Abstract
The control of DNA topology by DNA topoisomerases is essential for virtually all DNA transactions in the cell. These enzymes, present in every organism, exist as several non-homologous families. We previously identified a small group of atypical type IIB topoisomerases, called Topo VIII, mainly encoded by plasmids. Here, taking advantage of the rapid expansion of sequence databases, we identified new putative Topo VIII homologs. Our analyses confirm the exclusivity of the corresponding genes to mobile genetic elements (MGE) and extend their distribution to nine different bacterial phyla and one archaeal superphylum. Notably, we discovered another subfamily of topoisomerases, dubbed ‘Mini-A’, including distant homologs of type IIB topoisomerases and encoded by extrachromosomal and integrated bacterial and archaeal viruses. Interestingly, a short, functionally uncharacterized motif at the C-terminal extremity of type IIB topoisomerases appears sufficient to discriminate between Mini-A, Topo VI and Topo VIII subfamilies. This motif could be a key element for understanding the differences between the three subfamilies. Collectively, this work leads to an updated model for the origin and evolution of the type IIB topoisomerase family and raises questions regarding the role of topoisomerases during replication of MGE in bacteria and archaea.
INTRODUCTION
In bacteria and archaea, adaptation and genetic diversification take advantage of the acquisition of exogenous DNA via horizontal gene transfer (HGT) (1). Interestingly, the diversity of genes found in a specific bacterial species, e.g. the pan-genome, was shown to exceed by far the amount of genes found in a single bacteria (2,3). Considering that HGT can include DNA transfer across species, the term super-genome was used to describe the whole set of genes that a prokaryotic ‘individual’ can exploit in its environment (4,5). Importantly, the adaptation of prokaryotic organism is associated with antibiotic resistance and pathogenicity acquisition with major health implications (6,7). Understanding HGTs has also large implications in evolution, since cells and viruses are likely to have co-evolved with extensive HGT since the dawn of evolution (8,9).
Diverse mobile genetic elements (MGE) are involved in HGT, using several mechanisms. Conjugative plasmids are known to play a major role in HGT, as conjugation can cross species barrier, and can even spread between different domains of life (10,11). Conjugative plasmids show large diversity, in their size, and in the composition of genes encoded (12). In particular, these mobile elements possess distinct mechanisms regarding their replication and their mobility (12,13). In 2014, an atypical topoisomerase, the topoisomerase VIII (Topo VIII), was found encoded by a couple of free and integrated plasmids, including conjugative ones, raising questions about the function(s) of this protein in the replication or in the mobility of the MGE (14).
DNA topoisomerases are essential enzymes that can modify the number of topological links between the two strands of a DNA molecule. They introduce either single or double-strand breaks in the DNA, and move a DNA strand or a duplex through the breaks before closing them (15–17). DNA topoisomerases are classified into two major types (I and II), depending on the nature of the transient DNA break (single- or double-stranded) and have been grouped into five families (Topo IA, IB, IC, IIA and IIB), according to structural or sequence similarities. Some of these families have been further divided into subfamilies of homologous proteins and named either based on specific mechanistic features, e.g. gyrase (a subfamily of Topo IIA) or based on the timing of their discovery (Topo I, II, III, IV, V, VI, VIII). Notably, type I and type II DNA topoisomerases have uneven and even number in this historical nomenclature, respectively.
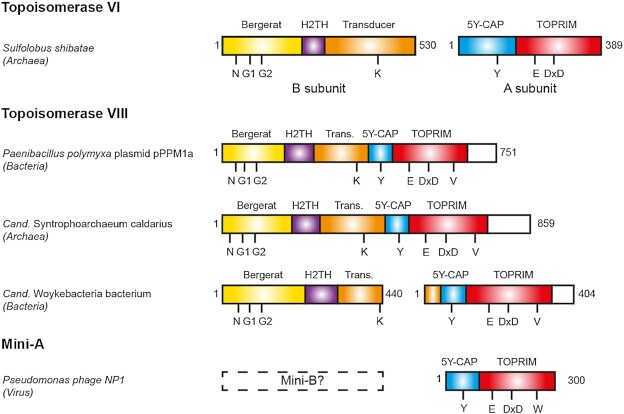
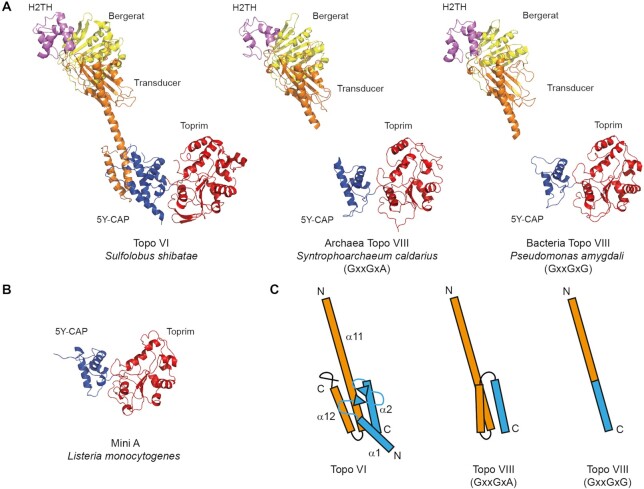
The type IIB topoisomerase family demonstrates complex distribution with diverse biochemical activity and cellular functions. The most understood member of this family is the topoisomerase VI (Topo VI), present in nearly all species of the Archaea domain, in which it functions as the canonical Type II topoisomerase, for transcription and replication (18). This enzyme is also found in several eukaryotes, including Viridiplantae (a eukaryotic division including plants, green and red algae) and in a small number of bacteria and protists (19). Topo VI is composed of two subunits termed Top6A and Top6B. Top6A contains the elements involved in DNA cleavage. This reaction requires a divalent metal ion, coordinated by the Toprim domain, and the nucleophilic attack of the catalytic tyrosine, within the 5Y-CAP domain, which covalently binds the 5′-end of DNA after cleavage (20). Top6B is involved in the binding and hydrolysis of adenosine triphosphate (ATP), which lead to the opening/closing of the topoisomerase gates, and enable the strand passage through the DNA double-strand break (21–23). This mechanism requires the conserved ATP-binding module called the Bergerat fold which is the hallmark of the GHKL protein family (24). The transducer domain, which bridges the ATPase and the Top6A (Figure 1) contains a highly conserved lysine, the switch-lysine, which side chain can interact with the γ-phosphate of a bound ATP. The hydrolysis of ATP and release of the inorganic phosphate (Pi) induces a rotation of the transducer. Current models suggest that this rotation promote the opening of a G-gate leading to the DNA strand passage (21,25).
Figure 1.
Schematic view of the domain composition of Topoisomerases VI, VIII and Mini-A. The Bergerat fold/GHKL, the H2TH, the Transducer, the 5Y-CAP and the TOPRIM domains are shown as yellow, purple, orange, blue and red boxes, respectively. The Topo VIII specific C-terminal region with unknown function is shown as white box. The position of the N, G1 and G2 boxes, the switch lysine (K), the catalytic tyrosine (Y), the Mg-binding residues (E and DxD), the RVELNAM sequence (V) and WELDAL signature (W) are indicated.
In nearly all eukaryotes, another type IIB enzyme, the Topo VI-like, is present (26–28). This enzyme is composed of two subunits Top6BL and Spo11. Spo11 is largely similar to Top6A, while Top6BL and Top6B display little sequence similarity, but similar predicted structures (18,26–27,29). Topo VI-like is important during meiosis, as it was shown to generate DNA double-strand breaks, essential for the correct chromosomal segregation, and to shape the genetic diversity through homologous recombination (18,29). Biochemical characterization of Topo VI-like has been difficult so far due to the poor solubility of the enzymes (30).
The other type IIB topoisomerase subfamily, Topo VIII, appears to be exclusively encoded by free or integrated MGE, mostly present in Bacteria. The only exception was encoded by the plasmid 5 from the archaeon Halalkalicoccus jeotgali B3 (14). Topo VIII proteins contain the different domains found in the other type IIB topoisomerases, with one major difference. Unlike Topo VI and Topo VI-like, which comprise two subunits, most Topo VIII such as Paenibacillus polymyxa, are composed of a single polypeptide, in which the N-terminal and C-terminal moieties are homologous to the B and A subunits of Topo VI, respectively (Figure 1) (14). Biochemical assays using purified Topo VIII revealed that some of the enzymes retain the topoisomerase activity, while another tested Topo VIII was generating double-strand breaks in vitro (14).
In this work, we have extended the number of Topo VIII genes from 21 to 77 in Bacteria, including two new sequences inside extrachromosomal conjugative plasmids. We also found 3 new Topo VIII in archaeal Metagenome-Assembled Genomes (MAGs). These genomes correspond to members of the Euryarchaeota superphylum, namely CandidatusSyntrophoarchaeum caldarius (Methanosarcinales), Candidatus division MSBL1 archaeon (Persephonarchaea) and Archaeglobales archaeon (Archaeoglobales) (31–33). Analysis of the genomic context of these new Topo VIII resulted in the identification of new MGEs, including free and integrated plasmids. Unexpectedly, we also identified a new family of proteins homologous to the A subunits of Topo IIB, that are present in the genome of archaeoviruses and bacterioviruses (bacteriophages). We called these proteins ‘Mini-A’, since they are shorter than other A subunits. Interestingly, Mini-A proteins are distantly related from both Topo VI and Topo VIII, suggesting a very ancient divergence between these three groups of enzymes and raising new question about the emergence and evolution of type IIB DNA topoisomerases.
MATERIALS AND METHODS
Sequence database search
We used several previously known Topoisomerase VIII as queries to carry out psi-BLAST and translated BLAST (tblastn) searches of different databases on the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Only sequences which display the conserved elements in the Bergerat Fold, the transducer, the Toprim and the 5Y-CAP domains were kept for further analysis.
Network analysis
All-against-all BLASTp analyses were performed on the set of Topo VIII proteins. The all-against-all results were grouped using the SiLiX (for SIngle LInkage Clustering of Sequences) package v1.2.8 (http://lbbe.univ-lyon1.fr/ SiLiX) (34). This approach for the clustering of homologous sequences, is based on single transitive links with alignment coverage constraints. Several different criteria can be used separately or in combination to infer homology separately (percentage of identity, alignment score or E-value, alignment coverage). For the Topo VIII protein dataset, we used the additional thresholds of 30 and 45% for the identity percentage and the query coverage, respectively.
Phylogenetic analyses
Alignments and trimming: each alignment used for phylogenetic analyses was performed using MAFFT v7 with default settings (35) and trimmed with BMGE (36) with a BLOSUM30 matrix and the -b 1 parameter.
Maximum likelihood trees: the IQ-TREE v1.7 software (http://www.iqtree.org/) (37) was used to calculate maximum likelihood (ML) trees with the best substitution model as suggested by the ModelFinder option (38). Branch robustness was estimated by several approaches as the non-parametric bootstrap procedure (100 replicates) (39), the SH-aLRT approximate likelihood ratio test (40), the ultrafast bootstrap approximation (1000 replicates) (41), and with the transfer bootstrap expectation approaches recently developed (42).
Protein structure predictions
Template-based protein structure were predicted using the RaptorX web server (http://raptorx.uchicago.edu/). The N-terminal half of Pseudomonas amygdali topoisomerase VIII was modeled using Sulfolobus shibatae Topo VI B (PDB 1MX0 chain A and 2ZBK chain B, score 261/406) (25,43) and its C-terminal half was modeled using Methanocaldococcus jannaschii Topo VI A subunit (PDB 1D3Y chain A, score 164/263) (20). The N-terminal half of S. caldarius topoisomerase VIII was modeled using Sulfolobus shibatae Topo VI B (PDB 1MX0 chain A and 2ZBK chain B, score 216/430) (25,43) and its C-terminal half was modeled using M. jannaschii Topo VI A subunit (PDB 1D3Y chain A, score 150/274) (20). Listeria monocytogenes mini-A was modeled using M. jannaschii Topo VI A subunit (PDB 1D3Y chain A, score 117/259) (20). Comparisons between different topoisomerases models were conducted with the PyMOL Molecular Graphics System (http://www.pymol.org).
Determination of integrated mobile genetic elements
Integrated MGE (iMGE) encoding topoisomerases were identified as described previously (44). The precise boundaries of integration were defined based on the presence of direct repeats corresponding to attachment sites or target site duplications. The direct repeats were searched for using Unipro UGENE (45).
RESULTS AND DISCUSSION
Identification of new type IIB topoisomerases: Topo VIII and Mini-A
In order to identify new Topo VIII sequences, we screened the NCBI non-redundant protein and environmental sequence databases by PSI-Blast and tblastn algorithms using several known Topo VIII sequences as queries. Using this approach, we identified a total number of 82 putative Topo VIII in bacterial and archaeal genomes, as well as in one unclassified contig (Table 1 and Supplementary Table S1).
Table 1.
Number of Topo VIII sequences in the different archaeal and bacterial phyla
| Domain | Phylum | Identified sequences |
|---|---|---|
| Total | 82 | |
| Bacterial | 77 | |
| Firmicutes | 40 | |
| Proteobacteria | 19 | |
| Actinobacteria | 6 | |
| Bacteroidetes | 5 | |
| Planctomycetes | 3 | |
| Chloroflexi | 1 | |
| Cyanobacteria | 1 | |
| Chisholmbacteria | 1 | |
| Woykebacteria | 1 | |
| Archaeal | 4 | |
| Euryarchaeota | 4 | |
| Unclassified contig | 1 |
Within Archaea, in addition to the previously reported plasmid-encoded Topo VIII (H. jeotgali, Halobacteriales) (14), we detected three new Topo VIII sequences in Metagenome-Assembled Genomes (MAGs): a Archaeoglobales archaeon, S. caldarius (Methanosarcinales) and in a Candidatus division MSBL1 archaeon (31–33). It has been proposed recently to rename archaea of the Candidatus division MSBL1 to Persephonarchaea, because they are a sister group of Hadesarchaea (Persephon being the wife of Hades in Greek mythology) (46). We will use this name thereafter. The Archaeoglobales archaeon protein is significantly shorter than other Topo VIII, missing the Bergerat fold and the H2TH domains, which may be due to the position of the topo VIII gene at the very 5′ extremity of the corresponding genomic contig. Further sequencing would be required to clarify the size of this protein.
In Bacteria, we identified 77 Topo VIII homologs encoded by both chromosomes and extrachromosomal plasmids. The latter included the two previously identified P. polymyxa and Paenibacillus alvei plasmids, and two new plasmids: Pseudomonas sp. Leaf58 pBASL58 (904 kb) and P. amygdali pMPPIa107 (972 kb). The identification of new Topo VIII sequences significantly expands the number of bacterial phyla, from 3 (Bacteroidetes, Proteobacteria and Firmicutes) to 9 phyla, including Planctomycetes, Chloroflexi, Cyanobacteria and two recently described phyla, Chisholmbacteria and Woykebacteria (Table 1) (47).
In addition to these Topo VIII, we found hundreds of shorter proteins that contain only a 5Y-CAP and a Toprim domains, domains which are essential for catalyzing the DNA cleavage (Figure 1). In this work, we chose a representative panel of 23 sequences, corresponding to 11 bacterial, 3 archaeal and 9 viral sequences (Supplementary Table S2). These proteins are notably different from previous known protein families with a Toprim domain (48). Indeed, only topoisomerases were known so far to contain both a Toprim and a 5Y-CAP domain. Since these proteins are homologs with the A subunits of Topo VI while significantly shorter (300 residues instead of 400 residues), we named these elements ‘Mini-A’. Since Top6B and Top6A encoding genes are invariably present as a tandem in archaeal Topo VI, we hypothesized that a Top6B subunit homologs (‘Mini-B’ proteins?) could be also encoded in the genome of organisms encoding a Mini-A gene. Our analysis of the genomic context surrounding the Mini-A genes indicated the absence of a Mini-B (e.g. containing the conserved Bergerat fold). It is therefore unclear whether Mini-A proteins are active alone, or whether they associated with another protein to carry their function.
Mini-A and Topo VIII are both encoded by mobile genetic elements
One characteristic feature of Topo VIII, is that it is the only know topoisomerase subfamily which seems dedicated to MGE, more specifically to free and integrated plasmids, including conjugative elements (14). This feature was conserved with the new Topo VIII sequences, which include 3 additional sequences on free plasmids. We further analyzed the genomic neighborhoods of the 8 topo VIII genes, including two new Topo VIII from archaea (Ca. Persephonarchaea, Ca.S. caldarius) and Topo VIII from different bacterial phyla: Actinobacteria (Kitasatospora aureofaciens), Cyanobacteria (Synechococcus sp. 1G10), Ca. Chisholmbacteria (Ca. Chisholmbacteria bacterium) and Planctomycetes (Two different Fimbriiglobus ruber genomes). Consistently, these sequences were found to be present in integrated elements, similar to what was already observed in the case of Topo VIII from Firmicutes and Proteobacteria (Supplementary Table S3) (14). Unfortunately, it was not possible to determine the genomic context of most new Topo VIII genes that were mostly retrieved from metagenome-assembled genomes (MAGs).
While Topo VIII enzymes were encoded by free and integrated plasmids, the Mini-As are encoded in the genomes of bacteria, archaea, archaeoviruses and bacterioviruses (bacteriophages), including pathogens such as Listeria, Pseudomonas, Clostridium and Salmonella. In the case of bacterial and archaeal sequences, we found viral elements at the vicinity of 9 out of 14 Mini-As (Supplementary Table S2). The remaining 5 Mini-As were found in small contigs, limiting the possibility of such genomic context analysis. Therefore, the Mini-A seems associated either with free viral sequences or with integrated proviral sequences. As a result our analysis reveals an additional level of diversity of type IIB topoisomerases in viruses.
Topo VIII exhibit two types of ATPase boxes
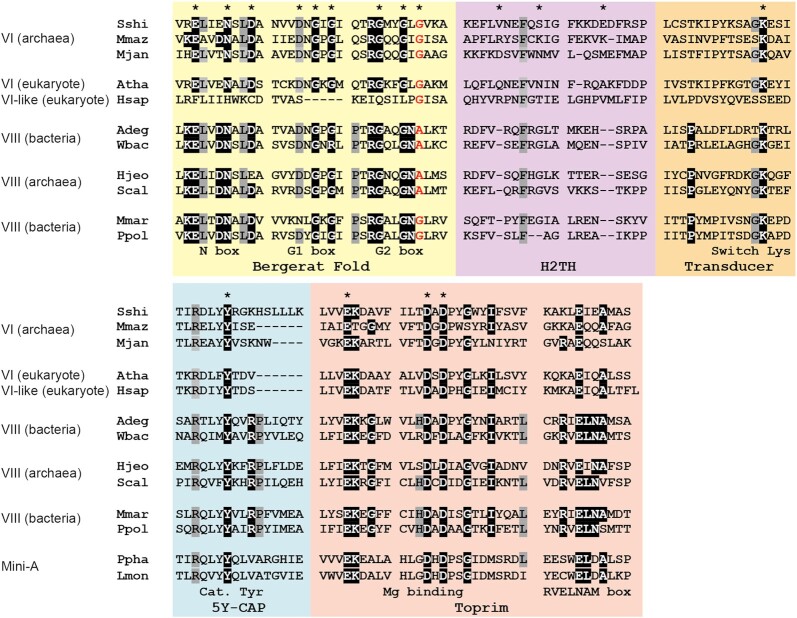
The B subunit of Topo VI is involved in the ATP binding and hydrolysis, thanks to one characteristic ATPase fold called the Bergerat fold or GHKL domain (18,24). In Topo VIII, the Bergerat fold is well conserved among the different sequences we identified (38% sequence identity). In particular, key residues involved in ATP-binding within the N-, the G1-and the G2-boxes are conserved in most Topo VIII (93%) (Figure 2). We nonetheless found two distinct G2-box sequences (GxxGxG and GxxGxA) in Topo VIII. The last residue of these G2-boxes are known to interact directly with ATP, so that GxxGxG Topo VIII and GxxGxA Topo VIII may display substantial activity differences. Importantly, both GxxGxG and GxxGxA sequences have been associated with functional ATPase, respectively in Topo VI and in MutL (24,49). Consistently, both GxxGxG and GxxGxA Topo VIII were associated with a functional switch lysine, in the transducer domain (81 out of 82 sequences).
Figure 2.
Conservation of Topo VIII. Alignment of Topo VIII, Topo VI, Topo VI-like and Mini-A proteins from Sulfolobus shibatae (Sshi), Methanosarcina mazei (Mmaz), Methanocaldococcus jannaschii (Mjan), Arabidopsis thaliana (Atha), Homo sapiens (Hsap), Ammonifex degensii (Adeg), Ca.Woykebacteria bacterium, Halalkalicoccus jeotgali (Hjeo), Ca.Syntrophoarchaeum caldarius, Microscilla marina (Mmar), Paenibacillus polymyxa plasmid pPPM1a (Ppol), Pseudomonas phage NP1 (Ppha) and Listeria monocytogenes (Lmon). More than 95% conserved residues are highlighted in black and >90% conserved residues are highlighted in gray. The last residue of the G2 box, either a glycine or an alanine is shown in red. Residues involved in ATP binding (Bergerat Fold and Transducer), the DNA binding (H2TH), divalent ion binding (Toprim) and the catalytic tyrosine are indicated by an asterisk.
Previously, a GxxGxA Topo VIII encoded by Ammonifex degensii was shown to linearize supercoiled plasmids, suggesting that the enzyme could cleave DNA without religation (14). Such an activity could mirror eukaryotic Topo VI-like enzymes, which are known to catalyze programmed double-strand breaks during meiosis (26,27). Interestingly, in these enzymes, the G2-box is largely degenerated, with only one G present (Gxxxxx), raising doubts about the ATPase activity (26,27). By contrast, GxxGxG Topo VIII encoded by P. polymyxa and Microscilla marina were able to relax supercoiled plasmids, with no detectable double-strand breaks, indicating that the DNA breaks were religated (14). Thus, the GxxGxA/GxxGxG signature may be important for the activity and, therefore, the function of topoisomerases.
Residues involved in DNA cleavage are conserved among type IIB topoisomerases
Contrary to the ATPase domains, the elements involved in DNA cleavage, more specifically within the 5Y-CAP and the Toprim domains are mostly conserved among the different type IIB topoisomerases. The main difference is the absence, in every Topo VIII and Mini-A, of the N-terminal part of the 5Y-CAP, which is involved in heterodimeric interaction between Top6A and Top6B (43,50). Overall, the 5Y-CAP domain is well conserved among different Topo VIII (40% sequence identity) and the catalytic tyrosine, found in that domain, is conserved not only in every Topo VIII but also in every Mini-A proteins (Figure 2). Consistently, the Toprim domain is well conserved in Topo VIII (44% sequence identity), and the divalent cation-binding region is similar between Topo VI and Topo VIII (Figure 2). More specifically, the residues involved in this interaction (E followed by DxDxxG) are conserved in every Topo VIII and Mini-A. This overall conservation suggests that both Mini-A and Topo VIII are likely able to cleave DNA similarly to Topo VI.
We identified several differences between Topo VI, Topo VIII and Mini-A. Both Topo VI and Mini-A contain one highly conserved arginine residue before the catalytic tyrosine, which seems to interact with DNA (Figure 2). In addition, the residue located immediately upstream of the catalytic tyrosine is often a tyrosine or phenylalanine which likely contributes to the respective positioning of the catalytic tyrosine and the DNA. Similarly, the majority of Topo VIII (59/82) displays the RxxY/FY sequence. However, a significant proportion of Topo VIII (18/82) have the RxxMY sequence while a few Topo VIII (5/82) displayed the HxxY/FY sequence. In addition, Topo VIII possess an arginine conserved in every sequences, three residues after the catalytic tyrosine, often followed by a proline residue, which are found in neither Topo VI nor in Mini-A proteins (Figure 2). Overall, the Topo VIII CAP-5Y domain shows an unexpected diversity surrounding the catalytic tyrosine with additional conserved residues compared to Mini-A and Topo VI. These conserved residues, at the vicinity of the catalytic tyrosine, are mostly arginine residues, known to interact with DNA. It is therefore possible that Topo VIII recognizes different DNA sequences and/or structures than Topo VI and Mini-A. Further functional studies will be important to dissect the precise function(s) of Topo VIII and Mini-A.
One major limitation to understand Topo VI, Topo VIII and Mini-A interplay with nucleic acids, is the absence of structure of any complexes between these proteins and DNA. A short domain, called the H2TH, located between the Bergerat fold and the transducer domain, was previously shown to bind DNA (23). This biochemical work indicated that the DNA was bend inside Topo VI catalytic site, thanks to the action of H2TH. Several basic residues from the H2TH seemed involved in DNA binding. In Topo VIII, this short domain (63–93 residues) is poorly conserved (28% sequence identity) and no basic residues seem to align with those identified in the archaeon Methanosarcina mazei Topo VI (Figure 2) (23). Basic residues at different positions might substitute for this DNA binding property, but the function of Topo VIII H2TH cannot be extrapolated from Topo VI at the moment.
Topo VIII show large diversity in the transducer domain
In Topo VI and Topo VIII, the transducer domain connects the ATPase region to the DNA cleavage region. While the switch lysine which interacts with ATP is conserved, the rest of the transducer domain sequence is considerably more divergent in different Topo VIII (28% sequence identity). In particular, the length of this domain varies from 135 residues for several Topo VIII from Proteobacteria to 204 residues for the Topo VIII from Persephonarchaea. There is a significant (Mann-Whitney U test (51) P < 10−10) difference between transducer domain lengths in GxxGxA Topo VIII (median 146 residues) and GxxGxG Topo VIII (median 179 residues) (Supplementary Table S1). The difference between Topo VIII transducers may affect the way the ATPase activity (in the N-terminal region) is connected with the transesterase activity (in the C-terminal region), and may also influence Topo VIII activity.
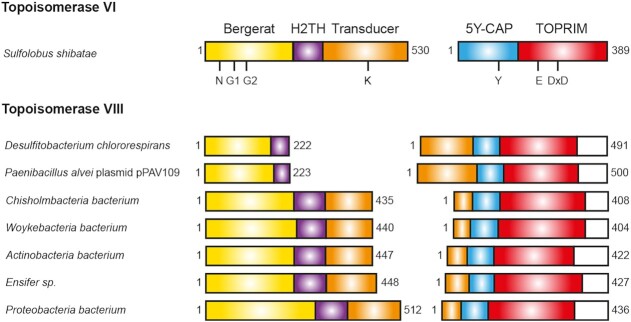
In seven sequences, we found the Topo VIII encoded by two adjacent genes, and the separation between the two subunits always occurs in the transducer domain, at two different positions (Figure 3). In the case of Desulfitobacterium chlororespirans (Clostridiales, Firmicutes) and P. alvei (Baciliales, Firmicutes), the Topo VIII separations occur at the limit between the H2TH and the transducer domain. Therefore, the position of the separations should not affect the folding of functional domains, and would be compatible with a functional enzyme. The Topo VIII sequences found in Candidatus Chisholmbacteria bacterium (Chisholmbacteria), Woykebacteria bacterium (Woykebacteria), Actinobacterium bacteria (Actinobacteria), Ensifer sp. (Proteobacteria) and Proteobacteria bacterium (Proteobacteria), are separated at the end of the transducer domain, just before the long α-helix, which connects the N-terminal and the C-terminal moieties of the protein. Similarly to the other 2-subunit Topo VIII, this separation should be compatible with the functionality and folding of the enzyme.
Figure 3.
Schematic view of the two-subunit Topoisomerases VIII. Position of the domains on two-subunit Topo VIII are shown, and compared with Topo VI domains organization. The coloring scheme is the same as that in Figure 1.
Therefore, the transducer domain is highly versatile, with different lengths, being even split into two subunits in several Topo VIII. Such split events have been also observed in other topoisomerase families, such as reverse gyrases and Topo IB, which can be functional either as a single polypeptide or as two subunits (52,53). One hypothesis is that the transducer domain could accommodate larger number of mutations compared to other domains that contain many conserved and indispensable residues. In spite of that, the observed differences may still affect the function of the Topo VIII.
Identification of a new conserved box in the different Type IIB topoisomerase subfamilies
As described previously (14), Topo VIII contain one particular RVELNAM sequence motif at the end of the Toprim domain. Consistently, the newly identified Topo VIII contain this conserved signature sequence confirming this observation. Interestingly, this region corresponds to a less conserved region in Top6A and Spo11 (R/KxExxA/Sx) in which, the arginine is either conserved or substituted to lysine, and the glutamate is conserved (Figure 2). The alanine is often either conserved or substituted to a serine residue, with the notable exception of Saccharomyces cerevisiae Spo11 in which this short residue is substituted by glutamate. Remarkably, Mini-A proteins also display their own characteristic signature sequence at this region, the xWELDAL sequence.
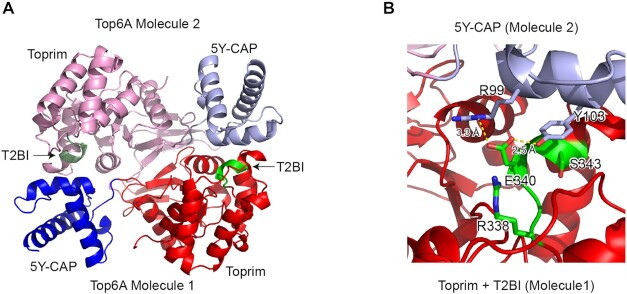
In Topo VI, the corresponding motif seems involved in the interaction between the different subunits inside the complex. Topo VI functional enzyme is composed of two B and two A subunits. In the crystal structures of the archaeal Topo VI and the crystal structure of Top6A subunits, the two A subunits dimerize and the (R/KxExxA/Sx) box is found at the interface between the two molecules (Figure 4) (20,43,50). We decided to call it the T2BI-box (Type IIB topoisomerases Interaction). In M. jannaschii Topo VI (20), the conserved T2BI glutamate residue (E340) forms hydrogen bonds with the conserved arginine (R99) and the catalytic tyrosine (Y103) in the 5Y-CAP. The aromatic ring of the catalytic tyrosine (Y103) also fits in a hydrophobic pocket centered on the T2BI serine (S343) (Figure 4). Notably, in Topo VI, Topo VIII and mini-A, the latter serine residue is often replaced by an alanine (Figure 2); nevertheless, both residues appear to be compatible with the formation of the hydrophobic pocket. Consistently, point mutations of the conserved lysine and glutamate in the T2BI-box have been previously generated in yeast Spo11, and the resulting mutants showed decreased DSBs formation, and subsequent meiotic recombination and spores formation (54).
Figure 4.
Structure of Methanocaldococcus jannaschii Top6A dimer (PDB ID: 1D3Y). (A) Cartoon Representation of two Top6A molecules in the dimers. The coloring scheme is the same as that in Figure 1. In addition, T2BI boxes are shown in green. (B) Detailed view of the interaction between T2BI and 5Y-CAP. Side chains of T2BI conserved residues (R338, E340 and S343) and 5Y-CAP (R99 and Y103) are shown. Relevant distances are shown in yellow dashed lines.
Surprisingly, none of the three Topo VI apo-form structures presently available are compatible with the presence of a DNA molecule, since the T2BI-box covers the catalytic tyrosine involved in the transesterase reaction. Thus, this box may be important for regulating type IIB enzymes activity, preventing uncontrolled cleavage of DNA. Both Topo VIII and Mini-A proteins possess additional conserved residues in the T2BI-box (resp. RVELNAM and xWELDAL sequences) which may reinforce their binding to the catalytic tyrosine region. Obtaining the structure of a type IIB topoisomerase (Topo VI, Topo VIII or Mini-A) with a DNA molecule will be determinant for understanding how the T2BI box can regulate the DNA cleavage. We therefore identify one motif which seems sufficient to discriminate between Topo VI, Topo VIII and Mini-A, and be therefore one key element distinguishing between the function of these three subfamilies of type IIB topoisomerases.
Structural prediction of Topo VIII and Mini-A proteins
The previously identified Topo VIII were predicted to fold similarly to the two known crystal structures of Topo VI (43,50), although the connection between the N-terminal and C-terminal moieties of the protein could not be modeled (14). Consistently, homology-based structure predictions indicate that newly identified Topo VIII should fold similarly to Topo VI. In particular, the overall protein architecture was very similar between archaeal Topo VIII from CandidatusS. caldarius (GxxGxA) and bacterial enzyme from P. amygdali plasmid (GxxGxG) (Figure 5). The N-terminal moiety of both Topo VIII superposes well with the Bergerat fold, the H2TH and the first half of the transducer domain of Topo VI, while the C-terminal moiety of these enzymes as well as Mini-A proteins superpose with the second half of the 5Y-CAP and the Toprim domains (Figure 5A).
Figure 5.
Homology modeling of Topo VIII and Mini-A structures. (A) Comparison of Topo VI crystal structure (PDB ID: 2ZBK) with Syntrophoarchaeum caldarius Topo VIII (GxxGxG) and Pseudomonas amygdali Topo VIII (GxxGxA). (B) Structure prediction of Listeria monocytogenes Mini-A. The coloring scheme is the same as that in Figure 1. (C) Modeling of Topo VIII junction between the transducer domain (orange) and the 5Y-CAP domain (blue) and comparison with Sulfolobus shibatae Topo VI known structure.
Neither Mini-A nor Topo VIII proteins contain the first part of the 5Y-CAP, which is involved in the interaction with Topo VI B subunit (Figure 5A) (43,50). This feature is consistent with the fact that both sides of the proteins are connected within one single polypeptide chain in the case of one-subunit Topo VIII but, at the same time, it raises question about how the two subunits associate in the case of split Topo VIII. The absence of this interacting region in Mini-A is one indication that these proteins may not be associated with a Mini-B.
Although the folding of the connections between N and C-terminal moieties cannot be modeled by homology, this region is predicted to fold as a single α-helix in the case of GxxGxG Topo VIII such as P. amygdali Topo VIII. As a consequence, in the case of this GxxGxG Topo VIII, the transducer may connect the ATPase region to the 5Y-CAP catalytic domain by a single α-helix (Figure 5B). By contrast, the corresponding region in GxxGxA Topo VIII such as Ca.S. caldarius Topo VIII are longer and contains two additional α-helices (Figure 5B). It is not clear how such three-helical connection folds, but this difference between GxxGxG and GxxGxA Topo VIII should impact the relationship between the ATPase region and the DNA cleavage region and, consequently, the Topo VIII activity.
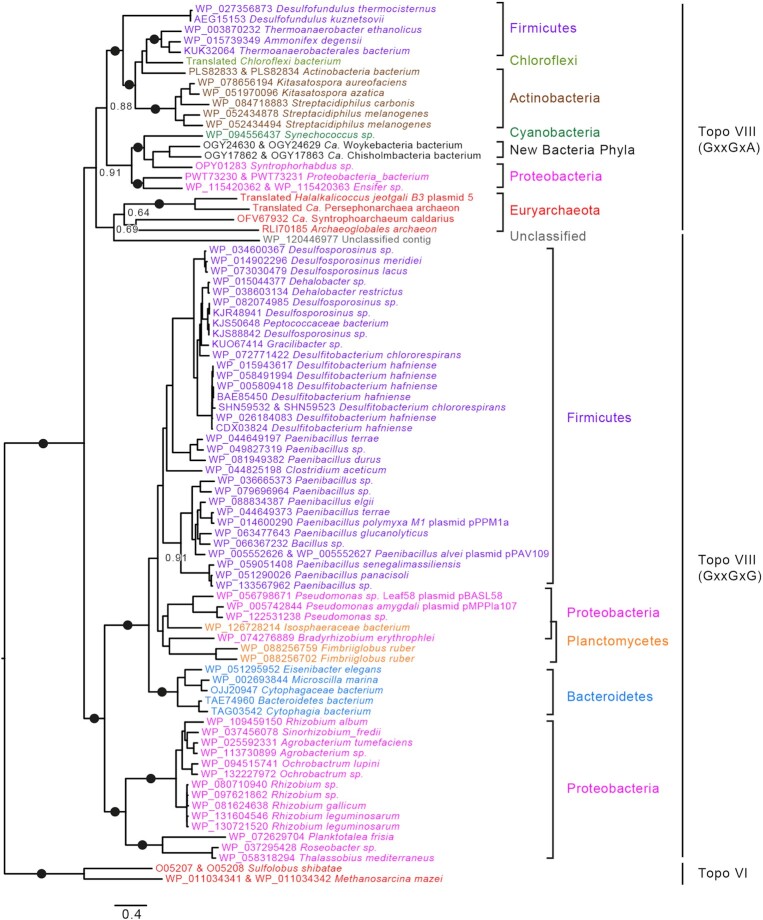
Topo VIII phylogeny indicates a separation between two groups of enzymes
We further aligned the 82 Topo VIII sequences together with the Topoisomerase VI B and A subunits from the archaea M. mazei and Sulfolobus shibatae using MAFFT (35). In the resulting maximum likelihood phylogenetic tree, we could identify two major monophyletic groups (BV = 100%), that corresponds to the GxxGxG and GxxGxA sequences with two exceptions (Figure 6). Archaeoglobales archaeon Topo VIII, which groups with GxxGxA Topo VIII, is truncated so that the sequence of the G2-box is unknown. The Topo VIII from one unclassified contig harbors the GxxGxG sequence while grouping with GxxGxA Topo VIII. One cannot exclude that the position of this Topo VIII corresponds to one long branch attraction, since this sequence displays the longest branch in Topo VIII phylogenetic tree (Figure 6).
Figure 6.
Phylogeny of Topoisomerases VIII. IQ Tree of the 82 topoisomerases VIII. Topo VIII sequences are color-coded corresponding to the taxonomy: Firmicutes in purple, Chloroflexi in light green, Actinobacteria in brown, Cyanobacteria in dark green, Proteobacteria in pink, Planctomycetes in orange, other bacterial phyla/MAG in black and archaea in red. Relevant booster values above 95% are represented as black closed circle, while other values are shown. The scale-bars represent the average number of substitutions per site.
The divergence between the GxxGxA and GxxGxG motifs may indicate an ancient event which led to two different subgroups of Topo VIII. Topo VIII may be further divided into additional subgroups, but they are not supported by statistical analyses and additional Topo VIII sequences would be required to confirm the relevance of these groups. For instance, the residues surrounding the catalytic tyrosine (mainly RxxY/FY) is found with variations in several monophyletic groups (Supplementary Figure S1).
GxxGxG Topo VIII comprises 21 sequences from seven different bacterial phyla and from archaea, most of which live in extreme environment (high salt, high temperature, acidic soil) while the 60 GxxGxA Topo VIII sequences are encoded by organisms living in ambient environments. Thus, Topo VIII diversification seems partly associated with environmental factors, indicating horizontal spread. In addition, sequences from the same bacterial phyla usually form monophyletic groups within a given Topo VIII subgroup, suggesting vertical descent. Nonetheless Firmicutes and Proteobacteria are present in both Topo VIII groups, suggesting that both groups originated from a gene duplication that could have occurred at the onset of bacterial evolution and later on co-evolved with their hosts (Figure 6). Interestingly, the four archaeal Topo VIII form a monophyletic clade inside the GxxGxA group. This suggests that Topo VIII were transferred from Bacteria to Archaea after the divergence between the two groups of Topo VIII. These results are consistent with the mobile character of Topo VIII found on plasmid and integrated mobile elements.
The seven two-subunit Topo VIII are found at different positions in the phylogenetic tree (Figure 6), and do not form monophyletic groups, although one subgroup includes larger proportion of the split Topo VIII variants. This distribution is consistent with multiple and independent and recent split events that produces functional enzyme.
Network analysis of the Topo VIII sequences is consistent with the results of phylogenetic analysis (Supplementary Figure S2). 59 out of 60 GxxGxG Topo VIII sequences gather into one group in this network, while 19 out of 23 GxxGxA sequences form a distinct group. The two halophilic archaeal Topo VIII (from H. jeotgali and Persephonarchaea archaeon), the truncated Archaeoglobales archaeon and the Topo VIII from the unclassified contig are excluded from this group, consistent with the phylogeny, in which these four sequences display long branches (Figure 6). In the corresponding phylogeny, branch supports are low, indicating that the identification of more archaeal Topo VIII is necessary for interpretation of the evolution of this family of topoisomerases.
Mini-A, Topo VI and Topo VIII form three distinct subfamilies of the Type IIB family
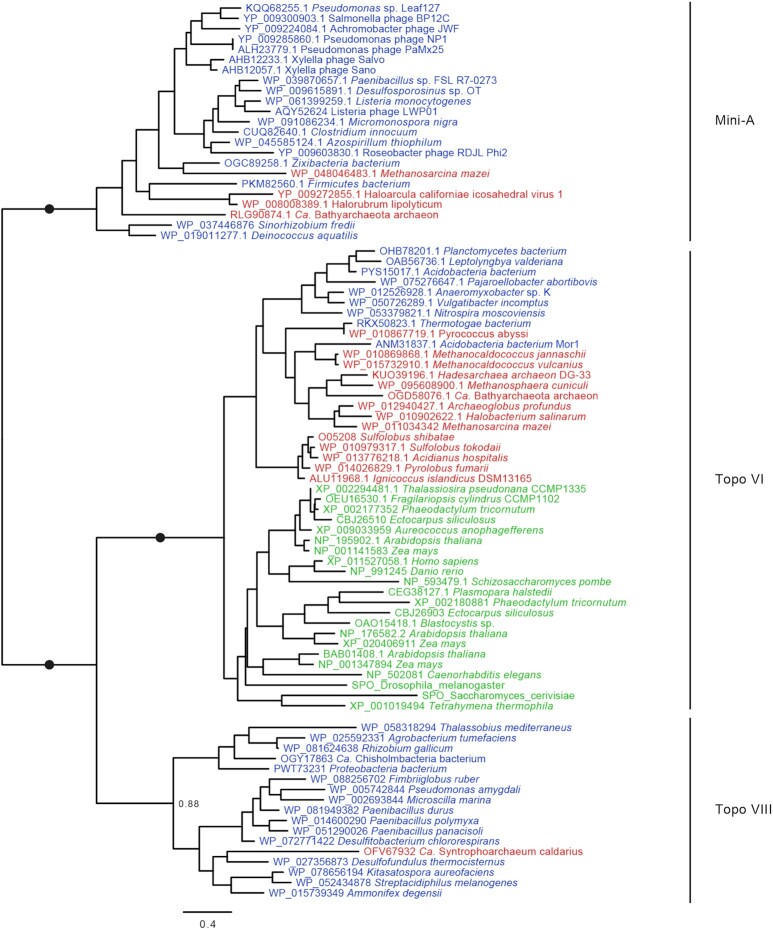
In order to investigate the evolutionary relationships between viral Mini-A proteins, plasmids Topo VIII and the chromosomal Topo VI, we selected 85 sequences representative of Mini-A, Topo VI A subunits, and the C-terminal moiety of Topo VIII, for phylogenetic analysis. The resulting maximum likelihood tree (Figure 7) shows that Mini-A, Topo VI and Topo VIII proteins forms three highly supported (bootstrap value = 100%) monophyletic groups, distinct from each other. Therefore, Mini-A represents a new subfamily of Type IIB topoisomerase, encoded by free and integrated virus. Type IIB topoisomerases family is therefore present as plasmids (Topo VIII), virus (Mini-A) and as non-mobile chromosomal (Topo VI) genes.
Figure 7.
Phylogeny of Type IIB Topoisomerases. IQ Tree of the 85 proteins, from Mini-A, Topo VI and Topo VIII subfamilies. Sequences are color-coded corresponding to domain of life: Archaea in red, Bacteria in Blue and Eukarya in green. Relevant booster values above 95% are represented as black closed circle, while other values are shown. The scale-bars represent the average number of substitutions per site.
At the moment, it is not possible to root the phylogeny, leading to two main alternatives regarding the evolution of type IIB enzymes. (i) The ancestor of the three A domains was already associated to a B subunit, and one lineage lost the B subunit leading to Mini-A. In that case, Mini-A ancestor protein would have emerged after the uptake of one fragment of Topo VI or Topo VIII gene by one virus. In that scenario, the B subunit was either lost from the viral genome or underwent major modifications leading to a non-canonical B-subunit (Mini-B) that is yet to be discovered. (ii) The ancestor of the three A domains was a stand-alone A protein, similarly to Mini-A. Thereafter, two of its descendants became associated to homologous B subunit in one or two events, to become the Topo VI and the Topo VIII. Since Mini-As are only found in viruses, it would be more likely, under this hypothesis, that the ancestor of Type IIB topoisomerases was encoded in viral genome. In addition, this protein would predate Topo VI, found in almost every archaeon and several eukaryotes, and thereafter predate LACA (Last Archaeal Common Ancestor) and possibly LEACA (Last Eukaryal and Archaeal Common Ancestor). Understanding the origins and the functions of these enzymes will require further investigation, including the determinations of the in vitro activity of Mini-A enzymes, and the in vivo functions of the protein during viral cycle.
CONCLUSION
Overall, our work confirms the main observation concerning Topo VIII: the 82 Topo VIII representatives seem to be encoded by MGE. Topo VIII is therefore the only known topoisomerase subfamily which is exclusive to MGE. Despite a major role of plasmids in HGT, driving bacterial adaptation and resistance to antibiotics (55,56), many aspects of plasmid replication remain poorly understood. In particular, there is little knowledge about how topoisomerases participate in plasmid replication, expression and conjugation. Plasmid type IA topoisomerase (Topo IA) has been previously shown to be encoded by the conjugative plasmid RPA, with many homologs in other plasmids from Proteobacteria and Firmicutes (57). Interestingly, both Topo VIII-encoding plasmids, pPPM1A of P. polymyxa and pAV109 of P. alvei, also encode Topo IA, suggesting that several topoisomerases may contribute to the replication/propagation of conjugative plasmids (14). In the new Topo VIII plasmids, pMPP1a107 and pBASL58, we found two (RefSeq WP_082476939 and WP_005742386) and one (RefSeq WP_082476939) Topo IA homologs, respectively. In addition, both plasmids encode the two subunits of Topo IV (WP_056799298 and WP_056799295 for pBASL58 and WP_114805133 and WP_005742002 for pMPP1a107). These plasmids, therefore, encode 3 and 4 different topoisomerases, respectively, highlighting the importance of these enzymes for the replication, expression and/or mobility of large plasmids.
The function of viral Mini-As also remains to be elucidated. Toprim domains consist of about 100 amino acids present in DnaG-type primases, type IA and type II topoisomerases, nucleases of the OLD family and bacterial DNA repair proteins of the RecR/M family (48). The absence of an ATPase domain in Mini-As could suggest that these proteins have a different activity than Type IIB topoisomerases. However, the pairing of Mini-A with other proteins, either virus of host encoded, cannot be excluded at the moment. Several Mini-As were found encoded by viruses infecting pathogenic bacteria, notably, Listeria monocytogenes and Salmonella enterica, the causative agents of listeriosis and salmonellosis, respectively (58,59). In the context of phage therapy advances, understanding the function(s) of these new viral topoisomerase-like proteins could bring important insight into the DNA transactions during the viral cycle. Future analyses of the prokaryotic mobilome are most likely to uncover novel (sub)families of topoisomerases and other genome replication enzymes.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Jacques Oberto, Dr Ryan Catchpole and Dr Stephanie Petrella for valuable discussions.
Contributor Information
Diane T Takahashi, Institut de Biologie Intégrative de la Cellule, CNRS, Université Paris-Saclay, 91198 Gif sur Yvette Cedex, France; Unité de Microbiologie Structurale, Institut Pasteur, CNRS, F-75015 Paris, France.
Violette Da Cunha, Institut de Biologie Intégrative de la Cellule, CNRS, Université Paris-Saclay, 91198 Gif sur Yvette Cedex, France.
Mart Krupovic, Institut Pasteur, Archaeal Virology Unit, Department of Microbiology, 75015 Paris, France.
Claudine Mayer, Unité de Microbiologie Structurale, Institut Pasteur, CNRS, F-75015 Paris, France; Université de Paris, Paris Diderot, F-75013 Paris, France.
Patrick Forterre, Institut de Biologie Intégrative de la Cellule, CNRS, Université Paris-Saclay, 91198 Gif sur Yvette Cedex, France; Institut Pasteur, F-75015 Paris, France.
Danièle Gadelle, Institut de Biologie Intégrative de la Cellule, CNRS, Université Paris-Saclay, 91198 Gif sur Yvette Cedex, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
European Research Council (ERC) Grant from the European Union's Seventh Framework Program (FP/2007-2013) (Project EVOMOBIL-ERC) [340440 to V.D.C., P.F.]; l’Agence Nationale de la Recherche [Project ESSPOIR ANR-17-CE12-0032 to D.T., D.G., C.M.; Project ENVIRA ANR-17-CE15-0005-01 to M.K.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Polz M.F., Alm E.J., Hanage W.P.. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. 2013; 29:170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R.. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005; 15:589–594. [DOI] [PubMed] [Google Scholar]
- 3. Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S.et al.. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc. Natl. Acad. Sci. U.S.A. 2005; 102:13950–13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Norman A., Hansen L.H., Sørensen S.J.. Conjugative plasmids: vessels of the communal gene pool. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2009; 364:2275–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Puigbò P., Lobkovsky A.E., Kristensen D.M., Wolf Y.I., Koonin E. V. Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol. 2014; 12:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davies J., Davies D.. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010; 74:417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyd E.F., Brüssow H.. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002; 10:521–529. [DOI] [PubMed] [Google Scholar]
- 8. Durzyńska J., Goździcka-Józefiak A.. Viruses and cells intertwined since the dawn of evolution. Virol. J. 2015; 12:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forterre P. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:3669–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dodsworth J.A., Li L., Wei S., Hedlund B.P., Leigh J.A., de Figueiredo P.. Interdomain conjugal transfer of DNA from bacteria to archaea. Appl. Environ. Microbiol. 2010; 76:5644–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cury J., Oliveira P.H., de la Cruz F., Rocha E.P.C.. Host range and genetic plasticity explain the coexistence of integrative and extrachromosomal mobile genetic elements. Mol. Biol. Evol. 2018; 35:2230–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smillie C., Garcillán-Barcia M.P., Francia M.V., Rocha E.P.C., de la Cruz F.. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010; 74:434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Orlek A., Stoesser N., Anjum M.F., Doumith M., Ellington M.J., Peto T., Crook D., Woodford N., Walker A.S., Phan H.et al.. Plasmid classification in an era of Whole-Genome sequencing: Application in studies of antibiotic resistance epidemiology. Front. Microbiol. 2017; 8:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gadelle D., Krupovic M., Raymann K., Mayer C., Forterre P.. DNA topoisomerase VIII: a novel subfamily of type IIB topoisomerases encoded by free or integrated plasmids in Archaea and Bacteria. Nucleic Acids Res. 2014; 42:8578–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vos S.M., Tretter E.M., Schmidt B.H., Berger J.M.. All tangled up: How cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011; 12:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Forterre P. Pommier Y Introduction and historical perspective. DNA Topoisomerases and Cancer. 2012; NY: Springer; 1–52. [Google Scholar]
- 17. Wang J.C. Untangling the Double Helix: DNA Entanglement and the Action of the DNA Topoisomerases. 2009; Cold Spring Harbor Laboratory Press. [Google Scholar]
- 18. Bergerat A., de Massy B., Gadelle D., Varoutas P.C., Nicolas A., Forterre P.. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997; 386:414–417. [DOI] [PubMed] [Google Scholar]
- 19. Forterre P., Gadelle D.. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009; 37:679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nichols M.D., DeAngelis K., Keck J.L., Berger J.M.. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 1999; 18:6177–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Corbett K.D., Berger J.M.. Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI. Structure. 2005; 13:873–882. [DOI] [PubMed] [Google Scholar]
- 22. Bates A.D., Berger J.M., Maxwell A.. The ancestral role of ATP hydrolysis in type II topoisomerases: prevention of DNA double-strand breaks. Nucleic Acids Res. 2011; 39:6327–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wendorff T.J., Berger J.M.. Topoisomerase VI senses and exploits both DNA crossings and bends to facilitate strand passage. Elife. 2018; 7:e31724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dutta R., Inouye M.. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 2000; 25:24–28. [DOI] [PubMed] [Google Scholar]
- 25. Corbett K.D., Berger J.M.. Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution. EMBO J. 2003; 22:151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vrielynck N., Chambon A., Vezon D., Pereira L., Chelysheva L., De Muyt A., Mezard C., Mayer C., Grelon M.. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science. 2016; 351:939–943. [DOI] [PubMed] [Google Scholar]
- 27. Robert T., Nore A., Brun C., Maffre C., Crimi B., Guichard V., Bourbon H.-M., de Massy B.. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science. 2016; 351:943–949. [DOI] [PubMed] [Google Scholar]
- 28. Ramesh M.A., Malik S.-B., Logsdon J.M.. A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr. Biol. 2005; 15:185–191. [DOI] [PubMed] [Google Scholar]
- 29. Keeney S., Giroux C.N., Kleckner N.. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997; 88:375–384. [DOI] [PubMed] [Google Scholar]
- 30. Yeh H.-Y., Lin S.-W., Wu Y.-C., Chan N.-L., Chi P.. Functional characterization of the meiosis-specific DNA double-strand break inducing factor SPO-11 from C. elegans. Sci. Rep. 2017; 7:2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mwirichia R., Alam I., Rashid M., Vinu M., Ba-Alawi W., Kamau A.A., Ngugi D.K., Göker M., Klenk H.-P., Bajic V.et al.. Metabolic traits of an uncultured archaeal lineage -MSBL1- from brine pools of the Red Sea. Sci. Rep. 2016; 6:19181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laso-Pérez R., Wegener G., Knittel K., Widdel F., Harding K.J., Krukenberg V., Meier D. V., Richter M., Tegetmeyer H.E., Riedel D.et al.. Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature. 2016; 539:396–401. [DOI] [PubMed] [Google Scholar]
- 33. Becker E.A., Seitzer P.M., Tritt A., Larsen D., Krusor M., Yao A.I., Wu D., Madern D., Eisen J.A., Darling A.E.et al.. Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic Osmo-response. PLoS Genet. 2014; 10:e1004784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miele V., Penel S., Duret L.. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics. 2011; 12:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Katoh K., Standley D.M.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Criscuolo A., Gribaldo S.. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010; 10:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q.. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015; 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S.. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017; 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Efron B. Bootstrap methods: another look at the jackknife. Ann. Stat. 1979; 7:1–26. [Google Scholar]
- 40. Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O.. New algorithms and methods to estimate Maximum-Likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010; 59:307–321. [DOI] [PubMed] [Google Scholar]
- 41. Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S.. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018; 35:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lemoine F., Domelevo Entfellner J.-B., Wilkinson E., Correia D., Dávila Felipe M., De Oliveira T., Gascuel O.. Renewing Felsenstein's phylogenetic bootstrap in the era of big data. Nature. 2018; 556:452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Graille M., Cladière L., Durand D., Lecointe F., Gadelle D., Quevillon-Cheruel S., Vachette P., Forterre P., van Tilbeurgh H.. Crystal structure of an intact type II DNA topoisomerase: insights into DNA transfer mechanisms. Structure. 2008; 16:360–370. [DOI] [PubMed] [Google Scholar]
- 44. Krupovic M., Makarova K.S., Wolf Y.I., Medvedeva S., Prangishvili D., Forterre P., Koonin E. V.. Integrated mobile genetic elements in Thaumarchaeota. Environ. Microbiol. 2019; 21:2056–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Okonechnikov K., Golosova O., Fursov M., Varlamov A., Vaskin Y., Efremov I., German Grehov O.G., Kandrov D., Rasputin K., Syabro M.et al.. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012; 28:1166–1167. [DOI] [PubMed] [Google Scholar]
- 46. Adam P.S., Borrel G., Brochier-Armanet C., Gribaldo S.. The growing tree of Archaea: new perspectives on their diversity, evolution and ecology. ISME J. 2017; 11:2407–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Anantharaman K., Brown C.T., Hug L.A., Sharon I., Castelle C.J., Probst A.J., Thomas B.C., Singh A., Wilkins M.J., Karaoz U.et al.. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 2016; 7:13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aravind L., Leipe D.D., Koonin E. V. Toprim–a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998; 26:4205–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ban C., Yang W.. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998; 95:541–552. [DOI] [PubMed] [Google Scholar]
- 50. Corbett K.D., Benedetti P., Berger J.M.. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat. Struct. Mol. Biol. 2007; 14:611–619. [DOI] [PubMed] [Google Scholar]
- 51. Mann H.B., Whitney D.R.. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947; 18:50–60. [Google Scholar]
- 52. Davies D.R., Mushtaq A., Interthal H., Champoux J.J., Hol W.G.J.. The structure of the transition state of the heterodimeric topoisomerase i of leishmania donovani as a vanadate complex with Nicked DNA. J. Mol. Biol. 2006; 357:1202–1210. [DOI] [PubMed] [Google Scholar]
- 53. Krah R., Kozyavkin S.A., Slesarev A.I., Gellert M.. A two-subunit type I DNA topoisomerase (reverse gyrase) from an extreme hyperthermophile. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Diaz R.L., Alcid A.D., Berger J.M., Keeney S.. Identification of residues in yeast Spo11p critical for meiotic DNA double-strand break formation. Mol. Cell Biol. 2002; 22:1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Frazão N., Sousa A., Lässig M., Gordo I.. Horizontal gene transfer overrides mutation in Escherichia coli colonizing the mammalian gut. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:17906–17915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ochman H., Lawrence J.G., Groisman E.A.. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000; 405:299–304. [DOI] [PubMed] [Google Scholar]
- 57. Li Z., Hiasa H., Kumar U., DiGate R.J.. The traE gene of plasmid RP4 encodes a homologue of Escherichia coli DNA topoisomerase III. J. Biol. Chem. 1997; 272:19582–19587. [DOI] [PubMed] [Google Scholar]
- 58. Phothaworn P., Dunne M., Supokaivanich R., Ong C., Lim J., Taharnklaew R., Vesaratchavest M., Khumthong R., Pringsulaka O., Ajawatanawong P.et al.. Characterization of flagellotropic, Chi-Like salmonella phages isolated from thai poultry farms. Viruses. 2019; 11:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Glaser P., Frangeul L., Buchrieser C., Rusniok C., Amend A., Baquero F., Berche P., Bloecker H., Brandt P., Chakraborty T.et al.. Comparative genomics of Listeria species. Science. 2001; 294:849–852. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.