Abstract
Characterizing genes that are critical for the survival of an organism (i.e. essential) is important to gain a deep understanding of the fundamental cellular and molecular mechanisms that sustain life. Functional genomic investigations of the vinegar fly, Drosophila melanogaster, have unravelled the functions of numerous genes of this model species, but results from phenomic experiments can sometimes be ambiguous. Moreover, the features underlying gene essentiality are poorly understood, posing challenges for computational prediction. Here, we harnessed comprehensive genomic-phenomic datasets publicly available for D. melanogaster and a machine-learning-based workflow to predict essential genes of this fly. We discovered strong predictors of such genes, paving the way for computational predictions of essentiality in less-studied arthropod pests and vectors of infectious diseases.
INTRODUCTION
The vinegar fly (Drosophila melanogaster) is a well-established model organism used to investigate insect biology and genetics as well as a range of molecular processes in metazoans such as development and inheritance (1,2). The availability of a high-quality genome for D. melanogaster (3) and molecular tools that allow transcriptional perturbation (e.g. RNAi (4–7)), genomic disruption (e.g. chemical/transposon mutagenesis (8–10) and/or site-directed methods such as CRISPR/Cas9 (11) have enabled the detailed elucidation of the functions of individual genes in this fly. These efforts have also allowed the discovery of genes that are critical for the survival of the organism, referred to as ‘essential’ genes. Extensive genomic-phenomic data and information as well as results from multiple ‘omics studies have been curated, integrated and catalogued in reference databases, such as FlyBase (12), FlyVar (13), modENCODE (14), Ensembl (15) and GenomeRNAi (16). Despite these efforts, the results on gene essentiality for individual genes from multiple functional genomics experiments can vary (17). Sources of such variability or ambiguity can relate to experimental or environmental conditions, developmental stage, sex, strain and/or experimental biases and/or errors (17,18). Defining essential genes in D. melanogaster and their characteristics might identify factors or features that define essentiality in other taxa (19), which suggest that computational tools would find applicability to predict gene essentiality in lesser studied organisms such as arthropod vectors and pests which cause substantial economic losses to agricultural industries as well as disease burdens in animals, humans and plants worldwide (20–23). In the absence of functional genomics platforms for most arthropods, computational methods capable of exploiting extensive ‘omics datasets to predict essential genes are desirable.
Despite the importance of D. melanogaster as a model organism, the abundance of publicly available ‘omics datasets for this insect has not been fully explored for the discovery of predictors of gene essentiality, and reliable computational approaches for the genome-wide prediction of essential genes of the vinegar fly are lacking. A range of genomic features, such as gene size, evolutionary rate, phyletic retention, transcription level, connectivity in protein–protein interaction (PPI) networks, cellular or subcellular localization, and/or sequence-derived features (24–27) have been linked to essentiality in eukaryotes. For D. melanogaster, studies have sought to infer essential genes computationally using features based on gene homology/orthology/ontology, PPI/co-expression networks, sequence-derived (nucleotide/protein), or combinations thereof (24,26–27). However, there are limitations in genome-wide predictions, particularly using PPI data and/or sequence-based features. In particular, PPI experiments may contain marked levels of false-positive and negative results (28,29), significantly affecting predictions. In addition, such datasets are limited or unavailable for non-model organisms. Additionally, although sequence-derived features alone have proven useful for essential gene predictions, their performance is still suboptimal, even if combined with PPI network features (26,27).
Here, we employed a novel computational approach that employs a large-scale feature extraction, engineering and selection procedure that takes into account variability in phenomic datasets for the inference of essential genes on a genome-wide scale. Using this approach, we built and systematically evaluated machine-learning (ML) models for the genome-wide prediction of essential genes in D. melanogaster.
MATERIALS AND METHODS
Datasets
We obtained comprehensive genomic-phenomic data and associated annotations from three sources: FlyBase (12), the Ensembl database (15) and/or peer-reviewed publications. Datasets derived from functional genomics (phenomic) as well as from genomic, transcriptomic, proteomic and epigenetic in GFF linked to the D. melanogaster genome were from FlyBase (version r6.30/FB2019_05). Data from RNAi experiments were obtained from the database GenomeRNAi (16). From Ensembl, we obtained genomic, coding sequences (CDSs) and proteins (canonical) data. For D. melanogaster, we also obtained gene transcription data for different tissues (30); multi-cell or single-cell transcriptomic data from embryo (31), gonads (GEO accession: GSE125947), testis (32), brain (33) and wing disc (34); Ribo-seq annotations (35); proteomic data (36); ATAC-seq peaks (37), and variomic data (genome-wide SNPs) (13,38).
Annotation of gene essentiality using phenomic data
Using phenomic data from FlyBase, we established a scoring system to provisionally annotate genes of D. melanogaster as essential (see ‘Data Availability’ section). Initially we extracted all mutant allele identifiers associated with ‘lethal’ or ‘viable’ phenotypes from the ‘allele_phenotypic_data_fb_2019_05.tsv’ file. Then, we used these allele identifiers to determine corresponding genes and count the number of ‘lethal’ or ‘viable’ entries per gene in the ‘fbal_to_fbgn_fb_2019_05.tsv’ file. For each gene, we then calculated an essentiality score (ES), defined as the total number of alleles linked to essential/lethal (E) terms squared divided by the total number of experiments linked to essential/lethal plus non-essential/viable terms (T) squared (E2/T2). A gene was designated as ‘essential’ (ES > 0.9), ‘non-essential’ (ES < 0.1) or ‘conditional-essential’ (0.1 ≤ ES ≤ 0.9).
Feature extraction or engineering
For individual genes, features were extracted from six (i.e. genomic, CDSs, overlapping-gene, transcriptomic, protein and ‘variome’) datasets derived from FlyBase, Ensembl and/or published studies; see ‘Datasets’ section above).
From genomic data, we extracted features including length, number of exons, distance from the chromosome center (average distance between start codon of the first gene and the stop codon of the last gene in a chromosome), number of isoforms and presence/absence of associated Pfam-domains using ‘biomaRt’ for R. From CDSs, we extracted nucleotide composition and correlation features using rDNAse for R (https://cran.r-project.org/web/packages/rDNAse) as well as codon usage features using CodonW (http://codonw.sourceforge.net).
For datasets associated with genomic coordinates (e.g. FlyBase annotations, TSS, Ribo-seq, proteomic and ATAC-seq), we engineered novel features by identifying and counting annotations whose genomic coordinates overlapped gene locations using the program BEDTools. For example, this approach was used to identify and count features in the GFF file obtained from FlyBase (column 2) which overlap with coordinates of genes.
For ‘pooled’ transcriptomic data from distinct tissues, we used the expression levels of individual genes for every tissue and experimental condition as features. For single-cell transcriptomic data, we obtained the transcription level for each gene in each cell and enumerated the cells transcribing a particular gene.
From protein sequences, we extracted features using ‘protr’ utilizing all descriptors defined in this package (https://cran.r-project.org/web/packages/protr) as well as the numbers of predicted transmembrane domains and signal peptides per protein employing TMHMM (39) and SignalP (40), respectively. We also obtained features from predicted protein subcellular localizations using WolfPsort (41) and DeepLoc (42) as well as protein disorder features employing DisEMBL (43).
For the variome of D. melanogaster, we calculated the numbers of SNPs in individual genes using BEDTools (44) and inferred the effect(s) of individual SNPs on gene function using SnpEff (45)—and used these data as features. The datasets and code used to extract or engineer features are in the ‘R Markdown’ script available at https://bitbucket.org/tuliocampos/essential_melanogaster (commit tag: NARGAB).
Feature sets
We combined all extracted/engineered features with essentiality annotations for respective genes and stacked this information into a matrix using R. In this feature matrix, each line represented a gene, each column represented an extracted feature and the last column represented the essentiality annotation (‘essential’ or ‘non-essential’); this matrix contained all data (‘FULL’). To create a non-redundant (NR) set of features, we first clustered protein sequences using USEARCH (parameters: -cluster_fast -centroids) (46), obtained gene identifiers and then removed genes and associated features if multiple amino acid sequences had ≥25% identity, retaining only the centroid sequences of all individual clusters. Subsequently, we removed features with low variance (i.e. when the percentage of unique values was <10%, or when the frequency of the commonest value divided by the frequency of the second commonest value was >19) from both the ‘FULL’ and ‘NR’ feature sets using the nearZeroVar method in ‘caret’. For the ‘FULL’ dataset, we also assessed statistical differences in the features between ‘essential’ and ‘non-essential’ using two-tailed pairwise t-tests (95% confidence interval) in R (t-test), recording p-values and Holm–Bonferroni-corrected (p.adjust) values.
Feature selection, ML training and performance assessment
Features were selected by random subsampling from 10 to 90% of data representing ‘essential’ or ‘non-essential’’ genes (in 10% stepwise increments) based on a consensus between elasticNet (alpha = 0.5) and ensemble Sparse Partial Least Squares (SPLS) methods using ‘glmnet’ and ‘enspls’ in R, respectively (26). The individual feature values were then normalized by subtracting the mean and dividing by the standard deviation calculated for each feature column. Normalized features were used to train each of six ML-models (GBM (Gradient Boosting Machine), GLM (Generalized Linear Model), NN (Neural Network—perceptron), Random Forest (RF), SVM (Support-Vector Machine) (26) and XGB (eXtreme Gradient Boosting—xgbTree) in the ‘caret’ R-package. During the training process, we employed parameter-tuning and 5-fold cross-validation, ultimately selecting the models with highest ROC-AUC. Following subsampling, we employed the remaining data (90 to 10%) to evaluate the performance of the final models using ROC-AUC and PR-AUC.
Subsequently, we trained each of the six ML-models with 100% of each feature set, and calculated the ‘importance’ of each feature for each ML algorithm and feature set using the varImp method in the ‘caret’ package. For each ML-model, we calculated ROC-AUCs using 5-fold cross-validation and plotted them against the parameters tested. We ranked the predictors according to the median feature-importance for the best three ML-models and selected 40 consensus-features that were highly predictive of gene essentiality employing the ‘FULL’ or ‘NR’ dataset. Then, we assessed whether these consensus-features correlated with essentiality using ‘correlationfunnel’ (https://cran.r-project.org/web/packages/correlationfunnel), and evaluated pairwise correlations among features using ‘corrplot’ (R). Using this reduced set of consensus-features (NR_SELECTED), we then trained the ML-methods and evaluated their prediction-performance using ROC-AUC and PR-AUC and used the final models to predict essentiality of all genes (n = 11 580) included in the present study. Finally, we assessed variation in these metrics using bootstrapping (1000-times) employing 90% of the consensus-features for training and the remaining 10% for testing.
Distribution of gene and SNPs on chromosomes
We counted the number of SNPs per each 1000 bp-window on each chromosome using published variomic data (13). We established the locations of genes that has been provisionally annotated as ‘essential’, ‘non-essential’ or ‘conditional-essential’ using the FlyBase annotation file (GFF format), and generated individual density plots to show the distributions of genes for each chromosome (‘ggplot’ for R). We compared the distributions of genes based on essentiality annotations conducted using Kolmogorov–Smirnov tests (ks.test in R).
Gene ontology (GO), transcription and tissue enrichment analyses
Using the GBM, RF and XGB methods trained with NR_SELECTED data, we identified D. melanogaster genes with the highest median probabilities (>0.7) of being essential, and then conducted gene ontology (GO) enrichment analysis. For these genes, GO enrichment (for biological process, molecular function or cellular component) was carried out using the database DAVID (47).
Independent validation of ML predictions using RNAi data
To validate the final ML-based predictions using the NR_SELECTED set, we queried individual genes predicted to be more likely essential (probability > 0.7) or non-essential (probability < 0.1) against the GenomeRNAi database, identifying matches to experiments linked to ‘lethal’ phenotypes (i.e. lethal, decreased cell number, decreased viability, decreased cell number, decreased viability, decreased cell viability or low cell number). As there was no clear description of ‘viable’ phenotypes in the GenomeRNAi database, we investigated the ratios of genes with at least one hit to a ‘lethal’ phenotype in this database. We queried from one to all genes, starting with genes with the highest probability of being essential, then adding an individual gene with a lower probability at a time, recalculating the ratios until all genes used in this study were queried. The same analysis was done, starting from a single gene with the lowest probability of being essential, then adding one gene with a higher probability at a time, recalculating the ratios as each gene were included, until all genes were represented. Pearson's correlation coefficients were calculated using cor.test in R to assess the correlation between the ratios and the ML probabilities. We also tested whether the ML probabilities could be used to predict the ratio of genes found with a ‘lethal’ phenotype by RNAi using a linear model (lm) in R.
RESULTS
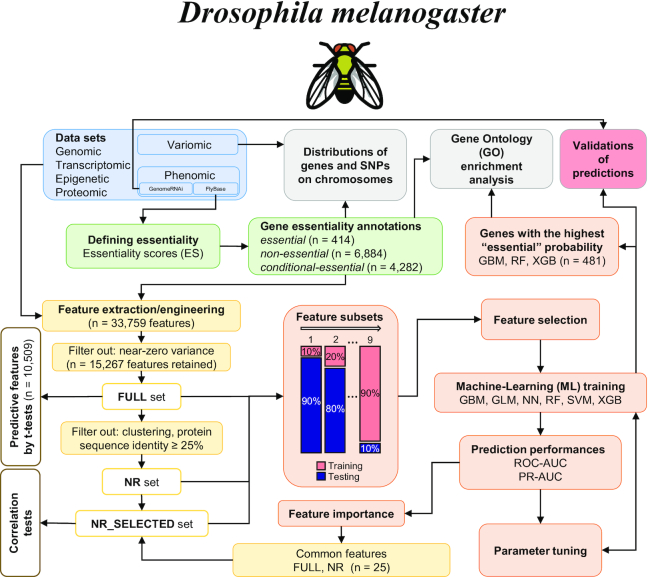
We implemented a workflow (Figure 1) that comprises: (i) the assignment of genes as essential based on genomic-phenomic data; (ii) the extraction of features while employing methods to identify predictors of essentiality on a genome-wide scale; (iii) the systematic training and evaluation of ML using subsets of features; (iv) the validation of gene essentiality predictions using independent experimental (gene knockdown) datasets; and (v) the inference of chromosome locations linked to SNPs and genes predicted to be essential; and (vi) the assignment of GO terms enriched for essential genes.
Figure 1.
Workflow employed in the present study. First, a wealth of publicly available ‘omics datasets for Drosophila melanogaster were obtained (blue). Then, we employed a ‘scoring system’ to annotate D. melanogaster genes for essentiality (green) using phenomic data. Next, we extracted or engineered features (yellow) from the datasets to establish feature sets (FULL—all features; NR—all features from sequences containing <25% amino acid identity; NR_SELECTED—25 highly predictive features of essentiality, selected from the NR dataset). These feature sets were used for a systematic evaluation of ML approaches for essential gene predictions (orange). Statistical significance (t-tests) and correlation tests were performed on the FULL and NR_SELECTED sets, respectively. The performances of the individual ML models, and the importance of the selected features for essentiality predictions were calculated and evaluated (orange). Independent validations of the ML predictions using knockdown (RNAi) data was also performed (red). Finally, GO enrichment and preferential genomic locations of SNPs and genes by essentiality annotations were evaluated (gray).
Provisional essentiality annotations
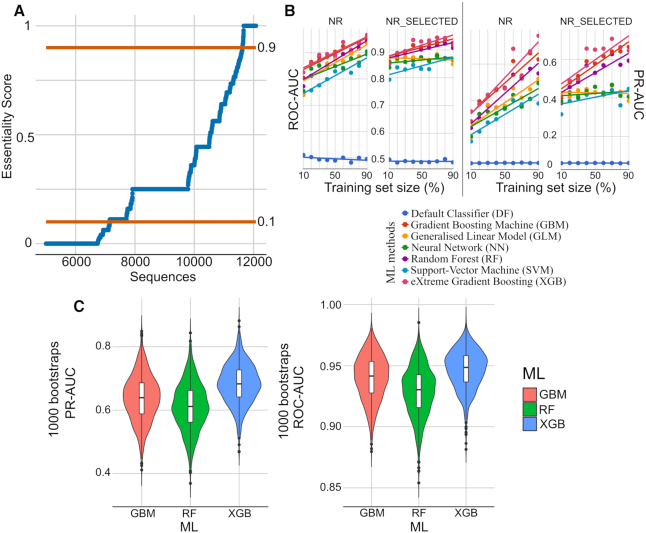
From phenomic data, we provisionally annotated genes as ‘essential’, ‘non-essential’ or ‘conditional-essential’ by applying an ES to each D. melanogaster gene (Figure 2A). For each gene, ES (E2/T2) was calculated using published genomic-phenomic data within FlyBase—the number of functional genomics experiments which recorded ‘lethal’ phenotypes (E) and the total number of experiments (T) reporting ‘lethal’ or ‘viable’ phenotypes (Supplementary Table S1). Based on ES and defined thresholds (0.1 and 0.9; cf. Figure 2A), we provisionally annotated 414 protein-coding genes as essential, 6884 as non-essential and 4282 as conditional-essential, with 158 (38.1%) essential, 946 (13.7%) non-essential and 2708 (63.2%) conditional-essential genes being supported by at least three experiments inferring ‘lethal’ and/or ‘viable’ phenotypes (Supplementary Tables S2–4). Of all of these 11 580 genes, 4920 genes were listed FlyBase as having both ‘lethal’ and ‘viable’ phenotypes, of which 87% (n = 4282) were annotated as conditional-essential.
Figure 2.
Annotation of essential genes from phenotype data, and performance of ML methods for essentiality predictions. (A) Drosophila melanogaster genes were curated for essentiality using phenotype data available in FlyBase. For each gene, an ES was calculated (y-axis) and ordered using the formula E2/T2, were ‘E’ is the number of entries relating to lethality/essentiality and ‘T’ is the total number of entries (‘lethal’ + ‘viable’) reported. Genes were annotated as ‘essential’ if ES was >0.9, or ‘non-essential’ if was ES < 0.1, or ‘conditional-essential’ otherwise. (B) In the systematic evaluation of gene essentiality predictions (‘essential’ versus ‘non-essential’) the performance of six ML algorithms and a default classifier were assessed, initially with a dataset (FULL) containing all genes curated previously and their features (not shown). In addition, a non-redundant (NR) dataset was created, containing all features from genes whose amino acid sequence identities were <25%. Another dataset containing the NR genes and a selection of 25 best-predictive features (NR_SELECTED) was also evaluated. For each dataset, random subsets of genes (10–90%, 10% increments) were used as training sets (x-axis), and the remaining 90–10% used as independent test sets. At each step, the prediction performance was evaluated using the test set using ROC-AUC (right) and PR-AUC (left) metrics. (C) Violin and box plots of ROC-AUC and PR-AUC from 1000 bootstraps of RF, XGB and GBM, with random sampling of 90% of the NR_SELECTED used for training and the remaining 10% of this feature set used for testing.
Predictors of essentiality identified from ‘omics data
For each D. melanogaster gene initially annotated for essentiality, we extracted 33 759 features from multiple (genomic, transcriptomic, variomic, proteomic and epigenetic) ‘omics datasets (Supplementary Table S5). The removal of features that exhibited low variance (deviation from the mean), left 15 267 for subsequent analyses (see Supplementary Table S5). We performed t-tests to compare these features between gene sets annotated as essential and non-essential, and identified 10 509 features with significant Holm–Bonferroni-corrected P-values (P < 0.05); 10 149 (96.6%) of these features were derived from single-cell RNA-seq data (scRNA-seq), 273 (2.6%) from sequence (nucleotide or protein), 53 (0.5%) from genomic annotations in FlyBase, 25 (0.2%) from RNA-seq data, 6 (0.06%) from subcellular localization predictions, 2 (0.02%) from genomic annotations (Ensembl) and 1 (0.01%) from proteomic data (see Supplementary Table S6).
Systematic feature selection and ML approaches
First, we used the complete (FULL) set of features (n = 15 267) obtained for genes annotated previously as ‘essential’ and ‘non-essential’. Briefly, subsets of the FULL set ranging from 10 to 90% (in 10%-increments) were used to select features and train six distinct ML algorithms (Gradient Boosting Machine, GBM; Generalized Linear Model, GLM; Neural Network, NN; Random Forest, RF; Support-Vector Machine, SVM; and eXtreme Gradient Boosting, XGB). The prediction performances (ROC-AUC and PR-AUC) of the models to predict the training sets was consistently ∼1 for both RF and XGB. For GBM and SVM, the ROC-AUC improved from 0.9 or 0.8 to 1, while the PR-AUC increased from 0.8 to 1 for both. ROC-AUC ranged between 0.93 and 0.95 for GLM and NN, whereas PR-AUC increased from 0.8 to 0.9 for NN, and decreased from 0.7 to 0.6 for GLM. For the predictions of independent test sets (with 90 to 10% of the data not being used for training), ROC-AUC for GBM, RF and XGB increased from >0.8 to 0.95, while PR-AUC improved from 0.3 to ∼0.65–0.7. Similar performances were observed for GLM and NN (ROC-AUC from 0.8 to ∼0.87 and PR-AUC from 0.3 to ∼0.5). For SVM, ROC-AUC improved from 0.75 to 0.9 and PR-AUC from 0.2 to 0.55. Following the final selection of features using the FULL set, a total of 200 essentiality predictors were identified, and their relevance in each ML model was recorded (Supplementary Table S7).
Second, we defined a non-redundant (NR) dataset by removing features of genes whose proteins contained ≥25% amino acid identity based on clustering analysis, retaining the features pertaining to a single representative gene (centroid). The NR dataset contained sets of 402 essential and 6138 non-essential genes, with 15 267 features for each gene. For the NR set, we employed the same systematic approach for data-partitioning and ML training/evaluation as used for the FULL set. When predicting the training sets, ROC-AUC was consistently ∼1 for GBM, RF, SVM and XGB, whereas it was ∼0.92 for GLM and NN. The PR-AUC was ∼1 for RF, ∼0.97 for SVM and XGB, and ∼0.87 for NN, but increased from 0.87 to 0.95 for XGB, and decreased from 0.75 to 0.6 for GLM. When predicting the test sets (Figure 2B), the ROC-AUC metric improved from 0.84 to 0.96 for GBM and XGB, from 0.8 to 0.94 for GLM and RF, from 0.8 to 0.9 for NN, and from 0.75 to 0.88 for SVM. PR-AUC increased from 0.35 to ∼0.65–0.7 for GBM and XGB, from 0.3 to 0.6 for RF, from 0.3 to ∼0.5 for GLM and NN, and from 0.2 to 0.45 for SVM. The final selection of features using the NR set identified 115 gene essentiality predictors (Supplementary Table S8).
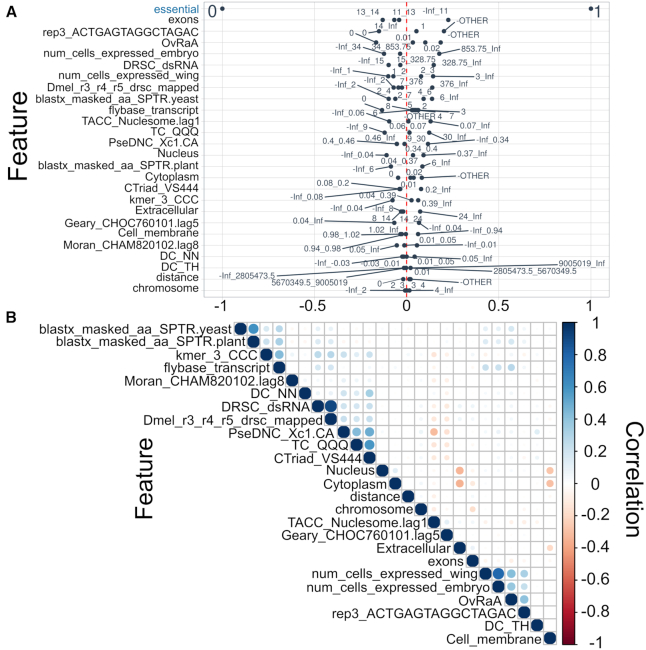
Third, we observed that 40 features from the FULL or NR sets contributed most to essentiality prediction (Supplementary Tables S7 and 8) and that 25 of these features were common to both sets. These 25 ‘strong predictors’ of gene essentiality were features from: (i) genomic annotations (e.g. number of ‘exons’, ‘chromosome’ location, ‘distance’ from the chromosome center); (ii) derived from composition or autocorrelation of nucleotide or amino acid sequences (e.g. ‘DC_NN’, ‘DC_TH’, ‘TC_QQQ’, ‘CTriad_VS444’, ‘PseDNC_Xc1.CA’, ‘Moran_CHAM820101.lag8’, ‘Geary_CHOC760101.lag5’, ‘kmer_CCC’, ‘TACC_Nucleosome.lag1’—obtained using ‘protr’ or ‘rDNAse’ for R); (iii) related to similarity to genes of other organisms (e.g. ‘blastx_masked_aa_SPTR.yeast’ and ‘blastx_masked_aa_SPTR.plant’—from FlyBase annotations); (iv) obtained from subcellular localization predictions (e.g. ‘Nucleus’, ‘Cytoplasm’, ‘Extracellular’, ‘Cell_membrane’); and (v) linked to transcription (e.g. ‘flybase_transcript’—represents annotations of transcripts from modENCODE within FlyBase; ‘DRSC_dsRNA’ and ‘Dmel_r3_r4_r5_drsc_mapped’—RNAi probes mapped to genes in FlyBase; ‘rep3_ACTGAGTAGGCTAGAT’—transcription levels of genes in a gonad cell (scRNA-seq); ‘num_cells_expressed_wing’ and ‘num_cells_expressed_embryo’—number of cells where a gene is transcribed in wing disc and embryo based on single cell data; and ‘OvRaA’—RNA-seq levels of transcription of genes in the ovary following treatment with rapamycin).
Fourth, we evaluated the correlations of each of these 25 strong predictors with gene essentiality, and then assessed pairwise correlations among them. The correlations with essentiality ranged from near zero (e.g. ‘chromosome’ and ‘distance’) to ∼0.25 (e.g. ‘exons’, ‘rep3_ACTGAGTAGGCTAGAC’, ‘OvRaA’) (Figure 3A). The number of exons (<11), transcription levels in a gonad cell (‘rep3_ACTGAGTAGGCTAGAC’; >1) and in the ovary treated with rapamycin (‘OvRaA’; >854.75) were each inferred to be linked to essential genes. Upon pairwise comparison (Figure 3B), there were weak or no correlations (−0.3 to 0.3) among >98% of predictors, moderate negative correlations (−0.3 to −0.5) between ‘extracellular’ and either ‘nucleus’ or ‘cytoplasm’, and moderate positive correlations (0.3 to 0.5) between ‘OvRaA’ and either ‘num_cells_expressed_wing’ or ‘num_cells_expressed_embryo’; between ‘kmer_CCC’ and ‘flybase_transcript’; and between the sequence features ‘PseDNC_Xc1.CA’ and ‘TC_QQQ’. Strong correlations (>0.5) were observed between ‘DRSC_dsRNA’ and ‘Dmel_r3_r4_r5_drsc_mapped’ (FlyBase features—RNAi probes), between ‘PseDNC_Xc1.CA’ and both ‘TC_QQQ’ and ‘CTriad_VS444’, between ‘num_cells_expressed_wing’ and ‘num_cell_expressed_embryo’ (scRNA-seq), and between ‘blastx_masked_aa_SPTR.yeast’ and ‘blastx_masked_aa_SPTR.plant’ (FlyBase annotations—protein sequence similarity to other organisms).
Figure 3.
Correlations of features. (A) The correlations (x-axis) of 25 highly predictive features (y-axis) with gene essentiality, generated using ‘correlationfunnel’ for R. Individual dots represent value ranges from i to j (i_j); Inf means any value above i; -Inf means any value below j; ‘OTHER’ means any other value. (B) The pairwise correlation among these 25 predictors.
Fifth, we carried out the systematic training and evaluation of each of the six ML algorithms using a subset of the NR set (402 essential and 6138 non-essential genes; the NR_SELECTED set) containing the 25 most highly predictive features identified in both the FULL and NR sets (Figure 2B). Using the NR_SELECTED set to predict the training sets, the ROC-AUC was ∼1 for GBM and RF and ∼0.9 for GLM and NN, and increased from 0.95 to ∼1 for XGB and from 0.92 to ∼1 for SVM. The PR-AUC was >0.97 for GBM, RF and XGB, increased from ∼0.65 to ∼1 for GBM and SVM, and remained relatively constant at ∼0.5 for GLM, and decreased from 0.8 to 0.7 for NN. When evaluating the prediction performances for the independent test sets, the ROC-AUC increased from 0.87 to between 0.93 and 0.96 for GBM, RF and XGB, from 0.82 to 0.87 for SVM, whereas it remained consistently at ∼0.87 for GLM and NN. The PR-AUC improved from ∼0.45 to 0.65 for GBM and XGB, and from 0.4 to 0.6 for RF, and remained at ∼0.4 for GLM, NN and SVM. Following the calculation of the median relative importance values for each of the six ML models, the five best of the 25 predictors for NR_SELECTED were ‘exons’ (importance = 100), ‘OvRaA’ (77.74), ‘num_cells_expressed_wing’ (53.54), ‘Nucleus’ (48.51) and ‘num_cells_expressed_embryo’ (Supplementary Table S9). Using the NR_SELECTED set, we assessed the variation in ROC-AUC and PR-AUC by employing a bootstrapping approach (n = 1000), where most (90%) of the data was randomly selected for ML training (GBM, RF and XGB), leaving the remaining 10% for testing and for the calculation of performance. Violin and box plots (Figure 2C) show that the ROC-AUC ranged between ∼0.85 and 1 for each of the three ML algorithms, with medians being between 0.93 and 0.95. Regarding the PR-AUC, the metric ranged from 0.4 to 0.8, and medians from 0.60 to 0.68. Overall, XGB exhibited the best performance for both metrics.
Finally, the ML models trained with the 25 features (i.e. ‘strongest predictors’) were each used to predict essentiality for all 11 580 genes included in this study, and the essentiality probabilities for each model was determined (Supplementary Tables S10 and 11). Considering the median probabilities of the best-performing models (i.e. GBM, RF and XGB), 482 genes had a high probability (>0.7) of being essential. Moreover, for each of the feature sets (i.e. FULL, NR and NR_SELECTED), we evaluated the effect of parameter-tuning on the ROC-AUC for each of the six ML algorithms. Regarding ML parameter tuning, the ROC-AUC achieved a higher performance in most cases using: a regularization-parameter value of <0.001 for GLM; ≥1000 boosting iterations and max-tree-depth of 10 or 20 for GBM; ≥200 boosting iterations and max-tree-depth of ≥10 for XGB; variable numbers of hidden-layer units for NN, depending on the dataset used; 50 randomly selected predictors for RF using the FULL or NR set, or 10 using NR_SELECTED; sigma-parameter of ≤0.01 for SVM using FULL or NR, or ≥0.1 using NR_SELECTED.
Independent validation of essential gene predictions using RNAi data
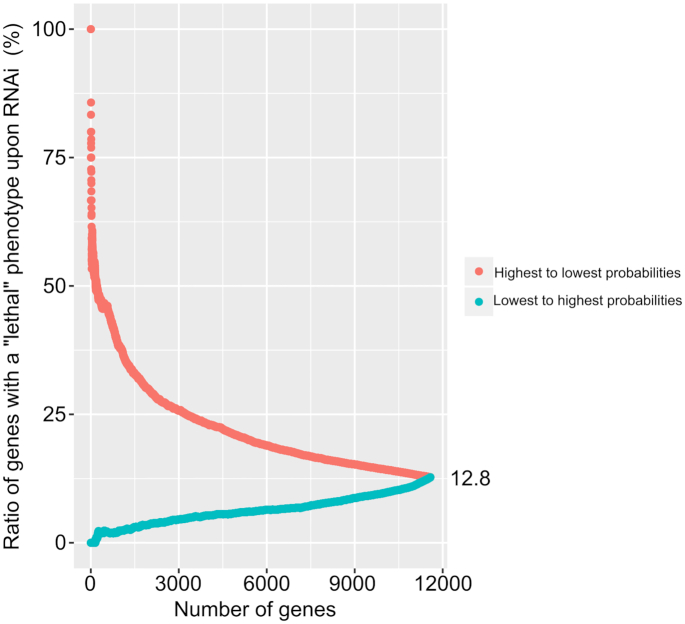
We validated the gene essentiality predictions using independent functional genomics (RNAi) data from the GenomeRNAi database (see Figure 4). The total number of genes with at least one ‘lethal’ phenotype was 1478, corresponding to a ratio of 12.8% of all 11 580 genes included in the present study. On the one hand, considering the genes predicted as most likely being essential by the ML approach (n = 482; probability of >0.7), 48.8% (n = 235) were supported by a ‘lethal’ phenotype from a least one experiment. On the other hand, of the genes predicted as most likely being non-essential (n = 9577; probability of <0.1), 9.6% (n = 918) had a ‘lethal’ RNAi phenotype. Next, we evaluated the relationship between the ML prediction probabilities and the ratios of genes with a ‘lethal’ RNAi phenotype. We serially queried individual genes against the GenomeRNAi database, from highest to lowest ML probability of being essential, and cumulatively re-calculated the ratios of genes linked to ‘lethal’ phenotypes. We then used the same approach, proceeding from the gene with the lowest probability (based on ML predictions) to that with the highest probability. The ratio decreased from 100% to 12.8% as more genes were included—from the highest to lowest probabilities (Figure 4; light red). Conversely, the ratio increased from zero to 12.8%, when including genes from the lowest to the highest probabilities (Figure 4). Interestingly, the Pearson's correlation coefficient between the ratios and the ML probabilities was ∼0.89 (P < 2.2e-16). Moreover, we tested whether the ML probabilities could be used as a cut-off to predict the ratio of genes showing a ‘lethal’ phenotype by RNAi. The adjusted R2 for the linear model was ∼0.80 (P < 2.2e-16).
Figure 4.
Independent validation of ML essentiality predictions using functional genomics data (RNAi). Initially, genes predicted for essentiality using ML approaches (GBM, RF and XGB; NR_SELECTED feature set) were ranked by probability of being essential. Then, from the highest to the lowest probability (light red) and from the lowest to the highest (light blue), these genes were cumulatively searched against the GenomeRNAi database and the ratios of genes with at least one ‘lethal’ phenotype were calculated.
Distribution of SNPs and genes along the chromosomes of D. melanogaster
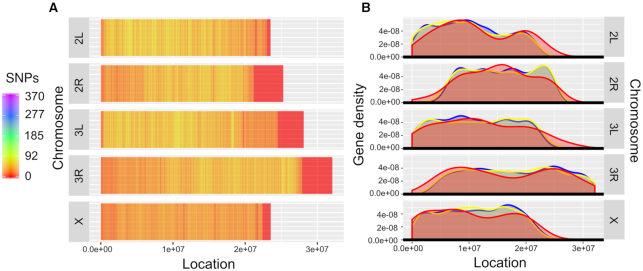
We generated a plot of the number of SNPs per 1000 bp-window for each of the largest chromosomes (i.e. 2, 3 and X; Figure 5A). The average number of SNPs per window was usually within the range of 0–92, with a maximum of 370, with reduced numbers being near the centromeric (autosomes) and telomeric (autosomes and X) regions (edges of chromosome segments; Figure 5A) compared with other areas of the genome. Fewer SNPs were found on the X chromosome compared with autosomal chromosomes. Chromosomal regions on the right of centromeres (i.e. 2R and 3R) exhibited wider genomic regions with low SNP density compared with the left.
Figure 5.
Distributions of single nucleotide polymorphisms (SNPs) and genes along Drosophila melanogaster chromosomes. (A) The distribution of SNPs (1000 bp-windows) along D. melanogaster chromosomes, based on a variant-call file (VCF) derived from whole-genome sequencing of D. melanogaster populations. (B) Density plots showing the distributions of genes along D. melanogaster chromosomes, stratified by essentiality annotations (red: ‘essential’; blue: ‘non-essential’; yellow: ‘conditional-essential’).
We assessed the distribution of genes along chromosomes (density plots), stratified by their essentiality annotations (Figure 5B). We observed that gene distributions on individual chromosomes were similar, irrespective of their annotation. Using Kolmogorov–Smirnov tests to compare these distributions, there was no significant difference in distribution of essential and non-essential (P = 0.16), or essential and conditional-essential (P = 0.39), or non-essential and conditional-essential (P = 0.19) genes. As for the SNP distributions, we observed a reduction in gene density near centromeric and telomeric regions. Based on Hartigans’ dip tests, the distributions of essential genes were unimodal for 2R (P = 0.48), 3L (P = 0.61) and X (P = 0.1), and non-unimodal for 2L (P = 0.043) and 3R (P = 0.01). Thus, essential genes were inferred to be preferentially located left on chromosome regions 2L (bimodal-low) and 3L, left on chromosome X; central on region 2R; and bimodal-even on region 3R. The numbers of essential genes on chromosomal regions were 73 (2L), 82 (2R), 79 (3L), 107 (3R), 2 (4), 71 (X) and none (Y), suggesting an overall preference for the right-hand side of chromosome 3 (3R).
Gene ontology (GO) terms enriched for essential genes
To obtain insight into the biological processes, cellular components and/or molecular functions in which essential genes are involved, we performed GO enrichment analysis using the database DAVID (47) (see Supplementary Table S12). For biological processes, the three most significantly enriched terms (P ≤ 1.9e-3) were ‘cytoplasmic translation’ (43 genes), ‘centrosome duplication’ (18) and ‘translation’ (47). For cellular components, highly enriched terms (P ≤ 1.7e-16) were ‘ribosome’ (43), ‘cytosolic small ribosomal subunit’ (23) and ‘cytosolic large ribosomal subunit’ (25). For molecular functions, the most predominant terms (P ≤ 1.7e-16) were ‘structural constituent of ribosome’ (47), ‘ATP binding’ (63), and ‘transcription factor activity sequence-specific DNA binding’ (37).
DISCUSSION
In the present work, we show that essential genes in D. melanogaster can be predicted with high confidence using ML methods. The prediction performance achieved here is attributed to: (i) a thorough annotation of essential genes using phenotypic data and development of a scoring system; and (ii) the discovery of informative predictive features (‘predictors’) from extensive ‘omics datasets publicly available for the vinegar fly.
The availability of the D. melanogaster genome (3) has enabled numerous functional investigations of genes in this fly (see (48)), allowing the experimental inference of essential and non-essential genes. However, it is evident that there is a subset of genes that has been classified as ‘essential’ (lethal phenotype) in particular knock-down experiments (4) and ‘non-essential’ (viable phenotype) in others (49). This ‘ambiguous’ subset, referred to here as conditional-essential, might be explained by the variable expression of a phenotype under distinct experimental conditions or in different life stages of the fly and/or an occasional mis-recording of a phenotype due to the large-scale nature of such experiments and the sheer number of flies being examined. In order to reduce possible bias or errors, in the present study, we established a system to separately score genes as essential, non-essential or conditional-essential. Establishing this system was crucial, so that we could set a threshold to infer essential and non-essential genes based on reliable phenotypic data as a basis to accurately train ML methods using feature engineering and selection.
We discovered strong predictors of essentiality, some of which were entirely novel (see Supplementary Table S6), and moved from using thousands of features to 25. In future work, a range of feature selection approaches could be used, including leave-one-out methods, to reduce this shortlist further, while assessing the loss in prediction performance. Among the 25 strongest predictors of essentiality inferred for D. melanogaster are features derived from scRNA-seq data for embryo, wing disc or gonads; transcripts defined by modENCODE; RNAi probes; and sequence similarity to genes of distantly related organisms (yeast and plants). Interestingly, we identified a feature—i.e. level of gene transcription in the ovary after rapamycin exposure in vitro—as one of the best predictors of essentiality in D. melanogaster. It is known that rapamycin extends the longevity of this fly via the modulation of mTOR pathway, and we propose that this modulation is tightly linked to essential genes in the reproductive tissues. We also found that the location (coordinates) of a gene on a chromosome, the exon number of a gene and selected nucleotide and protein sequence features (including ‘kmer_3_CCC’, and ‘TC_QQQ’, corresponding to the compositions of cytosine and glutamine trimers, respectively) as well as the inferred subcellular localization of a gene product were all strong predictors of essentiality; some of these features had been reported in previous studies (26–27,50). However, we showed that no single feature correlated perfectly with essentiality, although the features with the strongest predictive values were most significant based on t-test results. Therefore, we infer that the combination of individual features in the ‘strongest set’ provided incremental contributions, and was required to achieve a high performance of essentiality predictions by ML. This evidence indicates that the prediction of essentiality can be readily achieved without the need to include PPI data, which is costly, time-consuming and challenging to produce for non-model organisms such as parasites that cannot be maintained in culture in vitro.
Essentiality predictions were shown to be reliable based on threshold-independent metrics (ROC-AUC and PR-AUC), consistently achieving nearly perfect classification (∼1) by ROC-AUC and improving the PR-AUC by at least 2-fold compared with previously published studies (26,27). Overall, the ensemble-based ML methods (XGB, GBM and RF) achieved the best accuracy and consistency. Using these three methods, each trained with NR_SELECTED (a feature set less prone to sequence bias), we obtained final ML-based predictions of essentiality for all genes included in this study. Independent validation of these predictions using available RNAi data showed a strong correlation (0.89) of prediction probabilities with lethal phenotypes, such that these probabilities are considered excellent predictors of essentiality via RNAi (linear model, R2 ∼0.8). This analysis demonstrates the validity and ‘sensitivity’ of the scoring system and ML approaches. Complementary ontology enrichment analysis showed that protein processing was conspicuous for essential genes. However, the numbers of essential genes linked to each term was limited, supporting previous findings for model eukaryotes (26). For this reason, we elected not to use GO terms as features in the present study. In our opinion, the essential and non-essential gene sets inferred here provide a valuable resource to explore the functional roles of key subsets of genes using CRISPR/Cas9 and complementary experiments such as in situ-hybridization, proteomic and/or biochemical approaches (51–53).
A recent study (27) achieved an improved prediction of essential genes in D. melanogaster by integrating sequence, network and subcellular localization features. The genes included (441 essential and 11 788 non-essential) obtained from essentiality databases; 339 genes were from the Database of Essential Genes (DEG) (54) and 13 852 from the Online GEne Essentiality database (OGEE) (55). However, these two databases do not include RNAi data from GenomeRNAi (16) or mutant allele data from FlyBase (12). Indeed, all 339 genes of D. melanogaster listed in DEG had been characterized by transposon mutagenesis—a method reported to be error-prone and somewhat biased (56). An independent evaluation using our scoring system showed that most genes listed as essential (n = 202; 81.8%) in this database were categorized as conditional-essential. This result emphasizes that it is critical to infer confident (essential and non-essential) gene sets from genomic-phenomic data for the training of ML methods. As presently no gold-standards with unequivocal essentiality annotations exist, future efforts should focus on creating them, which would lead to enhanced ML predictions and analyses.
Similar numbers (n) of essential genes were localized to chromosomal regions 2L (n = 73), 2R (n = 82), 3L (n = 79) and chromosome X (n = 71) and ∼30% more genes were found on region 3R (n = 107), whereas only two were on chromosome 4 and none on Y. However, there was no statistical difference in chromosomal location among essential and non-essential genes and conditional-essential, although there were preferential locations on chromosomes for genes (Figure 5), irrespective of their classification. Although intuitively, one would expect essential genes to be within conserved regions of the genome, this was not supported by our analyses (Figure 5).
In conclusion, this study shows that a feature engineering/selection and ML-based workflow can identify novel predictors of gene essentiality of biological relevance and predict, with high confidence, essential genes in D. melanogaster. By using the vast genomic-phenomic datasets available for the vinegar fly, we demonstrate improved performance of gene essentiality predictions compared with previously published results (26,27) and without the inclusion of PPI network datasets. We believe that our workflow will be applicable to other arthropod species, provided that extensive, informative datasets are available. Presently, we hypothesize that features relating to sequence, subcellular localization and transcription data from reproductive tissues (scRNA-seq and RNA-seq) will be informative/useful predictors for Drosophila species more generally, but whether they will serve to predict gene essentiality for distantly related taxa remains to be explored in detail. Tackling this area would provide strength to the ML-based prediction of essentiality for a diverse range of species of invertebrates. Such a focus would be particularly beneficial for work on drug and vaccine target discovery in non-model eukaryotes, such as parasitic arthropods and worms, for numerous genomic, transcriptomic and/or proteomic datasets are available or can be produced, but for which the development of functional genomic tools may be unattainable or extremely challenging due to the intractability of such organisms to continuous in vitro-culture.
DATA AVAILABILITY
The data used in the present study, the ML code developed to perform the systematic ML approaches, and information on software versions and R libraries is available at: https://bitbucket.org/tuliocampos/essential_melanogaster. A static version of the package containing the code and data linked to this publication is available at: https://doi.org/10.6084/m9.figshare.12061815.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: Conceived and designed the study: T.L.C., N.D.Y. and P.K.K. Undertook the study and data analyses: T.L.C. Wrote the paper: T.L.C., N.D.Y. and R.B.G. Contributed to interpretation of findings: N.D.Y., R.B.G., P.K.K., A.H. Supervised the project: N.D.Y., R.B.G. and P.K.K. All authors read and approved the final version of the manuscript.
Contributor Information
Tulio L Campos, Department of Veterinary Biosciences, Melbourne Veterinary School, The University of Melbourne, Parkville, Victoria 3010, Australia; Bioinformatics Core Facility, Instituto Aggeu Magalhães, Fundação Oswaldo Cruz (IAM-Fiocruz), Recife, Pernambuco 50740-465, Brazil.
Pasi K Korhonen, Department of Veterinary Biosciences, Melbourne Veterinary School, The University of Melbourne, Parkville, Victoria 3010, Australia.
Andreas Hofmann, Department of Veterinary Biosciences, Melbourne Veterinary School, The University of Melbourne, Parkville, Victoria 3010, Australia; Griffith Institute for Drug Discovery, Griffith University, Brisbane, Queensland 4111, Australia.
Robin B Gasser, Department of Veterinary Biosciences, Melbourne Veterinary School, The University of Melbourne, Parkville, Victoria 3010, Australia.
Neil D Young, Department of Veterinary Biosciences, Melbourne Veterinary School, The University of Melbourne, Parkville, Victoria 3010, Australia.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
National Health and Medical Research Council (NHMRC), Australia (to R.B.G., P.K.K., N.D.Y.); Australian Research Council, Australia (to R.B.G., P.K.K., N.D.Y.); Yourgene Health and Melbourne Water Corporation (to R.B.G.); NHMRC Career Development Fellowship (to N.D.Y.); NHMRC Early Career Research Fellowship (to P.K.K.); Australian Government, Research Training Program Scholarship (to T.L.C.); Oswaldo Cruz Foundation (Fiocruz/Brazil) (to T.L.C.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Miklos G.L., Rubin G.M.. The role of the genome project in determining gene function: insights from model organisms. Cell. 1996; 86:521–529. [DOI] [PubMed] [Google Scholar]
- 2. Jennings B.H. Drosophila—a versatile model in biology & medicine. Mater. Today. 2011; 14:190–195. [Google Scholar]
- 3. Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G, Scherer S.E, Li P.W., Hoskins R.A., Galle R.F et al.. The genome sequence of Drosophila melanogaster. Science. 2000; 287:2185–2195. [DOI] [PubMed] [Google Scholar]
- 4. Boutros M., Kiger A.A., Armknecht S., Kerr K., Hild M., Koch B., Haas S.A., Paro R., Perrimon N.. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004; 303:832–835. [DOI] [PubMed] [Google Scholar]
- 5. Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. et al.. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007; 448:151–156. [DOI] [PubMed] [Google Scholar]
- 6. Boutros M., Ahringer J.. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 2008; 9:554–566. [DOI] [PubMed] [Google Scholar]
- 7. Heigwer F., Port F., Boutros M.. RNA interference (RNAi) screening in Drosophila. Genetics. 2018; 208:853–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spradling A.C., Stern D., Beaton A., Rhem E.J., Laverty T., Mozden N., Misra S., Rubin G.M.. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999; 153:135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bellen H.J., Levis R.W., Liao G., He Y., Carlson J.W., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M. et al.. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004; 167:761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blumenstiel J.P., Noll A.C., Griffiths J.A., Perera A.G., Walton K.N., Gilliland W.D., Hawley R.S., Staehling-Hampton K.. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics. 2009; 182:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bier E., Harrison M.M., O’Connor-Giles K.M., Wildonger J.. Advances in engineering the fly genome with the CRISPR-Cas system. Genetics. 2018; 208:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. dos Santos G., Schroeder A.J., Goodman J.L., Strelets V.B., Crosby M.A., Thurmond J., Emmert D.B., Gelbart W.M.. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015; 43:D690–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang F., Jiang L., Chen Y., Haelterman N.A., Bellen H.J., Chen R.. FlyVar: a database for genetic variation in Drosophila melanogaster. Database (Oxford). 2015; 2015:bav079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Washington N.L., Stinson E.O., Perry M.D., Ruzanov P., Contrino S., Smith R., Zha Z., Lyne R., Carr A., Lloyd P. et al.. The modENCODE Data Coordination Center: lessons in harvesting comprehensive experimental details. Database (Oxford). 2011; 2011:bar023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cunningham F., Achuthan P., Akanni W., Allen J., Amode M.R., Armean I.M., Bennett R., Bhai J., Billis K., Boddu S. et al.. Ensembl 2019. Nucleic Acids Res. 2019; 47:D745–D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmidt E.E., Pelz O., Buhlmann S., Kerr G., Horn T., Boutros M.. GenomeRNAi: a database for cell-based and in vivo RNAi phenotypes, 2013 update. Nucleic Acids Res. 2013; 41:D1021–D1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Caraus I., Alsuwailem A.A., Nadon R., Makarenkov V.. Detecting and overcoming systematic bias in high-throughput screening technologies: a comprehensive review of practical issues and methodological solutions. Brief. Bioinform. 2015; 16:974–986. [DOI] [PubMed] [Google Scholar]
- 18. Zhang Z., Ren Q.. Why are essential genes essential?—the essentiality of genes. Microb. Cell. 2015; 2:280–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhan T., Boutros M.. Towards a compendium of essential genes—from model organisms to synthetic lethality in cancer cells. Crit. Rev. Biochem. Mol. Biol. 2016; 51:74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Juroszek P., von Tiedemann A.. Plant pathogens, insect pests and weeds in a changing global climate: a review of approaches, challenges, research gaps, key studies and concepts. J. Agric. Sci. 2012; 151:163–188. [Google Scholar]
- 21. Anstead C.A., Batterham P., Korhonen P.K., Young N.D., Hall R.S., Bowles V.M., Richards S., Scott M.J., Gasser R.B.. A blow to the fly—Lucilia cuprina draft genome and transcriptome to support advances in biology and biotechnology. Biotechnol. Adv. 2016; 34:605–620. [DOI] [PubMed] [Google Scholar]
- 22. Bernigaud C., Samarawickrama G.R., Jones M.K., Gasser R.B., Fischer K.. The challenge of developing a single-dose treatment for scabies. Trends Parasitol. 2019; 35:931–943. [DOI] [PubMed] [Google Scholar]
- 23. Ahmed T., Hyder M.Z., Liaqat I., Scholz M.. Climatic conditions: conventional and nanotechnology-based methods for the control of mosquito vectors causing human health Issues. Int. J. Environ. Res. Public Health. 2019; 16:E3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Doyle M.A., Gasser R.B., Woodcroft B.J., Hall R.S., Ralph S.A.. Drug target prediction and prioritization: using orthology to predict essentiality in parasite genomes. BMC Genomics. 2010; 11:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dong C., Jin Y.T., Hua H.L., Wen Q.F., Luo S., Zheng W.X., Guo F.B.. Comprehensive review of the identification of essential genes using computational methods: focusing on feature implementation and assessment. Brief. Bioinform. 2018; 21:bby116. [DOI] [PubMed] [Google Scholar]
- 26. Campos T.L., Korhonen P.K., Gasser R.B., Young N.D.. An evaluation of machine learning approaches for the prediction of essential genes in eukaryotes using protein sequence-derived features. Comput. Struct. Biotechnol. J. 2019; 17:785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aromolaran O., Beder T., Oswald M., Oyelade J., Adebiyi E., Koenig R.. Essential gene prediction in Drosophila melanogaster using machine learning approaches based on sequence and functional Features. Comput. Struct. Biotechnol. J. 2020; 18:612–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuchaiev O., Rasajski M., Higham D.J., Przulj N.. Geometric de-noising of protein-protein interaction networks. PLoS Comput. Biol. 2009; 5:e1000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xiao Q., Wang J., Peng X., Wu F.-x, Pan Y.. Identifying essential proteins from active PPI networks constructed with dynamic gene expression. BMC Genomics. 2015; 16(Suppl. 3):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dobson A.J., He X., Blanc E., Bolukbasi E., Feseha Y., Yang M., Piper M.D.W.. Tissue-specific transcriptome profiling of Drosophila reveals roles for GATA transcription factors in longevity by dietary restriction. NPJ Aging Mech. Dis. 2018; 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karaiskos N., Wahle P., Alles J., Boltengagen A., Ayoub S., Kipar C., Kocks C., Rajewsky N., Zinzen R.P.. The Drosophila embryo at single-cell transcriptome resolution. Science. 2017; 358:194–199. [DOI] [PubMed] [Google Scholar]
- 32. Witt E., Benjamin S., Svetec N., Zhao L.. Testis single-cell RNA-seq reveals the dynamics of de novo gene transcription and germline mutational bias in Drosophila. Elife. 2019; 8:e47138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davie K., Janssens J., Koldere D., De Waegeneer M., Pech U., Kreft L., Aibar S., Makhzami S., Christiaens V., Bravo Gonzalez-Blas C. et al.. A single-cell transcriptome atlas of the aging Drosophila brain. Cell. 2018; 174:982–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bageritz J., Willnow P., Valentini E., Leible S., Boutros M., Teleman A.A.. Gene expression atlas of a developing tissue by single cell expression correlation analysis. Nat. Methods. 2019; 16:750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kiniry S.J., O’Connor P.B.F., Michel A.M., Baranov P.V.. Trips-Viz: a transcriptome browser for exploring Ribo-Seq data. Nucleic Acids Res. 2019; 47:D847–D852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deutsch E.W., Lam H., Aebersold R.. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008; 9:429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bozek M., Cortini R., Storti A.E., Unnerstall U., Gaul U., Gompel N.. ATAC-seq reveals regional differences in enhancer accessibility during the establishment of spatial coordinates in the Drosophila blastoderm. Genome Res. 2019; 29:771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Assaf Z.J., Tilk S., Park J., Siegal M.L., Petrov D.A.. Deep sequencing of natural and experimental populations of Drosophila melanogaster reveals biases in the spectrum of new mutations. Genome Res. 2017; 27:1988–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krogh A., Larsson B., von Heijne G., Sonnhammer L.E.. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001; 305:567–580. [DOI] [PubMed] [Google Scholar]
- 40. Petersen T.N., Brunak S., von Heijne G., Nielsen H.. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011; 8:785–786. [DOI] [PubMed] [Google Scholar]
- 41. Horton P., Park K.-J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K.. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007; 35:W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Almagro Armenteros J.J., Sonderby C.K., Sonderby S.K., Nielsen H., Winther O.. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017; 33:3387–3395. [DOI] [PubMed] [Google Scholar]
- 43. Linding R., Jensen L.J., Diella F., Bork P., Gibson T.J., Russell R.B.. Protein disorder prediction: implications for structural proteomics. Structure. 2003; 11:1453–1459. [DOI] [PubMed] [Google Scholar]
- 44. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M.. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010; 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- 47. Huang D.W., Sherman B.T., Tan Q., Collins J.R., Alvord W.G., Roayaei J., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A.. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007; 8:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mohr S.E., Hu Y., Kim K., Housden B.E., Perrimon N.. Resources for functional genomics studies in Drosophila melanogaster. Genetics. 2014; 197:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen S., Zhang E.Y., Long M.. New genes in Drosophila quickly become essential. Science. 2010; 330:1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kabir M., Barradas A., Tzotzos T.G., Hentges E.K., Doig J.A.. Properties of genes essential for mouse development. PLoS One. 2017; 12:e0178273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kanca O., Bellen H.J., Schnorrer F.. Gene tagging strategies to assess protein expression, localization, and function in Drosophila. Genetics. 2017; 207:389–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Korona D., Koestler S.A., Russell S.. Engineering the Drosophila genome for developmental biology. J. Dev. Biol. 2017; 5:E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Siddall N.A., Hime G.R.. A Drosophila toolkit for defining gene function in spermatogenesis. Reproduction. 2017; 153:R121–R132. [DOI] [PubMed] [Google Scholar]
- 54. Luo H., Lin Y., Gao F., Zhang C.-T., Zhang R.. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014; 42:D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen W.-H., Lu G., Chen X., Zhao X.-M., Bork P.. OGEE v2: an update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 2017; 45:D940–D944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. de Jong J., Akhtar W., Badhai J., Rust A.G., Rad R., Hilkens J., Berns A., van Lohuizen M., Wessels L.F., de Ridder J.. Chromatin landscapes of retroviral and transposon integration profiles. PLoS Genet. 2014; 10:e1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in the present study, the ML code developed to perform the systematic ML approaches, and information on software versions and R libraries is available at: https://bitbucket.org/tuliocampos/essential_melanogaster. A static version of the package containing the code and data linked to this publication is available at: https://doi.org/10.6084/m9.figshare.12061815.