Abstract
Lactobacillus crispatus is a common inhabitant of both healthy poultry gut and human vaginal tract, and the absence of this species has been associated with a higher risk of developing infectious diseases. In this study, we analyzed 105 L. crispatus genomes isolated from a variety of ecological niches, including the human vaginal tract, human gut, chicken gut and turkey gut, to shed light on the genetic and functional features that drive evolution and adaptation of this important species. We performed in silico analyses to identify the pan and core genomes of L. crispatus, and to reveal the genomic differences and similarities associated with their origins of isolation. Our results demonstrated that, although a significant portion of the genomic content is conserved, human and poultry L. crispatus isolates evolved to encompass different genomic features (e.g. carbohydrate usage, CRISPR–Cas immune systems, prophage occurrence) in order to thrive in different environmental niches. We also observed that chicken and turkey L. crispatus isolates can be differentiated based on their genomic information, suggesting significant differences may exist between these two poultry gut niches. These results provide insights into host and niche-specific adaptation patterns in species of human and animal importance.
INTRODUCTION
During the past decade, the composition and diversity of the human microbiota have been studied to understand how various members thereof impact human health and disease, and more recently special attention has been focused on microbiota-mediated animal health (1,2). The microbiota, defined as a complex microbial community, which exceeds the number of cells of the host and contains a gene pool that carries out many functions not encoded in the host genome (3), is now considered as another organ of a human being with significant roles in the physiology and metabolic pathways that impact our bodies (4,5). The gut microbiota has been of particular focus of interest with a clear dysbiosis described in certain inflammatory and immune diseases (6,7) but also linked with other diseases like autism (8) and with a key influence in the effect of drug metabolism (9). Recently, special attention has been focused on the other human microbiomes: oral, skin and vaginal microbiomes (10–12). Whereas the gut microbiome presents a high diversity of bacterial genera and species, the vaginal microbiome is typically low in diversity and commonly dominated by Lactobacillus genus. Among these, Lactobacillus crispatus has been described as one of the predominant species that plays a key role in women's health. Lactobacillus crispatus is also closely associated with animal health, particularly in poultry (chicken and turkey) gut health (13,14). The absence of L. crispatus has been correlated with higher risk of infectious diseases and sexually transmitted diseases in women (15,16). Remarkably, L. crispatus has become an emerging probiotic for both women and poultry health due to the capability to interfere with pathogenic bacteria, through the colonization, competitive exclusion and production of antimicrobial compounds and exopolysaccharides (EPSs) (17,18). However, only limited information has been elucidated on the genetic basis to explain how L. crispatus plays a key role in two distinctly different environments such as the poultry gut and the human vaginal niche.
Recent advances in next-generation sequencing technologies coupled with the development of bioinformatics tools have dramatically increased the capability of computational biology analyses to provide insights into the genetic contents of every way of life. In this regard, genomic comparative analyses can be performed on large-scale datasets, thanks to the notably increased number of bacterial genomes available, encouraging researchers to easily, speedily and conveniently analyze the data for a better understanding of the genomics and the biology thereof, allowing more accurate predictions to be made. Indeed, interrogating large datasets can provide insights into the genetic content and structure of bacterial genomes, together with the relationship within different isolates and with the ecological niche of isolation. In this regard, comparative genomic analyses, including pan genome reconstruction (19) at the species or genus level, have become crucial to undermine gene content, metabolomic capabilities, taxonomic relation and even the environmental niches of isolation and associated features.
Here, we performed comparative genomic analyses to investigate, in silico, how niche-specific adaption occurs across 105 L. crispatus strains isolated from different hosts (human and poultry) and body sites (mainly gut and vaginal isolates). We demonstrated that (i) the pan genome size varies between ecological niches, (ii) genomes cluster according to host and body site isolation source, and (iii) specific genetic features may affect colonization and probiotic efficacy with tissue specificity.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The L. crispatus strains from our North Carolina Klaenhammer (NCK) collection (Supplementary Table S1) were propagated from −80°C MRS glycerol (15%, v/v) stocks in MRS (de Man Rogosa and Sharpe, Difco) broth or in MRS agar (1.5%, w/v) plates, both at 37°C under anaerobic conditions.
DNA extraction, genome sequencing and assemblies
The DNA preparation, genome sequencing and genome assemblies were performed at CoreBiome (Saint Paul, MN, USA). Briefly, DNA extraction was performed from 16 h broth culture (see earlier) using MO Bio PowerFecal (Qiagen) automated for high throughput on QiaCube (Qiagen), with bead beating using 0.1 mm glass bead plates. DNA quantification was performed on Qiant-iT Picogreen dsDNA Assay (Invitrogen). Library preparation was performed with a procedure adapted from the Nextera Library Prep Kit (Illumina). Then, libraries were sequenced on Illumina NextSeq using paired-end 2 × 150 reads with a NextSeq 500/550 High Output v2 Kit (Illumina). The resulting DNA sequences were filtered for low quality (Q-score <20) and length (<50), and adapter sequences were trimmed using Cutadapt (v.1.15). For genome assemblies, the fastq files of the filtered pair-end reads obtained for each bacterial strain were used as input for genome assemblies using SPAdes v3.11.0. Contigs >1000 bases in length were used in a QUAST v4.5 analysis.
Genome annotation
We retrieved complete and partial sequences of L. crispatus genomes available at NCBI database in May 2019 avoiding any repeated genome (n = 88), in addition to the private sequenced strains of our NCK collection (n = 17), to reach a total of 105 strains for the comparative genomic analyses (Supplementary Table S1). For consistency in the annotations and open reading frame (ORF) predictions during the analyses, the 105 genomes were reannotated using Prokka v1.13.3 (20).
Pan genome analyses
The aforementioned set of 105 L. crispatus genomes was subjected to pan genome analyses using Roary v3.12.0 (19). Briefly, the predicted ORFs of each genome were used to perform the pan genome analyses to identify the total genes present in the pan genome, core genes, unique genes and new genes. The functional analyses of the core genes and unique genes were performed using eggNOG 5.0 (http://eggnog5.embl.de/#/app/home) (21). The resulting pan genome analysis output files were used to depict the corresponding plots in RStudio v1.1.463 (22).
Phylogenomic analyses
The 465 core genes shared among the 105 L. crispatus strains were aligned using the PRANK algorithm implemented in Roary v3.12.0 (19). The resulting multi-FASTA alignment file was used as input in ClustalW v2.1 (23) to infer the phylogenomic tree using the neighbor-joining clustering algorithm, including Lactobacillus acidophilus NCFM as an outgroup to root the tree. The core genome tree was depicted with FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Functional genomic analysis
RPS-BLAST was performed using conserved domain database (CDD) to identify the protein domains present in each genome of the L. crispatus strains (24). The identified protein domains were then classified into different functional clusters of orthologous groups (COGs) (25). The COG information of each stain was used to perform a discriminant analysis of principal components (DAPC) (26) with prior group information (based on isolation sources) implemented in the R package adegenet v2.0.1 (27). The most contributing COGs (contributions above a threshold of 0.0025) to the group membership were extracted and displayed in a heatmap.
Prophage identification
Prediction of prophage genes and regions in the 105 L. crispatus genomes was performed using PHASTER (www.phaster.ca) (28,29). The prophage regions were detected by blasting query sequences against bacterial and phage/prophage databases in GenBank. A completeness score was assigned to each detected prophage region based on whether (i) the region contains known phage sequence; (ii) >50% of the proteins in the detected regions are associated with known phage sequences; and (iii) <50% of the proteins in the detected regions are associated with known phage sequences. The predicted prophage region is considered intact if the score is >90, questionable if the score is between 60 and 90, and incomplete if the score is <60. For the detailed explanation of the score system, please refer to original PHASTER publication (28,29). The schematic representation of intact prophage region of the genomes NCK2514 and CTV-05 was directly exported from PHASTER website. Then, the signature proteins in the prophage regions were exported and depicted using ‘pheatmap’ package V1.0.12 in RStudio v1.1.463 (22).
CRISPR–Cas system identification
CRISPR–Cas systems were detected using CRISPRdisco using the 105 annotated genomes as input (30). After identification and manual curation of each CRISPR–Cas locus, the immediate downstream and upstream regions were extracted for each strain to check for conservation of nucleotide sequences. Then, certain strains were selected as representatives for each CRISPR subtype to manually depict the CRISPR loci. The CRISPR spacers were automatically extracted, for each strain and CRISPR subtype, and aligned using CRISPRviz for genotyping purposes (31).
Identification of features of interest
Different features of interest within L. crispatus genomes were investigated using local BLAST v2.7.1. Briefly, a local blastx was performed using previously identified amino acid sequences as queries. The results were hand curated to omit results with <40% identity at amino acid level. Finally, the number of positive hits for each protein was used to depict the heatmaps in RStudio v1.1.463 (22).
The ‘glycogen synthesis’ proteins were screened using proteins identified in L. acidophilus NCFM as database. The ‘glycogen hydrolase’ glgX amino acid sequence in RL03 was used as the database.
The ‘autolysin’ proteins were screened using 11 previously identified autolysins in L. acidophilus NCFM (32).
The ‘trehalose’ operon was screened using three previously identified proteins in L. acidophilus (33).
The ‘EPS’ operon was screened using 16 proteins identified in the strain NCK1350 in this study.
RESULTS AND DISCUSSION
Lactobacillus crispatus pan genome determination
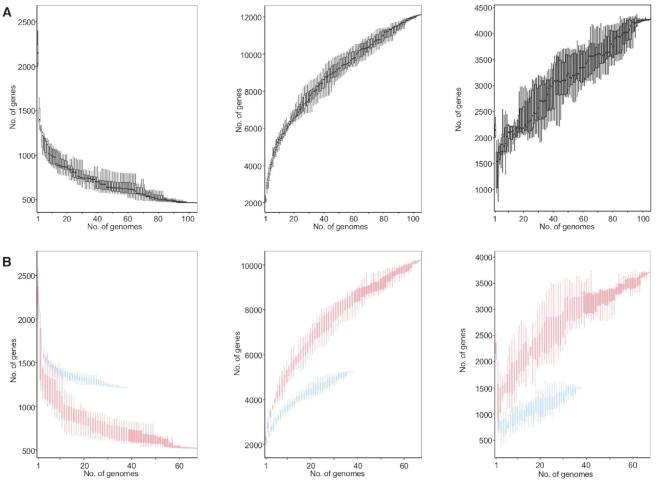
We first determined the pan genome across 105 L. crispatus genomes (Supplementary Table S1), including 88 publicly available genomes at NCBI (March 2019) and 17 newly sequenced from our bacterial collection (NCK series). From this large dataset, 67 of the genomes correspond to human isolates, whereas 13 correspond to chicken isolates, and 25 to turkey isolates (Supplementary Table S1). The initial genome screening did not display significant differences in the size (2.21 ± 0.18 Mb) (mean ± SD) or the GC content (36.95 ± 0.27%) related to the isolation source of the strains (Supplementary Figure S1A and B, Supplementary File S1). The number of genes detected in each genome varies by 9.5% (2144 ± 204.35) (mean ± SD), but no correlation was observed between the number of genes and the number of contigs or the contig length, indicating the genome quality was not an influencing factor in the analyses (Supplementary Figure S1C and D). It is worth noting that the L. crispatus genome is bigger than other well-known vaginal Lactobacillus species such as Lactobacillus gasseri (2 Mb), Lactobacillus iners (1.3 Mb) and Lactobacillus jensenii (1.7 Mb) (34). The comparative genomic analyses resulted in the identification of 12 114 genes that represent the pan genome of the 105 L. crispatus genomes (Figure 1A, middle panel). Moreover, we identified 465 core (conserved) genes that were shared among the strains and 4275 unique genes that are strain specific (Figure 1A, right panel). Both core and pan genomes are significantly larger than those previously reported for smaller datasets of L. crispatus analyses (35). However, the pan genome representation against the number of L. crispatus genomes revealed that it did not reach the plateau, as the addition of the last genome still increased the number of total genes (Figure 1A). According to these data, the pan genome of L. crispatus cannot be considered closed. The number of genomes included in this study is certainly biased by the number of isolates corresponding to each ecological niche of isolation. Then, to further analyze this dataset, we performed pan genome analyses in L. crispatus human isolates and poultry isolates independently, to understand the pan genome size regarding the isolation source and the number of genomes involved. The analyses of L. crispatus human isolates (n = 67) displayed a pan genome that is more complete (red) than the poultry pan genome (n = 38) (blue) (Figure 1B, middle panel), with 10 198 and 5250 genes, respectively. Indeed, the number of unique genes was distinct with 3706 and 1499 unique genes identified for human and poultry isolates, respectively (Figure 1B, right panel). The number of available genomes for each isolation host can partially account for these dramatic differences. However, with a cutoff of 38 genomes, the pan genome size of human L. crispatus isolates (8402 genes) was significantly bigger than the aforementioned poultry pan genome constituted by 5250 genes, partially accounted for by the higher number of unique genes present in human isolates.
Figure 1.
The core and pan genomes of Lactobacillus crispatus. The core genome decreases and the pan genome increases as a function of the number of genomes included in the analysis. (A) The core and pan genomes of 105 L. crispatus genomes. (B) The overlay of the core and pan genomes of L. crispatus human isolates (red, n = 67) and poultry isolates (blue, n = 38).
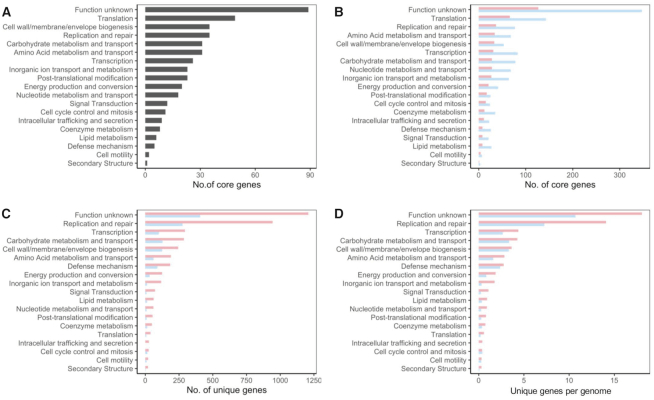
The functional categories of the COGs displayed that, besides the uncharacterized proteins, the majority of the core genes for the 105 strains were related to basic biological function such as translation, replication and repair, and cell wall/envelope biosynthesis, as expected (Figure 2A). When the isolation host (human versus poultry) is considered, the number of core genes in each functional category was different, mainly as a consequence of the number of genomes used for each host (Figure 2B). The unique genes identified during the pan genome analyses drive the differentiation between strains. In this regard, the number of unique genes on each functional category varied among the isolation host, with a higher number of unique genes on the human isolates (Figure 2C). To avoid misinterpretation due to the different number of genomes available for each isolation source, we calculated a ratio that represents the relative number of unique genes per genome (number of unique genes in specific category/number of genomes). Indeed, L. crispatus strains isolated from human origin tend to higher number of unique genes for every single functional category analyzed (Figure 2D), even if there are no differences in genome size (Supplementary Figure S1). The large repertoire of novel genes in L. crispatus indicated by this study along with other studies illustrated an abundance of undiscovered opportunities of applying L. crispatus for human and poultry health.
Figure 2.
Functional category of genes of interest. (A) Number of core genes corresponding to each functional category, out of the 465 core genes shared among the 105 L. crispatus strains. (B) Number of core genes corresponding to each functional category for L. crispatus human isolates (red) and poultry isolates (blue) in independent analyses. (C) Number of unique genes corresponding to each functional category. (D) Ratio of unique genes per genome.
The ecological niche of L. crispatus is relatively restricted to the female lower genital tract and poultry gut, with few exceptions of human isolates from human eye, human gut and human oral cavity. This is the largest comparative genomic analysis for this species to date, but the genomic diversity in L. crispatus has not been fully described. The pan genome for L. crispatus remains open in our analyses; the results indicated that pan genome analyses with larger dataset would not introduce dramatic changes to the pan genome of L. crispatus described here, unless genomes of new L. crispatus strains isolated from novel ecological niches are added.
Phylogenomic analyses
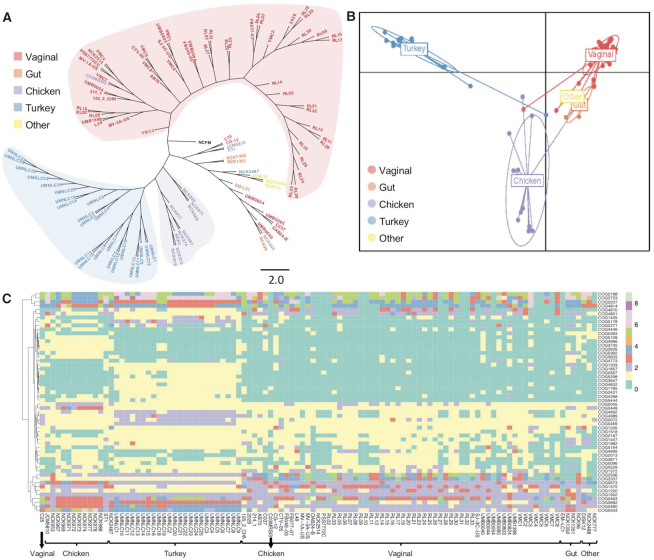
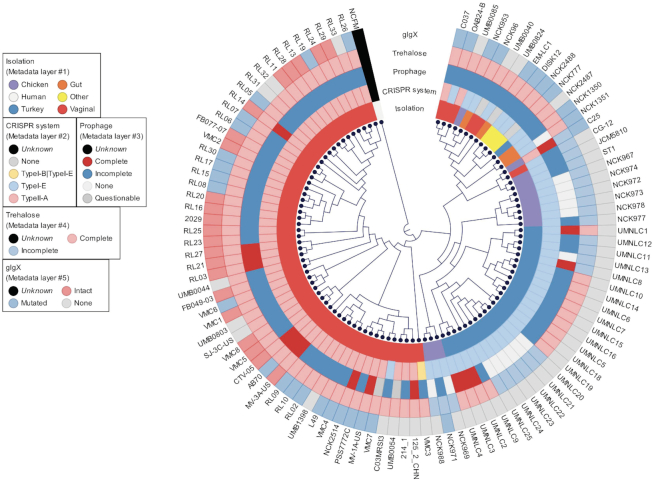
Historically, 16S rRNA gene sequence has been used to identify bacterial species and to perform phylogenetic comparative analyses. However, 16S rRNA provides relatively little or no information to distinguish closely related strains within the same species. Other conserved genes such as glycolysis enzyme(s) have been proposed as alternative for more accurate phylogenetic analyses (36,37). The recent democratization of next-generation sequencing technologies along with the rapid development of bioinformatics tools enables the use of pan genome analyses and thereof core genome-based phylogenomic trees to unearth the taxonomy. The phylogenomic tree based on the 465 core genes of the 105 L. crispatus displayed a clear segregation between the poultry isolates (blue-violet shadow) and the human vaginal isolates (red shadow) (Figure 3A). However, this phylogenomic tree also displayed a nonclearly delimited group with strains belonging to different isolation sources (according to the origin disclosed) combining human vaginal isolates, human gut isolates, and the unique human eye and unique human oral isolate together with two poultry isolates (Figure 3A). This mixed group can be related to cross-contaminations, misannotation on the origin of isolation or that the origin of isolation is not the real ecological niche of the strain. In this regard, Lactobacillus plantarum food isolates clustered closely together and mixed with human fecal isolates, suggesting these human fecal isolates may truly originate from food ingested (38). It is worth noting that phylogenomic analyses have been previously applied to understand the evolutionary history of bacterial isolates and to perform taxonomy analyses focused on strain and species level, as previously reported for commensal bacteria such as Lactobacillus (39), Bifidobacterium (40) and also for the main human pathogens such as Salmonella or Clostridium (41–43), among others. Our results showed that phylogenomic analyses represent a powerful methodology to elucidate the real ecological niche of the strains, being able to differentiate among isolates from closely related niches (e.g. different body sites in human isolates). Understanding the origin of isolation of each strain and their niche-specific adaptation can be of particular relevance for their further applications to improve probiotic efficacy and industrial workhorses. When a comprehensive dataset has been generated, like in this case, new isolates can be included in the pipeline, and based on their clustering on the phylogenomic analyses, their real ecological niche can be elucidated and their performance under certain environments can be predicted. Moreover, comparative genomics, pan genome analysis and phylogenomic analysis could help battle with mislabeling and misidentification of probiotic strains in commercial products, which are common problems in the probiotic industry (44). Indeed, these analyses will also help with a better understanding of microbial pathogens to elucidate their origin and evolution, specially at infection outbreaks.
Figure 3.
Lactobacillus crispatus strains can be distinguished based on phylogenomic tree or functional protein domain analysis. (A) Phylogenomic tree based on the 465 core genes of 105 L. crispatus genomes. (B) Discriminant analysis of principal components using the functional protein domains in L. crispatus. Each dot represents an L. crispatus strain, and the ecological niche is color coded: purple (chicken, n = 13), blue (turkey, n = 25), red (vaginal, n = 60), orange (gut, n = 4) and yellow (other, n = 3). (C) COGs of protein domains that contributed the most (threshold >0.0025) to the group differentiation.
Diverse protein domain occurrence in L. crispatus
Protein function characterization in silico is usually performed by comparing the conserved protein domains against NCBI’s CDD (24). In fact, mere presence or absence of protein domains in sequenced genomes can provide information regarding the phylogeny and evolutionary changes (45). To investigate the diversity of the protein function present in L. crispatus strains and their correlation with the isolation sources, we compared the protein domains from the 105 L. crispatus genomes used in this study. A clear distinction between human and poultry isolates was identified based on the protein functional domains using DAPC (Figure 3B), being consistent with the phylogenomic analysis performed with core genes (Figure 3A). Moreover, the L. crispatus isolates from different human body sites grouped closer and clearly distinct from the turkey and chicken isolates. Previous genomic and protein domain comparison analysis revealed that the functional capacities of microbiota present in the bladder and vaginal tract were distinctively different from gastrointestinal microbiota (46). There were only four human gut isolates included in our analysis. We believe that a larger number of human gut isolates will significantly increase the separation between the vaginal and gut L. crispatus isolates, regarding their protein functions. Nonetheless, the vaginal and gut isolates demonstrated clear separation in protein functions, although not to the extent present between poultry and human strains (Figure 3B). The DAPC analysis revealed that functional capacity of chicken isolates was distinctly different from that of turkey isolates. Chicken and turkey are believed to share similar physiology and similar gut microbiota containing Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria (47). There is an increase, but still scarcity of information regarding poultry microbiome composition, complexity and diversity especially in the case of turkey, despite its key role in animal health and growth efficiencies (48). However, the current common practices in poultry probiotic industry do not distinguish between the species of the birds (49). The differential protein function profile between chicken and turkey gut L. crispatus isolates shown in our DAPC analysis may suggest that the gut environments between these two birds are different and tailored probiotic formulation should be recommended for each individual poultry species for enhanced probiotic efficacy.
Then, a deep analysis was performed to elucidate the main protein functional domains that drive the differentiation among the L. crispatus isolates (Figure 3C, Supplementary File S2). We noticed that chicken isolates contained a few unique COGs related to surface structure encoding a Flp pilus assembly protein CpaB (COG3745), a predicted periplasmic lipoprotein (COG5633) and a Tfp pilus assembly protein PilW (COG4966), which were absent in the rest of L. crispatus strains. These enriched surface protein domains in L. crispatus chicken isolates suggest that they have evolved to develop unique surface structures to interact with and colonize the chicken gut. Likewise, two uncharacterized membrane proteins (COG3647 and COG2431) were mainly found in the turkey isolates, indicating adaptation of turkey L. crispatus to the turkey gut. The surface proteome of the strains is of particular interest since some of the niche-specific COGs driving the differentiation are related to bacterial surface. In a previous study, the noncovalently bound exoproteomes of three L. crispatus strains from different isolation sources were found to be significantly different (50). Defense mechanisms including a Na+-driven multidrug efflux pump (COG0534) and an McrBC 5-methylcytosine restriction system component (COG4268) were also enriched in the poultry isolates. Regarding carbohydrate transport and metabolism, a β-glucosidase (COG2723) that hydrolyzes the glycosidic bonds in β-d-glucosides and oligosaccharides commonly present in gut environment was found to be more enriched in poultry strains and human gut strains. Overall, these results implicated that the gut environment, both human and poultry, seemed to be more competitive and complex than the other niches in which L. crispatus resides, leading gut isolates to acquire unique stress coping mechanisms, surface structure aiding colonization and more diverse carbohydrate metabolism pathways.
Occurrence and diversity of CRISPR–Cas systems in L. crispatus
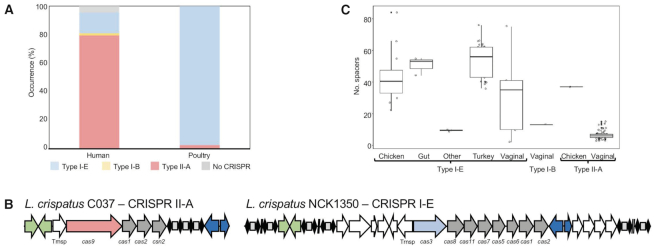
We investigated the occurrence and diversity of L. crispatus CRISPR–Cas systems in the aforementioned 105 genomes and characterized their distribution among the different niches of isolation using CRISPRdisco (30). Overall, we identified CRISPR loci in 97% (102/105) of the genomes and detected different systems including Type I-B (0.95% occurrence), Type I-E (43.8%) and Type II-A (52.4%) (Figure 4A) (Supplementary File S1). This high level of occurrence was previously described by our group in a smaller dataset (51) and it is consistent with the increased number of genomes used in this study. This is a significantly higher prevalence rate compared to the CRISPR occurrence described for other lactic acid bacteria like Lactobacillus (63%), Bifidobacterium (77%) (39,52) and Streptococcus thermophilus (53,54), even more drastically higher compared to the CRISPR distribution in nature, an estimated average of 46% in bacteria (55). As previously described, Type II-A are uniquely present in the human vaginal isolates (with the exception of the chicken isolate C25) of L. crispatus, while Type I systems are present in both human and poultry isolates, being the unique CRISPR subtype found in the poultry isolates (Figure 4). Generally, CRISPR Type I are the most common in nature (50%) with low abundance of Type II systems (56); however, that occurrence rate is biased in L. crispatus genomes due to higher number of vaginal isolates, all presenting subtype II-A systems (Supplementary File S1). Moreover, often flanked by the transposases, the CRISPR loci displayed a highly conserved architecture on the upstream and downstream regions across strains for each subtype, being a conserved feature of this particular species even at nucleotide level (Supplementary Figure S2). The CRISPR spacers represent the vaccination record of the strain and their evolution under selective pressure from invasive DNA, with higher number of spacers related to more predatory attacked by phages or other invasive nucleic acid elements. In L. crispatus, the repeat-spacer array presented variable size among the strains, depending on the CRISPR subtype and the isolation source of the strain. High abundance spacer content was displayed for the strains isolated from the gut, either human or poultry species, and very low spacer abundance for the human vaginal tract isolates with CRISPR subtype II-A but higher for vaginal isolates with CRISPR subtype I-E (Figure 4C). These spacer distributions reflect the high prevalence of phages in the gut environment (57) and also in the urogenital tract. Indeed, other commensal bacterial species isolated from gut environment present high occurrence of CRISPR–Cas systems with high number of spacers like Bifidobacterium longum (58,59). Interestingly, VMC3, a strain isolated from human vaginal tract in bacterial vaginosis state with a higher spacer content, is the unique strain carrying the CRISPR I-B system.
Figure 4.
CRISPR–Cas systems in L. crispatus. (A) Occurrence and diversity of CRISPR–Cas systems in 105 L. crispatus strains, color coded by CRISPR subtype. (B) CRISPR–Cas locus of subtype II-A (left) and subtype I-E (right), with signature cas gene color coded. (C) Box and whisker representation of the number of spacers detected for each CRISPR subtype and isolation source.
Moreover, CRISPR spacers have been used previously for genotyping strains in starter cultures (53), probiotics, commensal bacteria (59,60) and pathogens (61–66), studying the divergence and evolution of the strains among a common ancestor. The spacer content in L. crispatus strains displayed high heterogenicity among the origin of the strains, with highly diverse groups and absence of a common ancestor (Supplementary Figure S3). Interestingly, there is a clear ancestor as reflected by ancestral spacer conservation, for 21 strains, some of them isolated under the same project (Supplementary Figure S3).
Prophage occurrence and distribution among L. crispatus strains
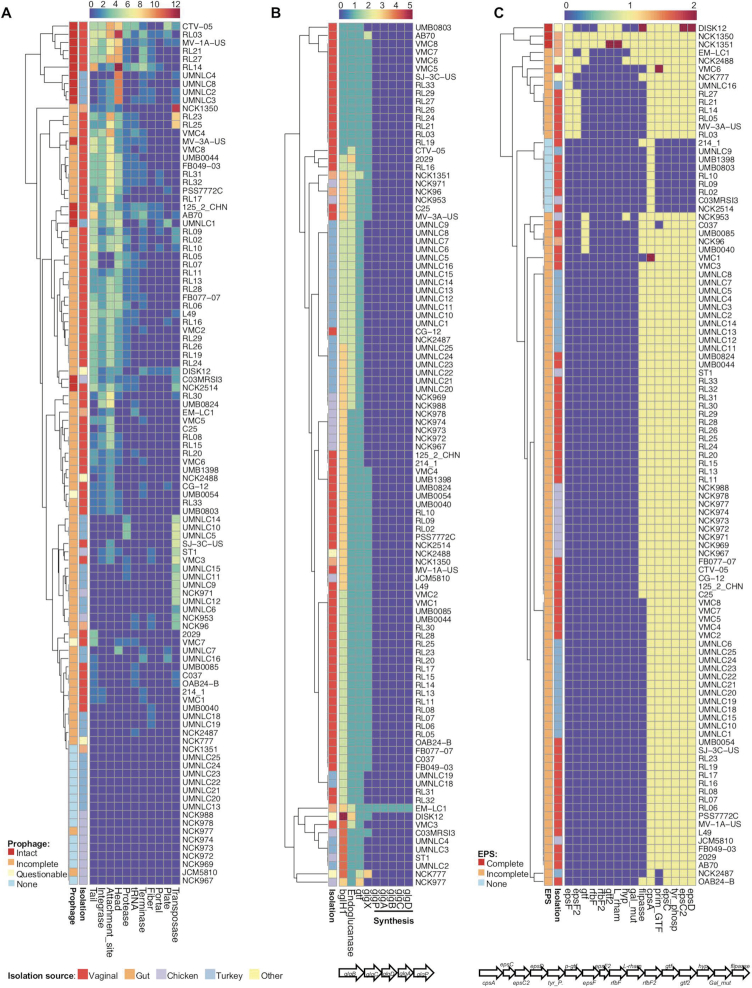
A significant portion of bacterial genomes (can be >20%) is composed of bacteriophage genes that can be either functional or nonfunctional (67). Prophage is the dormant stage of virulent lytic phages that got inserted in the bacterial chromosome and are transmitted to the next generation. Although little is known about the benefit for the bacteria to keep these big structures (even 100 kb) in their chromosomes, it has been illustrated that they may confer antibiotic resistance (68,69) and virulence (70), which can further increase their survivals in specific environmental niches. Prophage sequences were detected in the 90 out of the 105 L. crispatus genomes (Figure 5A, Supplementary File S1). Interestingly, out of the 15 L. crispatus strains that had no detectable prophage regions, only 1 strain (NCK1351) was isolated from human gut with the other 14 corresponding to poultry isolates, suggesting a higher occurrence of prophage in L. crispatus strains from human origin. Moreover, human strains are more likely to contain complete (intact) prophages (including attachment sites, integrase protein, protease protein, phage-like proteins, coat protein, tail protein and transposases) or prophage regions with higher completeness scores (Figure 5A). In this regard, the human gut isolate NCK1350 harbors a functional prophage that excised from the chromosome when induced with mitomycin C (51), although it was predicted as incomplete with the pipeline used in this characterization, reflecting that phenotypic assays need to be performed to verify functionality. An inverse correlation between the abundance of CRISPR spacers and prophage sequences was observed. The L. crispatus genomes with >10 spacers displayed an average of 1.4 prophage regions, whereas the genomes with <10 spacers presented an average of 2.7 prophage regions, as previously reported for other Lactobacillus spp. (39). Domestication of defective prophage genes by the bacteria can aid bacterial niche colonization (71). In this regard, Streptococcus mitis and Enterococcus faecalis platelet binding was enhanced by prophage tail proteins after mitomycin C or UV light treatment (72,73). We suspect that the various prophage structural proteins present in L. crispatus genomes may enhance the bacterial physiology for colonization or adaptation, although further investigations need to be done to unravel this intriguing correlation.
Figure 5.
Distribution of relevant genomic features in 105 L. crispatus strains. (A) Signature prophage proteins. (B) Glycogen hydrolysis (glgX) and glycogen synthesis operon. (C) EPS synthesis operon. The isolation source of each strain is indicated with a side color bar along each heatmap and the legend is on the bottom left side of the figure.
Glycogen metabolism
At the onset of puberty, the microbiota in female lower genital tract typically shifts to a Lactobacillus-dominated state that coincides with a drop in vaginal pH (pH 3.5 to 4.5), a spike in estrogen level and an increase in glycogen concentration in the vaginal epithelial cells (74). An abundance of free glycogen (around 3%) can be detected in female genital fluid, considered as the main carbohydrate that supports the growth of the vaginal microbiota, although controversial opinions exist (75). Therefore, the capability to metabolize glycogen as a carbon source will enhance bacterial competitiveness in the urogenital tract increasing their survival, growth and possible colonization. The glgx gene is responsible for glycogen degradation, debranching the polysaccharide outer chains by selectively hydrolyzing α-1,6-glycosidic linkages of chains containing three or four glucose residues, and has been well characterized in Escherichia coli (76,77). GlgX protein is an extracellularly anchored debranching enzyme with thermostability and higher activity in acidic pH (78), which corresponds to the environment in female lower genital tract. Interestingly, glgX gene was present in 61 out of 67 human strains, but was only detected in 4 chicken strains and zero turkey strain out of the 38 poultry isolates (Figure 5B). The glgX amino acid comparison among L. crispatus strains revealed significant sequence differences among the isolation source of the strains, even within human body sites. Indeed, the vaginal isolates present higher sequence similarity for GlgX protein (at least 99% similarity using Blosum62) compared to human gut or chicken isolates (Supplementary Figure S4). The high occurrence of glgx in L. crispatus vaginal isolates together with the highly conserved sequence, and the low abundance of poultry isolates, clearly represents a niche-specific feature that may enable the strains to metabolize the highly abundant glycogen in the vaginal tract, enhancing their colonization abilities. In this regard, a previous study also reported that growth pattern of L. crispatus on glycogen was correlated with the presence of glgX (pullulanase type I) (79). Moreover, van der Veer and colleagues reported that mutation in the N-terminal sequence, encoding the signal peptide involved in extracellular localization, was associated with poor growth or no growth on glycogen (79).
Perhaps the presence of specific genes involved in the uptake and catabolism of carbohydrates in general and the metabolism of glycogen in particular is correlated with the presence of specific substrates in a particular niche or host. Presumably, L. crispatus strains adapted to the human environment in which glycogen is relatively more abundant than in poultry are incentivized to maintain and/or acquire glycogen-debranching enzymes that could originate from other members of this community. Future metagenomic studies should investigate whether genomic differences might be due to functions encoded by other members of the same community.
Glycogen synthesis has been linked with increased survival and colonization persistence on the gut of certain Lactobacillus species such as L. acidophilus, as it can be used as a reservoir of energy during starvation and stressed conditions (80). The glg operon is responsible for glycogen biosynthesis, and it is known to be associated with certain bacterial species adapted to mammalian hosts (81). However, this operon was not detected in our L. crispatus genomic analysis except in the human gut isolate EM-LC1 (Figure 5B). Interestingly, a deep analysis revealed that this operon was likely acquired from Lactobacillus casei through horizontal transfer, supported by the fact that there is a MobA/MobL-like gene essential for plasmid transfer, upstream the glg operon.
Trehalose metabolism
Supplementation of prebiotics such as trehalose in poultry feed has been linked with multiple health benefits such as pathogens’ inhibition through competitive exclusion and stimulation of host immune system (82). Moreover, the capability to utilize a wide variety of carbon sources increases the adaptation and growth abilities of bacterial strains. The trehalose hydrolase operon in L. acidophilus NCFM encompasses a transcriptional regulator coupled with a trehalose-6 phosphate hydrolase in frame under the same promoter and a trehalose-specific phosphoenolpyruvate transferase system transporter (PTS) that is in the opposite direction as a part of the locus (33). Interestingly, our results showed that the presence of the trehalose operon in L. crispatus is strain dependent and directly related to the origin of isolation. In this regard, all the L. crispatus human isolates (gut and vaginal) displayed a complete operon (except vaginal isolates C25 and CG-12), whereas none of the chicken isolates presented a complete operon (unless NCK953 and C03MRSI3) (Supplementary Figure S5A). The turkey isolates present a mixed population where some of the isolates displayed a complete cluster and others lack the regulator or the regulator and the PTS. Then, the presence of trehalose operon is widely distributed among the L. crispatus vaginal isolates, being more absent in the poultry isolates; however, trehalose has not been described as a carbon source on the human urogenital tract. Nevertheless, the presence of the trehalose operon suggests the capability of the strains to internalize trehalose through the PTS system, releasing trehalose-6-phosphate into the cell for further hydrolyzation with the trehalose-6 phosphate hydrolase enzyme into glucose and glucose-6-phosphate that can be used as substrate for the glycolysis (83). The presence of this genetic feature will increase the wide range of carbohydrate sources that the strains will be able to metabolize favoring their growth and adaption to different ecological niches.
EPS cluster
EPSs are carbohydrate polymers present in the outer layer of bacteria belonging to a wide range of species and genus. EPS-producing strains have received special attention due to their industrial and medical applications (84). Moreover, the EPSs play a key role in the cross-talk interaction with the host being able to increase the colonization and modulate the immune system, while providing protection for the bacteria under stress conditions (85). Interestingly, among the 105 L. crispatus genomes, the human gut isolates NCK1350 and NCK1351 were the only strains displaying a complete eps cluster constituted by 16 genes. This eps operon includes the priming glycosyltransferase (p-gtf), involved in the starting step of EPS synthesis, glycosyltransferases, flipase, tyrosine kinase (epsC-D), tyrosine phosphatase, capsular polysaccharide gene (cpsA), rhamnose and membrane transporters, among others. However, most of the genomes screened (91/105) displayed an incomplete eps cluster as they harbored the p-gtf gene, together with other genes but not a complete cluster. Moreover, 12 strains did not present the required p-gtf, neither the others require EPS genes (Figure 5C). Nonetheless, truncated or incomplete eps clusters seem to be widely distributed among the 105 L. crispatus genomes, which correlates with the abundance of transposase and mobile elements flanking the complete eps locus of NCK1350, suggesting a potential acquisition through horizontal gene transfer. Up to date, there is a unique reference in the literature where EPS has been isolated from L. crispatus L1, a human vaginal isolate, able to counteract the adherence of Candida albicans (17) although the genome was not included in this study as it not was not publicly available.
Autolysins
Autolysins are enzymes responsible for cell division and separation by cleaving peptidoglycan, and are promising targets to develop novel antibiotics due to their cleavage activity (86). Autolysin has been shown to be involved in pathogenesis of Listeria monocytogenes and Staphylococcus epidermidis as a virulence factor for invasion and niche colonization (86,87). However, how autolysins assist with colonization of commensal bacteria has not been looked at extensively. Autolysins are commonly bound to the cell wall through noncovalent association in S-layer-forming Lactobacillus species such as L. acidophilus. Using the 11 predicted autolysins from L. acidophilus NCFM as reference, we observed heterogenicity in the presence of autolysins in L. crispatus isolates, with no clear association with the isolation source of the strain (Supplementary Figure S5B). It is worth noting that the human isolates tend to have a higher abundance of autolysin proteins than the poultry isolates, especially regarding the autolysins LBA1351 (containing a β-N-acetylmuramidase), AcmB (containing a β-N-acetylglucosaminidase) and LBA1140 (containing a β-N-acetylmuramidase) (Supplementary Figure S5B). AcmB has been linked with binding capacity of L. acidophilus NCFM to a number of extracellular matrices such as mucin, fibronectin, laminin and collagen in vitro (32) and therefore it may play a key role in L. crispatus adherence and colonization. The differential profile of autolysins likely suggests that human and poultry L. crispatus isolates interact distinctly with their native environments with different binding capacities, which can be related to the strong differences among ecological environments and tissue cell properties, between human and poultry species, and gut and vaginal niches.
CONCLUSIONS
Recent advances in next-generation sequencing have rapidly increased the availability of bacterial genomes, providing a vast amount of information for genetic content of bacteria of interest. In this study, we conducted a comprehensive comparative genomic analysis using 105 L. crispatus strains, isolated from either humans or poultry, to characterize their niche-specific adaptation based on pan genome analyses along with other genetic features. The core and pan genomes described here are significantly larger than those previously reported in smaller datasets (comparative genomics of L. crispatus suggests novel mechanisms for the competitive exclusion of Gardnerella vaginalis) displaying the need of larger amount of data to obtain concise conclusions. The phylogenomic analyses performed with core genes and the functional protein domains displayed a clear separation of the strain regarding their isolation source (Figure 3). Moreover, strains isolated from same host (human) but different body sites (gut versus vaginal) tend to cluster separately. It is worth noting that this niche-specific adaptation was also observed for some of the genetic features analyzed, like the CRISPR system subtype, the glycogen hydrolase (glgx) and trehalose operon (Figure 6). Nevertheless, this is the largest comparative genomic analysis for this species to date, although the genomic diversity in L. crispatus has not been fully described, as the number of samples was biased by the isolation source.
Figure 6.
Phylogenomic tree of 105 L. crispatus genomes with genetic features of interest. Phylogenomic tree was created based on the 465 core genes of L. crispatus (rooted with L. acidophilus NCFM as outgroup. NCFM was not included in the genetic features analyses). The genetic features of interest are displayed as different layers and color coded based on their occurrence and completeness.
Overall, this study provided new insights into the genomic content and variability of L. crispatus, which shed light on its genetic and functional attributes, with potential applications as probiotics for human and poultry health. Presumably, fundamental differences in the genetic content can translate to a better performance of specific strains in a particular ecological niche, increasing survival, colonization and functionalities. Indeed, the diverse genetic content, which varies and correlates with isolation source, is useful for rational design and formulation of probiotics and industrial workhorses, for host and body site-specific applications.
DATA AVAILABILITY
The chromosomal genome sequences of the newly sequenced L. crispatus strains reported in this manuscript have been deposited in the NCBI database under the BioProject ID PRJNA563077. The accession numbers for each strain are shown in Supplementary Table S1.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank their colleagues, especially Yong Jun Goh for her insights and Stefanie Andersen for bioinformatic support.
Author contributions: M.P. and C.H.-C. designed the study, performed bioinformatic analyses and participated in the manuscript writing. R.B. coordinated and supervised the study, and edited the manuscript. All authors approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
NC State University; DuPont; North Carolina Agricultural Foundation.
Conflict of interest statement. R.B. is a co-inventor on several patents related to probiotics and their uses. He is also a shareholder of Caribou Biosciences, Intellia Therapeutics, Locus Biosciences and Inari Ag, and a co-founder of Intellia Therapeutics and Locus Biosciences. C.H.-C. is an advisor and co-founder of Microviable Therapeutics.
REFERENCES
- 1. Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012; 486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Integrative HMP Research Network Consortium The Integrative Human Microbiome Project: dynamic analysis of microbiome–host omics profiles during periods of human health and disease. Cell Host Microbe. 2014; 16:276–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marchesi J.R., Ravel J.. The vocabulary of microbiome research: a proposal. Microbiome. 2015; 3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tremaroli V., Backhed F.. Functional interactions between the gut microbiota and host metabolism. Nature. 2012; 489:242–249. [DOI] [PubMed] [Google Scholar]
- 5. Sommer F., Backhed F.. The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 2013; 11:227–238. [DOI] [PubMed] [Google Scholar]
- 6. Palm N.W., de Zoete M.R., Flavell R.A.. Immune–microbiota interactions in health and disease. Clin. Immunol. 2015; 159:122–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Planer J.D., Peng Y., Kau A.L., Blanton L.V., Ndao I.M., Tarr P.I., Warner B.B., Gordon J.I.. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature. 2016; 534:263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mangiola F., Ianiro G., Franceschi F., Fagiuoli S., Gasbarrini G., Gasbarrini A.. Gut microbiota in autism and mood disorders. World J. Gastroenterol. 2016; 22:361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A.L.. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019; 570:462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y.E., Fischbach M.A., Belkaid Y.. Skin microbiota–host interactions. Nature. 2018; 553:427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jia G., Zhi A., Lai P.F.H., Wang G., Xia Y., Xiong Z., Zhang H., Che N., Ai L.. The oral microbiota—a mechanistic role for systemic diseases. Br. Dent. J. 2018; 224:447–455. [DOI] [PubMed] [Google Scholar]
- 12. Ravel J., Gajer P., Abdo Z., Schneider G.M., Koenig S.S., McCulle S.L., Karlebach S., Gorle R., Russell J., Tacket C.O. et al.. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:4680–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wei S., Morrison M., Yu Z.. Bacterial census of poultry intestinal microbiome. Poult. Sci. 2013; 92:671–683. [DOI] [PubMed] [Google Scholar]
- 14. Dec M., Nowaczek A., Stepien-Pysniak D., Wawrzykowski J., Urban-Chmiel R.. Identification and antibiotic susceptibility of lactobacilli isolated from turkeys. BMC Microbiol. 2018; 18:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu M.B., Xu S.R., He Y., Deng G.H., Sheng H.F., Huang X.M., Ouyang C.Y., Zhou H.W.. Diverse vaginal microbiomes in reproductive-age women with vulvovaginal candidiasis. PLoS One. 2013; 8:e79812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arokiyaraj S., Seo S.S., Kwon M., Lee J.K., Kim M.K.. Association of cervical microbial community with persistence, clearance and negativity of human papillomavirus in Korean women: a longitudinal study. Sci. Rep. 2018; 8:15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Donnarumma G., Molinaro A., Cimini D., De Castro C., Valli V., De Gregorio V., De Rosa M., Schiraldi C.. Lactobacillus crispatus L1: high cell density cultivation and exopolysaccharide structure characterization to highlight potentially beneficial effects against vaginal pathogens. BMC Microbiol. 2014; 14:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nardini P., Nahui Palomino R.A., Parolin C., Laghi L., Foschi C., Cevenini R., Vitali B., Marangoni A.. Lactobacillus crispatus inhibits the infectivity of Chlamydia trachomatis elementary bodies, in vitro study. Sci. Rep. 2016; 6:29024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T., Fookes M., Falush D., Keane J.A., Parkhill J.. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015; 31:3691–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014; 30:2068–2069. [DOI] [PubMed] [Google Scholar]
- 21. Huerta-Cepas J., Szklarczyk D., Heller D., Hernandez-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen L.J. et al.. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019; 47:D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. RStudio Team 2015; RStudio: integrated development environment for R. RStudio, Boston, MA.
- 23. Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. et al.. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 24. Marchler-Bauer A., Anderson J.B., Derbyshire M.K., DeWeese-Scott C., Gonzales N.R., Gwadz M., Hao L.N., He S.Q., Hurwitz D.I., Jackson J.D. et al.. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007; 35:D237–D240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V.. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000; 28:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jombart T., Devillard S., Balloux F.. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010; 11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008; 24:1403–1405. [DOI] [PubMed] [Google Scholar]
- 28. Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D.S.. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016; 44:W16–W21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arndt D., Marcu A., Liang Y., Wishart D.S.. PHAST, PHASTER and PHASTEST: tools for finding prophage in bacterial genomes. Brief. Bioinform. 2017; 20:1560–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crawley A.B., Henriksen J.R., Barrangou R.. CRISPRdisco: an automated pipeline for the discovery and analysis of CRISPR–Cas systems. CRISPR J. 2018; 1:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nethery M.A., Barrangou R.. CRISPR Visualizer: rapid identification and visualization of CRISPR loci via an automated high-throughput processing pipeline. RNA Biol. 2019; 16:577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson B.R., Klaenhammer T.R.. AcmB is an S-layer-associated beta-N-acetylglucosaminidase and functional autolysin in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2016; 82:5687–5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duong T., Barrangou R., Russell W.M., Klaenhammer T.R.. Characterization of the tre locus and analysis of trehalose cryoprotection in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2006; 72:1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mendes-Soares H., Suzuki H., Hickey R.J., Forney L.J.. Comparative functional genomics of Lactobacillus spp. reveals possible mechanisms for specialization of vaginal lactobacilli to their environment. J. Bacteriol. 2014; 196:1458–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ojala T., Kankainen M., Castro J., Cerca N., Edelman S., Westerlund-Wikstrom B., Paulin L., Holm L., Auvinen P.. Comparative genomics of Lactobacillus crispatus suggests novel mechanisms for the competitive exclusion of Gardnerella vaginalis. BMC Genomics. 2014; 15:1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brandt K., Barrangou R.. Phylogenetic analysis of the Bifidobacterium genus using glycolysis enzyme sequences. Front. Microbiol. 2016; 7:657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brandt K., Barrangou R.. Using glycolysis enzyme sequences to inform Lactobacillus phylogeny. Microb. Genom. 2018; 4:doi:10.1099/mgen.0.000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siezen R.J., Tzeneva V.A., Castioni A., Wels M., Phan H.T., Rademaker J.L., Starrenburg M.J., Kleerebezem M., Molenaar D., van Hylckama Vlieg J.E.. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ. Microbiol. 2010; 12:758–773. [DOI] [PubMed] [Google Scholar]
- 39. Sun Z., Harris H.M., McCann A., Guo C., Argimon S., Zhang W., Yang X., Jeffery I.B., Cooney J.C., Kagawa T.F. et al.. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015; 6:8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lugli G.A., Milani C., Duranti S., Mancabelli L., Mangifesta M., Turroni F., Viappiani A., van Sinderen D., Ventura M.. Tracking the taxonomy of the genus Bifidobacterium based on a phylogenomic approach. Appl. Environ. Microbiol. 2018; 84:e02249-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chapeton-Montes D., Plourde L., Bouchier C., Ma L., Diancourt L., Criscuolo A., Popoff M.R., Bruggemann H.. The population structure of Clostridium tetani deduced from its pan-genome. Sci. Rep. 2019; 9:11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Knight D.R., Kullin B., Androga G.O., Barbut F., Eckert C., Johnson S., Spigaglia P., Tateda K., Tsai P.J., Riley T.V.. Evolutionary and genomic insights into Clostridioides difficile sequence Type 11: a diverse zoonotic and antimicrobial-resistant lineage of global one health importance. mBio. 2019; 10:e00446-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Laing C.R., Whiteside M.D., Gannon V.P.J.. Pan-genome analyses of the species Salmonella enterica, and identification of genomic markers predictive for species, subspecies, and serovar. Front. Microbiol. 2017; 8:1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morovic W., Hibberd A.A., Zabel B., Barrangou R., Stahl B.. Genotyping by PCR and high-throughput sequencing of commercial probiotic products reveals composition biases. Front. Microbiol. 2016; 7:1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang S., Doolittle R.F., Bourne P.E.. Phylogeny determined by protein domain content. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomas-White K., Forster S.C., Kumar N., Van Kuiken M., Putonti C., Stares M.D., Hilt E.E., Price T.K., Wolfe A.J., Lawley T.D.. Culturing of female bladder bacteria reveals an interconnected urogenital microbiota. Nat. Commun. 2018; 9:1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilkinson T.J., Cowan A.A., Vallin H.E., Onime L.A., Oyama L.B., Cameron S.J., Gonot C., Moorby J.M., Waddams K., Theobald V.J. et al.. Characterization of the microbiome along the gastrointestinal tract of growing turkeys. Front. Microbiol. 2017; 8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Danzeisen J.L., Calvert A.J., Noll S.L., McComb B., Sherwood J.S., Logue C.M., Johnson T.J.. Succession of the turkey gastrointestinal bacterial microbiome related to weight gain. Peer J. 2013; 1:e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kabir S.M.L. The role of probiotics in the poultry industry. Int. J. Mol. Sci. 2009; 10:3531–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Johnson B.R., Hymes J., Sanozky-Dawes R., Henriksen E.D., Barrangou R., Klaenhammer T.R.. Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming lactobacilli. Appl. Environ. Microbiol. 2016; 82:134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hidalgo-Cantabrana C., Goh Y.J., Pan M., Sanozky-Dawes R., Barrangou R.. Genome editing using the endogenous type I CRISPR–Cas system in Lactobacillus crispatus. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:15774–15783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Briner A.E., Lugli G.A., Milani C., Duranti S., Turroni F., Gueimonde M., Margolles A., van Sinderen D., Ventura M., Barrangou R.. Occurrence and diversity of CRISPR–Cas systems in the genus Bifidobacterium. PLoS One. 2015; 10:e0133661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Horvath P., Romero D.A., Coute-Monvoisin A.C., Richards M., Deveau H., Moineau S., Boyaval P., Fremaux C., Barrangou R.. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008; 190:1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Magadan A.H., Dupuis M.E., Villion M., Moineau S.. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3–Cas system. PLoS One. 2012; 7:e40913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grissa I., Vergnaud G., Pourcel C.. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007; 35:W52–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Koonin E.V., Makarova K.S., Zhang F.. Diversity, classification and evolution of CRISPR–Cas systems. Curr. Opin. Microbiol. 2017; 37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stern A., Mick E., Tirosh I., Sagy O., Sorek R.. CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome. Genome Res. 2012; 22:1985–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gogleva A.A., Gelfand M.S., Artamonova I.I.. Comparative analysis of CRISPR cassettes from the human gut metagenomic contigs. BMC Genomics. 2014; 15:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hidalgo-Cantabrana C., Crawley A.B., Sanchez B., Barrangou R.. Characterization and exploitation of CRISPR loci in Bifidobacterium longum. Front. Microbiol. 2017; 8:1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Horvath P., Coute-Monvoisin A.C., Romero D.A., Boyaval P., Fremaux C., Barrangou R.. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 2009; 131:62–70. [DOI] [PubMed] [Google Scholar]
- 61. Andersen J.M., Shoup M., Robinson C., Britton R., Olsen K.E., Barrangou R.. CRISPR diversity and microevolution in Clostridium difficile. Genome Biol. Evol. 2016; 8:2841–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Freidlin P.J., Nissan I., Luria A., Goldblatt D., Schaffer L., Kaidar-Shwartz H., Chemtob D., Dveyrin Z., Head S.R., Rorman E.. Structure and variation of CRISPR and CRISPR-flanking regions in deleted-direct repeat region Mycobacterium tuberculosis complex strains. BMC Genomics. 2017; 18:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sola C., Abadia E., Le Hello S., Weill F.X.. High-throughput CRISPR typing of Mycobacterium tuberculosis complex and Salmonella enterica serotype Typhimurium. Methods Mol. Biol. 2015; 1311:91–109. [DOI] [PubMed] [Google Scholar]
- 64. Sun H., Li Y., Shi X., Lin Y., Qiu Y., Zhang J., Liu Y., Jiang M., Zhang Z., Chen Q. et al.. Association of CRISPR/Cas evolution with Vibrio parahaemolyticus virulence factors and genotypes. Foodborne Pathog. Dis. 2015; 12:68–73. [DOI] [PubMed] [Google Scholar]
- 65. Xie X., Hu Y., Xu Y., Yin K., Li Y., Chen Y., Xia J., Xu L., Liu Z., Geng S. et al.. Genetic analysis of Salmonella enterica serovar Gallinarum biovar Pullorum based on characterization and evolution of CRISPR sequence. Vet. Microbiol. 2017; 203:81–87. [DOI] [PubMed] [Google Scholar]
- 66. Xu X.Q., Xin Y.Q., Li X., Zhang Q.W., Yang X.Y., Jin Y., Zhao H.H., Jin X., Qi Z.Z.. [Genotyping by CRISPR and regional distribution of Yersinia pestis in Qinghai-plateau from 1954 to 2011]. Zhonghua Yu Fang Yi Xue Za Zhi. 2017; 51:237–242. [DOI] [PubMed] [Google Scholar]
- 67. Casjens S. Prophages and bacterial genomics: what have we learned so far. Mol. Microbiol. 2003; 49:277–300. [DOI] [PubMed] [Google Scholar]
- 68. Wang X., Wood T.K.. Cryptic prophages as targets for drug development. Drug Resist. Updat. 2016; 27:30–38. [DOI] [PubMed] [Google Scholar]
- 69. Wang X., Kim Y., Ma Q., Hong S.H., Pokusaeva K., Sturino J.M., Wood T.K.. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010; 1:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fortier L.C., Sekulovic O.. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013; 4:354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bobay L.M., Touchon M., Rocha E.P.. Pervasive domestication of defective prophages by bacteria. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:12127–12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bensing B.A., Siboo I.R., Sullam P.M.. Proteins PblA and PblB of Streptococcus mitis, which promote binding to human platelets, are encoded within a lysogenic bacteriophage. Infect. Immun. 2001; 69:6186–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Matos R.C., Lapaque N., Rigottier-Gois L., Debarbieux L., Meylheuc T., Gonzalez-Zorn B., Repoila F., Lopes Mde F., Serror P.. Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet. 2013; 9:e1003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hickey R.J., Zhou X., Settles M.L., Erb J., Malone K., Hansmann M.A., Shew M.L., Van Der Pol B., Fortenberry J.D., Forney L.J.. Vaginal microbiota of adolescent girls prior to the onset of menarche resemble those of reproductive-age women. mBio. 2015; 6:e00097-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mirmonsef P., Hotton A.L., Gilbert D., Burgad D., Landay A., Weber K.M., Cohen M., Ravel J., Spear G.T.. Free glycogen in vaginal fluids is associated with Lactobacillus colonization and low vaginal pH. PLoS One. 2014; 9:e102467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dauvillee D., Kinderf I.S., Li Z., Kosar-Hashemi B., Samuel M.S., Rampling L., Ball S., Morell M.K.. Role of the Escherichia coliglgX gene in glycogen metabolism. J. Bacteriol. 2005; 187:1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Song H.N., Jung T.Y., Park J.T., Park B.C., Myung P.K., Boos W., Woo E.J., Park K.H.. Structural rationale for the short branched substrate specificity of the glycogen debranching enzyme GlgX. Proteins. 2010; 78:1847–1855. [DOI] [PubMed] [Google Scholar]
- 78. Moller M.S., Goh Y.J., Rasmussen K.B., Cypryk W., Celebioglu H.U., Klaenhammer T.R., Svensson B., Abou Hachem M.. An extracellular cell-attached pullulanase confers branched alpha-glucan utilization in human gut Lactobacillus acidophilus. Appl. Environ. Microbiol. 2017; 83:e00402-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. van der Veer C., Hertzberger R.Y., Bruisten S.M., Tytgat H.L.P., Swanenburg J., de Kat Angelino-Bart A., Schuren F., Molenaar D., Reid G., de Vries H. et al.. Comparative genomics of human Lactobacillus crispatus isolates reveals genes for glycosylation and glycogen degradation: implications for in vivo dominance of the vaginal microbiota. Microbiome. 2019; 7:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Goh Y.J., Klaenhammer T.R.. Insights into glycogen metabolism in Lactobacillus acidophilus: impact on carbohydrate metabolism, stress tolerance and gut retention. Microb. Cell Fact. 2014; 13:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Goh Y.J., Klaenhammer T.R.. A functional glycogen biosynthesis pathway in Lactobacillus acidophilus: expression and analysis of the glg operon. Mol. Microbiol. 2013; 89:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kikusato M., Nanto F., Mukai K., Toyomizu M.. Effects of trehalose supplementation on the growth performance and intestinal innate immunity of juvenile chicks. Br. Poult. Sci. 2016; 57:375–380. [DOI] [PubMed] [Google Scholar]
- 83. Steen J.A., Bohlke N., Vickers C.E., Nielsen L.K.. The trehalose phosphotransferase system (PTS) in E. coli W can transport low levels of sucrose that are sufficient to facilitate induction of the csc sucrose catabolism operon. PLoS One. 2014; 9:e88688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Inturri R., Molinaro A., Di Lorenzo F., Blandino G., Tomasello B., Hidalgo-Cantabrana C., De Castro C., Ruas-Madiedo P.. Chemical and biological properties of the novel exopolysaccharide produced by a probiotic strain of Bifidobacterium longum. Carbohydr. Polym. 2017; 174:1172–1180. [DOI] [PubMed] [Google Scholar]
- 85. Hidalgo-Cantabrana C., Sanchez B., Milani C., Ventura M., Margolles A., Ruas-Madiedo P.. Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl. Environ. Microbiol. 2014; 80:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Asano K., Sashinami H., Osanai A., Asano Y., Nakane A.. Autolysin amidase of Listeria monocytogenes promotes efficient colonization of mouse hepatocytes and enhances host immune response. Int. J. Med. Microbiol. 2011; 301:480–487. [DOI] [PubMed] [Google Scholar]
- 87. Kohler T.P., Gisch N., Binsker U., Schlag M., Darm K., Volker U., Zahringer U., Hammerschmidt S.. Repeating structures of the major staphylococcal autolysin are essential for the interaction with human thrombospondin 1 and vitronectin. J. Biol. Chem. 2014; 289:4070–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The chromosomal genome sequences of the newly sequenced L. crispatus strains reported in this manuscript have been deposited in the NCBI database under the BioProject ID PRJNA563077. The accession numbers for each strain are shown in Supplementary Table S1.