Abstract
Objective(s):
Pseudomonas aeruginosa and Acinetobacter baumannii resist antibiotics by different intrinsic and acquired mechanisms. This study aims to define various carbapenem-resistant mechanisms of isolated P. aeruginosa and A. baumannii from nine different provinces of Iran.
Materials and Methods:
In this cross-sectional study, all carbapenem-resistant P. aeruginosa and A. baumannii samples from nine provinces of Iran on a one-year time horizon were gathered. Modified Hedge Test (MHT) and Carba NP-Test were applied to the identification of producing-carbapenemase strains. The most important carbapenemase genes recognized by PCR and gene overexpression of the efflux pump were surveyed by efflux pump inhibitors (EPIs) and confirmed by Real-Time PCR.
Results:
Twenty-one percent and 43.5% of P. aeruginosa and A. baumannii isolates were resistant to carbapenem, respectively. MHT and Carba-NP tests identified 21% and 11% carbapenemase-producing strains in these Gram-negative bacteria, respectively. NDM-1 was the most prevalently detected carbapenemase in P. aeruginosa; OXA-51 and OXA-23 were the most significant genes in A. baumannii. EPIs identified active efflux pumps in 20% and 28% of P. aeruginosa and A. baumannii, respectively. Real-time PCR confirmed gene overexpression of efflux pumps in 54% and 30% of positive EPIs in P. aeruginosa and A. baumannii, respectively.
Conclusion:
P. aeruginosa and A. baumannii may become multi-drug-resistant (MDR) and Extensively Drug-Resistant (XDR) strains and cause a high rate of mortality and morbidity. Thus, it is of necessity to prohibit the spread of antibiotic-resistant strains in hospitals.
Key Words: Acinetobacter baumannii, Carbapenems, Drug resistance, Iran, Pseudomonas aeruginosa
Introduction
Carbapenems are broad-spectrum beta-lactam antibiotic agents. They are usually considered the last choice for antibiotic therapy, especially in combatting Extended-Spectrum Beta-Lactamase (ESBL) producing microorganisms (1). Although some alternative antibiotics such as tigecycline and colistin can be used in case of carbapenem resistance, these are characterized by low effectiveness and/or high toxicity (1). The rate of carbapenem resistance to Gram-negative bacteria, especially in nosocomial pathogens such as P. aeruginosa and Acinetobacter baumannii, is high and increasing steadily (1-3). Nosocomial isolated P. aeruginosa and A. baumannii may be resistant to most of the available antibiotics and act as Multi-Drug Resistant (MDR) and Extensive Drug-Resistant (XDR) strains (3-5). The presence of carbapenem-resistant bacteria can be quite considerable because they enjoy the chance to shift to MDR strains commonly (1). P. aeruginosa and A. baumannii can become resistant to carbapenem through various mechanisms (2, 3, 6). The most important mechanism is the potential to produce carbapenemase because most of the carbapenemase genes can be found on the transferable genetic elements and they spread rapidly among bacteria (3, 5, 7). Different classes of carbapenemase can be detected in Gram-negative bacteria including Ambler classes A, B, and D β-lactamases (8). One of the inherent resistant mechanisms of carbapenems is the presence of active efflux pumps. It is important because it can cause cross-resistance to other antibiotic families (2, 9). Resistance to most of the available antibiotics in P. aeruginosa and A. baumannii can become a complex challenge for physicians due to the limited number of choices left for antibiotic therapy. This study is a multicenter research that aims to evaluate different mechanisms of carbapenem-resistant P. aeruginosa (CRPA) and A. baumannii (CRAB) through phenotypic and molecular techniques.
Materials and Methods
Setting and bacterial isolates
In this cross-sectional study, P. aeruginosa and A. baumannii strains were collected from nine provinces of Iran from September 2016 up to September 2017.
Antibiotic susceptibility testing
Carbapenem susceptibility was evaluated according to CLSI guidelines (10). P. aeruginosa ATCC 27853 was adopted as the control strain. All Carbapenems-resistant strains were included in the study.
Phenotypic screening of carbapenemase-producing strains
Phenotypic screening of carbapenemase-producing strains was carried out by the Modified Hodge Test (MHT) (10) and CarbaAcineto NP test according to CLSI guidelines (2016)(11).
Modified Hodge Test (MHT)
MHT was accomplished to identify carbapenemase-producing A. baumannii by using E. coli ATCC 25922 and ertapenem disc (10 µg). Strains with cloverleaf images of inhibition zone were considered as carbapenemase-producing strains according to the CLSI guidelines (2016)(10).
Carba NP test and CarbaAcineto NP test
THE CarbaAcineto NP test method has been described previously (11). In brief, one loop of a suspected strain was suspended in Tris-HCL mmol/l (5 M NaCl in CarbaAcineto NP Test) as a lysis buffer from antibiogram plates, vortexed for one min, and then incubated at room temperature for 30 min. The bacterial suspension was centrifuged at 10,000 xg at room temperature for 5 min. Next, 30 µl of the supernatant was mixed in 96 wells with 100 µl of imipenem monohydrate solution (3 mg per ml) pH 7.8, phenol red solution, and 0.1 mmol/l ZnSO4 (11).
Molecular detection of carbapenemase genes
The most prevalent carbapenemase genes were detected by conventional PCR. These genes included VIM, IMP, NDM-1, SPM-1, KPC, GES, and OXA-48 in P. aeruginosa and A. baumannii and OXA-23, OXA-40, OXA-24, OXA-58, and OXA-51 only in A. baumannii. Table 1 lists primers and Table 2 shows the previously described PCR conditions (12-18).
Table 1.
Primers used in this study for detection of resistance genes among P. aeruginosa and A. baumannii isolates
| Gene | Primer sequencing 5’ 3’ | PCR product size (bp) | Tm (oC) | Reference |
|---|---|---|---|---|
| kpc-F | CTGTCTTGTCTCTCATGGCC | 636 | 57.98 | [12] |
| kpc-R | CCTCGCTGTGCTTGTCATCC | 61.36 | ||
| ges -F | GTTTTGCAATGTGCTCAACG | 371 | 57.09 | [13] |
| ges-R | TGCCATAGCAATAGGCGTAG | 57.54 | ||
| vim -F | GATGGTGTTTGGTCGCATA | 390 | 55.61 | [14] |
| vim -R | CGAATGCGCAGCACCAG | 59.54 | ||
| imp -F | TTGACACTCCATTTACDG a | 139 | 48.56 | [14] |
| imp-R | GATYGAGAATTAAGCCACYCT a | 51.92 | ||
| NDM-1 -F | CCCGGCCACACCAGTGACA | 129 | 64.73 | [14] |
| NDM-1-R | GTAGTGCTCAGTGTCGGCAT | 60.11 | ||
| SPM-1- F | GGGTGGCTAAGACTATGAAGCC | 447 | 60.49 | [14] |
| SPM-1-R | GCCGCCGAGCTGAATCGG | 63.90 | ||
| oxa-48-F | CCAAGCATTTTTACCCGCATCKACC | 389 | 63.21 | [15] |
| oxa-48-R | GYTTGACCATACGCTGRCTGCG | 62.30 | ||
| oxa-23- F | GATGTGTCATAGTATTCGTCGT | 1058 | 55.86 | [6] |
| oxa-23- R | TCACAACAACTAAAAGCACTGT | 56.69 | ||
| oxa-40- F | GGAATTCCATGAAAAAATTTATACTTCC | 846 | 56.44 | [17] |
| oxa-40 - R | CGGGATCCCGTTAAATGATTCCAAGATTTTCTAGCG | 68.57 | ||
| oxa-24- F | GGTTAGTTGGCCCCCTTAAA | 246 | 57.39 | [18] |
| oxa-24- R | AGTTGAGCGAAAAGGGGATT | 57.41 | ||
| oxa-58- F | AAGTATTGGGGCTTGTGCTG | 598 | 58.45 | [18] |
| oxa-58- R | CCCCTCTGCGCTCTACATAC | 59.68 | ||
| oxa-51- F | TAATGCTTTGATCGGCCTTG | 353 | 56.48 | [18] |
| oxa-51- R | TGGATTGCACTTCATCTTGG | 56.01 | ||
| b AdeB - F | AACGGACGACCATCTTTGAGTATT | 84 | 60.32 | [36] |
| b AdeB - R | CAGTTGTTCCATTTCACGCATT | 58.36 | ||
| b16srRNA-F | CAGCTCGTGTCGTGAGATGT | 151 | 60.11 | [37] |
| b 16srRNA-R | CGTAAGGGCCATGATGACTT | 57.67 | ||
| b MexX-F | TGAAGGCGGCCCTGGACATCAGC | 326 | 69.22 | [2] |
| b MexX-R | GATCTGCTCGACGCGGGTCAGCG | 69.78 | ||
| b MexA-F | CGACCAGGCCGTGAGCAAGCAGC | 316 | 70. 52 | [2] |
| b MexA-R | GGAGACCTTCGCCGCGTTGTCGC | 70.42 | ||
| b MexC-F | GTACCGGCGTCATGCAGGGTTC | 164 | 65.93 | [2] |
| b MexC-R | TTACTGTTGCGGCGCAGGTGACT | 67.14 |
Table 2.
PCR conditions used in this study for detection of carbapenem-resistant genes
| Number of cycles | Time | Temperature ( o C) | Cycle |
|---|---|---|---|
| 1 | 1-10a min | 94 | Initial denaturation |
| 30-40 | 30- 45a sec | 94 | Denaturation Annealing Extension |
| 30- 40a sec | 54- 63a | ||
| 30 sec-1a min | 72 | ||
| 1 | 1-7a min | 72 | Final extension |
a based on each gene
Phenotypic screening of active efflux pumps
Treatment of the efflux pump by inhibitor
Phenotypic discovery of active efflux pumps was facilitated by detecting Minimum Inhibitory Concentration (MIC) of imipenem ranging between 2-256 µg/ml with and without Cyanide 3-Chlorophenylhydrazone (CCCP) as an EPI. The final concentration of CCCP (C2759 Sigma-Aldrich, France) was 25 µg/ml, simultaneously (19). The positive condition for the presence of active efflux pumps in the isolates was, at least, the 4-fold reduction of MIC in the presence of CCCP. A. baumannii ATCC 19606 was used as the control strain.
Relative gene expression by real-time PCR
RNA extraction was carried out by the Thermo RNA extraction kit (cat. No. K0732) according to the manual’s instructions.
We used an RNeasy Mini Kit with 1 hr on-column DNase digestion (Qiagen NV, Venlo, The Netherlands) for purification of total RNA. Total RNA was quantified using a spectrophotometer (WPA Biowave II Nanospectrophotometer, USA) and ratio of absorbance at 260 nm vs 280 nm was used to assess RNA purity. Moreover, extracted RNA was screened on a 3% agarose gel.
At the next step, cDNA synthesis was executed by the Thermo kit (cat. No. K1622). Finally, the gene overexpression of MexX, MexC, and MexA in P. aeruginosa and of adeB in A. baumannii from RND-type efflux systems, involved in carbapenem resistance, was prepared. 16srRNA was used as a house-keeping gene and P. aeruginosa ATCC 27853 and A. baumannii ATCC 19606 were considered as reference strains. The primers are shown in Table 1. Gene overexpression was calculated by the 2-ΔΔct formula (20).
a Y=T or C; D=A or G or T
b The relative gene expression was calculated for this gene by Real-Time PCR. Corbett Rotor-Gene 6000
Statistical analysis
SPSS 22.0 statistical software (IBM Corp., Armonk, USA) was utilized to conduct data analysis. Mean, Confidence Interval (CI), etc. were analyzed by the Explore test in SPSS version 22.0 software. Sensitivity and specificity of phenotypic methods were calculated through the following formula (21):
Sensitivity= (a/(a+c))×100
Specificity= (d/(b+d))×100
Positive predictive value (PPV)= (a/(a+b))×100
Negative predictive value (NPV)=(d/(c+d))×100
Results
In this cross-sectional study, 675 P. aeruginosa and 869 A. baumannii remained definite throughout the study, and 140 (20.7%) and 383 (44%) of them, respectively, were resistant to carbapenem. The results of MHT and Carba NP tests used to identify carbapenemase-producing strains are shown in Table 3.
Table 3.
Results of MHT of carbapenem-resistant strains
| MHT & Carba NP negative (%) | Carba NP positive (%) | MHT positive (%) | Bacteria |
|---|---|---|---|
| 102 (73) | 17 (12) | 21 (15) | P. aeruginosa |
| 258 (67) | 38 (10) | 87 (23) | A. baumannii |
MHT: Modified Hodge Test
According to the results from molecular detection of carbapenemase by PCR, NDM-1 was the most prevalent enzyme in CRPA and OXA-51 and OXA-23 were the most prevalent genes in CRAB. SPM-1, KPC, GES, and OXA-58 were not observed in any of the strains (Figures 1 and 2) (Tables 4 and 5).
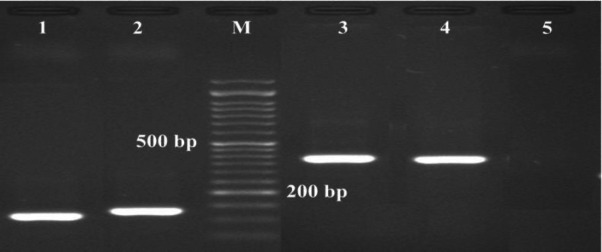
Figure 1.
PCR based identification of the NDM-1, imp, vim, and oxa-48 genes, using species primer pairs in P. aeruginosa and A. baumannii isolates
M: marker 50bp (SMO321, Fermantas). 1: positive NDM-1: 129bp, 2: positive imp: 139bp, 3: positive vim:390bp, 4: positive oxa-48:389 bp, and 5: negative control
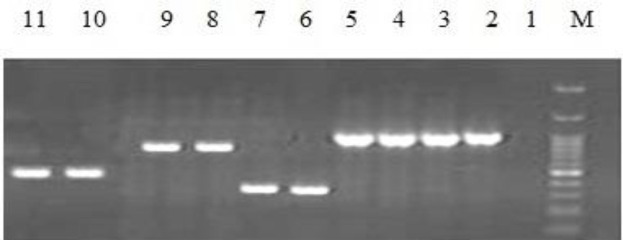
Figure 2.
PCR based identification of the oxa-23, oxa-24, oxa-51 and oxa-40 genes, using species primer pairs in P. aeruginosa and A. baumannii isolates
M: marker 100bp (SMO321, Fermantas). 1: negative control. 2- 5: positive oxa-23: 1058 bp. 6, 7: positive oxa-24: 246 bp. 8,9: positive oxa-40: 846 bp. 10, 11: positive oxa-51: 353 bp
Table 4.
Number (%) of detected carbapenemase genes in carbapenem-resistant Pseudomonas aeruginosa
| OXA_48 | GES | KPC | SPM-1 | NDM-1 | IMP | VIM | |
|---|---|---|---|---|---|---|---|
| 6 (4) | _ | _ | _ | 10 (7) | 8 (6) | _ | P. aeruginosa |
Table 5.
Number (%) of detected carbapenemase genes in carbapenem-resistant Acinetobacter baumannii
| OXA-58 | OXA-24 | OXA-40 | OXA-51 | OXA-23 | OXA_48 | GES | KPC | SPM-1 | NDM-1 | IMP | VIM | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| _ | 252 (65.8%) | 85 (22.3%) | 383 (91.6%) | 290 (76.5%) | 59 (15.4%) | _ | _ | _ | 36 (9.4%) | 1 (0.3%) | 7 (2%) | A. baumannii |
Sensitivity and specificity of MH and Carba NP tests concerning two non-fermentative Gram-negative bacteria are shown in Table 6.
Table 6.
Evaluation of MH and Carba NP test in detecting carbapenemase
| Carba NP test (%) |
MHT (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | NPV** | PPV* | Specificity | Sensitivity | Accuracy | NPV** | PPV* | Specificity | Sensitivity | Bacteria |
| 9 | 92 | 23 | 92 | 23 | 76 | 89 | 5 | 83 | 7 | P. aeruginosa |
| 16 | 3 | 97 | 89 | 14 | 21 | 3 | 98 | 89 | 20 | A. baumannii |
* Positive predictive value ; ** Negative predictive value; MH: Modified Hedge Test
According to phenotypic evaluations, it was found that 28 (20%) CRPA and 108 (28%) CRAB had active efflux pumps by adding CCCP. In the process of the real-time PCR assay, 15 (54%) P. aeruginosa with positive IEPs showed overexpression of MexX, MexC, and MexA (Figure 3). The mean gene expression of MexX was 8.34 with CI 95%: 1 to 17.15. The mean gene expression of MexA was 67.91 with CI 95%: 16.33 to 119.50. The mean gene expression of MexC was 2.73 with CI 95%: 1 to 5.7. As a common efflux pump, AdeB gene overexpression was detected in 32 (30%) positive EPI tests of A. baumannii with a mean gene expression of AdeB leveled at 9.81, CI 95%: 3.37 to 16.25.
Figure 3.
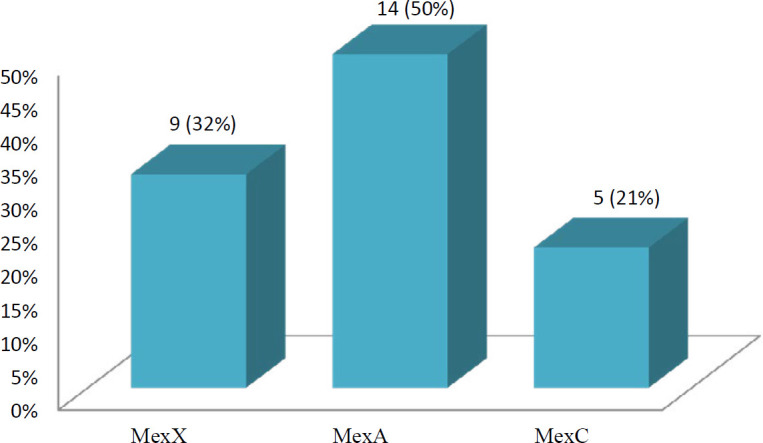
Results of gene expression of each efflux pump in Efflux Pump Inhibitors (EPIs) positive Pseudomonas aeruginosa
Discussion
In recent decades, CRPA and CRAB have been two of the most critical nosocomial pathogens threatening public health and the World Health Organization has included them in a global priority pathogens list of antibiotic-resistant bacteria (2, 4, 5, 7). Rapid horizontal spread of plasmid-borne carbapenemase in these bacteria can be one of the reasons that there is large-scale spread of carbapenem-resistant bacteria. According to the results of the conducted antibiotic susceptibility testing, 21% of the collected P. aeruginosa isolates were resistant to carbapenem. A research group from Brazil worked on P. aeruginosa isolated from blood (22). Their results confirmed that 44% of the mentioned isolates were resistant to carbapenems (22). The frequency of CRPA in the Brazilian study is higher than that in our results; therefore, the sources of collected bacteria may justify this difference in frequency. Ghasemian et al. (2019) published a review article about the frequency of CRPA and analyzed 36 studies from Iran (23). They reported the detection of CRPA in 55% of the studied isolates (22). We had access to materials and methods (the same as those of other studies) at 10 lab centers and different sorts of clinical specimens. Thus, the discrepancy between our proposed results and those in other studies may correspond to different specimens, materials, and methods (22, 23).
Production of carbapenemase is one of the significantly responsible mechanisms. NDM-1 is the most frequent carbapenemase in CRPA, as confirmed by the results of PCR and sequencing in the current study. NDM-1 is found on the plasmid and can carry other antibiotic-resistant genes (24). Therefore, the presence and identification of NDM-1 positive strains are quite important and the top priority for control by the nosocomial infection committee of each hospital. The results of a published study revealed that NDM-1-producing P. aeruginosa was not detected in or reported from Iran (25). Hence, the detection of NDM-1-producing P. aeruginosa in the current study is a very alarming sign for the health care system and it needs a significant approach.
Some phenotypic tests have been proposed so far to detect carbapenemase-producing organisms. MHT and Carba NP tests are two challenging methods. In the current study, the sensitivity and specificity of MHT and Carba NP tests to detecting carbapenemase in both of the bacteria under study are low and reasonable, respectively. In addition, other studies reported acceptable specificity but low sensitivity for MHT (26-28).
Efflux pumps are the other important carbapenem-resistant mechanisms that can cause the appearance of MDR and XDR strains because they can reject a different family of antibiotics, simultaneously (9). In the current study, 20% of CRPA showed active efflux pumps by the EPIs method. The results obtained in other studies showed the role of active efflux pumps in 18% of CRPA by EPIs (2), similarly to our findings. The results of Real-Time PCR confirmed 54% gene overexpression of the Mex family of efflux pumps in EPIs positive CRPA in this study. However, Azimi et al. reported gene overexpression in the Mex family of efflux pumps in 100% CRPA with the CCCP positive test (29). They used different methods for detecting the MIC method and EPI, which may explain the dissimilar results.
In the current study, 44% of the collected A. baumannii were resistant to at least one member of the carbapenem class. El Kettani A et al. (2017) showed that 76% of A. baumannii isolated from blood cultures were resistant to carbapenem (30). In 2018, researchers reported that 80% of A. baumannii isolated from wound burn specimens were imipenem-resistant (31). In the above two studies, strains were isolated from blood culture (30) and wounds burn (31); however, the current study evaluated the A. baumannii isolated from different clinical samples. We believe that the source of collected specimens and the use of different antibiotic discs in the brand (from different companies) can justify the divergence of our results from those in other studies. The concentration of more than one carbapenemase was observed in 1% and 2% of P. aeruginosa and A. baumannii, respectively. In addition, gene overexpression of the efflux pump was combined with carbapenemase in 1% of P. aeruginosa and all A. baumannii.
According to reports of other researchers, NDM-1-producing bacterium is one of the threatening isolates, while we found that 9% of CRAB pathogens were NDM-1 positive. Unfortunately, these results should be disturbing for Iran’s health system. Obtained results show that OXA-51 and OXA-23 are the most prevalent carbapenemase in isolated A. baumannii, as confirmed by other studies (5, 17, 32). Another responsible resistance mechanism is the efflux pump. According to real-time PCR results from evaluating AdeB gene expression, 29% of CRAB pathogens use the efflux pump mechanism. In previously published studies, several researcher groups worked on CRAB’s efflux pump mechanism and reported similar results to the findings of the current study (33-35).
Conclusion
The existence of different antibiotic-resistant mechanisms of P. aeruginosa and A. baumannii can cause cross antibiotic resistance, lead to the appearance of MDR and/or strains, and make the treatment difficult. The increasing number of NDM-1-producing bacteria is a very serious problem to combat in terms of antibiotic resistance. Therefore, finding a way to inhibit efflux pumps is quite essential for controlling the cross-resistance and appearance of MDR strains of bacteria.
Acknowledgment
The research reported in this publication was supported by the Elite Researcher Grant Committee (award no. (940290)) from the National Institutes for Medical Research Development (NIMAD), Tehran, Iran. The authors are grateful to the entire staff of the Pediatric Infectious Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Conflicts of Interest
This manuscript does not have any conflicts of interest.
References
- 1.Amini A, Ebrahimzadeh Namvar A. Antimicrobial resistance pattern and presence of beta-lactamase genes in Pseudomonas aeruginosa strains isolated from hospitalized patients, Babol-Iran. J Med Bacteriol. 2019;8:45–50. [Google Scholar]
- 2.Bijari A, Azimi L, Fallah F, Ardebili A, Lari ER, Lari AR. Involvement of the multidrug efflux pumps in betalactams resistant Pseudomonas aerugionsa clinical isolates collected from burn patients in Iran. Infect Disord Drug Targets. 2016;16:172–177. doi: 10.2174/1871526516666160829154302. [DOI] [PubMed] [Google Scholar]
- 3.Lari AR, Azimi L, Soroush S, Taherikalani M. Low prevalence of metallo-beta-lactamase in Pseudomonas aeruginosa isolated from a tertiary burn care center in Tehran. Int J Immunopathol Pharmacol. 2015;28:384–389. doi: 10.1177/0394632015578343. [DOI] [PubMed] [Google Scholar]
- 4.Armin S, Fallah F, Azimi L, Samadi Kafil H, Ghazvini K, Hasanzadeh S, et al. Warning: spread of NDM-1 in two border towns of Iran. Cell Mol Biol. 2018;64:125–129. [PubMed] [Google Scholar]
- 5.Azimi L, Talebi M, Pourshafie MR, Owlia P, Rastegar Lari A. Characterization of carbapenemases in extensively drug resistance Acinetobacter baumannii in a burn care center in Iran. Int J Mol Cell Med. 2015;4:46–53. [PMC free article] [PubMed] [Google Scholar]
- 6.Mobasseri P, Azimi L, Salehi M, Hosseini F, Fallah F. Multi-drug resistance profiles and expression of adeijk and abem in Acinetobacter baumannii collected from humans by Real-time PCR. J Med Bacteriol. 2018;7:50–56. [Google Scholar]
- 7.Owlia P, Azimi L, Gholami A, Asghari B, Lari AR. ESBL- and MBL-mediated resistance in Acinetobacter baumannii: a global threat to burn patients. Infez Med. 2012;20:182–187. [PubMed] [Google Scholar]
- 8.Karbasizade V, Heidari L, Jafari R. Detection of oxa-type carbapenemase genes in Acinetobacter baumannii isolates from nosocomial infections in Isfahan hospitals, Iran. J Med Bacteriol. 2015;4:31–36. [Google Scholar]
- 9.Nikaido H, Pagès JM. Broad-specificity efflux pumps and their role in multidrug resistance of gram-negative bacteria. FEMS Microbiol Rev. 2012;36:340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical laboratory standards institute. Performance standards for antimicrobial susceptibility testing: nineteenth informational supplement M100–S21. CLSI, Wayne, PA; 2016. [Google Scholar]
- 11.Dortet L, Poirel L, Errera C, Nordmann P. CarbAcineto NP test for rapid detection of carbapenemase-producing Acinetobacter spp. J Clin Microbiol. 2014;52:2359–2364. doi: 10.1128/JCM.00594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azimi L, Rastegar Lari A, Talebi M, Ebrahimzadeh Namvar AM, Soleymanzadeh Moghadam S. Evaluation of phenotypic methods for detection of Klebsiella Pneumoniae carbapenemase-producing K Pneumoniae in Tehran. J Med Bacteriol. 2013;2:26–31. [Google Scholar]
- 13.Gheorghe I, Czobor I, Chifiriuc MC, Borcan E, Ghiţă C, Banu O, et al. Molecular screening of carbapenemase-producing gram-negative strains in Romanian intensive careunits during a one year survey. J Med Microbiol. 2014;63:1303–1310. doi: 10.1099/jmm.0.074039-0. [DOI] [PubMed] [Google Scholar]
- 14.Lowings M, Ehlers MM, Dreyer AW, Kock MM. High prevalence of oxacillinases in clinical multidrug-resistant Acinetobacter baumannii isolates from the Tshwane region, South Africa – an update. BMC Infect Dis. 2015;14:2–10. doi: 10.1186/s12879-015-1246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamazaki Y, Funaki T, Yasuhara T, Sugano E, Ugajin K, Tahara S, et al. Molecular characteristics of a carbapenemase-producing Enterobacter species and Klebsiella species outbreak in a Japanese university hospital. Showa Univ J Med Sci. 2017;29:163–172. [Google Scholar]
- 16.Zhou H, Pi BR, Yang Q, Yu YS, Chen YG, Li LJ, et al. Dissemination of imipenem-resistant Acinetobacter baumannii strains carrying the ISAba1 blaOXA-23 genes in a Chinese hospital. J Med Microbiol. 2007;56:1076–1080. doi: 10.1099/jmm.0.47206-0. [DOI] [PubMed] [Google Scholar]
- 17.Héritier C, Poirel L, Lambert T, Nordmann P. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2005;49:3198–3202. doi: 10.1128/AAC.49.8.3198-3202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joshi PR, Acharya M, Kakshapati T, Leungtongkam U, Thummeepak R, Sitthisak S. Co-existence of blaOXA-23 and blaNDM-1 genes of Acinetobacter baumannii isolated from Nepal: antimicrobial resistance and clinical significance. Antimicrob Resist Infect Control. 2017;7:6–21. doi: 10.1186/s13756-017-0180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ardebili A, Talebi M, Azimi L, Rastegar Lari A. Effect of efflux pump inhibitor carbonyl cyanide 3-chlorophenylhydrazone on the minimum inhibitory concentration of ciprofloxacin in Acinetobacter baumannii clinical isolates. Jundishapur J Microbiol . 2014;7:e8691. doi: 10.5812/jjm.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azimi L, Namvar AE, Jamali S, Lari AR, Bijari A, Lari AR. Relative expression of efflux pumps in multi drug resistant Pseudomonas aeruginosa. Roum Arch Microbiol Immunol. 2015;74:86–90. [PubMed] [Google Scholar]
- 21.Trevethan R. Sensitivity, specificity, and predictive values: foundations, pliabilities, and pitfalls in research and practice. Front Public Health. 2017;5:1–7. doi: 10.3389/fpubh.2017.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadjadj L, Shoja S, Diene SM, Rolain JM. Dual infections of two carbapenemase-producing Acinetobacter baumannii clinical strains isolated from the same blood culture sample of a patient in Iran. Antimicrob Resist Infect Control. 2018;7:39–42. doi: 10.1186/s13756-018-0329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe M, Ehlers MM, Ismail F, Peirano G, Becker PJ, Pitout JDD, et al. Acinetobacter baumannii: epidemiological and beta-lactamase data from two tertiary academic hospitals in Tshwane, South Africa. Front Microbiol. 2018;9:1280–1289. doi: 10.3389/fmicb.2018.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramaniyan JS, Sundaram JM. Occurrence of bla genes encoding carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii from intensive care unit in a tertiary care hospital. J Lab Physicians. 2018;10:208–213. doi: 10.4103/JLP.JLP_108_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghasemian A, Salimian Rizi K, Rajabi Vardanjani H, Nojoomi F. Prevalence of clinically isolated metallo-beta-lactamase-producing Pseudomonas aeruginosa, coding genes, and possible risk factors in Iran. Iran J Pathol. 2018;13:1–9. [PMC free article] [PubMed] [Google Scholar]
- 26.Kuchibiro T, Komatsu M, Yamasaki K, Nakamura T, Nishio H, Nishi I, et al. Evaluation of the modified carbapenem inactivation method for the detection of carbapenemase-producing Enterobacteriaceae. J Infect Chemother. 2018;24:262–266. doi: 10.1016/j.jiac.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Davoudi-Monfared E, Khalili H. The threat of carbapenem-resistant gram-negative bacteria in a Middle East region. Infect Drug Resist. 2018;17:1831–1880. doi: 10.2147/IDR.S176049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azimi L, Talebi M, Owlia P, Pourshafie MR, Najafi M, Lari ER, et al. Tracing of false negative results in phenotypic methods for identification of carbapenemase by Real-time PCR. Gene. 2016;576:166–170. doi: 10.1016/j.gene.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 29.De Kievit TR, Parkins MD, Gillis RJ, Srikumar R, Ceri H, Poole K, et al. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2001;45:1761–1770. doi: 10.1128/AAC.45.6.1761-1770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El Kettani A, Maaloum F, Diawara I, Katfy K, Harrar N, Zerouali K, et al. Prevalence of Acinetobacter baumannii bacteremia in intensive care units of Ibn Rochd university hospital, Casablanca. Iran J Microbiol. 2017;9:318–323. [PMC free article] [PubMed] [Google Scholar]
- 31.Reza H. The frequency of multidrug-resistance and extensively drugresistant Acinetobacter baumannii in west of Iran. J Clin Microbiol Infect Dis. 2018;1:4–8. [Google Scholar]
- 32.Sarikhani Z, Nazari R, Nateghi Rostami M. First report of OXA-143-lactamase producing Acinetobacter baumannii in Qom, Iran. Iran J Basic Med Sci. 2017;20:1282–1286. doi: 10.22038/IJBMS.2017.9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hawkey J, Ascher DB, Judd LM, Wick RR, Kostoulias X, Cleland H, et al. Evolution of carbapenem resistance in Acinetobacter baumannii during a prolonged infection. Microb Genom. 2018;4:e000165. doi: 10.1099/mgen.0.000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dou Q, Zou M, Li J, Wang H, Hu Y, Liu W. AdeABC efflux pump and resistance of Acinetobacter baumannii against carbapenem. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2017;28;42:426–433. doi: 10.11817/j.issn.1672-7347.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Dias VC, Resende JA, Bastos AN, De Andrade Bastos LQ, De Andrade Bastos VQ, Bastos RV, et al. Epidemiological, physiological, and molecular characteristics of a brazilian collection of carbapenem- resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Microb Drug Resist. 2017;23:852–863. doi: 10.1089/mdr.2016.0219. [DOI] [PubMed] [Google Scholar]
- 36.Owrang M, Karimi A, Azimi L, Motaghi Nezhad R, Fallah F. Relative gene expression RND type efflux pumps in tigecycline resistant Acinetobacter baumannii isolated from training hospitals in Tehran, Iran. Int J Pediatr. 2018;6:8669–8674. [Google Scholar]
- 37.Peleg AY, Adams J, Paterson DL. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:2065–2069. doi: 10.1128/AAC.01198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]