Abstract
In recent years, eukaryotic long non-coding RNAs (lncRNAs) have been identified as important factors involved in a wide variety of biological processes, including histone modification, alternative splicing and transcription enhancement. The expression of lncRNAs is highly tissue-specific and is regulated by environmental stresses. Recently, a large number of plant lncRNAs have been identified, but very few of them have been studied in detail. Furthermore, the mechanism of lncRNA expression regulation remains largely unknown. Arabidopsis HISTONE DEACETYLASE 6 (HDA6) and LSD1-LIKE 1/2 (LDL1/2) can repress gene expression synergistically by regulating H3Ac/H3K4me. In this research, we performed RNA-seq and ChIP-seq analyses to further clarify the function of HDA6-LDL1/2. Our results indicated that the global expression of lncRNAs is increased in hda6/ldl1/2 and that this increased lncRNA expression is particularly associated with H3Ac/H3K4me2 changes. In addition, we found that HDA6-LDL1/2 is important for repressing lncRNAs that are non-expressed or show low-expression, which may be strongly associated with plant development. GO-enrichment analysis also revealed that the neighboring genes of the lncRNAs that are upregulated in hda6/ldl1/2 are associated with various developmental processes. Collectively, our results revealed that the expression of lncRNAs is associated with H3Ac/H3K4me2 changes regulated by the HDA6-LDL1/2 histone modification complex.
INTRODUCTION
High-throughput sequencing technologies have been widely used in the transcriptome analysis of eukaryotic systems. In previous studies, only ∼2% of eukaryotic genomes can be considered protein-coding regions. Many remaining transcripts with no protein-coding capacity are referred to as non-coding RNAs (ncRNAs) (1,2). Short-non-coding RNAs (<200 bp) comprise diverse groups, including transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), microRNA (miRNA) and small interfering RNA (siRNA). In recent years, long non-coding RNAs (lncRNAs, longer than 200 bp) have been identified as important regulators involved in diverse processes, including chromatin remodeling, histone modification, alternative splicing and transcription activation (3–7). Generally, the expression of lncRNAs is lower than that of protein-coding genes and tends to be more tissue specific (8–12). In the human genome, only ∼11–29% of lncRNAs are ubiquitously expressed in all tissues, compared to 65% of protein-coding genes (1,13–14). Previous research has also revealed that the expression of plant lncRNAs is lower than that of animal lncRNAs (10). High diversity of lncRNA expression in different tissues and under different growth conditions has also been observed in plants (10,15).
LncRNAs have been identified as being associated with cancer and other diseases and are involved in various developmental processes in animals (4,8–9,11). In plants, it has been reported that lncRNAs are involved in root, flower and fruit development, light responses, vernalization, auxin transport and stress responses (15–29). For example, the Arabidopsis lncRNAs cold-induced long antisense intragenic RNA (COOLAIR) and cold-assisted intronic non-coding RNA (COLDAIR) are both transcribed from flowering locus C (FLC) and can be induced by cold treatment. Both COOLAIR and COLDAIR can repress the floral repressor FLC by interacting with Polycomb Repressive Complex 2 (PRC2) to increase the histone H3 lysine 27 trimethylation (H3K27me3) level of FLC (24–29). Another Arabidopsis lncRNA, auxin-regulated promoter loop (APOLO), can be activated by auxin and regulates the formation of a chromatin loop encompassing the promoter of the neighboring gene Pinoid (PID), which encodes an essential regulator of auxin polar transport (30). Although a large number of lncRNAs have been identified in Arabidopsis, very few of them have been studied in detail. It was found that decreased FLC expression is associated with an increased level of the repression marker H3K27me3 at the FLC locus during vernalization. In contrast, although H3K27me3 is increased near the chromatin locations of COOLAIR and COLDAIR during vernalization, their expression is not decreased but is increased (26–28), implying that H3K27me3 may function differently in FLC and COOLAIR/COLDAIR. In humans, it has been suggested that the general expression pattern of lncRNAs is not sensitive to changes in DNA methylation and H3K9me2 (31). These results suggest that the expression regulation of lncRNAs may be different from that of protein-coding genes. However, the mechanism of lncRNA expression regulation is rarely discussed and remains elusive in plants.
In yeast and animal systems, histone deacetylases (HDACs or HDAs) and lysine-specific demethylase 1 (LSD1) are the core components of several multiprotein HDAC complexes, such as Mi2/NuRD and CoREST. Class I HDACs and LSD1 act as the core factors in these complexes to repress gene expression by removing the methylation of histone H3 lysine 4 (H3K4me) and histone acetylation, both of which are active histone marks for transcription (32–36). Arabidopsis Histone Deacetylase 6 (HDA6) is an RPD3-like class I HDAC involved in various transcriptional regulation processes, including transposon repression and the regulation of ribosomal RNA (37–42). Furthermore, HDA6 is involved in flowering, leaf development, senescence, abiotic stress responses and circadian rhythms (43–47). There are four LSD1 homologs in Arabidopsis: LSD1-LIKE 1 (LDL1), LDL2, LDL3 and Flowering Locus D (FLD) (48). LDL1 and LDL2 act redundantly to repress the expression of FLC by H3K4 demethylation. ldl1/ldl2 double mutants also show reduced DNA methylation at the FWA locus, which represses the floral transition (48,49). Our recent studies have demonstrated that Arabidopsis HDA6 directly interacts with LDL1 and LDL2 in the same protein complex (46) and that they function synergistically to regulate histone H3 acetylation (H3Ac) and H3K4me in the core circadian clock genes (46,47).
In this research, we further analyzed the functions of HDA6 and LDL1 by characterizing the global H3Ac/H3K4me2 profiles and transcriptomes of wild-type (WT), hda6, ldl1/ldl2 (ldl1/2) and hda6/ldl1/ldl2 (hda6/ldl1/2). Furthermore, we compared the genome-wide binding of HDA6 and LDL1 with these global H3Ac/H3K4me2 profiles and transcriptomes. Our results provide further details about the function of the Arabidopsis HDA6-LDL1/2 complex and reveal its involvement in the regulation of lncRNAs.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana) was grown in growth chambers under 12/12 h light/dark conditions at 22°C. In this study, the WT Arabidopsis Columbia (Col-0) ecotype was used. The mutants used in this research, including ldl1/ldl2 (48), hda6 (axe1-5) (45), hda6/ldl1/2, LDL1pro::LDL1:GFP/ldl1 and HDA6pro::HDA6:GFP/hda6 (46), were previously described.
RNA-seq analyses
For genome-wide expression analysis, RNA from 14-day-old seedlings of WT, hda6, ldl1/2 and hda6/ldl1/2 plants was isolated using the AllPure Plant RNA Purification Kit (Allbio, ABTGNA020-50) and treated with RNase-free DNase. RNA from at least three biological replicates was sequenced separately. Sequencing libraries were built using the Illumina TruSeq RNA library preparation protocol. The libraries were sequenced on the Illumina HiSeq 2500 platform using a paired-end scheme (2 × 150 bp) with TruSeq v3 chemistry. Reads were mapped to the TAIR10 Arabidopsis genome (50) using Qiagen CLC Genomics Workbench (QIAGEN, https://www.qiagenbioinformatics.com) with the default settings. The procedure of CLC Genomics Workbench data-processing was listed as following: Step 1, import fastaq.gz RAW data of the sequenced reads by Standard Import tool. Step 2, generate metadata to match samples and groups. Step 3, mapping reads to TAIR10 Arabidopsis genome (50) by RNA-Seq Analysis tool with default settings. Step 4, compare the reads with each gene between groups by Differential Expression for RNA-Seq tool with default settings. Step 5, export the computed reads and relative gene expression levels for further analysis. Genes exhibiting at least a 1.5-fold change in expression with a P-value < 0.05 were considered to be differentially expressed (Supplementary Table S1). The computed reads of all genes and the random selected coding gene lists were also listed in Supplementary Table S2 and 3.
To generate strand-specific bigWig files for visualization with the Integrated Genome Viewer (IGV) (51), raw sequencing reads were mapped to the Arabidopsis genome by usingTopHat2. The mapped reads were filtered with SAMtools (-F 0x4 -f 0x2), and only properly mapped read pairs were retained. To generate the strand-specific coverage files, the filtered reads were processed by bamCoverage with the following parameters: -bs 10, –normalizeUsing RPKM, reverse strand –samFlagInclude 64, –samFlagExclude 16; forward strand –samFlagInclude 80.
Quantitative reverse transcription PCR analysis
Total RNA was isolated using TRIzol reagent (Invitrogen, 15596026) according to the manufacturer's instructions. Two micrograms of DNase (Promega, RQ1 #M6101)-treated total RNA was used to synthesize cDNA (Promega, #1012891). RT-qPCR (real-time quantitative PCR) was performed using iQ SYBR Green Supermix solution (Bio-Rad, #170-8880). The CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) was used with the following cycling conditions: 95°C for 10 min, followed by 45 cycles of 95°C for 15 s, 60°C for 30 s and fluorescence detection. This was immediately followed by a melting curve (65–95°C in increments of 0.5°C for 5 s each with plate reading at each step). The melting curve analysis confirmed the absence of non-specific products. Each sample was quantified at least in triplicate and normalized by calculating the delta Cq (quantification cycle) value relative to the expression of the internal control Ubiquitin10 (UBQ10). The Cq and relative expression values were calculated with Bio-Rad CFX Manager 3.1 based on the MIQE guidelines. The standard deviations were determined from at least three technical and two biological replicates. The variance of the average data is represented by the SEM (standard error of the mean). SDs (standard deviations), SEMs and P-values were calculated using Student's paired t-test. The gene-specific primers used for qRT-PCR are listed in Supplementary Table S4.
Chromatin immunoprecipitation assays
ChIP assays were performed as previously described (45). Whole 14-day-old seedlings grown under long-day conditions were collected and treated with 1% formaldehyde for chromatin-DNA extraction. The obtained chromatin DNA was sheared to a mean length of 500 bp by sonication, and proteins and DNA fragments were then immunoprecipitated using antibodies against acetylated histone H3K9K14 (Millipore, catalog no. 06-599), dimethylated histone H3K4 (Diagenode, catalog no. C15410035) or GFP (Abcam, catalog no. ab290). DNA crosslinking to the immunoprecipitated proteins was reversed, and the DNA was then analyzed by real-time PCR using specific primers (Supplementary Table S4). The percent input was calculated as follows: 2∧(Cq(IN)-Cq(IP))X100, where Cq is the quantification cycle, as calculated by Bio-Rad CFX Manager 3.1 based on the MIQE guidelines. The SD were obtained from at least three technical and two biological replicates. The variance of the average data is represented by the SEM. SDs, SEMs and P-values were calculated using Student's paired t-test.
ChIP-seq and data analyses
ChIP-seq assays were performed based on previous research (46,52–55). The HDA6 ChIP-seq data (GSE132563) used in this study were proceed together with previously published LDL1 ChIP-seq data (GSE118025) (46). Samples of 5 ng of DNA from at least 5 ChIP replicates were pooled to ensure that there was enough starting DNA for library construction. Two biological replicates were prepared and sequenced for each ChIP-seq experiment. The ChIP DNA was first tested by qRT-PCR and then used to prepare ChIP-seq libraries. End repair, adaptor ligation and amplification were carried out using the Illumina Genomic DNA Sample Prep kit according to the manufacturer's protocol. An Illumina HiSeq 2500 instrument was used for the high-throughput sequencing of the ChIP-seq libraries. The raw sequence data were processed using the GAPipeline Illumina sequence data analysis pipeline. Bowtie2 was then employed to map the reads to the Arabidopsis genome (TAIR10) (50). Only perfectly and uniquely mapped reads were retained for further analysis. To determine the correlation between biological repeats, the Pearson correlation was computed using R statistical software according to the normalized signal intensity for ChIP binding peaks. The alignments were first converted to Wiggle (WIG) files using deepTools. The data were then imported into the Integrated Genome Viewer (IGV) (51) for visualization. The distribution of the ChIP binding peaks was analyzed with ChIPseeker (56), and a high-read random Arabidopsis genomic region subset (1 350 000 regions) was used to represent the ratio of the total Arabidopsis genomic regions. To identify DNA motifs enriched at LDL1-associated sites, 400-bp sequences encompassing each peak summit (200 bp upstream and 200 bp downstream) were extracted and searched for enriched DNA motifs using MEME-ChIP (57). Searches were performed using the default parameters. HDA6/LDL1-targeted lncRNAs were identified by comparing the HDA6/LDL1 binding sites with the lncRNA-associated genomic regions (15) (Supplementary Table S5). To compare the correlation between RNA expression and histone modifications, the ChIP-seq results as well as previously published Arabidopsis H3K4me3, H3K9me2, H3K27me3, H3K36me3, Pol II and Pol V ChIP-seq data (52,58–60) were analyzed by using deepTools to generate a correlation matrix among the different gene groups.
RESULTS
Genome-wide occupancy profiles of HDA6 and LDL1
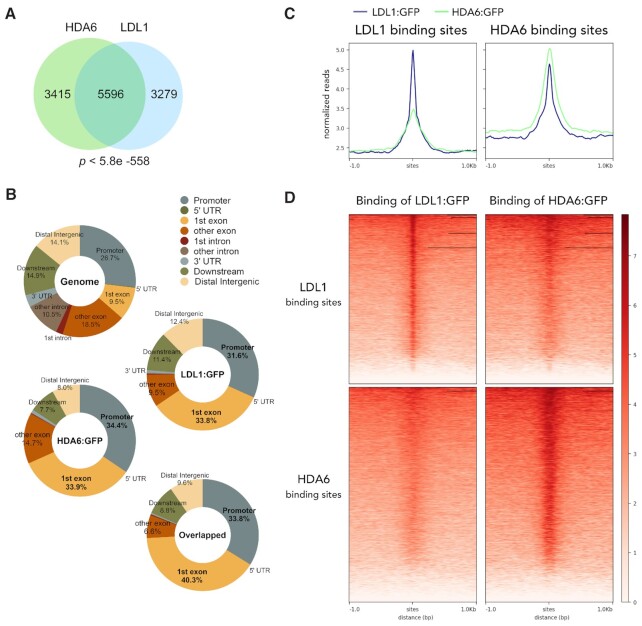
To further investigate the function of the HDA6-LDL1/2 complex, we mapped the genome-wide occupancy of HDA6 by chromatin immunoprecipitation followed by sequencing (ChIP-seq). ChIP-seq analysis was carried out using transgenic plants expressing the HDA6:GFP fusion protein driven by the native HDA6 promoter (HDA6pro::HDA6:GFP) in the hda6 mutant background. The HDA6:GFP transgene can fully rescue the late-flowering phenotype of the hda6 mutant, indicating that it is functional in vivo (Supplementary Figure S1). The ChIP-seq analyses identified 9,011 HDA6-targeted genomic regions (Figure 1A), including several regions located close to the direct targets of HDA6 identified in previous studies (41–45). We previously reported the genome-wide occupancy profile of LDL1 (46). We further compared the genome-wide occupancy profiles of HDA6 and LDL1. We found that HDA6 and LDL1 cooccupy 5596 targeted genomic regions, indicating significant overlap between HDA6- and LDL1-occupied genomic regions (hypergeometric test, P < 5.8e-558) (Figure 1A). Compared to the Arabidopsis genomic region distribution, we found that the binding of HDA6 and LDL1 was more enriched in the first exon and promoter regions but less enriched in UTR and intron regions (Figure 1B). Approximately 30–35% of HDA6- and LDL1-occupied sites are located in the promoters of protein-coding genes and 30-40% are located in the first exons of gene bodies (Figure 1B). Furthermore, the HDA6-enriched sites were located precisely at the LDL1-occupied sites, indicating that HDA6 and LDL1 show high similarity in their binding regions (Figure 1C and D). In addition, many similar cis-elements were found to be enriched within the HDA6 and LDL1 binding sites, including (GA/TC)n repeats, G-boxes and TCP binding sites (TBSs) (Supplementary Figure S2). These results are consistent with the previous finding that HDACs and LSD1 function cooperatively in the same protein complex in Arabidopsis (33–36,46).
Figure 1.
HDA6 and LDL1 show similar binding patterns in Arabidopsis. (A) A Venn diagram showing statistically significant overlap between HDA6 binding peaks and LDL1 binding peaks (5596 peaks; P < 5.8e -558, hypergeometric test). (B) Pie charts showing the distribution of HDA6 and LDL1 in annotated genic and intergenic regions in the genome. (C) The mean density of HDA6 and LDL1 occupancy at all LDL1-associated sites or HDA6-associated sites. The average HDA6 and LDL1 binding signal within 1 kb genomic regions flanking the center of LDL1 or HDA6 peaks is shown. (D) Heat map representation of the cooccupancy of HDA6 and LDL1 in the genome. Each horizontal line represents an LDL1- or HDA6-bound region, and the signal intensity is shown for LDL1 binding and HDA6 binding (center). Columns show the genomic region surrounding each LDL1 or HDA6 peak. Signal intensity is indicated by the shade of red.
By comparing the genome-wide occupancy profiles of HDA6 and LDL1, we found that LDL1 is highly associated with HDA6-binding sites, but the binding of HDA6 at LDL1-binding sites is not as strong as that of LDL1 itself (Figure 1C and D). These results implied that although the function of LDL1 is highly correlated with that of HDA6, LDL1 may not always be associated with HDA6, suggesting that in addition to HDA6, other HDACs may also functionally associate with LDL1/2 in Arabidopsis.
The mechanism of expression regulation may be different between protein-coding genes and lncRNAs
We analyzed the distribution of the expression levels of lncRNAs and protein-coding genes in Arabidopsis and found that the expression of lncRNAs was lower than that of the protein-coding genes (Supplementary Figure S3A), which is consistent with previous studies in animals and plants (8–12). To eliminate analysis error, we created random expression-matched coding gene subsets for further analysis (Supplementary Figure S3A).
Histone modification marks are removable and may therefore provide a flexible strategy for gene repression or activation. H3Ac, H3K4me2/3 and H3K36me3 are active marks for transcription; in contrast, H3K9me2 and H3K27me3 are signals for transcriptional silencing (35). The data presented in recent studies have implied that the effect of H3K9me and H3K27me3 on lncRNAs may be different from that of protein-coding genes (26–28,31). However, the mechanism of lncRNA expression regulation is rarely discussed and remains elusive in plants. To further investigate the role of histone modifications in the regulation of lncRNA in Arabidopsis, we compared the H3Ac and H3K4me2 ChIP-seq results with previously published ChIP-seq results for H3K4me3, H3K9me2, H3K27me3 and H3K36me3 in WT Arabidopsis (52,58). We found that the expression of lncRNAs was positively correlated with H3Ac, H3K4me2/3 and H3K36me3 (Supplementary Figure S3B). However, the expression of lncRNAs was less correlated with H3K9me2 and H3K27me3 (Supplementary Figure S3B). Nevertheless, the expression of protein-coding genes still presented a negative correlation with H3K27me3 (Supplementary Figure S3B). These results indicated that the general expression of lncRNAs is less affected by H3K9me2 and H3K27me3 in the gene body of lncRNAs, implying that the effect of histone modifications may be different between protein-coding genes and lncRNAs.
RNA polymerase V (Pol V) is a plant-specific RNA polymerase that has an independent function from Pol II. Pol II is the most active RNA polymerase for protein-coding genes, small RNAs and rRNAs. In contrast, Pol V is highly associated with other non-coding RNA-mediated gene silencing processes, such as RNA-directed DNA methylation (RdDM) (61,62). A previous study suggested that the transcripts of specific lncRNAs are associated with Pol V (63). To further investigate the function of RNA polymerases in the regulation of lncRNA in Arabidopsis, we also compared the expression of lncRNAs with previously published ChIP-seq results for Pol II and Pol V in Arabidopsis (59,60). We found that the expression of lncRNAs was positively correlated with the enrichment of both Pol II and Pol V, in which the correlation between lncRNA expression and the enrichment of Pol V was relatively minor (Supplementary Figure S4). In contrast, the expression of protein-coding genes was positively correlated with the enrichment of Pol II but was not positively correlated with the enrichment of Pol V (Supplementary Figure S4). These results indicated that the regulation may be different between protein-coding genes and lncRNAs in Arabidopsis.
HDA6 and LDL1/2 are involved in the regulation of lncRNAs in Arabidopsis
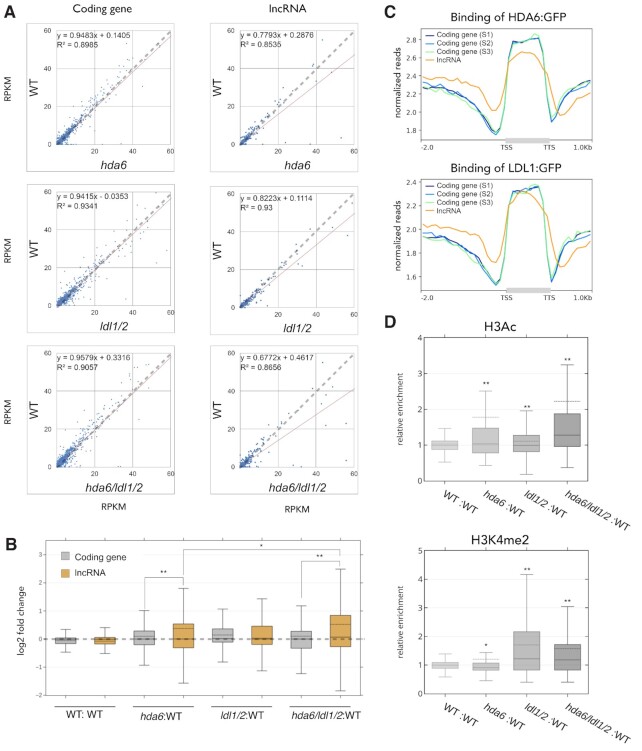
To further investigate the regulatory targets of HDA6-LDL1/2, we performed RNA-seq analyses in WT, hda6, ldl1/ldl2 (ldl1/2) and hda6/ldl1/ldl2 (hda6/ldl1/2) mutant plants and identified differentially expressed genes (DEGs) from the TAIR10 Arabidopsis gene annotation. The upregulated and downregulated DEGs in hda6, ldl1/2 and hda6/ldl1/2 were analyzed among different gene types. Interestingly, we found that among all of the differentially expressed protein-coding genes identified in hda6, ldl1/2 and hda6/ldl1/2, ∼60% were upregulated and ∼40% were downregulated. However, among all of the differentially expressed lncRNAs in hda6 and ldl1/2, more than 74% were upregulated. Furthermore, more than 83% of the differentially expressed lncRNAs were upregulated in hda6/ldl1/2 (Supplementary Figure S5), suggesting that HDA6-LDL1/2 may have greater effects on the expression of lncRNAs than that of protein-coding genes. The overall expression of all lncRNAs was higher in hda6, ldl1/2 and hda6/ldl1/2 than in WT (Figure 2A). In contrast, the expression patterns of the protein-coding genes were not substantially changed in hda6, ldl1/2 and hda6/ldl1/2 compared to WT (Figure 2A and Supplementary Figure S6A). We also compared the relative expression of protein-coding genes and lncRNAs. The fold change of lncRNAs was higher than that of protein-coding genes in hda6 and hda6/ldl1/2 compared to WT, and the largest increase in the expression of lncRNAs was observed in hda6/ldl1/2 (Figure 2B). The general expression pattern showed no substantial difference between all coding genes and the expression-matched coding gene subsets (Supplementary Figure S6). However, the mean fold change observed for the total lncRNAs in hda6/ldl1/2 was not greatly increased (Figure 2B), suggesting that HDA6-LDL1/2 may not regulate all of the lncRNAs, but only a specific fraction of them.
Figure 2.
LDL1/2 and HDA6 regulate lncRNA expression by modulating H3K4me2 and H3Ac levels.(A) X-Y scatter plot and abline of WT-hda6, WT-ldl1/2 and WT-hda6/ldl1/2 showing the expression patterns of the protein-coding gene subset and lncRNAs. (B) Boxplots showing the relative expression of the protein-coding gene subset and lncRNAs in hda6, ldl1/2 and hda6/ldl1/2 compared to those in WT. *P < 0.01, **P < 0.001 (Student's t-test). (C) Metagene ChIP-seq binding profiles of HDA6 and LDL1 in lncRNAs and the protein-coding gene subsets (S1–S3). The profile is from 2 kb upstream of the TSS to 1 kb downstream of the TTS, and the gene lengths were scaled to the same size. (D) Boxplots showing the relative enrichment of H3Ac and H3K4me2 in lncRNAs in hda6, ldl1/2 and hda6/ldl1/2 compared to those in WT. The dotted line indicates the mean value of each group. *P < 0.01, **P < 0.001 (Student's t-test).
Interestingly, we also found that the levels of HDA6 and LDL1 binding within the promoters of lncRNAs were higher than within the protein-coding genes (Figure 2C). These results suggested that HDA6 and LDL1/2 may be highly associated with the regulation of lncRNAs. In addition, we performed H3Ac and H3K4me2 ChIP-seq analysis in WT, hda6, ldl1/2 and hda6/ldl1/2 to identify the effects of HDA6-LDL1/2 on histone modification and found that the relative H3Ac and H3K4me2 enrichment levels of the lncRNAs were also increased in hda6, ldl1/2 and hda6/ldl1/2 (Figure 2D), indicating that LDL1, LDL2 and HDA6 are involved in the regulation of lncRNAs by affecting H3K4me2 and H3Ac levels.
To determine whether the regulation of lncRNA is associated with other HDACs in Arabidopsis, we also analyzed the H3Ac ChIP-seq profiles of WT and hda9 (64) to compare the changes in H3Ac in lncRNAs. We found that the relative H3Ac level of the lncRNA in hda9 was not significantly increased compared with that in hda6 (Supplementary Figure S7A), indicating that lncRNAs are not universally regulated by all HDACs in Arabidopsis.
The binding of HDA6 and LDL1 to lncRNAs is associated with increased H3Ac and H3K4me2 levels
A recent study systematically annotated a total of 6510 lncRNAs in Arabidopsis thaliana via high-depth strand-specific RNA sequencing (15). These lncRNAs were used for further analysis in this study. We found that H3Ac and H3K4me2 levels in WT Arabidopsis were associated with the distance between neighboring genes, where the lncRNAs exhibiting higher H3Ac and H3K4me2 levels appeared to be located closer to their neighboring genes (Supplementary Figure S7B). However, we also found that lncRNAs that were located farther from their neighboring genes appeared to be more enriched in hda6/ldl1/2 (Supplementary Figure S7C). These results suggested that HDA6-LDL1/2 may only regulate the expression of specific lncRNAs.
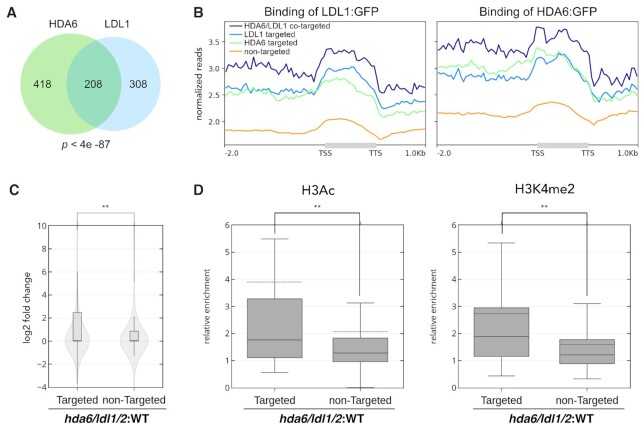
Among the annotated lncRNAs, 626 and 516 were directly targeted by HDA6 and LDL1, respectively (Figure 3A). A total of 208 lncRNAs were cotargeted by HDA6 and LDL1, indicating high overlap between HDA6- and LDL1-targeted lncRNAs (hypergeometric test, P < 4e -87) (Figure 3A). As we expected, the binding of HDA6 and LDL1 to the targeted lncRNAs was stronger than their binding to nontargeted lncRNAs (Figure 3B). Furthermore, the binding of HDA6 and LDL1 was increased in the HDA6/LDL1-cotargeted lncRNAs (Figure 3B). We further compared the relative expression of the HDA6/LDL1-cotargeted and non-targeted lncRNAs. The relative expression of the HDA6/LDL1-cotargeted lncRNAs in hda6/ldl1/2 was significantly increased compared to that of the non-targeted lncRNAs (Figure 3C). In addition, the relative H3Ac and H3K4me2 levels of the HDA6/LDL1-cotargeted lncRNAs in hda6/ldl1/2 were increased compared to those of the non-targeted lncRNAs (Figure 3D). These results indicated that the changes in the H3Ac and H3K4me2 levels of the lncRNAs are associated with the binding of HDA6 and LDL1.
Figure 3.
The expression of H3Ac and H3K4me2 in HDA6/LDL1-targeted lncRNAs is increased in hda6/ldl1/2 compared to WT. (A) A Venn diagram showing statistically significant overlap between lncRNAs targeted by HDA6 and those targeted by LDL1 (208 lncRNAs; P < 4e -87, hypergeometric test). (B) Metagene ChIP-seq binding profiles of HDA6 and LDL1 in HDA6-targeted lncRNAs, LDL1-targeted lncRNAs, HDA6/LDL1-cotargeted lncRNAs and lncRNAs not targeted by HDA6 or LDL1. The profile is from 2 kb upstream of the TSS to 1 kb downstream of the TTS, and the gene lengths were scaled to the same size. (C) Violin plots with boxplots showing the relative expression on the HDA6/LDL1-cotargeted and nontargeted lncRNAs in hda6/ldl1/2 compared to those in WT. **P < 0.001 (Student's t-test). (D) Boxplots showing the relative enrichment of H3Ac and H3K4me2 in the HDA6/LDL1-cotargeted and non-targeted lncRNAs in hda6/ldl1/2 compared to those in WT. The dotted line indicates the mean value of each group. **P < 0.001 (Student's t-test).
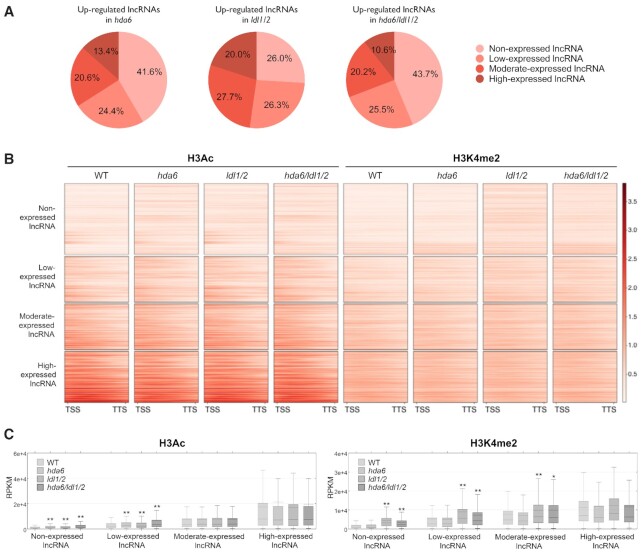
Among the 6510 annotated lncRNAs, 475, 284 and 526 were upregulated in hda6, ldl1/2 and hda6/ldl1/2, respectively (Supplementary Figure S8A). In addition, among the 526 lncRNAs that were upregulated in hda6/ldl1/2, 307 and 253 displayed increases in H3Ac (hypergeometric test, P < 6e-57) and H3K4me2 (hypergeometric test, P < 3.7e-20) , respectively, in hda6/ldl1/2 (Supplementary Figure S8B). We classified the 6510 annotated lncRNAs into non-expressed, low expression, moderate expression and high expression groups according to their expression levels in WT. Interestingly, we found that most of the lncRNAs that were upregulated in hda6, ldl1/2 and hda6/ldl1/2 were non-expressed or expressed at low levels in WT (Figure 4A and Supplementary Figure S8A). In addition, most of the HDA6/LDL1-targeted lncRNAs were also non-expressed or expressed at low levels (Supplementary Figure S8C and D). The levels of H3Ac and H3K4me2 among the lncRNAs that were non-expressed or showed low expression were lower than those in the highly expressed lncRNAs (Figure 4B and C), indicating that H3Ac and H3K4me2 levels are correlated with the expression levels of lncRNAs. Moreover, we found that the total H3Ac and H3K4me2 levels of the lncRNAs that were non-expressed or showed low expression in hda6, ldl1/2 and hda6/ldl1/2 were increased compared to those in WT (Figure 4B and C). In contrast, the total H3Ac and H3K4me2 levels of the highly expressed lncRNAs in hda6, ldl1/2 and hda6/ldl1/2 were not changed compared to those in WT (Figure 4B and C). These results suggested that HDA6-LDL1/2 played an important role in the repression of lncRNAs that were nonexpressed or showed low expression. It has been reported that the expression of lncRNAs is highly tissue specific and is regulated by environmental stresses (8–15). Thus, we extracted the ABA-, drought- and cold-induced lncRNAs as well as the lncRNAs showing high expression in inflorescences or siliques identified in a previous study (15) for further analysis. We found that these stress-induced or tissue-specific lncRNAs were mostly nonexpressed or showed low expression under regular treatment (Supplementary Figure S9). Altogether, these results implied that the regulation of lncRNAs that were nonexpressed or showed low expression may be essential for plant development and responses to the environment.
Figure 4.
The H3Ac and H3K4me2 levels of lncRNAs that are nonexpressed or show low expression are increased in hda6/ldl1/2. (A) Pie chart showing the distribution of lncRNAs that were non-expressed or showed low-, moderate- or high- expression among the lncRNAs that were upregulated in hda6, ldl1/2 and hda6/ldl1/2. (B) H3Ac and H3K4me2 plot heatmap of lncRNAs in WT, hda6, ldl1/2 and hda6/ldl1/2 arranged by their expression levels in WT. Each horizontal line represents one lncRNA, and the signal intensity is shown for H3Ac or H3K4me2 levels in WT, hda6, ldl1/2 and hda6/ldl1/2. Signal intensity is indicated by the shade of red. (C) Boxplots showing the H3Ac and H3K4me2 levels of lncRNAs that were non-expressed or showed low-, moderate- or high-expression in WT, hda6, ldl1/2 and hda6/ldl1/2. The dotted line indicates the mean value of each group. *P < 0.05, **P < 0.005 (Student's t-test).
The neighboring genes of the lncRNAs that are upregulated in hda6/ldl1/2 are associated with development in Arabidopsis
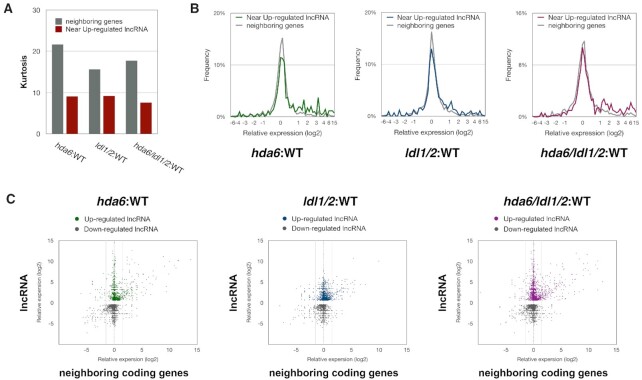
Previous studies have revealed that lncRNAs may positively or negatively regulate the expression of their neighboring genes (3–9,15,24–30). The kurtosis value is a measure of deviation, with lower kurtosis values indicating that there are more extreme values in the sample population. We calculated the kurtosis values of the relative expression levels of the neighboring genes near lncRNAs and found that the kurtosis values of the neighboring genes near the upregulated lncRNAs in hda6, ldl1/2 and hda6/ldl1/2 were lower than those of the overall neighboring genes (Figure 5A). In addition, the frequencies of extreme values among the neighboring genes near the upregulated lncRNAs in hda6, ldl1/2 and hda6/ldl1/2 were higher than those among the overall neighboring genes (Figure 5B). When the expression levels of the lncRNAs and their neighboring genes in hda6, ldl1/2 and hda6/ldl1/2 were compared to those in WT, we found that the expression variation of neighboring genes among the upregulated lncRNAs was higher than that among the downregulated lncRNAs (Figure 5C). These results suggested that the expression levels of neighboring genes are associated with the changes in the expression of lncRNAs.
Figure 5.
The expression changes in neighboring coding genes located near HDA6/LDL1/2-regulated lncRNAs. (A) The statistical kurtosis values of the relative expression levels in WT-hda6, WT-ldl1/2 and WT-hda6/ldl1/2. All annotated neighboring coding genes (gray) (15) and the neighboring coding genes located close to the upregulated lncRNAs (red) are presented. (B) The distribution of relative expression levels in WT-hda6, WT-ldl1/2 and WT-hda6/ldl1/2. Data is shown for all annotated neighboring coding genes (15) and the neighboring coding genes located close to upregulated lncRNAs. (C) X-Y scatter plot showing the relative expression levels of the lncRNAs and their neighboring genes in WT-hda6, WT-ldl1/2 and WT-hda6/ldl1/2.
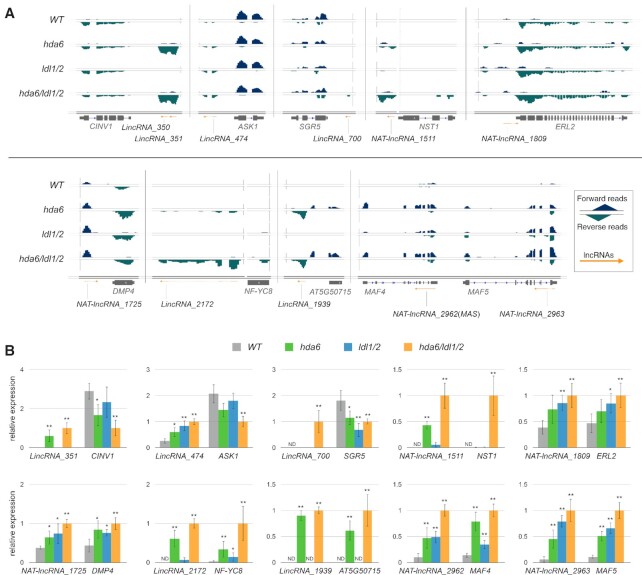
In addition, we found that the upregulation of lncRNAs in hda6, ldl1/2 and hda6/ldl1/2 was generally positively correlated with the expression of neighboring genes (Supplementary Figure S10A), which was consistent with the results of a previous study (15). The function of these neighboring genes near the upregulated lncRNAs in hda6/ldl1/2 was further analyzed according to Gene Ontology Biological Processes (GO-BP). These genes were shown to be involved in various developmental processes, including shoot and flower development (Supplementary Figure S10B and C). These results suggested that the neighboring genes of the lncRNAs that are upregulated in hda6/ldl1/2 are associated with various developmental processes in Arabidopsis. To further confirm the expression patterns of the lncRNAs showing upregulation in hda6/ldl1/2 and their neighboring genes, the RNA-seq reads for the forward strand and reverse strand were visualized separately for WT, hda6, ldl1/2 and hda6/ldl1/2. We found that the reads of the relevant lncRNAs exhibited substantially higher levels in hda6/ldl1/2 than in WT (Figure 6A and Supplementary Figure S11). Moreover, we found that the expression of the genes near or overlapping with these lncRNAs was substantially changed in hda6/ldl1/2 (Figure 6A and Supplementary Figure S11).
Figure 6.
The expression of HDA6-LDL1/2-regulated lncRNAs and their neighboring genes. (A) Integrated Genome Viewer showing forward- and reverse-transcript reads of the selected lncRNA genes and their neighboring genes in WT, hda6, ldl1/2 and hda6/ldl1/2. Orange arrows indicate transcripts of lncRNA genes from previously published data (15). Bars: normalized reads = 2. (B) Gene expression of the selected lncRNA genes and their neighboring genes in WT, hda6, ldl1/2 and hda6/ldl1/2. Gene expression was determined by qRT-PCR and normalized to UBQ10. N.D.: not detected. Error bars correspond to standard deviations from three biological replicates. *: P < 0.05, **: P < 0.005 (Student's t-test).
We selected 10 lncRNAs and their neighboring genes to further validate their expression by qRT-PCR (Figure 6B). These selected up- or downregulated neighboring genes, including cytosolic invertase 1 (CINV1), arabidopsis skp1 homologue 1 (ASK1), shoot gravitropism 5 (SGR5), nac secondary wall thickening promoting factor1 (NST1), rrecta-like 2 (ERL2), duf679 domain membrane protein 4 (DMP4), nuclear factor Y subunit C8 (NF-YC8), wrky DNA-binding protein 17 (WRKY17), mads affecting flowering 4 (MAF4) and mads affecting flowering 5 (MAF5), are reported to be involved in plant development. ASK1 is a scaffold linker protein of the SCF ubiquitin-protein E3 ligase complex that has been reported to be involved in multiple developmental processes, including early development, flowering time and the circadian clock (65–68). SGR5 is highly expressed in petioles, hypocotyls, roots, floral organs and especially inflorescence stems and is involved in shoot and flower development (69,70). ERL2 is associated with organ growth and flower development by promoting cell proliferation (71). NF-YC8 is a subunit of the NF-Y DNA-binding complex, and Arabidopsis NF-YC homologs act as important factors in flowering time control (72,73). In addition, NST1 and DMP4 have been reported to be involved in flower development (74,75), whereas CINV1, ASK1 and WRKY17 have been reported to be involved in root development and abiotic stress responses in Arabidopsis (76–78).
MAF4 and MAF5 are paralogs of FLC and act as floral repressors in Arabidopsis (79). MAS (NAT-lncRNA_2962) is an antisense lncRNA produced from the 3′ region of MAF4 that acts as a positive regulator of MAF4 (15). In our study, we found that the expression of MAS was increased in hda6, ldl1/2 and hda6/ldl1/2 (Figure 6A and B). MAF4 expression was also increased in hda6, ldl1/2 and hda6/ldl1/2 (Figure 6A and B). Similar to MAS, NAT-lncRNA_2963 is an antisense lncRNA transcribed from the 3′ region of MAF5 (15). The expression of both NAT-lncRNA_2963 and MAF5 was increased in hda6, ldl1/2 and hda6/ldl1/2 (Figure 6A and B).
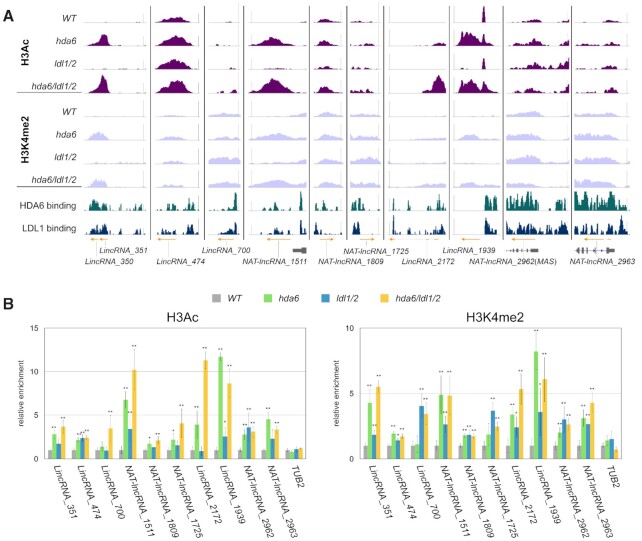
The HDA6/LDL1 binding peaks and H3Ac and H3K4me2 profiles of these lncRNAs in WT, hda6, ldl1/2 and hda6/ldl1/2 were viewed and curated in IGV. We identified specific binding of HDA6 and LDL1 to the promoter regions of these lncRNAs (Figure 7A; Supplementary Figures S12 and 13). Moreover, H3Ac and H3K4me2 levels were substantially increased in the downstream regions of HDA6 and LDL1 binding sites in hda6/ldl1/2, and the increased H3Ac and H3K4me2 peaks were located near the transcribed regions of these lncRNAs (Figure 7A; Supplementary Figure S12 and 13). The H3Ac and H3K4me2 levels of these lncRNAs were further validated by ChIP–qPCR (Figure 7B). It appears that HDA6 and LDL1 can bind to the promoters of specific lncRNAs to regulate their expression. Furthermore, we extracted all the neighboring genes of the lncRNAs that were upregulated in hda6/ldl1/2. The general binding of HDA6 and LDL1 to the promoters of these neighboring genes was reduced compared with that among all genes in Arabidopsis (Supplementary Figure S14), suggesting that these neighboring genes may not be directly regulated by HDA6 and LDL1.
Figure 7.
H3Ac and H3K4me2 in the lncRNAs regulated by HDA6-LDL1/2. (A) Integrated Genome Viewer showing H3Ac and H3K4me2 peaks in WT, hda6, ldl1/2 and hda6/ldl1/2 and the binding of HDA6 and LDL1 in the selected lncRNA genes. Orange arrows indicate transcripts of lncRNA genes from previously published data (15). Bars: normalized reads = 5. (B) ChIP analysis of H3Ac and H3K4me2 levels of the selected lncRNA genes. ChIP assays were performed with H3K9K14Ac and H3K4me2 antibodies. The relative enrichment was determined by qRT-PCR and normalized to ACT2. Error bars correspond to standard deviations from three biological replicates. *: P < 0.05, **: P < 0.005 (Student's t-test).
Among the neighboring genes located near the lncRNAs that were upregulated in hda6/ldl1/2, over 75% were protein-coding genes, whereas ∼19% were transposable element (TE) genes (Supplementary Table S2). We found that the neighboring genes of the lncRNAs that were upregulated in hda6/ldl1/2 may be associated with various developmental processes in Arabidopsis. Both hda6 and ldl1/2 mutants are late flowering (45,48). Interestingly, we found that hda6/ldl1/2 plants flowered later than hda6 and ldl1/2 plants (Supplementary Figure S15A and B). The enhanced late-flowering phenotype of hda6/ldl1/2 may be associated with the lncRNA-neighboring genes regulated by HDA6-LDL1/2.
DISCUSSION
HDACs and LSD1 have been identified as the core components of several multiprotein complexes, such as Mi2/NuRD and CoREST, that regulate gene expression cooperatively in yeast and animal systems (32–36). It has also been found that human HDAC1/2 and LSD1 are aberrantly highly expressed and positively regulate cell growth in many kinds of cancer cell lines (80–82), suggesting that the HDAC complex is important in cell growth and development. In this study, we found that HDA6 and LDL1 are associated with promoter and gene body regions. A previous global binding analysis of human HDACs also showed that HDAC2 and HDAC6 can bind to both promoter and gene body regions (83). The binding of human HDACs is associated with the function of RNA Pol II and the repression of transcription elongation in human cells (84–86). A recent study revealed that human HDACs are correlated with the positive regulation of transcription elongation (86). Furthermore, the binding of human HDACs to gene bodies is involved in the regulation of alternative splicing (87–90). The binding of Arabidopsis HDA6 and LDL1 in gene body regions implies that the HDA6-LDL1/2 complex may also be involved in transcription elongation and alternative splicing. In this study, we found that HDA6-LDL1/2 is associated with the regulation of lncRNAs. Because ∼30% of the annotated lncRNAs are antisense transcripts and overlap with protein-coding genes (15), it is possible that the gene body regions bound by HDA6 and LDL1 may act as the promoters of the overlapping lncRNAs.
In plants, lncRNAs have been implicated in development, light responses, vernalization, auxin transport and stress responses (15–28). However, only a few plant lncRNAs have been studied in detail. In addition, the expression of lncRNAs is lower than that of protein-coding genes, and they tend to be more tissue specific (8–12). Furthermore, the expression of plant lncRNAs is lower than that of animal lncRNAs (10,15). Recently, it was reported that the expression of the human lncRNAs TINCR and PLAC2 is regulated by histone acetylation (91,92). However, the mechanism of lncRNA expression regulation in plants is still largely unclear. In this study, we found that a subset of Arabidopsis lncRNAs that are nonexpressed or show low expression are regulated by HDA6-LDL1/2. Because the expression of lncRNAs is highly tissue specific and can be induced under certain environmental conditions (1,10,13–15), HDA6-LDL1/2 may play an important role in plant development by repressing lncRNAs.
lncRNAs can positively or negatively regulate the expression of their neighboring genes (3–9,15,24–30). We found that a subset of genes located near the HDA6-LDA1/2-regulated lncRNAs are involved in various plant developmental processes. These genes include CINV1, ASK1, SGR5, NST1, ERL2, NF-YC8, WRKY17, MAF4 and MAF5 (60–74). The misexpression of these neighboring genes may result in developmental defects in Arabidopsis. Previously, HDA6 was reported to repress the expression of MAF4 and MAF5 (45). The repression of MAF4 and MAF5 by HDA6 may be related to their nearby lncRNAs. Furthermore, HDA6 is involved in the regulation of TEs by interacting with DNA Methyltransferase1 (MET1) and Kryptonite/Suvh4 (KYP) (39–41). Interestingly, we found that ∼19% of the neighboring genes near the upregulated lncRNAs in hda6/ldl1/2 are TEs. It has been reported that TEs contribute to the tissue-specific and stress-induced expression of lncRNAs (93,94). Therefore, these TEs regulated by HDA6 may also be involved in the regulation of lncRNAs.
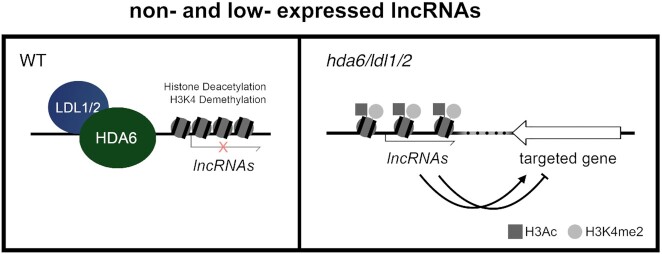
Collectively, our research revealed that the expression of lncRNAs that are non-expressed or show low expression is associated with H3Ac/H3K4me2 changes regulated by the HDA6-LDL1/2 histone modification complex (Figure 8). Furthermore, we found that a subset of the neighboring genes of the lncRNAs regulated by HDA6 and LDL1 are associated with various plant developmental processes. These results indicated that the expression of Arabidopsis lncRNAs can be suppressed by the HDA6-LDL1/2 complex and that HDA6-LDL1/2 may be associated with plant developmental processes by modulating the expression of lncRNAs.
Figure 8.
The expression of lncRNAs that are nonexpressed or show low expression is associated with H3Ac/H3K4me2 changes regulated by the HDA6-LDL1/2 histone modification complex. The HDA6-LDL1/2 histone modification complex can bind to the promoter region or gene body of lncRNAs that are non-expressed or show low expression to suppress their expression by histone deacetylation and H3K4 demethylation. The expression of protein-coding genes may be positively or negatively regulated by their neighboring lncRNAs; therefore, HDA6-LDL1/2 may affect the expression of protein-coding genes by regulating neighboring lncRNAs.
DATA AVAILABILITY
The RNA-seq and ChIP-seq short read data have been submitted to the NCBI Gene Expression Omnibus (GEO) database, including the previously published LDL1 ChIP-seq data (GSE118025) (46) and unpublished data from HDA6 ChIP-seq (GSE132563), H3Ac ChIP-seq of WT, hda6, ldl1/2 and hda6/ldl1/2 (GSE132636), H3K4me2 ChIP-seq of WT, hda6, ldl1/2 and hda6/ldl1/2 (GSE133005) and RNA-seq of WT, hda6, ldl1/2 and hda6/ldl1/2 (GSE132873).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Technology Commons, College of Life Science, National Taiwan University for the convenient use of the Bio-Rad real-time PCR system and CLC Genomics Workbench.
Authors’ Contributions: F.-Y. H.,Y.C. and K.W. designed the research, F.-Y. H., C.C., Y.-H.S. and F.-F.C. performed the research. F.-Y.H., C.C., C.L., M.-R.Y., J.-W.A.H., P.-Y.C and K.W. analyzed the data. F.-Y.H. and K.W. wrote the article.
Contributor Information
Fu-Yu Hung, Institute of Plant Biology, National Taiwan University, Taipei 10617 Taiwan; Agriculture and Agri-Food Canada, London Research and Development Centre, London, ON N5V 4T3 Canada.
Chen Chen, Agriculture and Agri-Food Canada, London Research and Development Centre, London, ON N5V 4T3 Canada; Department of Biology, Western University, London, ON N6A 3K7 Canada; Key Laboratory of South China Agricultural Plant Molecular Analysis and Genetic Improvement, Provincial Key Laboratory of Applied Botany, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China; Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Guangzhou 510650, China.
Ming-Ren Yen, Institute of Plant and Microbial Biology, Academia Sinica, Taipei 11529, Taiwan.
Jo-Wei Allison Hsieh, Institute of Plant and Microbial Biology, Academia Sinica, Taipei 11529, Taiwan; Genome and Systems Biology Degree Program, Academia Sinica and National Taiwan University, Taipei, Taiwan.
Chenlong Li, Agriculture and Agri-Food Canada, London Research and Development Centre, London, ON N5V 4T3 Canada; Department of Biology, Western University, London, ON N6A 3K7 Canada; State Key Laboratory of Biocontrol and Guangdong Key Laboratory of Plant Resource, School of Life Sciences, Sun Yat-sen University, Guangzhou, 510275 China.
Yuan-Hsin Shih, Institute of Plant Biology, National Taiwan University, Taipei 10617 Taiwan.
Fang-Fang Chen, Institute of Plant Biology, National Taiwan University, Taipei 10617 Taiwan.
Pao-Yang Chen, Institute of Plant and Microbial Biology, Academia Sinica, Taipei 11529, Taiwan; Genome and Systems Biology Degree Program, Academia Sinica and National Taiwan University, Taipei, Taiwan.
Yuhai Cui, Agriculture and Agri-Food Canada, London Research and Development Centre, London, ON N5V 4T3 Canada; Department of Biology, Western University, London, ON N6A 3K7 Canada.
Keqiang Wu, Institute of Plant Biology, National Taiwan University, Taipei 10617 Taiwan.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
Ministry of Science and Technology of Republic of China [108-2311-B-002-013-MY3, 108-2311-B-002-001 to K.W.]; NTU-Academia Sinica Joint grant [NTU-AS-108L104310 to K.W., P.-Y.C]; Agriculture and Agri-Food Canada; National Science and Engineering Research Council of Canada [RGPIN/04625-2017 to Y. C.). Funding for the open access charge: National Taiwan University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F.et al.. Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ariel F., Romero-Barrios N., Jegu T., Benhamed M., Crespi M.. Battles and hijacks: noncoding transcription in plants. Trends Plant Sci. 2015; 20:362–371. [DOI] [PubMed] [Google Scholar]
- 3. Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A.et al.. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011; 472:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Geisler S., Coller J.. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013; 14:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu X., Hao L.L., Li D.Y., Zhu L.H., Hu S.N.. Long non-coding RNAs and their biological roles in plants. Genome Proteomics Bioinformatics. 2015; 13:137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamada M. Functions of long intergenic non-coding (linc) RNAs in plants. J. Plant Res. 2017; 130:67–73. [DOI] [PubMed] [Google Scholar]
- 7. Bardou F., Ariel F., Simpson C.G., Romero-Barrios N., Laporte P., Balzergue S., Brown J.W.S., Crespi M.. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell. 2014; 30:166–176. [DOI] [PubMed] [Google Scholar]
- 8. Iwakiri J., Terai G., Hamada M.. Computational prediction of lncRNA-mRNA interactions by integrating tissue specificity in human transcriptome. Biol. Direct. 2017; 12:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Popadin K., Gutierrez-Arcelus M., Dermitzakis E.T., Antonarakis S.E.. Genetic and epigenetic regulation of human lincRNA gene expression. Am. J. Hum. Genet. 2013; 93:1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu J., Jung C., Xu J., Wang H., Deng S.L., Bernad L., Arenas-Huertero C., Chua N.H.. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012; 24:4333–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dinger M.E., Amaral P.P., Mercer T.R., Pang K.C., Bruce S.J., Gardiner B.B., Askarian-Amiri M.E., Ru K., Solda G., Simons C.et al.. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008; 18:1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lagarde J., Uszczynska-Ratajczak B., Carbonell S., Perez-Lluch S., Abad A., Davis C., Gingeras T.R., Frankish A., Harrow J., Guigo R.et al.. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat. Genet. 2017; 49:1731–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G.et al.. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dempsey J., Zhang A., Cui J.Y.. Coordinate regulation of long non-coding RNAs and protein-coding genes in germ-free mice. BMC Genomics. 2018; 19:834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao X.Y., Li J.R., Lian B., Gu H.Q., Li Y., Qi Y.J.. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 2018; 9:5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khemka N., Singh V.K., Garg R., Jain M.. Genome-wide analysis of long intergenic non-coding RNAs in chickpea and their potential role in flower development. Sci. Rep. 2016; 6:33297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kang C.Y., Liu Z.C.. Global identification and analysis of long non-coding RNAs in diploid strawberry Fragariavesca during flower and fruit development. BMC Genomics. 2015; 16:815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song X.B., Sun L., Luo H.T., Ma Q.G., Zhao Y., Pei D.. Genome-wide identification and characterization of long non-coding RNAs from mulberry (Morusnotabilis) RNA-seq data. Genes (Basel). 2016; 7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang H., Chung P.J., Liu J., Jang I.C., Kean M.J., Xu J., Chua N.H.. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014; 24:444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ben Amor B., Wirth S., Merchan F., Laporte P., d’Aubenton-Carafa Y., Hirsch J., Maizel A., Mallory A., Lucas A., Deragon J.M.et al.. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009; 19:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Di C., Yuan J.P., Wu Y., Li J.R., Lin H.X., Hu L., Zhang T., Qi Y.J., Gerstein M.B., Guo Y.et al.. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014; 80:848–861. [DOI] [PubMed] [Google Scholar]
- 22. Li S., Yamada M., Hang X.W., Ohler U., Benfey P.N.. High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev. Cell. 2016; 39:508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y.C., Liao J.Y., Li Z.Y., Yu Y., Zhang J.P., Li Q.F., Qu L.H., Shu W.S., Chen Y.Q.. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014; 15:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Swiezewski S., Liu F.Q., Magusin A., Dean C.. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009; 462:799–802. [DOI] [PubMed] [Google Scholar]
- 25. Heo J.B., Sung S.. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011; 331:76–79. [DOI] [PubMed] [Google Scholar]
- 26. Liu F.Q., Marquardt S., Lister C., Swiezewski S., Dean C.. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010; 327:94–97. [DOI] [PubMed] [Google Scholar]
- 27. Csorba T., Questa J.I., Sun Q.W., Dean C.. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:16160–16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heo J.B., Sung S.. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011; 331:76–79. [DOI] [PubMed] [Google Scholar]
- 29. Kim D.H., Sung S.. Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev. Cell. 2017; 40:302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ariel F., Jegu T., Latrasse D., Romero-Barrios N., Christ A., Benhamed M., Crespi M.. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell. 2014; 55:383–396. [DOI] [PubMed] [Google Scholar]
- 31. Sati S., Ghosh S., Jain V., Scaria V., Sengupta S.. Genome-wide analysis reveals distinct patterns of epigenetic features in long non-coding RNA loci. Nucleic Acids Res. 2012; 40:10018–10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khochbin S., Verdel A., Lemercier C., Seigneurin-Berny D.. Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 2001; 11:162–166. [DOI] [PubMed] [Google Scholar]
- 33. Lee M.G., Wynder C., Cooch N., Shiekhattar R.. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005; 437:432–435. [DOI] [PubMed] [Google Scholar]
- 34. Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L.et al.. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009; 138:660–672. [DOI] [PubMed] [Google Scholar]
- 35. Lan F., Nottke A.C., Shi Y.. Mechanisms involved in the regulation of histone lysine demethylases. Curr. Opin. Cell Biol. 2008; 20:316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y.. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004; 119:941–953. [DOI] [PubMed] [Google Scholar]
- 37. Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I.et al.. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell. 2004; 16:1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Earley K., Lawrence R.J., Pontes O., Reuther R., Enciso A.J., Silva M., Neves N., Gross M., Viegas W., Pikaard C.S.. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006; 20:1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu X.C., Yu C.W., Duan J., Luo M., Wang K.C., Tian G., Cui Y.H., Wu K.Q.. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 2012; 158:119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. To T.K., Kim J.M., Matsui A., Kurihara Y., Morosawa T., Ishida J., Tanaka M., Endo T., Kakutani T., Toyoda T.et al.. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLos Genet. 2011; 7:e1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu C.W., Tai R., Wang S.C., Yang P., Luo M., Yang S., Cheng K., Wang W.C., Cheng Y.S., Wu K.. HIstone Deacetylase6 acts in concert with histone methyltransferases SUVH4, SUVH5, and SUVH6 to regulate transposon silencing. Plant Cell. 2017; 29:1970–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu K., Zhang L., Zhou C., Yu C.W., Chaikam V.. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 2008; 59:225–234. [DOI] [PubMed] [Google Scholar]
- 43. Chen L.T., Luo M., Wang Y.Y., Wu K.. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010; 61:3345–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Luo M., Yu C.W., Chen F.F., Zhao L., Tian G., Liu X., Cui Y., Yang J.Y., Wu K.. Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in arabidopsis. PLos Genet. 2012; 8:e1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu C.W., Liu X.C., Luo M., Chen C.Y., Lin X.D., Tian G., Lu Q., Cui Y.H., Wu K.Q.. Histone Deacetylase6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 2011; 156:173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hung F.Y., Chen F.F., Li C.L., Chen C., Lai Y.C., Chen J.H., Cui Y.H., Wu K.Q.. The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res. 2018; 46:10669–10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hung F.Y., Chen F.F., Li C.L., Chen C., Chen J.H., Cui Y.H., Wu K.Q.. The LDL1/2-HDA6 histone modification complex interacts with toc1 and regulates the core circadian clock components in arabidopsis. Front. Plant Sci. 2019; 10:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jiang D., Yang W., He Y., Amasino R.M.. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell. 2007; 19:2975–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Greenberg M.V.C., Deleris A., Hale C.J., Liu A., Feng S.H., Jacobsen S.E.. Interplay between active chromatin marks and RNA-Directed DNA methylation in Arabidopsis thaliana. PLos Genet. 2013; 9:e1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lamesch P., Berardini T.Z., Li D.H., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M.et al.. The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012; 40:D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li C., Chen C., Gao L., Yang S., Nguyen V., Shi X., Siminovitch K., Kohalmi S.E., Huang S., Wu K.et al.. The Arabidopsis SWI2/SNF2 chromatin remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLos Genet. 2015; 11:e1004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li C., Gu L., Gao L., Chen C., Wei C.Q., Qiu Q., Chien C.W., Wang S., Jiang L., Ai L.F.et al.. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 2016; 48:687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen C., Li C.L., Wang Y., Renaud J., Tian G., Kambhampati S., Saatian B., Nguyen V., Hannoufa A., Marsolais F.et al.. Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat. Plants. 2017; 3:814–824. [DOI] [PubMed] [Google Scholar]
- 55. Chen C., Shu J., Li C., Thapa R.K., Nguyen V., Yu K., Yuan Z.C., Kohalmi S.E., Liu J., Marsolais F.et al.. RNA polymerase II-independent recruitment of SPT6L at transcription start sites in Arabidopsis. Nucleic Acids Res. 2019; 47:6714–6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu G., Wang L.G., He Q.Y.. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 57. Machanick P., Bailey T.L.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luo C.Y., Sidote D.J., Zhang Y., Kerstetter R.A., Michael T.P., Lam E.. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J. 2013; 73:77–90. [DOI] [PubMed] [Google Scholar]
- 59. Liu C., Xin Y., Xu L., Cai Z., Xue Y., Liu Y., Xie D., Liu Y., Qi Y.. Arabidopsis ARGONAUTE 1 binds chromatin to promote gene transcription in response to hormones and stresses. Dev. Cell. 2018; 44:348–361. [DOI] [PubMed] [Google Scholar]
- 60. Wierzbicki A.T., Cocklin R., Mayampurath A., Lister R., Rowley M.J., Gregory B.D., Ecker J.R., Tang H., Pikaard C.S.. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 2012; 26:1825–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wierzbicki A.T., Haag J.R., Pikaard C.S.. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008; 135:635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Haag J.R., Pikaard C.S.. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011; 12:483–492. [DOI] [PubMed] [Google Scholar]
- 63. Bohmdorfer G., Sethuraman S., Rowley M.J., Krzyszton M., Rothi M.H., Bouzit L., Wierzbicki A.T.. Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. Elife. 2016; 5:e19092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim Y.J., Wang R.Z., Gao L., Li D.M., Xu C., Mang H.G., Jeon J.E., Chen X.S., Zhong X.H., Kwak J.M.et al.. POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:14858–14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Deshaies R.J. SCF and cullin/RING H2-based ubiquitin ligases. Annu Rev. Cell Dev. .Bi. 1999; 15:435–467. [DOI] [PubMed] [Google Scholar]
- 66. Zhao D.H., Yang M., Solava J., Ma H.. The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev. Genet. 1999; 25:209–223. [DOI] [PubMed] [Google Scholar]
- 67. Song Y.H., Estrada D.A., Johnson R.S., Kim S.K., Lee S.Y., MacCoss M.J., Imaizumi T.. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:17672–17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu F.Q., Ni W.M., Griffith M.E., Huang Z.Y., Chang C.Q., Peng W., Ma H., Xie D.X.. The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell. 2004; 16:5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tanimoto M., Tremblay R., Colasanti J.. Altered gravitropic response, amyloplast sedimentation and circumnutation in the Arabidopsis shoot gravitropism 5 mutant are associated with reduced starch levels. Plant Mol. Biol. 2008; 67:57–69. [DOI] [PubMed] [Google Scholar]
- 70. Cui D.Y., Zhao J.B., Jing Y.J., Fan M.Z., Liu J., Wang Z.C., Xin W., Hu Y.X.. The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting Auxin biosynthesis and transport. PLos Genet. 2013; 9:e1003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shpak E.D., Berthiaume C.T., Hill E.J., Torii K.U.. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development. 2004; 131:1491–1501. [DOI] [PubMed] [Google Scholar]
- 72. Hou X.L., Zhou J.N., Liu C., Liu L., Shen L.S., Yu H.. Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 2014; 5:4601. [DOI] [PubMed] [Google Scholar]
- 73. Liu X., Yang Y.H., Hu Y.L., Zhou L.M., Li Y.G., Hou X.L.. Temporal-specific interaction of NF-YC and CURLY LEAF during the floral transition regulates flowering. Plant Physiol. 2018; 177:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M.. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005; 17:2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gao Z., Daneva A., Salanenka Y., Van Durme M., Huysmans M., Lin Z.C., De Winter F., Vanneste S., Karimi M., Van de Velde J.et al.. KIRA1 and ORESARA1 terminate flower receptivity by promoting cell death in the stigma of Arabidopsis. Nat.Plants. 2018; 4:365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lou Y., Gou J.Y., Xue H.W.. PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell. 2007; 19:163–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li C., Liu Z., Zhang Q., Wang R., Xiao L., Ma H., Chong K., Xu Y.. SKP1 is involved in abscisic acid signalling to regulate seed germination, stomatal opening and root growth in Arabidopsis thaliana. Plant Cell Environ. 2012; 35:952–965. [DOI] [PubMed] [Google Scholar]
- 78. Ali M.A., Azeem F., Nawaz M.A., Acet T., Abbas A., Imran Q.M., Shah K.H., Rehman H.M., Chung G., Yang S.H.et al.. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J. Plant Physiol. 2018; 226:12–21. [DOI] [PubMed] [Google Scholar]
- 79. Gu X.F., Jiang D.H., Wang Y.Q., Bachmair A., He Y.H.. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 2009; 57:522–533. [DOI] [PubMed] [Google Scholar]
- 80. Metzger E., Wissmann M., Yin N., Muller J.M., Schneider R., Peters A.H.F.M., Gunther T., Buettner R., Schule R.. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005; 437:436–439. [DOI] [PubMed] [Google Scholar]
- 81. Schulte J.H., Lim S.Y., Schramm A., Friedrichs N., Koster J., Versteeg R., Ora I., Pajtler K., Klein-Hitpass L., Kuhfittig-Kulle S.et al.. Lysine-Specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009; 69:2065–2071. [DOI] [PubMed] [Google Scholar]
- 82. Qin Y., Vasilatos S.N., Chen L., Wu H., Cao Z.S., Fu Y.M., Huang M., Vlad A.M., Lu B.F., Oesterreich S.et al.. Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene. 2019; 38:390–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang Z.B., Zang C.Z., Cui K.R., Schones D.E., Barski A., Peng W.Q., Zhao K.J.. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009; 138:1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kim Y.J., Greer C.B., Cecchini K.R., Harris L.N., Tuck D.P., Kim T.H.. HDAC inhibitors induce transcriptional repression of high copy number genes in breast cancer through elongation blockade. Oncogene. 2013; 32:2828–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bartholomeeusen K., Fujinaga K., Xiang Y.H., Peterlin B.M.. Histone deacetylase inhibitors (HDACis) that release the positive transcription Elongation Factor b (P-TEFb) from its inhibitory complex also activate HIV transcription. J. Biol. Chem. 2013; 288:14400–14407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Greer C.B., Tanaka Y., Kim Y.J., Xie P., Zhang M.Q., Park I.H., Kim T.H.. Histone deacetylases positively regulate transcription through the elongation machinery. Cell Rep. 2015; 13:1444–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hnilicova J., Hozeifi S., Duskova E., Icha J., Tomankova T., Stanek D.. Histone deacetylase activity modulates alternative splicing. PLoS One. 2011; 6:e16727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ye J.M., Llorian M., Cardona M., Rongvaux A., Moubarak R.S., Comella J.X., Bassel-Duby R., Flavell R.A., Olson E.N., Smith C.W.J.et al.. A pathway involving HDAC5, cFLIP and caspases regulates expression of the splicing regulator polypyrimidine tract binding protein in the heart. J. Cell Sci. 2013; 126:1682–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhou H.L., Luo G.B., Wise J.A., Lou H.. Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res. 2014; 42:701–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nakano Y., Kelly M.C., Rehman A.U., Boger E.T., Morell R.J., Kelley M.W., Friedman T.B., Banfi B.. Defects in the alternative Splicing-Dependent regulation of REST cause deafness. Cell. 2018; 174:536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dong H.Y., Hu J.G., Zou K.J., Ye M.L., Chen Y.W., Wu C.Y., Chen X., Han M.L.. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol. Cancer. 2019; 18:3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92. Chen F., Qi S., Zhang X., Wu J., Yang X., Wang R.. lncRNA PLAC2 activated by H3K27 acetylation promotes cell proliferation and invasion via the activation of Wnt/betacatenin pathway in oral squamous cell carcinoma. Int. J. Oncol. 2019; 54:1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang D., Qu Z.P., Yang L., Zhang Q.Z., Liu Z.H., Do T., Adelson D.L., Wang Z.Y., Searle I., Zhu J.K.. Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants. Plant J. 2017; 90:133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chishima T., Iwakiri J., Hamada M.. Identification of transposable elements contributing to Tissue-Specific expression of long Non-Coding RNAs. Genes (Basel). 2018; 9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and ChIP-seq short read data have been submitted to the NCBI Gene Expression Omnibus (GEO) database, including the previously published LDL1 ChIP-seq data (GSE118025) (46) and unpublished data from HDA6 ChIP-seq (GSE132563), H3Ac ChIP-seq of WT, hda6, ldl1/2 and hda6/ldl1/2 (GSE132636), H3K4me2 ChIP-seq of WT, hda6, ldl1/2 and hda6/ldl1/2 (GSE133005) and RNA-seq of WT, hda6, ldl1/2 and hda6/ldl1/2 (GSE132873).