Abstract
Heteroplasmy, the existence of multiple mitochondrial haplotypes within an individual, has been studied across different scientific fields. Mitochondrial genome polymorphisms have been linked to multiple severe disorders and are of interest to evolutionary studies and forensic science. Before the development of massive parallel sequencing (MPS), most studies of mitochondrial genome variation were limited to short fragments and to heteroplasmic variants associated with a relatively high frequency (>10%). By utilizing ultra-deep sequencing, it has now become possible to uncover previously undiscovered patterns of intra-individual polymorphisms. Despite these technological advances, it is still challenging to determine the origin of the observed intra-individual polymorphisms. We therefore developed a new method that not only detects intra-individual polymorphisms within mitochondrial and chloroplast genomes more accurately, but also looks for linkage among polymorphic sites by assembling the sequence around each detected polymorphic site. Our benchmark study shows that this method is capable of detecting heteroplasmy more accurately than any method previously available and is the first tool that is able to completely or partially reconstruct the sequence for each mitochondrial haplotype (allele). The method is implemented in our open source software NOVOPlasty that can be downloaded at https://github.com/ndierckx/NOVOPlasty.
INTRODUCTION
Mitochondria are small organelles involved in various cellular functions, with ATP production through respiration as their primary task. On average, one mitochondrion harbors between 2 and 10 mtDNA molecules, and each cell contains hundreds to thousands of mitochondria, depending on the cell function (1). When inherited or somatic mutations cause variation within one cell or among different cells of an organism, we speak of heteroplasmy. It is mainly caused, but not exclusively, by somatic mutations occurring within an organism, at a rate increasing with age (2). As mitochondrial genomes are maternally inherited in most animals, multiple variants can sometimes be inherited from the mother, if they are present in the egg cell, providing another source of heteroplasmy. Finally, in a few animal species, paternal leakage of mitochondria during fertilization has been reported to occur, generating heteroplasmy by combining alleles that originated in separate individuals (3,4).
In humans, multiple severe disorders like cancer (5,6), autism (7), mitochondrial encephalopathy (8) and late-onset neurodegenerative diseases (9) have already been associated with this phenomenon (10). Mitochondrial heteroplasmy is common in healthy individuals, although some mutations can become pathogenic after they reach a certain frequency threshold. This threshold is generally >50% and therefore relatively easy to discover, but it would be beneficial to detect these pathogenic mutations in an early phase, when present at a much lower frequency (11). There is also great interest from the field of forensic science, since these mutation patterns can enhance the likelihood ratio for a potential match (12–14). Considering that mitochondrial genomes are frequently used as markers in evolutionary studies, widespread occurrence of heteroplasmy within a species could have important implications for divergence times between populations or species (15).
Before the application of next-generation sequencing (NGS) technologies, heteroplasmy detection was achieved mostly by Sanger DNA sequencing or PCR-based approaches and most studies focused solely on the mtDNA control region (1,16). The detection limit of Sanger sequencing (10–20%) is higher than the frequency of most heteroplasmic mutations, while PCR-based approaches are only available for specific sites, which led to a limited view of the potential impact of heteroplasmy (17,18). These limitations were partially overcome by the rise of NGS technologies. These technologies are able to produce a very high coverage of the complete mitochondrial genome, making it possible to detect mitochondrial variants associated with a frequency as low as 1–2% (13,19). Despite these technological advances, it is still difficult to detect all mutations below a frequency of 5% while at the same time avoiding false positives (20). If we look at the Illumina technology, currently the most accurate next-generation sequencer, the error rate is on average between 0.01 and 0.1%. However, this error rate varies depending on the platform, chemistry, read length and experiment, reaching over 10% for some nucleotide positions. For example, sequences made of single-nucleotide repeats (SNR) are associated with much higher error rates; depending on the length of the repeat, error rates can exceed 20% (21). Besides limitations of the sequencing technology itself, nuclear copies of mitochondrial DNA segments (NUMTs) can be mistaken for real mitochondrial sequences, thereby creating the illusion of heteroplasmy. NUMTs have been reported in several species and can range in length from 40 bp to the entire mitochondrial genome (22). The interference of these NUMTs in heteroplasmy detection can be reduced by computational and molecular approaches, but it is difficult to completely exclude their influence from the analysis (18,23).
There are several strategies to detect heteroplasmy with NGS data. Laboratory enrichment of mitochondrial sequences is the most affordable method for achieving very deep coverage, which can be achieved by PCR amplification or mtDNA isolation. Methods of isolation focus on separating organelle genomes from the nucleus by centrifugation or capture array at an early stage, while amplification methods focus on increasing the proportion of mitochondrial DNA by polymerase chain reaction (PCR) with specific primers (18). Both strategies cannot avoid NUMT contamination completely and are often not successful in isolating the complete mitochondrial genome. A more recent and probably more direct approach lies with whole genome sequencing (WGS), where entire genomic DNA extracts are sequenced using NGS (24). While achieving a very deep coverage is more costly, it provides a dataset that can also be used for other genomic analyses. There are already large amounts of WGS datasets available online and the continuous decrease of NGS sequencing costs will make it more attractive to produce them in the future. For the detection of rare single-nucleotide substitutions (with frequencies <0.5%), there is a method named duplex sequencing, which has a reliable detection threshold of up to 0.01% (25). This method tags both strands of the DNA and will only identify SNPs when they are present on both strands, which strongly reduces false calls from random sequencing errors. However, duplex sequencing is less effective for indel detection and its high costs make it inapplicable for whole genome sequencing. This improved accuracy counts only for sequencing errors; the presence of NUMTs will still result in false positives.
Currently available heteroplasmy detection pipelines are based on aligning the reads to a mitochondrial reference genome. These algorithms use simple base-quality filtering or more complex statistical strategies to deal with sequencing errors (26,27). Depending on the chosen strategy and the quality of the dataset, it can decrease the detection threshold to 1%. NUMT interference can be reduced by filtering out these sequences before heteroplasmy detection, which can be achieved by aligning all reads to an assembly of the nuclear genome, if available. This works very well when the nuclear genome originates from the same individual. However, in most cases, a reference nuclear genome is used, which does not necessarily contain the same NUMTs as the sequenced individual.
Most tools for heteroplasmy detection are solely developed for the human mitochondrial genome (27,28) or not suitable for indel detection (28). The final output is at best a VCF (Variant Call Format) file that lists all possible heteroplasmic sites. To the best of our knowledge, there is no algorithm that outputs how different polymorphic sites are linked to each other. We present a novel method that detects low-frequency intra-individual polymorphisms during a de novo assembly of the mitochondrial genome. We extended the organelle assembler NOVOPlasty with this new heteroplasmy detection option that is also able to assemble the region around each detected polymorphic site and subsequently allows phasing of nearby SNPs. Depending on the detected SNP density, NOVOPlasty is able to assemble between 200 bp and the complete mitochondrial haplotype or NUMT sequence. This new method evaluates whether a haplotype is an NUMT or is of mitochondrial origin (i.e. true heteroplasmy). Finally, it outputs graphical views for user-friendly representations.
MATERIALS AND METHODS
Sequencing and data availability
The Gonioctena intermedia dataset (PCR-free) was sequenced on the Illumina HiSeq 2500 platform (250 bp paired-end reads). The mitochondrial reads of this dataset can be found in the European Nucleotide Archive (ENA) under accession number ERX3482826. The six human samples (PCR-free) were sequenced on the Illumina HiSeqX platform (150 bp paired-end reads). As these datasets are not publicly available, we also included three PCR-free WGS datasets from the ENA to the benchmark (SRR2098263, ERR3243159 and ERR1395547). The simulated datasets from the MToolBox publication (27) are publicly available on https://sourceforge.net/projects/mtoolbox/files/Simulations/. We also received one dataset from the University of Innsbruck, which was used in a benchmark for their scalable web server for the analysis of mtDNA studies (mtDNA-Server). This dataset was made available to us after contacting the authors of the mtDNA-Server publication (28). The dataset is a sample-mix up (1:100) of two human samples (HM625679.1 and KC286589.1) sequenced on the Illumina HiSeq platform.
Data preparation
Before heteroplasmy analysis, the G. intermedia mitochondrial genome was assembled by NOVOPlasty and submitted to Genbank under accession number NC_042500. As the mitochondrial genome of G. intermedia has a long repetitive control region (∼3500 bp) where heteroplasmy detection would not be reliable, we decided to exclude this region before analysis. To facilitate the comparison between our results and previously published heteroplasmy sites of G. intermedia, we chose the same reference start and end points (1–14 644) as in that study (29). This should be considered before each heteroplasmy analysis, as repetitive regions above >80% of the read length can cause false positives. These problematic regions can be found in the mitochondrial control region of some insects. The inverted repeat in chloroplast genomes does not have to be removed, though it is not possible to distinguish heteroplasmic point mutations between the two regions.
Depending on the size of the dataset and the detection method, original FASTQ files or filtered FASTQ files were used as input. As the mtDNA-Server and MToolBox datasets have file sizes <1 GB, the original FASTQ files were used as input for each detection method. The WGS datasets have a very large file size (>30 GB) and contain only a small fraction of mitochondrial sequencing reads (<1%). To facilitate the heteroplasmy detection on these large datasets, we filtered them for mitochondrial sequences by selecting sequencing reads that comprise any small fraction (16 bp) of the mitochondrial reference genome. This ‘filter’ script can be downloaded together with NOVOPlasty at https://github.com/ndierckx/NOVOPlasty.
All assemblies and heteroplasmy analyses were executed on both an AMD Opteron 6134 CPU machine of 2.3 GHz with a total of 1500 GB of RAM, and on a Intel(R) Core(TM) i7-4710HQ CPU of 2.50 GHz with 16 GB of RAM.
NOVOPlasty updates
NOVOPlasty is a de novo seed-extend based assembler for organelle genomes, which was published in 2016 (30). The assembly has to be initiated by a seed, which is iteratively extended in both directions. The seed sequence can be one sequence read, a conserved gene or even a complete organelle genome from a distant species. Since the initial release of NOVOPlasty, there have been several updates to improve its robustness and to include new functionalities. Some updates were suggested by users to improve user friendliness, such as the acceptance of zipped files as input, automatic subsampling, batch inputs and extraction of the assembled reads into a separate file. Besides these small improvements, three major updates broadened the capabilities of NOVOPlasty. The first one was a new assembly mode for plant mitochondrial genomes, which requires the chloroplast genome as an input to avoid chimeric assemblies that can be caused by duplicated chloroplast sequences inside the mitochondrial genome. The second large update was the introduction of reference-assisted assembly. Although the assembly will still be de novo, NOVOPlasty will use the reference genome to resolve ambiguous positions where more than one extension is possible. This method makes it possible to resolve the inverted repeat in chloroplast genomes, which reduces the post-processing time. Additional information about each update can be found under the ‘Wiki’ section of the NOVOPlasty Github page.
Heteroplasmy detection
The latest major update includes a variant caller and a heteroplasmy detector. The variant caller option generates a VCF file that includes all variants within the new assembly and a given reference. For heteroplasmy detection, it is necessary to first assemble the organelle genome with NOVOPlasty and then to use this assembly as a reference and seed input. It is also possible to use the rCRS sequence of the human mitochondrial DNA directly as reference and seed, this is recommended if you want the positions of the variants to be according to the human reference genome. All mutations that have a frequency above a specified minor allele frequency (MAF) will be detected. The MAF can be a value between 0.6% and 50%, although the actual detection limit will depend on the coverage available (e.g. to detect a variant associated with a frequency below 3%, a coverage >300× is needed). Since detected frequencies can vary around the average value, it is recommended to choose a value that is 0.3% to 1% lower than the expected MAF. Depending on the quality of the dataset, frequencies <1% can result in false positives originating from sequencing errors. In the heteroplasmy mode, NOVOPlasty will reassemble the mitochondrial genome, but this time considering the MAF during consensus calling of each base. When the allele frequency is above the given threshold (MAF), the position will be examined further with additional parameters described below. In contrast with the standard assembly mode of NOVOPlasty, the heteroplasmy mode uses the quality scores available in the FASTQ files to remove base calls with a score <21. Some reads can be over-represented as a result of duplications during PCR amplification, which can inflate sequencing error and mutation frequencies. We therefore apply a duplication normalization to reduce the duplication count when it exceeds the theoretical value by 3-fold. This theoretical value is calculated for each position in the mitochondrial genome by taking the median duplication count of each unique read alignment position. Mutations that still have a frequency above the MAF will be stored in a VCF (version 4.0) file, enclosing the frequency and total coverage of each mutation. This is the final output given by most heteroplasmy pipelines.
Mutation linkage
Because the heteroplasmy caller in NOVOPlasty is based on de novo assembly, it is able to use assemblies to infer linkage between variants and to produce phased haplotypes. While this information appears crucial for heteroplasmy analysis, to the best of our knowledge, no pipeline available to date is capable of phasing the polymorphic sites. NOVOPlasty uses a short sequence around each mutation as a seed for assembling its flanking sequences. The seed will only be extended with sequences that contain this mutation in one of the paired reads. Any variation to the major haplotype that is linked to the initial mutation can be categorized as ‘fully linked’ or ‘partially linked’. Only ‘fully linked’ positions are used to filter sequences for the local assembly, as ‘partially linked’ mutations have multiple alleles and can therefore be linked to multiple sequences (Figure 1). The length of each assembly depends on the read length and the distribution of intra-individual polymorphisms across the mitochondrial genome. NOVOPlasty will output a file with all assembled contigs and a file that lists all detected linkages for each intra-individual polymorphism from the VCF file. We make a distinction between fully linked, partially linked and zero linkage. Assemblies that contain nuclear sequences at either end will be automatically marked as NUMTs and outputted in a separate file.
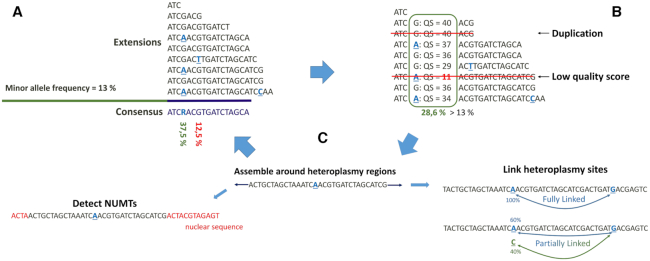
Figure 1.
Workflow of heteroplasmy detection with NOVOPlasty. For simplicity, the representation of the workflow is limited to unidirectional extension. (A) During consensus calling, all positions with a polymorphism higher than the given minor allele frequency (MAF) are marked for further analysis. (B) Each marked position will be subjected to additional filtering steps, low quality scores and over-represented reads (as a result of duplications during PCR) will be removed. (C) The remaining polymorphisms will be subjected to an additional assembly step to link to other mutations sites or to exclude NUMT sequences.
Supplementary Figure S1 depicts the different outputs from the heteroplasmy module. All intra-individual polymorphisms that are flagged as potential heteroplasmy will be outputted in a VCF file, together with a FASTA file with all assemblies and a TXT file with a table listing the linkage between each polymorphism. Similar files will also be outputted for the detected NUMTs. We also added an optional output to visualize the links between intra-individual polymorphisms with Circos (31). All necessary files to run Circos are generated by NOVOPlasty, but non-human mitochondrial genomes need manual correction if annotation is desired. Supplementary Figure S3 in the supplementary material is an example of a Circos output for a human mitochondrial genome.
Benchmark
We tested the heteroplasmy function of NOVOPlasty on one G. intermedia and ten human datasets. The mtDNA-Server and MToolBox datasets result from respectively targeted mtDNA sequencing and simulations by simNGS (http://www.ebi.ac.uk/goldman-srv/simNGS/), while the other datasets result from whole genome sequencing. The targeted mtDNA sequencing approach makes it possible to obtain a very high coverage of the mitochondrial genome with a minimum of nuclear sequences contamination. Generating a whole genome sequencing dataset is less laborious but will consist predominately of nuclear sequences (between 99.9% and 99.97% in the case of our datasets). As these datasets were meant for nuclear genome assembly, the overall high coverage resulted in enough mitochondrial genome coverage for low frequency (<3%) heteroplasmy detection. A summary of the characteristics of each dataset is presented in Table 1.
Table 1.
Summary of the characteristics of each dataset
| F1&F2 | mtDNA-server | MToolBox | G. intermedia | SRR2098263 | ERR3243159 | ERR1395547 | |
|---|---|---|---|---|---|---|---|
| Origin | WGS | Targeted | Simulated mtDNA | WGS | WGS | WGS | WGS |
| Platform | HiSeqX | HiSeq | / | HiSeq 2500 | HiSeq 2500 | NovaSeq 6000 | HiSeq 2000 |
| Read length | 151 bp | 101 bp | 101 bp | 251 bp | 100 bp | 150 bp | 100 bp |
| Insert size | 420 bp | 265 bp | 500 bp | 520 bp | 300 bp | 450 bp | 325 bp |
| Mitochondrial coverage | 3650 | 59 500 | 100–2000 | 4200 | 4000 | 20 000 | 5000 |
| Fraction of dataset | 0.04% | 97% | 99% | 0.13% | 0.08% | 0.25% | 0.08% |
The mtDNA-Server publication compared their web-based heteroplasmy caller with LoFreq (32), a software for sensitive variant detection. Four different sample-mix ups (1:2, 1:10, 1:50 and 1:100) were created from two human samples (HM625679.1 and KC286589.1). We can expect 27 heteroplasmic point mutations, excluding the sample-specific heteroplasmic mutations. We have limited our analysis to the 1:100 dataset, as it is the most challenging and the only one we were able to obtain.
System requirements (wall time and peak virtual memory) were compared between the four methods for the mtDNA-Server dataset and the six MToolBox datasets. Overall MToolBox requires the most computational power as it is the only tool that aligns all the reads back to the nuclear genome. Increasing sequencing depth of the mitochondrial genome will increase the wall time of each method, nevertheless the maximum wall time is still limited to 102 min for a mitochondrial coverage of 35 000. The complete results can be found as Supplementary Data in Supplementary Table S1.
RESULTS
Benchmark on the mtDNA-Server dataset
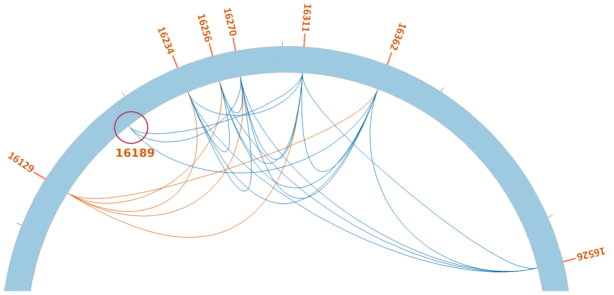
Three algorithms designed for the detection of low-frequency variants (mtDNA-Server (28), LoFreq (32), MToolBox (27)) were used in comparison to NOVOplasty to call heteroplasmic sites on the mtDNA-server dataset (see ‘Materials and Methods’ section). The mtDNA-Server was able to detect 24 out of 27 heteroplasmic sites, without any false positives. LoFreq achieved a higher sensitivity with 26 out of 27, but at the expense of specificity since it also detected 30 false positives. Without the mutation linkage module, NOVOPlasty was able to detect 26 out of 27 heteroplasmic sites, without any false positive. Both LoFreq and NOVOPlasty detected all 5 sample-specific heteroplasmic mutations of the majority sample, while mtDNA-Server only detected 3 out of 5. NOVOPlasty failed to detect one mutation that is located in a low complexity region (C-repeat), where read quality is generally lower. Although this SNP was not detected during the initial step of identifying polymorphic sites, it was detected during mutation linkage. The position of this SNP can be found in the linkage table output and is clearly visible in the Circos drawings of Figure 2 and Supplementary Figure S3. Figure 2 is a detailed view of the region around the missing SNP, which clearly shows linkage with nearby heteroplasmic positions.
Figure 2.
Detailed view of the control region from the Circos output of the mtDNA-Server dataset. Positions linked by blue lines are on the same sequence, while positions linked by red lines originate from different sequences. Position 16,129 is one of the 5 sample-specific heteroplasmic mutations and has no linkage with the other polymorphisms. The red circle indicates the missing SNP in the low complexity region that was not detected without the linkage module.
The benchmark study in the mtDNA-Server publication tested for the 27 SNPs, yet never mentioned a verified deletion between the two samples. We repeated the experiment and mtDNA-Server was not able to detect this deletion, while LoFreq detected 8 different indels around this position. NOVOPlasty successfully detected the deletion and linked it to adjacent heteroplasmic positions. There were also no false positives, which means that for this dataset, NOVOPlasty achieved a specificity and sensitivity of 100% by using both the heteroplasmy and haplotyping modules (Table 2). The average minor frequency of the 27 heteroplasmic sites detected by NOVOPlasty was 1.04%, compared to a theoretical value of 1%. The complete VCF output can be found as Supplementary Data (Supplementary Figure S2).
Table 2.
Benchmark results of the heteroplasmy analysis of the mtDNA-Server and MToolBox datasets
| NOVOPlasty | mtDNA-Server | LoFreq | MToolBox | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Correct calls | False positives | Correct calls | False positives | Correct calls | False positives | Correct calls | False positives | False positives >0.6% | |
| mtDNA-Server | 27/28* | 0 | 24/28 | 0 | 26/28 | 30 | 28/28 | 741 | 92 |
| mtDNA-Server (intra-individuel) | 5/5 | 0 | 3/5 | 0 | 5/5 | 30 | 5/5 | 741 | 92 |
| MToolBox (2000×, 25%) | 5/5 | 0 | 1/5 | 0 | 5/5 | 1 | 5/5 | 2 | 1 |
| MToolBox (2000×, 0.5%) | 0/5 | 0 | 0/5 | 0 | 2/5 | 0 | 5/5 | 0 | 0 |
| MToolBox (500×, 25%) | 5/5 | 0 | 1/5 | 0 | 5/5 | 0 | 5/5 | 0 | 0 |
| MToolBox (500×, 0.5%) | 1/5 | 0 | 0/5 | 0 | 1/5 | 0 | 0/5 | 0 | 0 |
| MToolBox (100×, 25%) | 3/3 | 0 | 1/3 | 0 | 3/3 | 0 | 3/3 | 0 | 0 |
| MToolBox (100×, 0.5%) | 0/3 | 0 | 0/3 | 0 | 0/3 | 0 | 0/3 | 0 | 0 |
*The missing heteroplasmic position was indirectly detected by the linkage module.
We also included MToolBox (27), a highly automated pipeline for heteroplasmy annotation, to the benchmark. Although MToolBox was able to detect all 28 heteroplasmic sites, it generated at the same time 741 false positives. The majority of these false positives have very low frequencies and are most likely to originate from sequencing errors. To obtain a more truthful assessment of MToolBox’s performance, we removed all polymorphisms associated with a frequency <0.6%, which reduced the number of false positives to 60 (Table 2). Besides the relatively low specificity, multiple incorrect alleles (1–4) were detected at the same positions than those of 4 true heteroplasmic sites. False positives are especially clustered in low-complexity regions (LCR), demonstrating the difficulty for MToolBox to resolve those regions.
The 5 sample-specific heteroplasmic mutations of the majority allele were excluded from the benchmark; nevertheless, some were also detected by each method and are in fact true heteroplasmic sites. Current methods are not able to detect that these 5 mutations originate from different sequences, while our new haplotyping assembly strategy confirmed that each of these 5 positions are not linked to any of the other 27 heteroplasmic positions (Figure 2 and Supplementary Figure S2).
MToolBox dataset
In the publication describing MToolBox (27), the authors assess its performance on simulated datasets with coverages ranging from 100× to 2000× and heteroplasmic frequencies from 0.5% to 75%. They inserted 5 heteroplasmic sites in each simulated dataset: 2 deletions, 2 insertions and 1 SNP. We ran NOVOPlasty, mtDNA-Server and LoFreq on 6 of these datasets (with frequencies of 0.5% and 25%) and compared our results to the ones of the MToolBox publication. NOVOPlasty, LoFreq and MToolBox detected all heteroplasmic sites across all 25% heteroplasmy datasets, while mtDNA-Server only detected the SNP for these datasets, as indels are problematic for this tool. Heteroplasmic frequencies of 0.5% are below the detection threshold of NOVOPlasty and mtDNA-Server and were therefore not capable of detecting any of the heteroplasmic positions in the 0.5% heteroplasmy datasets (with the exception of one heteroplasmic position with NOVOPlasty). From the three 0.5% datasets, LoFreq detected 2 heteroplasmic positions in the 2000× dataset and 1 in the 500× dataset. MToolbox was able to detect the 5 heteroplasmic positions in the dataset with the highest coverage (2000×), without any false positives. These results suggest that MToolBox is capable of accurately detecting low frequency heteroplasmy (∼0.5%) for high-coverage data. However, results of MToolBox on non-simulated datasets reveal a very high rate of false positives for low frequency variants.
We also wish to rectify an error in the benchmark of the MToolBox publication. The authors reported a 60% sensitivity for the 100× datasets, while we obtained a sensitivity of 100%. After close inspection of the 100× datasets, we confirm that they only contain 3 heteroplasmic positions. The 2 missing positions may have been accidentally omitted when creating the 100× simulated datasets.
Human WGS datasets
The nine human WGS datasets consisted out of two sets of related individuals (F1 and F2), one including a mother and her child; and three public datasets from the ENA. We compared our heteroplasmy analysis with the ones of LoFreq, MToolBox and mtDNA-Server (Table 3).
Table 3.
Benchmark results of the heteroplasmy analysis of 9 human WGS datasets
| NOVOPlasty | mtDNA-Server | LoFreq | MToolBox | ||||
|---|---|---|---|---|---|---|---|
| Before NUMT assembly | After NUMT assembly | Frequency Range | SNPs > 0.6% | SNPs > 0.6% | All SNPs | SNPs > 0.6% | |
| F1-P1 (Distant relative) | 3 | 2 | 0.75–0.87 % | 1535 | 1460 | 1372 | 119 |
| F1-P2 (Mother) | 4 | 3 | 0.7–6.4 % | 1601 | 1372 | 465 | 182 |
| F1-P3 (Son) | 1 | 1 | 1.10% | 1073 | 1236 | 2225 | 89 |
| F2-P1 (Daughter) | 75 | 9 | 0.8–1.4 % | 3045 | 2731 | 248 | 186 |
| F2-P2 (Distant relative) | 6 | 6 | 0.66–52 % | 978 | 817 | 1711 | 116 |
| F2-P3 (Father) | 71 | 3 | 0.6–1.2 % | 3245 | 2855 | 192 | 165 |
| SRR2098263 | 3 | 3 | 0.7–67 % | 1160 | 275 | 88 | 31 |
| ERR3243159 | 4 | 4 | 0.89–33% | / | 63 | 397 | 71 |
| ERR1395547 | 21 | 20 (6*) | 0.6–1.1% | / | 196 | 161 | 72 |
*After inspection of the linkage assemblies, 14 additional heteroplasmic positions were identified as NUMTs.
The most remarkable observation is the huge amount of detected intra-individual polymorphisms by LoFreq and mtDNA-Server compared to NOVOPlasty and MToolBox. NOVOPlasty detected no more than 9 polymorphisms per sample, compared to over a thousand by LoFreq and mtDNA-Server. These large amounts of heteroplasmic sites seem rather unusual when looking at previous studies on human samples (33–35). LoFreq and mtDNA-Server share 1032 polymorphisms for sample F1-P1, which means that approximately half of all positions are only detected by one of the two methods. The mtDNA-Server provides additional information for every detected polymorphism to interpret the results, including whether the polymorphism is linked to any known NUMT.
For a reason we cannot explain, LoFreq detected a significant lower amount of intra-individual polymorphisms in the three public datasets. While mtDNA-Server had a similar high number of intra-individual polymorphisms for SRR2098263 and two unsuccessful runs for ERR3243159 and ERR1395547 (caused by incompatible quality scores).
If we look at the NOVOPlasty results of F1, we found an identical position (8557G>A) in both mother and child with frequencies of 6.4% and 1.1%, respectively. The relatively high frequency associated with this polymorphism and the fact that the local assembly around this region did not expose any NUMT sequence make it highly probable that this is a true heteroplasmic mutation. NOVOPlasty, MToolBox and mtDNA-Server detected this polymorphism in F1-P2 (6.4%) and F1-P3 (1.1%), while LoFreq only detected it in F1-P2. mtDNA-Server categorized this polymorphism as a reliable heteroplasmic polymorphism but also linked it to an NUMT sequence. The fact that this NUMT sequence was not the origin of the 8557G>A mutation shows that the only reliable way to link an observed polymorphism to the presence of an NUMT sequence is to conduct a haplotyping assembly around that position. In the second family, father (F2-P3) and daughter (F2-P1) share a high presence of NUMT content. NOVOPlasty was able to reduce the detected polymorphisms from 75 to 9 (daughter) and from 71 to 3 (father) by local assembly around each position.
Although MToolBox is designed to detect very low frequencies, it produces a great amount of false positives for variant frequencies <0.6%. If we consider only values >0.6%, MToolBox performs significantly better than mtDNA-Server and LoFreq on the analyzed WGS datasets. This can be explained by the presence of an NUMT removal module in the MToolBox pipeline. This module will filter out sequences that align to a reference nuclear genome to reduce NUMT interference. Yet this will not completely eliminate the issue, as NUMT sequences can differ between individuals (i.e. some NUMT sequences in a sample could be absent from the reference nuclear genome assembly used). The additional local assembly module of NOVOPlasty is capable of filtering out false positives that originate from any NUMT sequence (i.e. without the need for a reference nuclear genome assembly). We compared the linked mutations in the NUMT assemblies of F2-P1 and F2-P3 produced by NOVOPlasty to the results generated by MToolBox; our program was able to identify nine false positives that originate from NUMT sequences that are absent from the human reference genome and thus non-eliminated (i.e. considered to be true heteroplasmic variants) by MToolBox. While NOVOPlasty did not assemble any NUMT sequence in datasets ERR3243159 and SRR2098263, it did detect one in dataset ERR1395547 automatically; plus 14 additional intra-individual polymorphisms were linked to NUMTs after manual inspection of the linkage assemblies. Six out of these fifteen variants that were linked to NUMT sequences were still detected as true heteroplasmic positions by LoFreq. MToolBox did not identified any of these 15 positions as heteroplasmic as the corresponding NUMT sequences can be recovered from the human reference genome. In conclusion, the haplotyping module allows NOVOplasty to be the most sensitive and specific heteroplasmy caller for any variant >0.6% frequency.
Gonioctena intermedia dataset
The widespread occurrence of heteroplasmy within a population of G. intermedia was confirmed in a previous study (29). WGS datasets of 2 individuals were used for heteroplasmy detection with BWA and SAMtools and revealed possible heteroplasmy in both cases. This observation was confirmed by sequencing (Sanger) an 814 bp fragment of the COI gene for 24 individuals. Eleven individuals out of twenty-four were clearly heteroplasmic for two distant mtDNA haplotypes. For heteroplasmy detection of the complete mitochondrial genome (minus the control region), a subset (2.5%) of the G. intermedia dataset (Table 1) was used as input for SAMtools. This resulted in the detection of 169 intra-individual polymorphisms with an average frequency around 20%. We repeated this experiment with SAMtools and LoFreq on the subsampled dataset and on the complete dataset. Similar results were obtained for the subsampled dataset with SAMtools, although when using the complete dataset, SAMtools only detected 8 intra-individual polymorphisms. We believe that the 2.5% subset is not sufficient to provide an objective estimation of the frequency of each mutation. SAMtools only detects frequencies >10%, which makes it more sensitive for over-represented sequences. LoFreq, which detects frequencies up to 0.6%, detected 7826 intra-individual polymorphisms for the complete dataset and 1370 for the subsampled dataset. NOVOPlasty detected 0 polymorphisms with a MAF of 10%, 3 with a MAF of 5%, 94 with a MAF of 3% and 350 with a MAF of 1%. These results are inconsistent with the previously estimated average frequency of 20% for the low frequency allele. An explanation can be found in the assembled NUMT sequences by NOVOPlasty. Every assembled NUMT sequence that were composed of COI gene sequence included the variants from the low frequency haplotype. This probably means that all COI NUMTs originated from that allelic version of the mitochondrial genome. Since the initial analysis did not specifically eliminate NUMT sequences to identify polymorphic sites in the mitochondrial genome, and did not attempt to directly phase them based on an assembly, inferred frequencies of heteroplasmic mutations are probably based on a combination of true alternative mitochondrial haplotype variants and NUMT sequences.
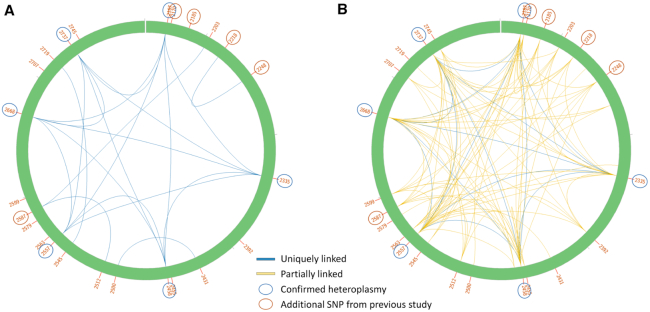
Compared to the human datasets, NOVOPlasty was not able to automatically exclude all NUMT-related polymorphisms, as most polymorphisms were linked to multiple sequences. There are several factors that make this dataset more complex to analyze than the other datasets we tested. The combination of a long repetitive control region (which is too complex to assemble) with long NUMT sequences (in some cases their length spanned the entire mitochondrial genome), means NOVOPlasty will sometimes stop the assembly before reaching the nuclear region within which it is inserted (e.g. because the assembly has reached the control region first), and will thus not catalogue this sequence as a NUMT. Besides, most NUMT sequences seem to have originated from the low frequency haplotype. This complicates the assembly of the low frequency haplotype, as many partially linked variants that originate from NUMT sequences interfere during mutation linkage. We attempted to visualize this with the Circos files that were generated by NOVOPlasty for the run with a MAF of 1% (Figure 3). We limit the visualization to the COI fragment, as Sanger sequencing of 24 individuals identified a haplotype that comprises 6 polymorphisms in this region. Figure 3A connects SNPs that are fully linked to each other, which implies that these SNPs can always be found together. The six confirmed SNPs (indicated by a blue circle) are clearly all fully linked to each other. There is some linkage between the other positions, but none is fully linked to any of the six confirmed positions. These results confirm the presence of the second haplotype that was found within the 24 individuals. Partially linked connections were included to obtain a more complete picture (Figure 3B). This image reveals that the six variants inferred previously for the low frequency COI haplotype are linked to most of the assembled sequences, including the confirmed NUMT sequences, demonstrating the difficulty to resolve all possible mitochondrial haplotypes and NUMTs. Even though it was not possible to assemble the complete minor haplotype of the mitochondrial genome, NOVOPlasty was still able to assemble >9500 bp of the minor haplotype and to link 76 intra-individual polymorphisms within this assembly.
Figure 3.
Circos output of the COI region generated by NOVOPlasty for the Gonioctena intermedia dataset with a MAF of 1%. All the detected SNPs for this region are indicated by their position in the complete mitochondrial genome. The 6 positions confirmed by Sanger in a previous study (29) are encircled in blue, the additional SNPs that were detected by SAMtools (MAF > 10%) in the same study are encircled in red. (A) Only SNPs that are fully linked to each other, which implies that these SNPs can always be found together, are connected with each other. (B) Fully linked (blue) and partially linked (yellow) SNPs are shown. Partially linked SNPs can be found together in some sequences, but not all.
DISCUSSION
Current methods for heteroplasmy detection rely on sequence alignments (BAM files) to identify SNPs. Alignment tools will try to align all sequencing reads to the reference sequence and in general do not consider allele frequencies below 10%. However, there are many cases of heteroplasmy in which the second allele is associated with a lower frequency; detection of heteroplamsy is difficult in these cases because it becomes necessary to discriminate between low frequency mitochondrial variants and nuclear sequences (NUMTS). Nuclear contamination is especially problematic with WGS datasets, as we can infer from the results of this study. We introduced here a new approach, in which heteroplasmy is detected during a de novo assembly of the mitochondrial genome and subsequently verified by local assembly and phasing around each potential heteroplasmic position.
To avoid a biased benchmark by overfitting to one type of dataset, we opted to compare our new method to previously published results. Despite the absence of NUMT sequences in these datasets, NOVOPlasty still generated the most accurate results and is the only method capable of distinguishing the sample-specific heteroplasmic mutations from the artificial mutations in the mtDNA-Server dataset. To demonstrate the full strength of this new method, we also compared the performance of each tool on 10 PCR-free WGS datasets. In general, the large interference of NUMT sequences can be reduced by aligning reads to the reference nuclear genome, yet will not exclude individual-specific NUMT sequences. NOVOPlasty is able to extend the sequence around each intra-individual polymorphism and accurately link nearby variants when there is no ambiguity for that position. Thanks to this strategy, it can detect haplotypes that actually belong to the nuclear genome (NUMTs), by identifying mitochondrial sequences that extend into nuclear sequence.
In our experience, heteroplasmy analysis is best conducted on data sets with sufficient coverage (>300x for MAF of 3% and >500x for MAF of 1%) and ideally obtained from PCR-free libraries, as recombination during PCR amplification generates chimeric sequences, which creates artificial linkages between mutations. Currently, heteroplasmy analysis with NOVOPlasty only works for sequencing reads that originate from Illumina technologies. The detection limit lays around 0.6%, any lower frequency will greatly increase the possibility of false positives by failing to eliminate sequencing errors. MToolBox is the only method that presented a successful benchmark on lower frequencies (0.5%, 2,000x dataset in Table 2), although only for the dataset that was simulated and MToolBox was not capable of achieving a similar performance with other datasets.
In conclusion, NOVOplasty was the most sensitive and specific mitochondrial heteroplasmy caller for simulated and real WGS datasets for any variant with an allelic frequency above 0.6%. Such property should be useful for medicine, forensic science and evolution research. The software is open source and can be downloaded at https://github.com/ndierckx/NOVOPlasty. Besides a standard Perl installation, there are no software or module requirements to run the script.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Sebastian Schoenherr for providing us the mtDNA-Server dataset.
Authors’ contributions: N.D. conceived, designed and scripted the heteroplasmy module. P.M. provided the G. intermedia sequences. G.S. provided the human sequences. N.D. wrote the manuscript. P.M. and G.S. provided guidance and reviewed the manuscript.
Notes
Present address: Nicolas Dierckxsens, State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-sen University, Guangzhou 510060, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
PhD bursary of the Belgian Kids Fund (to N.D.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Duan M., Tu J., Lu Z.. Recent advances in detecting mitochondrial DNA heteroplasmic variations. Molecules. 2018; 23:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang R., Wang Y., Ye K., Picard M., Gu Z.. Independent impacts of aging on mitochondrial DNA quantity and quality in humans. BMC Genomics. 2017; 18:890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kvist L., Martens J., Nazarenko A.A., Orell M.. Paternal leakage of mitochondrial DNA in the great tit (Parus major). Mol. Biol. Evol. 2003; 20:243–247. [DOI] [PubMed] [Google Scholar]
- 4. Bentley K.E., Mandel J.R., McCauley D.E.. Paternal leakage and heteroplasmy of mitochondrial genomes in Silene vulgaris: evidence from experimental crosses. Genetics. 2010; 185:961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kalsbeek A.M.F., Chan E.K.F., Corcoran N.M., Hovens C.M., Hayes V.M.. Mitochondrial genome variation and prostate cancer: a review of the mutational landscape and application to clinical management. Oncotarget. 2017; 8:71342–71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li H.L., Kang T., Chen L., Zhang W., Liao Y., Chen J., Shi Y.. Detection of mitochondrial DNA mutations by high-throughput sequencing in the blood of breast cancer patients. Int. J. Mol. Med. 2013; 33:77–82. [DOI] [PubMed] [Google Scholar]
- 7. Valiente-Pallejà A., Torrell H., Muntané G., Cortés M.J., Martínez-Leal R., Abasolo Y., Alonso Y., Vilella E., Martorell L.. Genetic and clinical evidence of mitochondrial dysfunction in autism spectrum disorder and intellectual disability. Human Mol.Genet. 2018; 27:891–900. [DOI] [PubMed] [Google Scholar]
- 8. Uusimaa J., Finnil S., Remes A.M., Rantala H., Vainionp L., Hassinen I.E., Majamaa K.. Molecular Epidemiology of Childhood Mitochondrial Encephalomyopathies in a Finnish Population: Sequence Analysis of Entire mtDNA of 17 Children Reveals Heteroplasmic Mutations in tRNA Arg, tRNA Glu, and tRNA Leu(UUR) Genes. Pediatrics. 2004; 114:443–450. [DOI] [PubMed] [Google Scholar]
- 9. Pinto M., Moraes C.T.. Mitochondrial genome changes and neurodegenerative diseases. Biochim. Biophys. Acta. 2014; 1842:1198–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ryzhkova A.I., Sazonova M.A., Sinyov V.V., Galitsyna E.V., Chicheva M.M., Melnichenko A.A., Grechko A.V., Postnov A.Y., Orekhov A.N., Shkurat T.P.. Mitochondrial diseases caused by mtDNA mutations: a mini-review. Ther. Clin. Risk Manag. 2018; 14:1933–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stewart J.B., Chinnery P.F.. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet. 2015; 16:530–542. [DOI] [PubMed] [Google Scholar]
- 12. Gallimore J.M., McElhoe J.A., Holland M.M.. Assessing heteroplasmic variant drift in the mtDNA control region of human hairs using an MPS approach. Forensic. Sci. Int. Genet. 2018; 32:7–17. [DOI] [PubMed] [Google Scholar]
- 13. Holland M.M., Makova K.D., McElhoe J.A. Deep-coverage MPS analysis of heteroplasmic variants within the mtgenome allows for frequent differentiation of maternal relatives. Genes. 2018; 9:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sultana G.N.N., Sultan M.Z.. Mitochondrial DNA and methods for forensic identification. J. Forensic. Sci. Crimin, Inves. 2018; 9:1. [Google Scholar]
- 15. Irwin J.A., Saunier J.L., Niedersttter H., Strouss K.M., Sturk K.A., Diegoli T.M., Brandsttter A., Parson W., Parsons T.J.. Investigation of heteroplasmy in the human mitochondrial DNA control region: a synthesis of observations from more than 5000 global population samples. J. Mol. Evol. 2009; 68:516–527. [DOI] [PubMed] [Google Scholar]
- 16. Ramos A., Santos C., Mateiu L., Gonzalez M., Alvarez L., Azevedo L., Amorim A., Aluja M.P.. Frequency and Pattern of Heteroplasmy in the Complete Human Mitochondrial Genome. PLoS One. 2013; 8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Just R.S., Irwin J.A., Parson W.. Mitochondrial DNA heteroplasmy in the emerging field of massively parallel sequencing. Forensic. Sci. Int. Genet. 2005; 18:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Duan M., Tu J., Lu Z.. Recent advances in detecting mitochondrial DNA heteroplasmic variations. Molecules. 2018; 23:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McElhoe J.A., Holland M.M., Makova K.D., Su M.S., Paul I.M., Baker C.H., Faith S.A., Young B.. Development and assessment of an optimized next-generation DNA sequencing approach for the mtgenome using the Illumina MiSeq. Forensic. Sci. Int. Genet. 2014; 13:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li M., Schnberg A., Schaefer M., Schroeder M., Nasidze I., Stoneking M.. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am. J. Hum. Genet. 2010; 87:237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schirmer M., DAmore R., Ijaz U.Z., Hall N., Quince C.. Illumina error profiles: resolving fine-scale variation in metagenomic sequencing data. BMC Bioinformatics. 2016; 17:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richly E., Leister D.. NUMTs in sequenced eukaryotic genomes. Mol. Biol. Evol. 2004; 21:1081–1084. [DOI] [PubMed] [Google Scholar]
- 23. Marquis J., Lefebvre G., Kourmpetis Y.A.I., Kassam M., Ronga F., De Marchi U., Wiederkehr A., Descombes P.. MitoRS, a method for high throughput, sensitive, and accurate detection of mitochondrial DNA heteroplasmy. BMC Genomics. 2017; 18:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang S., Huang T.. Characterization of mitochondrial DNA heteroplasmy using a parallel sequencing system. BioTechniques. 2010; 48:287–296. [DOI] [PubMed] [Google Scholar]
- 25. Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A.. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 2012; 36:14508–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo Y., Li J., Li C., Shyr Y., Samuels D.C.. MitoSeek: extracting mitochondria information and performing high-throughput mitochondria sequencing analysis. Bioinformatics. 2013; 29:1210–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Calabrese C., Simone D., Diroma M.A., Santorsola M., Gutt C., Gasparre G., Picardi E., Pesole G., Attimonelli M.. MToolBox: a highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing. Bioinformatics. 2014; 30:3115–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weissensteiner H., Forer L., Fuchsberger C., Schpf B., Kloss-Brandsttter A., Specht G., Kronenberg F., Schnherr S.. mtDNA-Server: next-generation sequencing data analysis of human mitochondrial DNA in the cloud. Nucleic Acids Res. 2016; 44:W64–W69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kastally C., Mardulyn P.. Widespread co-occurrence of two distantly related mitochondrial genomes in individuals of the leaf beetle Gonioctena intermedia. Biol. Lett. 2017; 13:20170570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dierckxsens N., Mardulyn P., Smits G.. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017; 45:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krzywinski M.I., Schein J.E., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A.. Circos: an information aesthetic for comparative genomics. Genome Res. 2009; 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilm A., Aw P.P.K., Bertrand D., Yeo G.H.T., Ong S.H., Wong C.H., Khor C.C., Petric R., Hibberd M.L., Nagarajan N.. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012; 40:11189–11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ye K., Lu J., Ma F., Keinan A., Gu Z.. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:10654–10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Newell C., Hume S., Greenway S.C., Podemski L., Shearer J., Khan A.. Plasma-derived cell-free mitochondrial DNA: A novel non-invasive methodology to identify mitochondrial DNA haplogroups in humans. Mol. Genet. Metab. 2018; 125:332–337. [DOI] [PubMed] [Google Scholar]
- 35. Zambelli F., Vancampenhout K., Daneels D., Brown D., Mertens J., Van Dooren S., Caljon B., Gianaroli L., Sermon K., Voet T. et al.. Accurate and comprehensive analysis of single nucleotide variants and large deletions of the human mitochondrial genome in DNA and single cells. Eur. J. Hum. Genet. 2017; 25:1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.