Abstract
Objective(s):
Tuberculosis (TB), caused by Mycobacterium tuberculosis, is a widespread infectious disease around the world. Early diagnosis is always important in order to avoid spreading. At present, many studies have confirmed that microRNA (miRNA) could be a useful tool for diagnosis. This study aimed to evaluate whether miRNAs could be regarded as a noninvasive diagnosis biomarker from sputum for pulmonary tuberculosis (PTB).
Materials and Methods:
The M. tuberculosis strain H37Rv was incubated and cultured with human macrophage line THP-1. The total RNA was extracted from the THP-1 cells for detection. Six increased expressions of miRNAs were selected by miRNA microarray chips and the miRNAs were confirmed by qRT-PCR in the M. tuberculosis infection cell model. At last, the efficiency of other methods was compared with using miRNA.
Results:
Only miR-155 showed a better diagnostic value for PTB than the other five miRNAs to distinguish PTB from non-PTB, including pneumonia, lung cancer, and unexplained pulmonary nodules. Next, we detected and analyzed the results of 68 PTB patients and 122 non-PTB, the sensitivity and specificity of miR-155 detection was 94.1% and 87.7%, respectively. It was higher than sputum smear detection and anti-TB antibody detection. But slightly lower than ELISpot (97%, P=0.404). Interestingly, the ranking of sputum smear by Ziehl-Neelsen staining had positive correlation with the expression level of miR-155 in smear-positive sputum (R2=0.8443, P<0.05).
Conclusion:
Our research suggested that miR-155 may be an efficiency biomarker for active PTB diagnosis and bacteria-loads evaluation.
Key Words: Biomarker, Diagnosis, microRNA-155, Pulmonary tuberculosis, Sputum
Introduction
Tuberculosis (TB), caused by various strains of mycobacteria, mainly Mycobacterium tuberculosis in humans, is a very contagious infectious disease (1). As one of the most deadly infectious diseases to seriously threaten human health, TB can attack multiple organs, especially the lungs causing pulmonary TB (PTB), but can also invade other parts of the body causing extra-PTB (2). Most infections are asymptomatic and latent, but about ten percent of PTB patients infected with M. tuberculosis eventually develop active disease which, in case of delayed treatment, kills over fifty percent of those infected (3). Therefore, early diagnosis of PTB is the key to reducing mortality; however, diagnosis remains a challenge.
At present, the methods of TB diagnosis mainly have direct sputum smear staining for acid-fast bacilli (AFB), sputum culture, tuberculin skin test (TST), and chest radiography in routine clinical practice (4). However, direct sputum smear staining has a low sensitivity and M. tuberculosis culture, the gold standard for the diagnosis of M. tuberculosis, has the drawbacks of being time-consuming and low success rate of culture (4-6). In infants of less than six years, the TST is used while this test is not specific for M. tuberculosis detection. The chest radiography has high technical requirements for operators and high cost and, it is not suitable for children and pregnant women. In view of the above, these conventional methods of TB diagnosis have certain disadvantages that cause diagnostic delays and misdiagnosis, increasing the rate of morbidity and mortality. Therefore, discovering new methods to effectively improve the diagnosis of PTB is urgently needed.
MicroRNAs (miRNAs) are small single-stranded noncoding RNAs (generally 19~25 nucleotides in length) that play a vital role in many important biological processes, such as cell proliferation, cell migration, cell invasion, cell apoptosis, cell death, and early embryonic development, through regulating the expression of their target mRNAs (7-9). Except for being highly conserved in variety of species, miRNAs have remarkable stability in different kinds of biological specimen, including serum, plasma, sputum, urine, fresh snap-frozen materials, formalin-fixed, and even in very harsh conditions, this is due to their resistance to extreme pH and temperatures, repeated freeze-thaw cycles, long storage in frozen conditions, and endogenous or exogenous RNA enzymes (10). Owing to these prominent characteristics, miRNAs can be considered as good candidates for use as biomarkers in human diseases. Recently, miRNAs as potential diagnostic biomarkers of multiple diseases have been widely studied (11-14).
Ndzi et al. reported that plasma miR-29a-3p showed good diagnostic performance (84.35%) between active TB and latent TB and good distinguishing performance (81.37%) between active TB and control groups (15). Serum and sputum miR-144 levels were up-regulated significantly in TB patients, the ROC analysis of serum miR-144 showed high sensitivity (99.19%) and specificity (94.87%), the ROC analysis of sputum miR-144 also showed high sensitivity (86.29%) and specificity (99.15%), indicating the potential application value of serum and sputum miR-144 levels in the diagnosis of TB (16). Zheng et al. reported that plasma miR-155 was significantly up-regulated in TB patients and showed a high area under curve (AUC) value of 0.976, obtaining a good diagnostic performance in discriminating PTB from healthy controls (17). Taken together, the above results indicated that fluid miRNAs show a good diagnostic performance in TB patients.
Compared with other samples, sputum is one of the most conveniently and noninvasively accessible biological fluids. Growing evidence has suggested that sputum miRNAs could be a promising diagnostic biomarker for early detection of many human diseases, such as chronic obstructive pulmonary disease and lung cancer (18-20). However, at present, there is little research on sputum miRNA in the diagnosis of tuberculosis. Therefore, considering the above advantages of sputum miRNAs, the study aimed to detect M. tuberculosis induced differentially expressed miRNAs from host macrophage and explore the potential diagnostic value of sputum miR-155 in the diagnosis of active PTB.
Materials and Methods
Sixty-eight patients with active PTB (39 males and 29 females) were enrolled from the Second Peoples’ Hospital of Wuhu City and the Second Affiliated Hospital of Wannan Medical College, China, from January 2017 to October 2018. The subjects ranged in age from 19 to 78 years (mean age 46.8±28.3 years). According to the revised international definitions in TB control of the World Health Organization (WHO) (21), the inclusion criteria of patients with active PTB were as follows: 1. typical PTB clinical symptoms such as fever and cough, 2. fibrocavitary lung infiltrate on chest computed tomography (CT) and chest X-ray, 3. at least one positive sputum examination results (sputum smear or sputum culture), 4. detection results of ELISpot was positive. Patients were excluded who had other coexisting lung diseases. During the same time period, 50 healthy volunteers (25 males and 25 females) who attended the physical examination, were recruited as healthy controls. The subjects ranged in age from 27 to 74 years (mean age 52.4±14.7 years). Healthy donors had no history of TB infection, no organic diseases, and no clinical symptoms of any infectious disease. Meanwhile, three pulmonary disease groups with total of 72 patients were included: pneumonia, lung cancer, unexplained pulmonary nodules. In conclusion, the three basic characteristic of 190 participants, including 122 non-PTB donors and 68 PTB patients, were described in this study (Table 1).
Table 1.
The basic information of 190 donors in this research for clinical verification
| N ame | P rimer sequence |
|---|---|
| hsa-miR-155 | Forward 5’-ACGCTCAGTTAATGCTAATCGTGATA-3’ |
| Reverse 5’-ATTCCATGTTGTCCACTGTCTCTG-3’ | |
| hsa-miR-29b-1* | Forward 5’-GGGTAGCACCATTTGAAATC-3’ |
| Reverse 5’-TTTGGCACTAGCACATT-3’ | |
| hsa-miR-150 | Forward 5’-TAGACGCGTCCGCGAGAGA-3’ |
| Reverse 5’-GTGCAGGGTCCGAGGT-3 | |
| hsa-miR-146a | Forward 5’-ACACTCCAGCTGGGTGAGAACTGAATTCCATG-3’ |
| Reverse 5’-TGTCGTGGAGTCGGCAATTC-3’ | |
| hsa-miR-212 | Forward 5’-GCGCTGGAATGTAAAGAAGT-3’ |
| Reverse 5’-GTGCAGGGTCCGAGGT-3’ | |
| hsa-miR-483-5p | Forward 5’-ACACTCCAGCTGGGAAGACGGGAGGAAAGAA-3’ |
| Reverse 5’-CTCAACTGGTGTCGTGGA-3’ |
F: female; M: male
The study was approved by the ethics committee of the Second Peoples’ Hospital of Wuhu City and the Second Affiliated Hospital of Wannan Medical College and carried out in compliance with the Helsinki Declaration. Written informed consent was obtained from all participants and their families before the commencement of the study.
Sputum samples collection
Early morning sputum samples, 1.5~2.5 ml sputum without saliva, were collected into a sterile plastic container (nuclease-free), and processed within 1 hr before anti-TB treatment and at 1 month after treatment. Obtained sputum was processed with 0.1% dithiothreitol and 20 U/ml RNase inhibitor for homogenization. The homogenized sputum samples were centrifuged at 4000 rpm at 4 °C for 20 to 30 min and the supernatant was collected, subpackaged into 1.5 ml Eppendorf tubes without RNase, and then stored in liquid nitrogen immediately for later testing. To exclude unqualified sputum specimens, at least two sputum smears were prepared for Wright-Giemsa staining. The cells in the sputum smears were counted and classified under a low-magnification microscope. The specimen qualified for the study if the percentage of squamous cells was less than 80% (22, 23).
THP-1 cells culture
Human macrophage cell line THP-1 (GDC100) was kindly provided by professor Xiao-Lian Zhang (Wuhan University, Wuhan, China). THP-1 cells were routinely grown in RPMI 1640 (Life Technologies) supplemented with 10% Fetal Bovine Serum (FBS, Wisent Inc., Quebec, Canada) and induced differentiation by the addition of 100 nM phorbol 12-myristate 13-acetate (PMA) (Beyotime Biotechnology, Shanghai, China) to the culture medium for 48 hr at 37 °C.
M. tuberculosis culture and infection model
M. tuberculosis H37Rv (strain American Type Culture Collection (ATCC) 93009) was purchased from the Beijing Biological Product Institute (Beijing, China) (24). 120 clinical M. tuberculosis strains were obtained from Hainan Agricultural Reclamation Hospital, Dongfang People’s Hospital, and Sanya People’s Hospital. M. tuberculosis strains were cultivated in Middlebrook 7H9 (Roche) containing 10% albumin dextrose catalase (ADC, Roche) for three weeks at 37 °C, and harvested while in the log phase of growth and resuspended with RPMI 1640 culture medium.
For the M. tuberculosis infection model, THP-1 cells were seeded and cultivated in 6-well plates to reach approximately 80% confluent monolayer cells. Then, the cells were washed in PBS and the fresh culture medium was added. THP-1 cells were infected with M. tuberculosis strains at a multiplicity of infection (MOI) of 100 (cells: bacteria=1:100) for 12 hr, 24 hr, and 48 hr, respectively as experimental groups and then washed by PBS three times to remove extracellular M. tuberculosis. Cells without M. tuberculosis strains infection were cultivated for indicated time periods as control groups. Then, cells were collected in TRIzol reagent (Invitrogen, California, USA) and preserved at -80 °C until RNA isolation.
RNA isolation
Total RNA was extracted from THP-1 cells with M. tuberculosis infection and sputum supernatant using TRIzol reagent (Invitrogen) and further purified with an RNeasy mini kit (Qiagen, USA) according to the manufacturer’s protocol. The quantity of the extracted RNA was detected by a NanoDrop2000 spectrophotometer (NanoDrop Technologies, USA). The purity and integrity were assessed using denaturing agarose gel electrophoresis.
Microarray analysis
miRNA expression profile in RNA samples from THP-1 cells with M. tuberculosis infection and THP-1 cells without M. tuberculosis infection was detected by Human miRNA Expression Microarray Release 14.0 (Agilent Technologies, Inc.). The experiment and quality control were performed by CapitalBio Corp (Beijing, China) according to the manual. Fluorescence images on the chips were obtained using luxScan10K Microarray Scanner (CapitalBio) and digitized using Array-Pro image analysis software (Media Cybernetics) for data analysis. Different expressions of miRNAs were selected based on a two-fold change of increase or decrease between TB patients group and non-TB participants group.
Quantitative real-time PCR (qRT-PCR)
Total RNA samples (1 μg each group) were reverse transcribed into cDNA using the miScript® II RT Kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s protocols. Specific primers for qRT-PCR amplification of different genes were purchased from Qiagen (Hilden, Germany). The primer sequences are listed in Table 2. qRT-PCR analysis was performed on ABI Step One Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) in a total volume of 20 μl, containing 2 μl of synthesized cDNA solution, 500 nM of each primer, 10 μl of 2× miScript SYBR-Green PCR Mix (Qiagen), and added sterile water to 20 μl. The amplification conditions included 95 °C for 5 min, followed by 42 cycles of 95 °C for 15 sec, and extension at 58 °C for 30 sec. Each assay was performed in duplicate, and the expression fold change of each miRNA was calculated according to the 2-ΔΔCt method (25) and normalized to the housekeeping U6 expression. Differences in expression levels between two groups (PTB and non-PTB group, patients with active PTB group and healthy group) were measured using the Student’s t-test using SPSS 20.0 with P<0.05 considering statistically significant.
Table 2.
The six pairs of primers sequence for qRT-PCR detection in this research
| miRNAs | 12 hr ( X±SD) | 24 hr ( X±SD) | 48 hr ( X±SD) | F , P- value |
|---|---|---|---|---|
| miR-155 | 6.25±1.23 | 13.56±2.31 | 22.71±5.22 | 17.95, 0.0029 |
| miR-29b-1* | 5.11±1.54 | 17.26±1.09 | 19.94±2.47 | 58.18, <0.000 |
| miR-150 | 4.62±0.94 | 4.28±1.41 | 6.01±1.62 | 1.376, 0.3223 |
| miR-146a | 3.26±0.88 | 3.81±1.02 | 4.29±0.83 | 0.954, 0.436 |
| miR-212 | 3.14±0.65, | 3.94±0.55 | 4.01±0.74 | 1.672, 0.264 |
| miR- 483-5p | 2.76±0.39 | 2.98±0.24 | 3.46±0.38 | 3.257, 0.110 |
ELISpot assay
Peripheral blood mononuclear cells (PBMCs) were separated by centrifugation from 5 ml heparinized blood samples and then added into the pre-coated ELISPOT plate. The experiment was performed using ELISpot kit (Dakewe Biotech Co., Ltd, Shenzhen, China) according to the manufacturer’s instructions. The number of spot-forming cells (SFCs) was counted by an automated ELISpot reader (Cellular Technology Ltd., Shaker Heights, OH) in each well.
Acid-fast staining test (Ziehl-Neelsen staining)
At least two sputum smears were prepared on clean glass slides and were subjected to acid-fast staining using an acid-fast staining kit (Y-S Biotechnology, Shanghai, China) according to the manufacturer’s instructions.
TB antibody test
The serum was separated by centrifugation (3500 rpm for 5 min ) from a 3 ml blood sample. The experiment was performed using a TB antibody test kit (Nattbio Co., Ltd, Guangzhou, China) according to the manufacturer’s instructions.
Statistical analysis
Data were recorded as the mean±standard deviation (±SD) and analyzed by IBM SPSS Statistics version 20.0 software (IBM Corp., Armonk, NY, USA). ANOVA test or Student’s t-test was used for statistical analysis. The chi-square test was performed for comparing results between the sputum miRNA-155 detection and other test methods. P<0.05 was regarded as significantly different.
Results
Different expression of miRNAs in THP-1 cells with H37Rv treatment
The miRNA microarray chips were used to analyze the expression of 36 miRNAs in THP-1 cells with M. tuberculosis H37Rv strain infection. The results showed that only two miRNAs were down-regulated, while most of miRNAs were up-regulated in THP-1 cells infected with H37Rv. Among them, the expression levels of six miRNAs had a significant up-regulation: miR-155, miR-29b-1*, miR-150, miR-146a, miR-212, and miR-483-5p in THP-1 cells with M. tuberculosis infection for 12 hr, 24 hr, and 48 hr. The up-regulation of these six miRNAs was time-dependent. Compared with the uninfected group, the up-regulation of miR-155 and miR-29b-1* was statistically significant (F=17.95 and 58.18, all P-values were <0.01) (Table 3).
Table 3.
Differential expression of six miRNAs in THP-1 cells with H37Rv infection at 3 time points
| Groups (n) |
P
ulmonary
tuberculosis (68) |
P
neumonia
(32) |
Lung cancer
(28) |
Unexplained pulmonary nodules
(12) |
Healthy donors
(50) |
|---|---|---|---|---|---|
| Age( X±S) | 46.8±28.3 | 56.9±29.4 | 61.5±20.6 | 41.8±17.6 | 52.4±14.7 |
| F/M (n/n) | 39/29 | 19/13 | 18/10 | 3/9 | 25/25 |
| Smoking (n) | 47 | 14 | 21 | 1 | 32 |
| BMI | 19.9±5.6 | 22.9±7.8 | 16.7±6.1 | 23.9±6.8 | 24.1±7.7 |
| HIV co-infection(n) | 1 | 0 | 0 | 0 | 0 |
Differential expression of miRNAs in THP-1 cells infected with clinical isolated M. tuberculosis strains
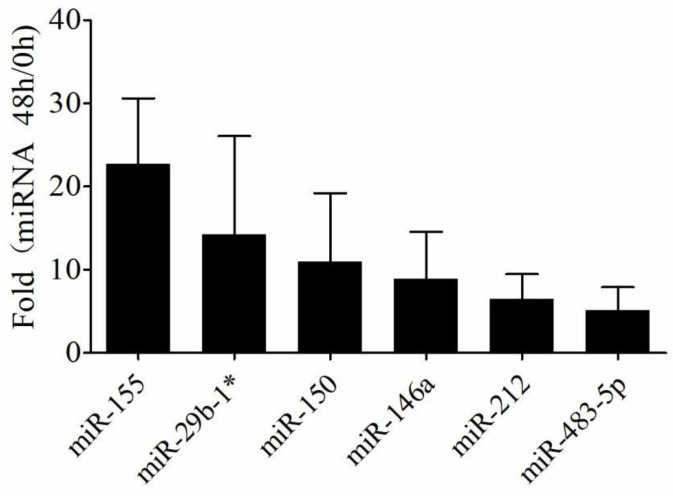
Expression levels of the above six miRNAs were analyzed by qRT-PCR in THP-1 cells infected with M. tuberculosis strains, isolated from 120 clinical sputum samples. The data showed that the expression levels of the six miRNAs (miR-155, miR-29b-1*, miR-150, miR-146a, miR-212, and miR-483-5p) were up-regulated, which was consistent with the above findings. In addition, the up-regulated multiple of miR-155 was higher than the other 5 miRNAs (all P-values were <0.01) (Figure 1). Thus, miR-155 was selected as the diagnostic target for further study.
Figure 1.
Differential expression of miRNAs in THP-1 cells infected with clinical isolated Mycobacterium tuberculosis strains
Expression levels of the above six miRNAs were analyzed by qRT-PCR in THP-1 cells infected with Mycobacterium tuberculosis strains, isolated from 120 clinical sputum samples, respectively. The data showed that the expression levels of the six miRNAs (miR-155, miR-29b-1*, miR-150, miR-146a, miR-212 and miR-483-5p) were up-regulated, which was consistent with the above finding. In addition, the up-regulated multiple of miR-155 was higher than other 5 miRNAs (all P-values were <0.01). Thus, miR-155 was selected as the diagnostic target for further study
The relative level of six kinds of miRNA in sputum from 190 donors
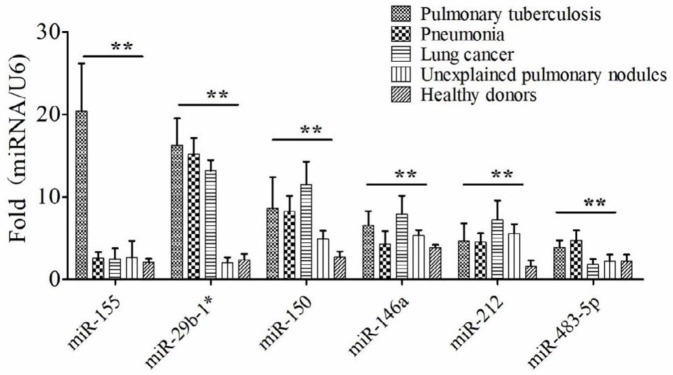
The total RNA was extracted from 190 sputa and the relative level of miRNAs was detected via qRT-PCR. The 190 donors were from five groups, 68 patients with PTB, 32 patients with pneumonia, 28 patients with lung cancer, 12 patients with unexplained pulmonary nodules, and 50 healthy donors (Table 1). The six kinds of miRNAs showed different detection performances among the five groups (Figure 2). miR-155 showed a better detection performance to distinguish PTB patients from non-PTB donors. Although there were statistical differences between PTB and non-PTB using the other five kinds of miRNAs, they had lower value than miRNA-155 for PTB diagnosis from sputum. So miRNA-155 was selected and used to be a detection tool in the next experiment.
Figure 2.
The relative level of miRNA-155 from six kinds of miRNAs be worthy for Pulmonary tuberculosis (PTB) diagnosis in clinical sputum
The relative level of six kinds of miRNAs from were detected from five groups’ sputum via qRT-PCR respectively, 68 patients with pulmonary tuberculosis (PTB), 32 patients with pneumonia, 28 patients with lung cancer, 12 patients with unexplained pulmonary nodules and 50 healthy donors. The miR-155 showed a better detection performance to distinguish PTB patients from non-PTB donors. Although there were statistical differences between PTB and non-PTB using other five kinds of miRNAs, them were less value than miRNA-155 for PTB diagnosis from sputum
Diagnostic value of sputum miR-155 for active PTB
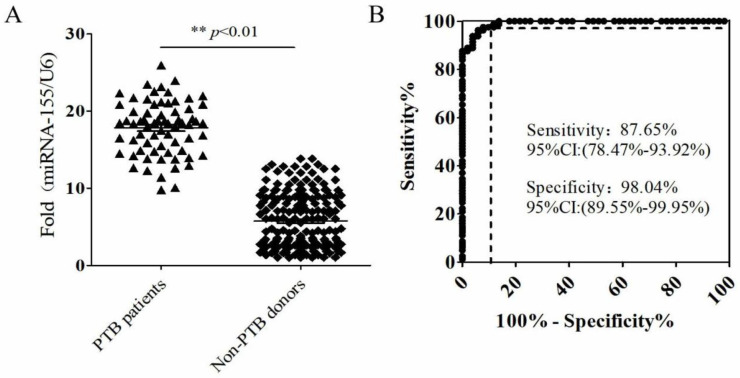
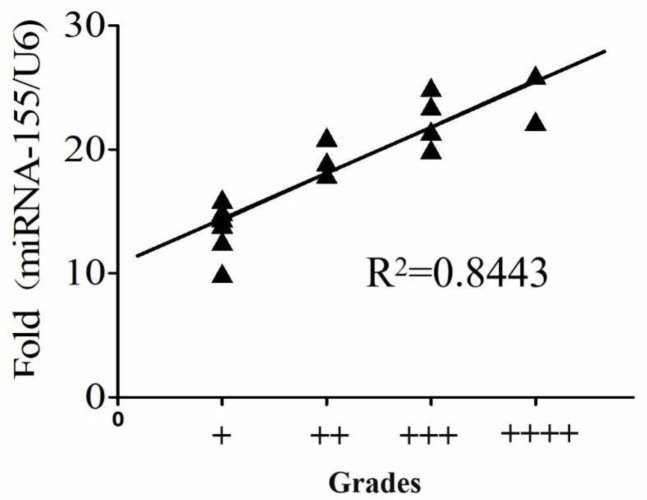
The qRT-PCR was used to analyze the expression levels of miR-155 in sputum specimens from 68 active PTB patients and 122 non-PTB patients (32 patients with pneumonia, 28 patients with lung cancer, 12 patients with unexplained pulmonary nodules, and 50 healthy donors) (Table 1 and Figure 3A). Then, we used the receiver operating characteristic curve (ROC curve) to evaluate the sensitivity and specificity of sputum miR-155 for active PTB diagnosis. Results showed that the sensitivity of sputum miR-155 detection was 94.1% (95% CI:86.18%-96.45%), the specificity was 87.7% (95% CI:76.13%-94.38%) (Figure 3B). The positive rate of sputum miR-155 detection was 94.1%, which was higher than sputum smear detection (22.1%, P<0.001) and anti-tuberculosis antibody detection (83.8%, P<0.05). But it was slightly lower than ELISpot (97%, P=0.404) (Table 4). Interestingly the ranking of sputum smear was positively correlated with the expression level of miR-155 in sputum (R2=0.8443, P<0.05) (Figure 4).
Figure 3.
MiR-155 showed a better diagnostic value for active PTB from sputum
The expression levels of miR-155 in sputum specimens from 68 active Pulmonary tuberculosis (PTB) patients and 122 non-PTB patients (32 patients with pneumonia, 28 patients with lung cancer, 12 patients with unexplained pulmonary nodules and 50 healthy donors) respectively. The receiver operating characteristic curve (ROC curve) we used to evaluate the sensitivity and specificity of sputum miR-155 for active PTB diagnosis. Results showed that the sensitivity of sputum miR-155 detection was 94.1% (95% CI:86.18%-96.45%), the specificity was 87.7% (95% CI:76.13%-94.38%)
Table 4.
Comparison of diagnostic values of different methods for active Pulmonary tuberculosis (PTB)
| M ethods | N umber of positive cases detected (n) | D etection rate (%) | 2, P-value |
|---|---|---|---|
| s putum miR-155 detection | 64 | 94.1 | — |
| s putum smears staining | 15 | 22.1 | 72.515,.000** |
| TB antibody test | 57 | 83.8 | 3.672, 0.045* |
| ELISpot | 66 | 97 | 0.697, 0.404 |
Figure 4.
The good correlation between the grade of Ziehl-Neelsen staining and the level of miR-155 in sputum smear-positive
The detection data of 15 patients with Pulmonary tuberculosis (PTB) were analyzed the correlation between Ziehl-Neelsen staining and miR-155. From this results, the expression level of miR-155 risen as the grade increasing. The four grades of smear detection had a good correlation with miRNA-155 (R2=0.8443)
Discussion
In some areas of China, TB epidemic has increased in recent years, due to the dual infection of TB and HIV, the spread of drug-resistant TB, and the expansion of the floating population, which has resulted in increased health care costs and other socioeconomic burdens (26, 27). Macrophages constitute the first line of immune defense against M. tuberculosis infection. Previous studies indicated that miRNA expression profiles of macrophages showed differences between M. tuberculosis infection groups and controls (28, 29). Consistent with the previous study, we found some differential expression miRNAs in macrophages with M. tuberculosis H37Rv strain infection. Among them, the expression levels of six miRNAs (miR-155, miR-29b-1*, miR-150, miR-146a, miR-212, and miR-483-5p) had a significant up-regulation, and the up-regulation of miR-155 and miR-29b-1* was statistically significant (F=17.95 and 58.18, all P-values were <0.01). Next, we used qRT-PCR to verify the differential expression of six miRNAs in THP-1 cells with clinical isolated M. tuberculosis strains. The data was consistent with the above finding.
In clinical practice, TB diagnosis usually depends on clinical symptoms, sputum culture, TST, and radiography. However, these routine clinical diagnostic methods have many disadvantages and make them difficult to meet clinical needs. Therefore, an easy, rapid, and accurate diagnostic method for tuberculosis diagnosis is urgently needed. Mounting studies have indicated that miRNA expression profiles in biological fluids can reflect altered physiological and/or pathological conditions (30, 31). Moreover, miRNAs are stably present in sputum (32, 33) and the differential expression of sputum miRNAs is closely associated with multiple human diseases. For instance, expression levels of miR-629-3p, miR-142-3p, and miR-223-3p are up-regulated in the sputum of patients with severe asthma and are related to neutrophilic airway inflammation, indicating that these sputum miRNAs contribute to this asthma inflammatory phenotype (34). miR-223 was increased in sputum samples of non-small cell lung cancer (NSCLC) patients compared to healthy controls. Sputum miR-223 detection showed 82% sensitivity and 95% specificity with AUC at 0.90 in the detection of NSCLC, suggesting that sputum miR-223 could be a useful biomarker for NSCLC diagnosis (35). Zheng et al. reported for the first time that miRNA expression profiles in sputum were dramatically different between PTB and healthy controls, which supports the potential application for improving the diagnosis of TB (36).
In this work, our data suggested that the expression levels of miR-155 were significantly up-regulated in THP-1 cells infected with M. tuberculosis H37Rv strain and clinical isolated M. tuberculosis strains, respectively. Previous studies also have shown that miR-155 was up-regulated in CD14+ or PBMCs cells from patients with active TB (37, 38). miR-155 as a crucial regulator of innate or adaptive immune responses plays an important role in host immune response against M. tuberculosis infection (39, 40). Considering the vital role of miR-155 in TB and prominent features of sputum miRNAs in the assessment of human diseases, we hypothesized that sputum miR-155 expression is altered in active PTB patients and could be the potential application for active PTB diagnosis. Consistent with the expected results, the expression of miR-155 was significantly increased in the sputum of patients with active PTB. ROC analysis showed that the sensitivity of sputum miR-155 detection was 94.1% and the specificity was 87.7%. Sputum miR-155 quantification obtained a good diagnostic performance in discriminating active PTB from healthy controls, which was higher than sputum smear detection (22.1%, P<0.001) and anti-TB antibody detection (83.8%, P<0.05). But it was slightly lower than ELISpot and the difference was not statistically significant (97%, P=0.404). In addition, interestingly the ranking of sputum smear was positively correlated with the expression level of miR-155 in sputum (R2=0.8443, P<0.05). There were some inadequacies in this study because of the limited number of donors, the 190 participants were distributed in the adult population and Han nationality. There was no verification for TB diagnosis at different ages and nationalities, especially ethnic minorities. Of course, we will expand the scope of research in the next experiment.
In conclusion, sputum miR-155 detection maybe a novel non-invasive biomarker for rapid and accurate diagnosis of 68 adult patients with active PTB.
Conclusion
Our research suggests that miR-155 is a potential biomarker for tuberculosis diagnosis in sputum. This miRNA detection method showed higher sensitivity and specificity of diagnostic potency than other traditional methods. Especially, the sputum sample was obtained easily and conveniently for most people, due to non-invasive sampling. Of course, this target will be detected using a rapid and visualized method in our next experiment.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (no. 81601806).
Conflicts of Interest
The authors declare that there are no potential conflicts of interest.
References
- 1.Forrellad MA, Klepp LI, Gioffre A, Sabio y Garca J, Morbidoni HR, de la Paz Santangelo M, et al. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4:3–66. doi: 10.4161/viru.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang Z, van der Merwe RG, Warren RM, Schubert WD, Gey van Pittius NC. Assessing the progress of Mycobacterium tuberculosis H37Rv structural genomics. Tuberculosis (Edinb) 2015;95:131–136. doi: 10.1016/j.tube.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. World Health Organization press release WHO sees surge in progress against tuberculosis on eve of global summit. Saudi Med J. 2004;25:1140–1141. [PubMed] [Google Scholar]
- 4.Wallis RS, Pai M, Menzies D, Doherty TM, Walzl G, Perkins MD, et al. Biomarkers and diagnostics for tuberculosis: progress, needs, and translation into practice. Lancet. 2010;375:1920–1937. doi: 10.1016/S0140-6736(10)60359-5. [DOI] [PubMed] [Google Scholar]
- 5.Magalhães J, Lima J, Araújo AA, Coutinho IO, Leal NC, Almeida A. Microscopic detection of Mycobacterium tuberculosis in direct or processed sputum smears. Rev Soc Bras Med Trop. 2018;51:237–239. doi: 10.1590/0037-8682-0238-2017. [DOI] [PubMed] [Google Scholar]
- 6.Lourens M, Philips L, Kleinhans CC, Friedrich SO, Martinson N, Venter A, et al. Liquid mycobacterial culture outcomes after different sputum collection techniques before and during treatment. Tuberculosis (Edinb) 2019;116:17–21,. doi: 10.1016/j.tube.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Zou X. MiR-652 serves as a prognostic biomarker in gastric cancer and promotes tumor proliferation, migration, and invasion via targeting RORA. Cancer Biomark. 2019;26:323–331. doi: 10.3233/CBM-190361. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Wang X, Jiang J, Cao Z, Yang B, Cheng X. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol Immunol. 2011;48:1084–1090. doi: 10.1016/j.molimm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Acunzo M, Croce CM. MicroRNA in Cancer and Cachexia--A Mini-Review. J Infect Dis. 2015;212 Suppl 1:S74–77. doi: 10.1093/infdis/jiv197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyu J, Zhao L, Wang F, Ji J, Cao Z, Xu H, et al. Discovery and validation of serum micrornas as early diagnostic biomarkers for prostate cancer in Chinese population. Biomed Res Int. 2019;2019:9306803. doi: 10.1155/2019/9306803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santamaria-Martos F, Benítez I, Ortega F, Zapater A, Giron C, Pinilla L, et al. Circulating microRNA profile as a potential biomarker for obstructive sleep apnea diagnosis. Sci Rep. 2019;9:13456. doi: 10.1038/s41598-019-49940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Záveský L, Jandáková E, Weinberger V, Minář L, Hanzíková V, Dušková D, et al. Ovarian cancer: Differentially expressed microRNAs in tumor tissue and cell-free ascitic fluid as potential novel biomarkers. Cancer Invest. 2019;18:1–13. doi: 10.1080/07357907.2019.1663208. [DOI] [PubMed] [Google Scholar]
- 14.Hofbauer SL, de Martino M, Lucca I, Haitel A, Susani M, Shariat SF, et al. A urinary microRNA (miR) signature for diagnosis of bladder cancer. Urol Oncol. 2018;36:531.e1–531. doi: 10.1016/j.urolonc.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Ndzi EN, Nkenfou CN, Mekue LM, Zentilin L, Tamgue O, Pefura E, et al. MicroRNA hsa-miR-29a-3p is a plasma biomarker for the differential diagnosis and monitoring of tuberculosis. Tuberculosis (Edinb) 2019;114:69–76. doi: 10.1016/j.tube.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Lv Y, Guo S, Li XG, Chi JY, Qu YQ, Zhong HL. Sputum and serum microRNA-144 levels in patients with tuberculosis before and after treatment. Int J Infect Dis. 2016;43:68–73. doi: 10.1016/j.ijid.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 17.Zheng ML, Zhou NK, Luo CH. MiRNA-155 and miRNA-132 as potential diagnostic biomarkers for pulmonary tuberculosis: A preliminary study. Microb Pathog. 2016;100:78–83. doi: 10.1016/j.micpath.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Lacedonia D, Palladino GP, Foschino-Barbaro MP, Scioscia G, Carpagnano GE. Expression profiling of miRNA-145 and miRNA-338 in serum and sputum of patients with COPD, asthma, and asthma-COPD overlap syndrome phenotype. Int J Chron Obstruct Pulmon Dis. 2017;12:1811–1817. doi: 10.2147/COPD.S130616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Wang Q, Zhang S. MicroRNAs in sputum specimen as noninvasive biomarkers for the diagnosis of nonsmall cell lung cancer: An updated meta-analysis. Medicine (Baltimore) 2019;98:e14337. doi: 10.1097/MD.0000000000014337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Y, Guarnera MA, Fang H, Jiang F. Small non-coding RNA biomarkers in sputum for lung cancer diagnosis. Mol Cancer. 2016;15:36. doi: 10.1186/s12943-016-0520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organization. International Union Against Tuberculosis and Lung Disease Royal Netherlands Tuberculosis Association. Revised international definitions in tuberculosis control. Int J Tuberc Lung Dis. 2001;5:213–215. [PubMed] [Google Scholar]
- 22.Sloane AJ, Lindner RA, Prasad SS, Sebastian LT, Pedersen SK, Robinson M, et al. Proteomic analysis of sputum from adults and children with cystic fibrosis and from control subjects. Am J Respir Crit Care Med. 2005;172:1416–1426. doi: 10.1164/rccm.200409-1215OC. [DOI] [PubMed] [Google Scholar]
- 23.Sagel SD, Kapsner R, Osberg I, Sontag MK, Accurso FJ. Airway inflammation in children with cystic fibrosis and healthy children assessed by sputum induction. Am J Respir Crit Care Med. 2001;164:1425–1431. doi: 10.1164/ajrccm.164.8.2104075. [DOI] [PubMed] [Google Scholar]
- 24.Tang XL, Yuan CH, Ding Q, Zhou Y, Pan Q, Zhang XL. Selection and identification of specific glycoproteins and glycan biomarkers of macrophages involved in Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 2017;104:95–106. doi: 10.1016/j.tube.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Dye C, Williams BG. The population dynamics and control of tuberculosis. Science. 2010;328:856–861. doi: 10.1126/science.1185449. [DOI] [PubMed] [Google Scholar]
- 27.Shi QN, Ma JQ. [Big data analysis of flow of tuberculosis cases in China, 2014] Zhonghua Liu Xing Bing Xue Za Zhi. 2016;37:668–672. doi: 10.3760/cma.j.issn.0254-6450.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 28.Sharbati J, Lewin A, Kutz-Lohroff B, Kamal E, Einspanier R, Sharbati S. Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS One. 2011;6:e20258. doi: 10.1371/journal.pone.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furci L, Schena E, Miotto P, Cirillo DM. Alteration of human macrophages microRNA expression profile upon infection with Mycobacterium tuberculosis. Int J Mycobacteriol. 2013;2:128–134. doi: 10.1016/j.ijmyco.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Backes C, Meese E, Keller A. Specific miRNA disease biomarkers in blood, serum and plasma: Challenges and prospects. Mol Diagn Ther. 2016;20:509–518. doi: 10.1007/s40291-016-0221-4. [DOI] [PubMed] [Google Scholar]
- 31.Dieter C, Assmann TS, Costa AR, Canani LH, de Souza BM, Bauer AC, et al. MiR-30e-5p and MiR-15a-5p expressions in plasma and urine of type 1 diabetic patients with diabetic kidney disease. Front Genet. 2019;10:563. doi: 10.3389/fgene.2019.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu Z, et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie Y, Todd NW, Liu Z, Zhan M, Fang H, Peng H, et al. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67:170–176. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maes T, Cobos FA, Schleich F, Sorbello V, Henket M, De Preter K, et al. Asthma inflammatory phenotypes show differential microRNA expression in sputum. J Allergy Clin Immunol. 2016;137:1433–1446. doi: 10.1016/j.jaci.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 35.Bagheri A, Khorram Khorshid HR, Mowla SJ, Mohebbi HA, Mohammadian A, Yaseri M, et al. Altered miR-223 expression in sputum for diagnosis of non-small cell lung cancer. Avicenna J Med Biotechnol. 2017;9:189–195. [PMC free article] [PubMed] [Google Scholar]
- 36.Zhengjun Yi, Yurong Fu, Rui Ji, Ruifang Li, Zhiyu Guan. Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PLoS One. 2012;7:e43184. doi: 10.1371/journal.pone.0043184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, Lu C, Diao N, Zhang S, Wang S, Wang F, et al. Analysis of microRNA expression profiling identifies miR-155 and miR-155* as potential diagnostic markers for active tuberculosis: a preliminary study. Hum Immunol. 2012;73:31–37. doi: 10.1016/j.humimm.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Huang J, Jiao J, Xu W, Zhao H, Zhang C, Shi Y, et al. MiR-155 is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO3. Mol Med Rep. 2015;12:7102–7108. doi: 10.3892/mmr.2015.4250. [DOI] [PubMed] [Google Scholar]
- 39.Dudda JC, Salaun B, Ji Y, Palmer DC, Monnot GC, Merck E, et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 2013;38:742–753. doi: 10.1016/j.immuni.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huffaker TB, Hu R, Runtsch MC, Bake E, Chen X, Zhao J, et al. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep. 2012;2:1697–1709. doi: 10.1016/j.celrep.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]