Abstract
Detection of structural variants (SVs) on the basis of read alignment to a reference genome remains a difficult problem. De novo assembly, traditionally used to generate reference genomes, offers an alternative for SV detection. However, it has not been applied broadly to human genomes because of fundamental limitations of short-fragment approaches and high cost of long-read technologies. We here show that 10× linked-read sequencing supports accurate SV detection. We examined variants in six de novo 10× assemblies with diverse experimental parameters from two commonly used human cell lines: NA12878 and NA24385. The assemblies are effective for detecting mid-size SVs, which were discovered by simple pairwise alignment of the assemblies’ contigs to the reference (hg38). Our study also shows that the base-pair level SV breakpoint accuracy is high, with a majority of SVs having precisely correct sizes and breakpoints. Setting the ancestral state of SV loci by comparing to ape orthologs allows inference of the actual molecular mechanism (insertion or deletion) causing the mutation. In about half of cases, the mechanism is the opposite of the reference-based call. We uncover 214 SVs that may have been maintained as polymorphisms in the human lineage since before our divergence from chimp. Overall, we show that de novo assembly of 10× linked-read data can achieve cost-effective SV detection for personal genomes.
INTRODUCTION
Cost-effective whole-genome sequencing has been revolutionized over the past decade by short-fragment approaches (1,2). Standard Illumina data support high-quality, read-mapping-based detection of single-nucleotide variants (SNVs) in ∼90% of the human genome (3–7). De novo assembly of Illumina data has been recognized to be an alternative way to generate comparable SNV and better small indel (insertion/deletion) calls (8). However, detection of structural variants (SVs) on the basis of short-fragment Illumina data alone continues to be challenging (9–11), and de novo assembly of anything but the simplest microbial genomes (12) does not yet generate usefully contiguous genome sequences unless Illumina data are supplemented with other data (13–15).
The lack of long-range contiguity in standard Illumina data has distinct consequences depending on the applications. For SV discovery, split reads and other mapping-based approaches can detect breakpoints but connecting them to call a specific SV remains extremely challenging (16–19). For haplotyping, variants can be phased by population-based methods (20,21) or family-based recombination inference (22,23), but such approaches are only feasible for common SNVs or large pedigrees. Finally, highly polymorphic regions such as the HLA in which the reference sequence does not adequately capture the diversity present in the population are refractory to mapping-based approaches and require de novo assembly (24). However, for de novo assembly, short-fragment data are challenged by interspersed repetitive sequences from mobile elements and by segmental duplications, and only support highly fragmented genome reconstruction (25,26).
In principle, many of the challenges of short-fragment approaches for comprehensive variant discovery can be overcome by long-fragment/read sequencing (27,28). Direct sequencing of long DNA fragments requires single-molecule approaches, such as Pacific Biosciences (PacBio) or Oxford Nanopore (ONT) (29,30). This is because no enzymatic technology exists that can reliably amplify long DNA fragments of arbitrary sequences. The main trade-offs between Illumina and single-molecule long read approaches can at present be characterized as low-cost, high base quality, short fragments (Illumina) versus higher cost, low raw base quality, long fragments (PacBio and ONT) (9,31). As a consequence, whole-genome sequencing technologies now tend to be deployed in highly specialized ways that emphasize different methodologies depending on the goal to be achieved: standard 30× Illumina sequencing for small variant detection and relatively low-power SV detection (7,32); mate-pair libraries or single-molecule approaches (i.e. long-fragment) for better SV detection and haplotyping (9,33), and hybrid approaches with more than one technology for de novo assembly (15,34).
Novel computational approaches leveraging the special characteristics of 10× Genomics data have already generated significant advances in power and accuracy of haplotyping (35,36), cancer genome reconstruction (37,38), metagenome assemblies (39) and de novo assembly of large genomes (14,40,41). 10× linked-read sequencing combines low per-base error and good small-variant discovery with long-range information for much improved SV detection (38,42), and the possibility of long-range contiguity in de novo assembly (40,41,43).
As assembly-based approaches become prevalent for SV detection (44,45), it becomes important to evaluate assembly quality and its dependence on library preparation parameters. We therefore assessed the ability of de novo 10× assemblies to support SV detection with different parameters in 10× linked-read libraries generation on two well-studied individual genomes. Our analyses are based on pairwise alignment of the assemblies' contigs to the reference genome and finding gaps, a procedure whose compelling simplicity is only possible with assembly-based approaches (8). We use three metrics (SVs shared between individuals, support by PacBio data, and alignment to Ape genomes) to assess the accuracy of our assembly-based SV calls. Additionally, we explore the difference between the SV calls and the molecular mechanism that produced the derived allele and are able to identify the true molecular event that brought about a subset of SVs. Finally, we uncover an unexpected number of SVs that have most likely been maintained as polymorphisms since before the last common ancestor of chimps and humans.
MATERIALS AND METHODS
DNA extraction, library construction and sequencing
We ordered NA12878 and NA24385 from Coriell Institute and sequenced them accordingly with a variety of parameters. These two cell lines were chosen because they have the most complete data from other sources to validate our variant calls. For library L1, genomic DNA was extracted from ∼1 000 000 cultured NA12878 cells using the Gentra Puregene Blood Kit following manufacturer’s instructions (Qiagen, Cat. No 158467). To generate longer DNA fragments (W  = 150 kb and longer) for L2 to L6, a modified protocol for DNA extraction was applied. Two-hundred thousand NA12878 or NA24385 cells of fresh culture were added to 1 ml cold 1× PBS in a 1.5-ml tube and pelleted for 5 min at 300 g. The cell pellets were completely resuspended in the residual supernatant by vortexing and then lysed by adding 200 μl Cell Lysis Solution and 1 μl of RNaseA Solution (Qiagen, Cat. No 158467), mixing by gentle inversion, and incubating at 37°C for 15–30 min. This cell lysis solution is used immediately as input for the 10× Chromium prep (ChromiumTM Genome Library & Gel Bead Kit v2, PN-120258; ChromiumTM i7 Multiplex Kit, PN-120262). Fragment size of the input DNA was controlled by gentle handling during lysis and DNA preparation for 10× Chromium system. Different amounts of input DNA (between 1.25 and 4 ng) were used to generate libraries with different CF. The 10× Chromium Controller was operated and the GEM prep was performed as instructed by the manufacturer. Individual libraries were then constructed by end repairing, A-tailing, adapter ligation and PCR amplification. Each library was sequenced with three lanes of paired-end 150 bp runs on the Illumina HiSeqX instrument to obtain high genomic coverage. The assembly-based SNVs and SVs from these libraries were analyzed and validated by a variety of strategies (Supplementary Figure S1).
= 150 kb and longer) for L2 to L6, a modified protocol for DNA extraction was applied. Two-hundred thousand NA12878 or NA24385 cells of fresh culture were added to 1 ml cold 1× PBS in a 1.5-ml tube and pelleted for 5 min at 300 g. The cell pellets were completely resuspended in the residual supernatant by vortexing and then lysed by adding 200 μl Cell Lysis Solution and 1 μl of RNaseA Solution (Qiagen, Cat. No 158467), mixing by gentle inversion, and incubating at 37°C for 15–30 min. This cell lysis solution is used immediately as input for the 10× Chromium prep (ChromiumTM Genome Library & Gel Bead Kit v2, PN-120258; ChromiumTM i7 Multiplex Kit, PN-120262). Fragment size of the input DNA was controlled by gentle handling during lysis and DNA preparation for 10× Chromium system. Different amounts of input DNA (between 1.25 and 4 ng) were used to generate libraries with different CF. The 10× Chromium Controller was operated and the GEM prep was performed as instructed by the manufacturer. Individual libraries were then constructed by end repairing, A-tailing, adapter ligation and PCR amplification. Each library was sequenced with three lanes of paired-end 150 bp runs on the Illumina HiSeqX instrument to obtain high genomic coverage. The assembly-based SNVs and SVs from these libraries were analyzed and validated by a variety of strategies (Supplementary Figure S1).
De novo diploid assembly
Scaffolds were generated by the ‘pseudohap2’ output style of Supernova2 (40), which explicitly generated scaffolds for two haplotypes, simultaneously. Pairs of scaffolds were extracted as the two haplotypes from the Supernova2 megabubble structures if they shared the same start and end nodes in the assembly graph. Diploid contigs were generated by breaking the candidate scaffolds at the sequences with least 10 consecutive ‘N’s and were aligned to human reference genome (hg 38) by Minimap2 (46). The genome was split into 500 bp windows and diploid regions were defined as the maximum extent of successive windows covered by two contigs, each from one haplotype (47).
SNV and SV calls from diploid contigs
We used Paftools (https://github.com/lh3/minimap2/tree/master/misc) to identify SNVs and SVs no shorter than 50 bp from the CS tags generated by Minimap2 alignment. A valid variant was covered by exactly two contigs with mapping quality >20, each from one haplotype. SVs were called as homozygous if the calls from the two allelic contigs were overlapping. SVs were considered shared among assemblies from the same individual if there was any overlap in coordinates.
Validation of SNV calls
We validated SNVs by comparison with the ‘gold standard’ GIAB (Genome in a Bottle) SNV call set (NA12878: ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/NA12878_HG001/latest/GRCh38/, NA24385: ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio/HG002_NA24385_son/latest/ GRCh38/). Any SNV calls were removed if they are outside of GIAB high-confidence regions or diploid regions. The SNVs were generated by freebayes (https://github.com/ekg/freebayes) from the barcode-aware alignments of Lariat (48).
Validation of SV calls
SVs were examined by three approaches: (i) we applied svviz2 (49) to analyze PacBio reads from NA12878 (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/NA12878/NA12878_PacBio_MtSinai/) and NA24385 (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/PacBio_MtSinai_NIST/). svviz2 aligned and compared the PacBio reads to the reference sequence and the reconstructed the alternative allele of candidate SVs. Genotypes 0/1 and 1/1 confirmed our SV calls; genotypes were also used to evaluate the genotype accuracy in the validated call set. (ii) We identified SVs called in both NA12878 and NA24385 and considered them reciprocally validated if their coordinates differed by fewer than 20 bp. We only considered the existence of SVs regardless of their genotype concordance. The complete set of SVs for each sample was the union of calls of the three libraries. (iii) We aligned each SV and 500 bp flanking sequence on either side from the involved contigs to their chimpanzee (chimp, reference genome Pan_tro_3.0) and orangutan (orang, reference genome PPYG2) orthologs. We defined the aligned distance between the end of the left flanking sequence and the start of the right flanking sequence as dis(align). For deletions, if dis(align) was <2 bp, then the derived allele was recognized as an insertion carried by the reference genome; if dis(align) was between 0.9 and 1.1 times of the SV length, then the derived allele was recognized as a deletion in the individual’s genome. For insertions, if dis(align) was <2 bp, then the derived allele was recognized as an insertion carried by the target genome; if dis(align) was between 0.9 and 1.1 times of the SV length, then the derived allele was recognized as a deletion carried by the reference genome.
Comparison to other callsets
The mapping-based SVs were called by Long Ranger 2.2.2 (https://support.10xgenomics.com/genome-exome/software/pipelines/latest/what-is-long-ranger). Truvari (50) was used to compare SV calls with Tier 1 benchmark of GIAB (50). The SVs from the other assemblies (ONT; 10x/Bionano) were called based on the same methods and parameters as we used for Supernova2.
Annotation of SV sequence
Deletions and insertions were annotated as Alu sequences if they were between 250 and 350 bp long and could be uniquely aligned to the Alu consensus sequence from the UCSC Genome Browser. We used Tandem Repeats Finder (51) to annotate tandem repeats.
Multiple sequence alignment to detect ancient polymorphism
We produced the four-way multiple sequence alignments using MUSCLE (52) from the SVs where orang and chimp differed in matching the reference sequence or the alternate allele. The sequences were (i) human reference sequence, (ii) assembled target sequence, (iii) orangutan reference sequence and (iv) chimpanzee reference sequence. We then examined all such alignments to verify whether the SV sequence was orthologous and if the breakpoints were identical.
RESULTS
Library preparation, physical parameters and sequence depth
We prepared and sequenced six whole-genome libraries with diverse total input DNA and fragment size distributions, three for NA12878 and NA24385 each (Materials and Methods section). Accordingly, the data varied in physical fragment coverage (CF), read coverage per fragment (CR) and average fragment size ( ) (Supplementary Table S1). We used Supernova2 for assembly, limiting the depth by subsampling to include 1200 M reads, corresponding to ∼56-fold sequence coverage (subsampled libraries from
) (Supplementary Table S1). We used Supernova2 for assembly, limiting the depth by subsampling to include 1200 M reads, corresponding to ∼56-fold sequence coverage (subsampled libraries from  to
to 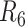 in Table 1). The contigs from the six assemblies were aligned against the human reference genome (hg38) to identify SNVs, and indels of 50 bp or greater (Materials and Methods section). We quantified the assembly qualities of libraries constructed and sequenced with different parameters (Table 1). CF between 800× and 1000× achieved the best contig N50 without sacrificing the fraction of the genome that was diploid, which suggested that the CF recommended by 10× Genomics is not always the optimal metric for 10× linked-read assembly. Furthermore, our assemblies suggested the optimal W
in Table 1). The contigs from the six assemblies were aligned against the human reference genome (hg38) to identify SNVs, and indels of 50 bp or greater (Materials and Methods section). We quantified the assembly qualities of libraries constructed and sequenced with different parameters (Table 1). CF between 800× and 1000× achieved the best contig N50 without sacrificing the fraction of the genome that was diploid, which suggested that the CF recommended by 10× Genomics is not always the optimal metric for 10× linked-read assembly. Furthermore, our assemblies suggested the optimal W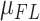 should be around 50–150 kb.
should be around 50–150 kb.
Table 1.
Summary of the assemblies of the six libraries from NA12878 and NA24385
| Library | Sample | Contig N50/NA50 (kb) | Scaffold N50/NA50 (Mb) | Coverage (%) | Diploid regions (%) | Haploid regions (%) |
|---|---|---|---|---|---|---|
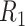
|
NA12878 | 141.2/116.8 | 27.86/13.43 | 91.9 | 58.9 | 27.7 |
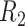
|
NA12878 | 114.9/100.4 | 17.22/6.96 | 91.1 | 73.3 | 11.3 |
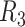
|
NA12878 | 99.4/86.3 | 7.93/4.77 | 91.7 | 77.2 | 9.2 |
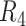
|
NA24385 | 101.2/89.2 | 8.76/4.66 | 91.3 | 73.4 | 12.2 |
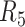
|
NA24385 | 58.4/54.2 | 2.85/1.94 | 91.7 | 79.2 | 5.8 |
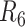
|
NA24385 | 129.2/110.3 | 48.66/12.57 | 91.7 | 78.1 | 7.9 |
Contigs are aligned to human reference genome (hg38) to calculate the overall genomic coverage and the genomic regions in diploid and haploid states.
Concordance and accuracy of assembly based SNV calls
We first analysed SNV calls from the pairwise alignments in order to assess the overall feasibility of assembly based variant calling. The number of SNV calls from five libraries ( to
to 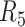 ) was comparable, around 3 000 000 (Supplementary Table S2). By contrast,
) was comparable, around 3 000 000 (Supplementary Table S2). By contrast,  covered the lowest percentage of diploid regions (58.9%) and generated the smallest SNV set (2 635 173; Table 1 and Supplementary Table S2). The assemblies of the libraries from the same individual shared >92% of SNVs with another, and 2–2.4 × 106 SNVs were shared by all the three (Supplementary Tables S3 and S4). Genotype concordances were high for those SNVs shared by all three assemblies of the same individual, >99.9% (Supplementary Table S5). These assembly based calls cover 92.4–93.6% (NA12878) and 95.1–96.5% (NA24385) of SNVs called by barcode-aware, mapping-based calls. Genotype concordance between assembly and mapping-based calls was high for all the libraries, around 99.8% (Supplementary Table S6). Furthermore, we compared assembly based calls with the ‘gold standard’ GIAB call set (53). We only evaluated the ‘gold standard’ SNVs that fell within the overlap of diploid regions of our assemblies and of high confidence regions from GIAB (Materials and Methods section). Around 93–97% of these SNVs could be detected by assembly based calls (Supplementary Tables S7–S12).
covered the lowest percentage of diploid regions (58.9%) and generated the smallest SNV set (2 635 173; Table 1 and Supplementary Table S2). The assemblies of the libraries from the same individual shared >92% of SNVs with another, and 2–2.4 × 106 SNVs were shared by all the three (Supplementary Tables S3 and S4). Genotype concordances were high for those SNVs shared by all three assemblies of the same individual, >99.9% (Supplementary Table S5). These assembly based calls cover 92.4–93.6% (NA12878) and 95.1–96.5% (NA24385) of SNVs called by barcode-aware, mapping-based calls. Genotype concordance between assembly and mapping-based calls was high for all the libraries, around 99.8% (Supplementary Table S6). Furthermore, we compared assembly based calls with the ‘gold standard’ GIAB call set (53). We only evaluated the ‘gold standard’ SNVs that fell within the overlap of diploid regions of our assemblies and of high confidence regions from GIAB (Materials and Methods section). Around 93–97% of these SNVs could be detected by assembly based calls (Supplementary Tables S7–S12).
We also investigated whether the parameters of library preparation and sequencing might explain some of the differences in SNVs detection between libraries (Supplementary Table S1). For NA12878 and NA24385, the two libraries with the lowest physical coverages of 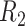 and
and 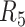 (CF = 123× and 208×) had the worst performance (highest false negative rates and lowest genotype concordance). Substantially greater CF had much better performance (Supplementary Table S13). We did not observe much difference between
(CF = 123× and 208×) had the worst performance (highest false negative rates and lowest genotype concordance). Substantially greater CF had much better performance (Supplementary Table S13). We did not observe much difference between 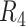 and
and 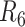 , suggesting the performance of SNV calls would not dramatically change if the physical coverage was sufficiently high (CF = 803×). The most common assembly based genotyping errors were heterozygous SNVs miscalled as homozygosity (Supplementary Table S13).
, suggesting the performance of SNV calls would not dramatically change if the physical coverage was sufficiently high (CF = 803×). The most common assembly based genotyping errors were heterozygous SNVs miscalled as homozygosity (Supplementary Table S13).
SV calls from diploid contigs
We inferred large and mid-size indels (≥50 bp) from the same contig-to-reference alignments that were used for SNV calling (Materials and Methods section). Two to three times more deletions than insertions were detected in the six assemblies (Supplementary Table S2). The size distributions of different libraries were comparable, with a peak near 300 bp. Most of the SVs in that peak are Alu sequences (Materials and Methods section and Figure 1; Supplementary Figures S2 and S3). We also observed peaks around 6 kb in deletions, corresponding to LINE1s (L1s) (Materials and Methods section and Figure 1; Supplementary Figures S2 and S3). SV calls in the three assemblies from the same individual differ somewhat with each assembly having around 30–40% unique calls, and overlapping calls also constitute similar proportion for each library (Supplementary Figure S4 and Supplementary Tables S14–S17).
Figure 1.

Deletion and Insertion size distributions of NA12878 for 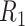 (A, B, G and H),
(A, B, G and H),  (C, D, I and G) and
(C, D, I and G) and 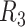 (E, F, K and I).
(E, F, K and I).
Comparison between Supernova-based and other SV calls
We compared the overlap in SVs between 10×-based calls from barcode-aware read mapping by Long ranger with our Supernova assemblies, using the same data from the six libraries. In order to compare to published work, we replicated a pipeline that had been used previously (Materials and Methods section) to highlight the potential of 10x-based SV calling (54). Regardless of library, Supernova assemblies generated more than twice the number of calls than mapping-based calls; a majority of the mapping-based calls were covered by the Supernova calls, and many calls were unique to the Supernova call set (Supplementary Tables S18 and S19).
We also compared our calls from NA12878 with callsets we generated from recently released assemblies of ONT (55) and 10×/Bionano data (14) (Materials and Methods section). Our Supernova-based SV calls had more overlap with the ones from ONT than 10x/Bionano (Supplementary Tables S20–S23). The SVs shared by at least two of our libraries were more likely to also be called by the other technology.
During preparation of our manuscript, the GIAB consortium released a preliminary callset of SVs in NA24385, v0.6 (50). We focused on Tier1, the most specific of the GIAB callsets. Overall, precision of the Supernova-based calls was ∼0.5 depending on the library, with recall being lower (Supplementary Table S24). Excluding tandem repeats increased precision to almost 80%, with recall below 0.2 (Supplementary Table S25).
SV set evaluation
For additional insight into the details of SV calling on the basis of Supernova assemblies, we designed three criteria to further evaluate our calls: supporting evidence from PacBio reads analyzed by svviz2 (49); overlap between the two individuals; and finally, by alignment to two ape genomes (chimp and orang; Materials and Methods section; Supplementary Figure S5). For these analyses, we pooled the non-redundant calls from the three libraries from each individual. This inflates the false positive rate but allows for a more comprehensible analysis. By using the union of the abovementioned three criteria, we could validate roughly half of the deletions (51.3% for NA12878 and 50.7% for NA24385) and almost 80% of the insertions (78.5% for NA12878 and 78.3% for NA24385; Figure 2).
Figure 2.

Three SV evaluation approaches: (i) overlap between NA12878 and NA24385 (both individuals, green), (ii) supported by any ape genome (Ape, blue), (iii) supported by PacBio reads (PacBio, red). Numbers are SV counts.
Overlaps of calls between the two individuals or between one individual and an ape are likely to be highly specific, but not sensitive: specific because it is extraordinarily unlikely to produce the same SV twice in two independent hominid lineages; not sensitive because the two individuals do not share all variants, but rather a fraction that depends on population genetic parameters and stochasticity. The PacBio reads, by contrast, are derived from the same individual and are therefore expected to be both sensitive and specific. Indeed, PacBio reads validated the largest fraction of our SV calls compared to the other methods (Figure 2). However, ∼20% of deletions with support from apes, and ∼18% of deletions with support from the other individual, were not validated by PacBio reads. This suggests that validation by PacBio is not fully sensitive either, and that some of the unvalidated deletion calls are in fact true positives. For insertions, the fraction of calls validated by the other individual but not by PacBio is considerably lower (∼4%), which is consistent with the idea that insertion calls are more specific than deletion calls, as also suggested by their lower number.
We next investigated whether the type of sequence influenced the validation rate. Classification of insertions and deletions into Alu, non-Alu repetitive, and non-repetitive sequences revealed considerably higher validation rates of Alu insertions than for the other two classes (Figure 3; Supplementary Figures S6 and S7). This is presumably because the assembly process is unlikely to produce a full-length Alu sequence erroneously, and so any insertion whose sequence matches an Alu is highly likely to be correct. Conversely, the fact that different assemblies produce a large number of unique Alu insertion calls that are likely correct again underscores that sensitivity of insertion detection is low, but specificity is high.
Figure 3.

Sensitivities of deletions (A, B, C and D) and insertions (E, F, G and H) for the three libraries of NA12878. Percentages denote the proportion of SVs from assembly based calls validated by any of the three evaluation approaches.
Finally, we examined whether the validation rate differed between SV calls unique or shared among the assemblies for each individual. As expected, the overall validation rate of SVs shared by all three libraries was >95%, whereas unique SVs reached ∼30% for deletions and ∼50% for all insertions (Figure 3).
SV call genotype accuracy and breakpoint precision
To further evaluate assembly based SV calls, we also assessed the accuracies of genotypes. As before, we validated unique and shared SV calls among the three libraries for each individual using PacBio reads. Overall, shared deletions reached above 68% genotype accuracy, with the subset that comprises Alus achieving 84%. Unique deletions reached above 40% accuracy. For insertions, accuracies for both shared and unique ones were significantly higher, above 92% and 75%, respectively. Shared Alu insertions achieved perfect accuracy (100%) (Supplementary Figures S8–S11).
Finally, to assess the base-pair level accuracy of the SV breakpoints we binned the SVs shared by both individuals based on their size differences between the two calls and evaluated their validation rates by PacBio reads and the alignments to ape genomes. If the SVs were validated in both of the individuals, >80% of the deletions and 70% of the insertions had size differences <2 bp. The rates were lower for calls not validated (60% for deletions, 40% for insertions; Supplementary Figure S12).
SV call versus actual molecular mechanism
SVs are called ‘insertions’ or ‘deletions’ by comparison to the reference sequence, but that call does not necessarily reflect the actual molecular mechanism that gave rise to the SV: if the reference sequence carries the derived allele and our sequenced individual carries the ancestral state, the call is the opposite of the molecular mechanism. For 12,537 SVs, 1 kb of flanking sequence (500 bp on either side) could be aligned to at least one of their ape orthologs (Materials and Methods section). On the basis of these alignments, assuming that the ape sequence represents the ancestral state, we thus classified each such SVs as either a true insertion or a true deletion (Figure 4A). As expected from population genetic principles, a large fraction (37%) of deletion calls were in fact derived insertions, and half of called insertions were in fact deletions.
Figure 4.
Classification of insertion and deletion calls into ancestral and derived state and inference of the originating molecular mechanism by comparison against ape genomes. (A) Inference of derived allele and molecular mechanism by alignment to ape sequences; colored circle on tree denotes the lineage in which the mutation occurred. (B) Derived allele insertion size distribution. (C) Derived allele deletion size distribution.
Evidence that the derived allele actually reflects the molecular mechanism that initially generated the variant can be found in the size distribution of the events. Insertions (Figure 4B) follow an exponential dropoff in frequency as a function of size, with the major exception being a peak at 310–330 base pairs, in which 96% of insertions are full-length Alu sequence. By contrast, the deletion size distribution (Figure 4C) exhibits two regions of deviation from an exponential distribution, from ∼110 to 150 bp and from 290 to 330 bp; the latter is somewhat enriched for Alu sequence, reflecting either (i) that we do not classify all called insertions correctly or (ii) that there is some propensity for Alu elements to be deleted across their full length. We also note that the vast majority of detected polymorphic L1 insertions were called as deletions in the assembled individual (i.e. the reference sequence carries the derived insertion allele), suggesting that SusperNova2 has a hard time assembling through young L1s that have not yet accumulated SNVs or other small variants.
Ancient SVs
For 5167 SVs, the two human sequences (reference and alternate allele plus 1 kb flanking sequence as above) could be aligned to both orang and chimp orthologs. The vast majority of alignments were consistent between the two apes, supporting either the reference allele or the alternate allele as being ancestral. However, there were 225 events for which the chimp aligned to one allele, and the orang to the other (Figure 5A). Such inconsistencies can only be explained by two possibilities: (i) two independent insertions or deletions, one having occurred in one of the ape lineages, and another of the same sequence and coordinates generating the human derived allele or (ii) an ancient polymorphism that arose before our last common ancestor with chimp and that has been maintained in the human population since.
Figure 5.

Ancient origin of SVs. (A) The four cases in which orang matches one human allele and chimp the other, and their count in our dataset. (B) Size distributions of the inferred 32 deletions and 182 insertions. Venn diagram indicates how many are shared between the two individuals and how many are unique to one of them. (C) Phasing the SVs with closely linked SNVs; counts in Venn diagram indicate the number of each configuration.
To distinguish between these two possibilities, we proceeded as follows. SVs in our data sets that aligned to both chimp and orang occur approximately once per half-megabase (5167/length of genome covered in diploid contigs), and they are not clustered anywhere in the genome. The evolutionary distance between the apes and human is quite close, and while no models exist from which the probability of a hypothetical co-occurrence of SVs could be predicted, the proportion of such events in our data set (225/5167 = 4%) seems quite high. We constructed multiple sequence alignments among the four sequences and visually inspected each of them. For 214 events, we verified that the ape and human breakpoints precisely aligned and that the sequence of the ape SV was identical (excepting an occasional SNV or small indel) to that of the human allele. Size, sequence and breakpoint locations of overlapping parallel events, by contrast, would be expected to vary independently in humans and apes. We did not observe any such variance for the vast majority of the shared events, strongly suggesting that each SV has a single evolutionary origin and represents an ancient polymorphism maintained in our population since our last common ancestor with chimp.
Assuming that the orang sequence represents the ancestral state, we classified the SVs according to molecular mechanism, yielding 182 derived insertions and 32 derived deletions (Figure 5A and B). This represents a highly significant (Chi-square test, P < 10E-24) deviation from expectation (108 deletions, 106 insertions, based on their proportion in the set of 5167 SVs that could be aligned to both apes). This deviation is consistent with the idea that insertion sequence is more likely than a deletion to produce evolutionary novelty and may be selected for. This finding represents indirect evidence for the selection (positive or balancing) that would be necessary to maintain these polymorphisms for such a long time.
Finally, the multiple alignments provided further opportunity to test the ancient polymorphism hypothesis by analysis of linked SNVs (Figure 5C). About 129 alignments had at least one SNP in the 1 kb of sequence surrounding the SVs; 94 of them were not informative, that is, both ape sequences had the same base, shared with either the reference or the individual. About 25 alignments had at least one SNV that was in phase with the SV; 13 alignments had 5 or more phased SNVs. Curiously, 15 alignments had SNVs that were out of phase with the SV, and 5 of these also had at least one SNV that was in phase. Four of these 5 were arranged such that the SNVs with consistent phase were closer to the SV and the SNV with inconsistent phase was further away, suggesting that these four alignments capture not only ancient polymorphisms (SVs and SNVs) but also ancient recombination events between the consistent and the inconsistent SNVs. The considerable fraction of alignments that contain phased SNVs in the immediate vicinity of an SV is perhaps the strongest evidence in favor of the ancient polymorphism hypothesis.
DISCUSSION
SVs are abundant and important but require long-range information for their detection; thus, they are not easily identified by standard (short-fragment) sequencing. We here explored the utility of assembly based approaches for SV detection, specifically by using de novo assembly on the basis of 10× Genomics data. Our study demonstrates the promising future of assembly based approaches to detect SVs in personal genomes, with reasonable sensitivity and genotype accuracy. Importantly, our pairwise-alignment based SV calls had remarkable breakpoint consistency and accuracy as evaluated by comparisons between the two individuals and with ape sequences.
Diploid assembly and variant detection
In the context of diploid assembly, which is the natural approach for assembly of genomes that harbor heterozygosity, the diploid fraction of the assembly is an important metric: it directly impacts variant discovery and genotyping, in that erroneously haploid regions will be missing all of their heterozygosity. The short input fragment length ( or W
or W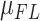 ) of
) of  resulted in roughly 20% less of the genome in a diploid state (Table 1 and Supplementary Table S1, <60% versus <80%) compared to the other libraries of the same individual. As a consequence, there were fewer SNV and SV calls in the analyses involving
resulted in roughly 20% less of the genome in a diploid state (Table 1 and Supplementary Table S1, <60% versus <80%) compared to the other libraries of the same individual. As a consequence, there were fewer SNV and SV calls in the analyses involving  (Supplementary Table S2).
(Supplementary Table S2).
Sensitivity of SNV detection is naturally limited by the fraction of the genome that is covered by the assembly; genotype accuracy evaluation is limited to the fraction of the assembly that is in a diploid state. Overall sensitivity of assembly based calls is ∼90% of that of mapping-based SNV calls and incorrect call rates in high-confidence regions of GIAB are also higher than with mapping-based calls. We conclude that at this point, assembly based SNV calls from Supernova2 are not competitive with barcode-aware read-mapping approaches. However, we note that this is not a compromise as exactly the same sequence data can be used for SNV detection (via barcode-aware mapping) and SV detection (via assembly). We estimate that the cost increase over standard Illumina sequencing is about 2×, given the 10× prep cost and the higher level of sequence coverage required. There may be many applications for which this combination of excellent SNV detection (via barcode-aware read-mapping) and highly precise SV discovery (via assembly), achieved by the same data set, is worth the cost.
Importance for de novo assembly based SV detection
Our study highlights two concepts that are important for SV science. First, the variation call that is based on comparison to reference is not the same as the allelic origin of the variant. Molecularly, that allelic origin is also the mechanism that gave rise to the variant as the initial single mutation that arose in an ancestral individual’s germline. In our individuals, very large fractions of deletion calls were actually insertions, and vice versa, as expected and as illustrated with hundreds of Alu insertions. The second concept is that there may be many more regions than previously thought in which heterozygosity has been maintained in our lineage since before our last common ancestor with chimp. Our results in this regard support the idea that there is distinct value in assembly based approaches for determining SVs in large numbers of individuals for population genetic questions as well.
DATA AVAILABILITY
The raw sequencing data are deposited in the Sequence Read Archive and the corresponding BioProject accession number is PRJNA527321.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Justin Zook, Marc Salit, Alex Bishara, Noah Spies, Nancy Hansen, David Jaffe, and Deanna Church for informative discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
This research was supported by training and research grants from the National Institute of Standards and Technology, Gaithersburg, MD, USA.
Conflict of interest statement. Arend Sidow is a consultant and shareholder of DNAnexus, Inc.
REFERENCES
- 1. Metzker M.L. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010; 11:31–46. [DOI] [PubMed] [Google Scholar]
- 2. Shendure J., Balasubramanian S., Church G.M., Gilbert W., Rogers J., Schloss J.A., Waterston R.H.. DNA sequencing at 40: past, present and future. Nature. 2017; 550:345–353. [DOI] [PubMed] [Google Scholar]
- 3. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nielsen R., Paul J.S., Albrechtsen A., Song Y.S.. Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 2011; 12:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zook J.M., Chapman B., Wang J., Mittelman D., Hofmann O., Hide W., Salit M.. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat. Biotechnol. 2014; 32:246–251. [DOI] [PubMed] [Google Scholar]
- 7. Rimmer A., Phan H., Mathieson I., Iqbal Z., Twigg S.R.F., Consortium W.G.S., Wilkie A.O.M., McVean G., Lunter G.. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 2014; 46:912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics. 2012; 28:1838–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaisson M.J., Huddleston J., Dennis M.Y., Sudmant P.H., Malig M., Hormozdiari F., Antonacci F., Surti U., Sandstrom R., Boitano M. et al.. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015; 517:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Merker J.D., Wenger A.M., Sneddon T., Grove M., Zappala Z., Fresard L., Waggott D., Utiramerur S., Hou Y., Smith K.S. et al.. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet. Med. 2018; 20:159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Onishi-Seebacher M., Korbel J.O.. Challenges in studying genomic structural variant formation mechanisms: the short-read dilemma and beyond. Bioessays. 2011; 33:840–850. [DOI] [PubMed] [Google Scholar]
- 12. Teeling H., Glockner F.O.. Current opportunities and challenges in microbial metagenome analysis–a bioinformatic perspective. Brief. Bioinform. 2012; 13:728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li X., Fan D., Zhang W., Liu G., Zhang L., Zhao L., Fang X., Chen L., Dong Y., Chen Y. et al.. Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat. Commun. 2015; 6:8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mostovoy Y., Levy-Sakin M., Lam J., Lam E.T., Hastie A.R., Marks P., Lee J., Chu C., Lin C., Dzakula Z. et al.. A hybrid approach for de novo human genome sequence assembly and phasing. Nat. Methods. 2016; 13:587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zimin A.V., Puiu D., Luo M.C., Zhu T., Koren S., Marcais G., Yorke J.A., Dvorak J., Salzberg S.L.. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res. 2017; 27:787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alkan C., Coe B.P., Eichler E.E.. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011; 12:363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Layer R.M., Chiang C., Quinlan A.R., Hall I.M.. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014; 15:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mohiyuddin M., Mu J.C., Li J., Bani Asadi N., Gerstein M.B., Abyzov A., Wong W.H., Lam H.Y.. MetaSV: an accurate and integrative structural-variant caller for next generation sequencing. Bioinformatics. 2015; 31:2741–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rausch T., Zichner T., Schlattl A., Stutz A.M., Benes V., Korbel J.O.. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012; 28:i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O’Connell J., Sharp K., Shrine N., Wain L., Hall I., Tobin M., Zagury J.F., Delaneau O., Marchini J.. Haplotype estimation for biobank-scale data sets. Nat. Genet. 2016; 48:817–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Delaneau O., Zagury J.F., Marchini J.. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013; 10:5–6. [DOI] [PubMed] [Google Scholar]
- 22. O’Connell J., Gurdasani D., Delaneau O., Pirastu N., Ulivi S., Cocca M., Traglia M., Huang J., Huffman J.E., Rudan I. et al.. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014; 10:e1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roach J.C., Glusman G., Hubley R., Montsaroff S.Z., Holloway A.K., Mauldin D.E., Srivastava D., Garg V., Pollard K.S., Galas D.J. et al.. Chromosomal haplotypes by genetic phasing of human families. Am. J. Hum. Genet. 2011; 89:382–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kajitani R., Toshimoto K., Noguchi H., Toyoda A., Ogura Y., Okuno M., Yabana M., Harada M., Nagayasu E., Maruyama H. et al.. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014; 24:1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alkan C., Sajjadian S., Eichler E.E.. Limitations of next-generation genome sequence assembly. Nat. Methods. 2011; 8:61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Treangen T.J., Salzberg S.L.. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 2011; 13:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huddleston J., Ranade S., Malig M., Antonacci F., Chaisson M., Hon L., Sudmant P.H., Graves T.A., Alkan C., Dennis M.Y. et al.. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Res. 2014; 24:688–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu H., Giordano F., Ning Z.. Oxford Nanopore MinION Sequencing and Genome Assembly. Genomics Proteomics Bioinform. 2016; 14:265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roberts R.J., Carneiro M.O., Schatz M.C.. The advantages of SMRT sequencing. Genome Biol. 2013; 14:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jain M., Olsen H.E., Paten B., Akeson M.. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016; 17:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sedlazeck F.J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., Schatz M.C.. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018; 15:461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. et al.. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011; 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cretu Stancu M., van Roosmalen M.J., Renkens I., Nieboer M.M., Middelkamp S., de Ligt J., Pregno G., Giachino D., Mandrile G., Espejo Valle-Inclan J. et al.. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 2017; 8:1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo J.F., Zhang L., Li K., Mei J.P., Xue J., Chen J., Tang X., Shen L., Jiang H., Chen C et al.. Coding mutations in NUS1 contribute to Parkinson's disease. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:11567–11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edge P., Bafna V., Bansal V.. HapCUT2: robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res. 2017; 27:801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patterson M., Marschall T., Pisanti N., van Iersel L., Stougie L., Klau G.W., Schonhuth A.. WhatsHap: Weighted haplotype assembly for future-generation sequencing reads. J. Comput. Biol. 2015; 22:498–509. [DOI] [PubMed] [Google Scholar]
- 37. Zheng G.X., Lau B.T., Schnall-Levin M., Jarosz M., Bell J.M., Hindson C.M., Kyriazopoulou-Panagiotopoulou S., Masquelier D.A., Merrill L., Terry J.M. et al.. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 2016; 34:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spies N., Weng Z., Bishara A., McDaniel J., Catoe D., Zook J.M., Salit M., West R.B., Batzoglou S., Sidow A.. Genome-wide reconstruction of complex structural variants using read clouds. Nat. Methods. 2017; 14:915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bishara A., Moss E.L., Kolmogorov M., Parada A.E., Weng Z., Sidow A., Dekas A.E., Batzoglou S., Bhatt A.S.. High-quality genome sequences of uncultured microbes by assembly of read clouds. Nat. Biotechnol. 2018; 36:1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weisenfeld N.I., Kumar V., Shah P., Church D.M., Jaffe D.B.. Direct determination of diploid genome sequences. Genome Res. 2017; 27:757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hulse-Kemp A.M., Maheshwari S., Stoffel K., Hill T.A., Jaffe D., Williams S.R., Weisenfeld N., Ramakrishnan S., Kumar V., Shah P. et al.. Reference quality assembly of the 3.5-Gb genome of Capsicum annuum from a single linked-read library. Hortic. Res. 2018; 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Elyanow R., Wu H.T., Raphael B.J.. Identifying structural variants using linked-read sequencing data. Bioinformatics. 2017; 34:353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones S.J., Haulena M., Taylor G.A., Chan S., Bilobram S., Warren R.L., Hammond S.A., Mungall K.L., Choo C., Kirk H. et al.. The Genome of the Northern Sea Otter (Enhydra lutris kenyoni). Genes (Basel). 2017; 8:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wong K.H.Y., Levy-Sakin M., Kwok P.Y.. De novo human genome assemblies reveal spectrum of alternative haplotypes in diverse populations. Nat. Commun. 2018; 9:3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chaisson M.J.P., Sanders A.D., Zhao X., Malhotra A., Porubsky D., Rausch T., Gardner E.J., Rodriguez O.L., Guo L., Collins R.L. et al.. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019; 10:1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pedersen B.S., Quinlan A.R.. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 2017; 34:867–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bishara A., Liu Y., Weng Z., Kashef-Haghighi D., Newburger D.E., West R., Sidow A., Batzoglou S.. Read clouds uncover variation in complex regions of the human genome. Genome Res. 2015; 25:1570–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Spies N., Zook J.M., Salit M., Sidow A.. svviz: a read viewer for validating structural variants. Bioinformatics. 2015; 31:3994–3996. [DOI] [PubMed] [Google Scholar]
- 50. Zook J.M., Hansen N.F., Olson N.D., Chapman L.M., Mullikin J.C., Xiao C., Sherry S., Koren S., Phillippy A.M., Boutros P.C. et al.. A robust benchmark for germline structural variant detection. 2019; 09 June 2019, preprint: not peer reviewed 10.1101/664623. [DOI]
- 51. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999; 27:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zook J., McDaniel J., Parikh H., Heaton H., Irvine S.A., Trigg L., Truty R., McLean C.Y., De La Vega F.M., Xiao C. et al.. Anopen resource for accurately benchmarking small variant and reference calls. Nat. Biotechnol. 2018; 37:561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Marks P., Garcia S., Barrio A.M., Belhocine K., Bernate J., Bharadwaj R., Bjornson K., Catalanotti C., Delaney J., Fehr A. et al.. Resolving the full spectrum of human genome variation using Linked-Reads. Genome Res. 2019; 29:635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T. et al.. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018; 36:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data are deposited in the Sequence Read Archive and the corresponding BioProject accession number is PRJNA527321.



