Abstract
Background
Fecal colonization by carbapenem-resistant Enterobacteriaceae (CRE) can be the main reservoir for transmission of these resistant organisms especially in the Intensive Care Units (ICUs).
Aim
This study was conducted to evaluate the rate of rectal carriage and molecular characterization of CRE in patients hospitalized in the ICUs of 2 major hospitals (Adan and Mubarak Al Kabeer Hospitals) in Kuwait.
Materials and methods
Rectal swabs were collected from all patients at admission, 48 h after admission and once weekly from April 2017- March 2018. Initial CRE screening was carried out on MacConkey agar on which meropenem disc 10μg was placed. Identification of isolates was by API 20E. Susceptibility testing was performed using the E-test method. Polymerase chain reaction (PCR) was used to detect the carbapenemase-encoding genes. Clonal relationship was investigated by pulsed-field electrophoresis (PFGE). Genes of blaOXA-181 and blaNDM-5–carrying plasmids were detected in some strains.
Results
A total of 590 patients were recruited into the study. Of these, 58 were positive for CRE, giving a prevalence of 9.8%; 25/320 (7.8%) in Adan and 33/270 (12.2%) in Mubarak Al Kabeer Hospitals. All isolates were resistant to multiple antibiotics. Resistance rates to colistin and tigecycline were 17% and 83%, respectively. Single genes of blaOXA-181 were detected in isolates from 38 (65.5%) out of 58 patients and in 5 patients colonized by blaOXA-48-positive CRE. A combination of 2 genes was detected in 12 isolates; 5 blaKPC-2 and blaOXA-181, 4 blaVIM-1 and blaOXA-181, and 3 blaNDM-5 and blaOXA-181. PFGE showed an overall level of similarity of 38%. Southern hybridization studies localized the blaOXA-181 and blaNDM-5 genes to a large plasmid of 200kb in 3 K. pneumoniae isolates and a small plasmid of 80kb in 2 E. coli isolates, respectively.
Conclusion
The prevalence of CRE colonization in the 2 hospital ICUs was relatively high and the emergence of blaOXA-181-mediated CRE is a cause for concern as there is the possibility of rapid horizontal spread among hospital patients in Kuwait. Active surveillance of CRE in the ICUs is highly recommended to stem its spread.
Introduction
Gut flora is a major reservoir for Enterobacteriaceae which are potentially pathogenic to hospitalized patients especially those in the Intensive Care Units (ICUs). These Enterobacteriaceae have acquired resistance to several antimicrobial agents over time including the cephalosporins due inactivation by extended-spectrum β-lactamases (ESBLs) and AmpC production [1, 2]. This has led to excessive use of carbapenems for treating documented infections as well as for empirical therapy of suspected ESBL-associated infections especially those acquired in the ICUs [3]. The consequence of this has been accumulation of selective pressure for carbapenem resistance and the emergence of carbapenem-resistant Enterobacteriaceae (CRE). CRE infections are among the most problematic clinical challenges due to limited therapeutic options, limited isolation facilities, and higher financial cost [4]. They are increasingly reported worldwide primarily due to cumulative awareness globally and better identification methods in the hospital diagnostic laboratories.
Fecal colonization by CRE has been rarely investigated compared with colonization by ESBLs-producing Enterobacteriaceae in non-outbreak settings. In Kuwait, the emergence of carbapenem resistance among members of the family Enterobacteriaceae was not reported until 2012 when 2 patients infected with nosocomial acquired NDM-1 producing Klebsiella pneumoniae were encountered in the adult ICU of Mubarak Al Kabeer hospital [5]. Since then more and more infected or colonized patients by CRE have been encountered in many hospitals in Kuwait in the subsequent years [6–8]. The isolates responsible for these infections/colonizations have been NDM-1, VIM-4, and OXA-48-producing strains. OXA-181-producing CRE have never been isolated before as clinical or colonizing strains anywhere in Kuwait. The enzyme is a variant of OXA-48 by 4 amino acid substitutions at the Thr104Ala, Asn110Asp, Glu175Gln, and Ser179Ala positions [9]. It was initially identified in Enterobacter cloacae and K. pneumoniae strains isolated from patients in India in 2007 [10]. Since then OXA-181-producing Enterobacteriaceae has been reported in many other countries around the world including Nepal, Sri Lanka, Bangladesh, Canada, France, The Netherlands, Oman, Romania, South Africa, and United Kingdom [11–13]. Controlling the spread of CRE isolates involves reduction in antibiotic use, adherence to the infection control guidelines, and early detection of rectal colonization. Early detection of carriers or colonizers allows for early and rapid establishment of contact precautions to prevent CRE transmission to other patients. Infection with CRE has important implications on the care of the patients especially those in the ICUs because of limited therapeutic options. There is also a risk of outbreaks which may be attributed to environmental contamination or break in the infection control guidelines [14, 15]. This study was designed to evaluate the rectal prevalence of and molecular characteristics of CRE among patients admitted to two adult ICUs in Adan and Mubarak Al Kabeer Hospitals.
Materials and methods
Study design: This study was conducted for a period of one year (April 2017 to March 2018) in the 32–bedded and 26-bedded medical/surgical ICUs of Adan and Mubarak Al Kabeer Hospitals, Kuwait, respectively. These hospitals are large University-affiliated secondary and tertiary hospitals with 935 and 830 beds, respectively. They are located about 16 km apart responsible for a catchment population of about 2.5 million people.
Patients
A total of 590 consecutive patients admitted to the ICUs of Adan Hospital (n = 320; 54.2%) and Mubarak Al Kabeer Hospital (n = 270; 45.8%) over a period of 12 months were recruited into this study. Rectal swabs were collected from these patients on the day of admission, after 48 hours, and then at weekly intervals, till the day of discharge or death. A colonized patient with CRE on admission was defined as a carrier from the community. Nosocomial ICU-acquired CRE isolates were defined as those acquired after 48 hours of admission. The acquisition rate was determined by follow-up rectal cultures obtained from those negative at admission every week until the patient was discharged or deceased. The definition of acquisition rate was the number of patients who were not carriers on admission but became carriers later, divided by the number of patients who are not carriers on admission [16]. Demographic bio-data, e.g. age, gender, sample type, nationality, travel history, previous hospitalization within one year of current admission, diagnosis, co-morbidity, and prior antibiotic use, was recorded.
Ethics statement
Institutional ethical approval was obtained from the authorizing Medical Ethics Committee of the Ministry of Health (permit number 2016/272). Collection of the specimens was conducted according to the Declaration of Helsinki and with particular institutional ethical and professional standards. Written informed consent was obtained from the patients or their guardians and no additional clinical specimens were collected for the purpose of the study.
Microbiological studies
Rectal swabs were inoculated directly onto MacConkey (MAC) agar (Oxoid, Basingstoke, Hants, UK) on which 10μg meropenem disk (Becton Dickinson Ltd., Research Park Drive, North Ryde, Australia) was placed which allowed the establishment of optimal zone diameters for the screening of carbapenem-resistant Gram-negative bacteria. After incubation for 24 h at 37°C in an incubator (GallenKamp Economy Incubator size 2, Akribis Scientific Limited, Common Farm, Frog Lane, Pickmere, Knutsford, Cheshire), a reduced zone of inhibition of ≤19 mm around the meropenem disc was considered a potential CRE [17–19]. After the growth on MAC, identification of the isolates to the species level was done using the API 20E (bioMérieux, Marcy, L’Etoile, France).
Antibiotic susceptibility testing
The minimum inhibitory concentrations (MICs) of the antibiotics tested were determined by using E-test (bioMérieux) according to the manufacturer’s instruction. Antibiotics tested were the following: amikacin, cefoxitin, ciprofloxacin, colistin, ertapenem, imipenem, meropenem, and tigecycline. Interpretation of the results was carried out according to the Clinical and Laboratory Standards Institute [17] interpretative criteria. Tigecycline and colistin susceptibility were interpreted according to the EUCAST guidelines [19]. The following control strains, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and E. coli ATCC 35218 were included in each run. To compare the susceptibilities of different groups, the MICs that inhibited 90% (MIC90) and 50% (MIC50) of the isolates were calculated. CRE was defined as an Enterobacteriaceae isolate that showed decreased susceptibility to ertapenem (MIC>0.5μl/ml), imipenem (MIC>1μl/ml), and/or meropenem (MIC>1μl/ml) regardless of carbapenemase production [17].
Phenotypic and molecular characterization
The phenotypic production of carbapenemase by the isolates was investigated by using PCR and sequencing. PCR assays were done using previously described primers designed to detect Ambler class A (blaKPC), class B (blaVIM, blaIMP, blaNDM) and class D (blaOXA-48 and blaOXA-like) carbapenemase genes [20–22]. Sequencing was performed using the GeneAmp PCR system 9700 by cycle sequencing with BigDye termination (AB Applied Biosystems, Foster City, California, USA). The sequencing results were analyzed using software from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov).
Pulsed-field gel electrophoresis (PFGE)
Clonality of the CRE isolates was investigated by macrorestriction analysis of the genomic DNA with XbaI (New England Biolabs, Ipswich, MA, USA). The genomic DNA fragments were separated by PFGE in the CHEF-DR III system (Bio-Rad, Hemel Hempstead, UK). Electrophoresis conditions were pulsed-times ranging from 5 to 45 sec for 20 h at 6 V/cm at 14°C. Restriction patterns were analyzed according to previously described criteria [23]. In the PFGE analysis, the isolates were considered as highly related (100–97.5%), related (97.5–80%) and unrelated (<80%).
Plasmid analysis
Plasmids carrying blaOXA-181 and blaNDM-5 were detected and characterized in 3 K. pneumoniae (MK-18, MK-19, Adan-42) and 2 E. coli (MK-3, MK-6,) isolates by alkaline lysis method using episomes in E. coli 39R861 and E. coli V517 as molecular mass controls according to the previously described method of Kado and Liu [24]. It was followed by hybridization with blaOXA-181 and blaNDM-5 probes, using the ECL Random-Prime Labelling and detection system (Amersham Pharmacia Biotech, Little Chalfont, UK). The NDM-5 containing plasmids were conjugally transferred into E. coli J53RAZ [25].
Statistical analysis
The Epical 2000, version 1.02 (Brixton Health, Llanidloes, Powys, Wales, UK) was used to compare 2 proportions—percentages with 95% confidence interval and one-sided P-value. A Pearson’s Chi-squared test (IBM SPSS Statistics 26) was carried out to assess whether nationality and comorbidity were related significantly. P value of ≤0.05 was considered as statistically significant.
Results
Demographic characteristics
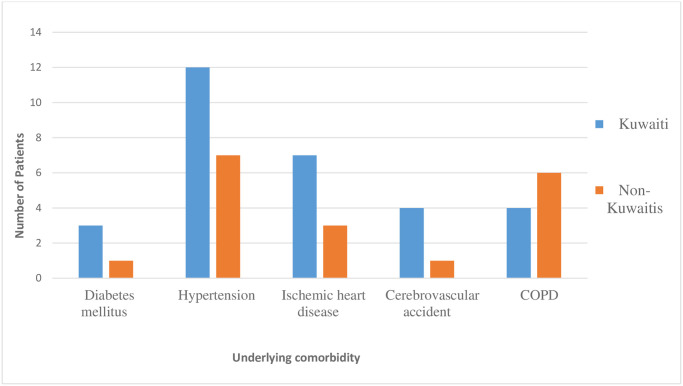
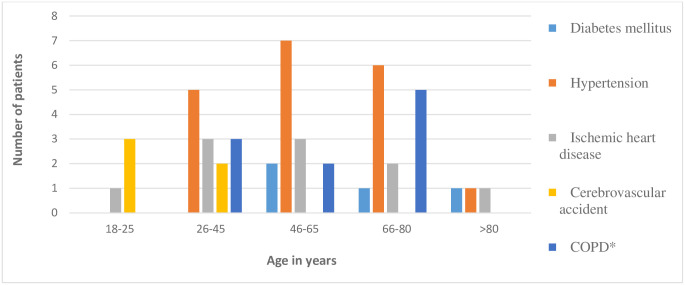
The colonization rates of the ICU patients during the study period were 7.8% (25 / 320) at Adan Hospital, and 12.2% (33/270) at Mubarak Al Kabeer Hospital. The demographic characteristics of all CRE-positive patients were generally comparable and they are summarized in Table 1. None of the general characteristics of the patients attained statistical significance between the two hospitals, except chronic obstructive pulmonary disease (COPD) comorbidity (P = 0.007). Of the 58 colonized patients, 33 (56.9%) were males and 25 (43.1%) females, with a male-to-female ratio of 1:0.8. The ages ranged from 18 to 89 years (mean = 50.4 years). Approximately 70% of the patients were over the age of 45 years in both hospitals; 20/25 (80%) in Adan versus 23/33 (69.7%) Mubarak Al Kabeer, (P>0.05). Nine (36%) versus 12 (36.4%), respectively were in the age group 46–65 and 10 (40%) versus 10 (30.3%), respectively in the 66–80 age group, (P>0.05). Thirty-nine (67.2%) were Kuwaitis, followed by 10 (17.2%) Indians and seven (12.1%) Egyptians. Intra-hospital, but not inter-hospital, differences between Kuwaitis and non-Kuwaitis was statistically significant (P<0.0001) in both hospitals. The most prominent comorbidities were hypertension (19/58: 32.7%), ischemic heart diseases (10/58: 17.2%), and COPD (10/58: 17.2%); none of these, except COPD (P<007), attained inter-hospital statistical significance. Other underlying conditions included cerebrovascular accident (5/58: 8.6%) and diabetes mellitus (4/58: 6.9%). A total of 22 (37.9%) gave history of previous treatment with a carbapenem and 10 (17.2%) with intravenous cephalosporins prior to admission into the ICU. Figs 1 and 2 show the inter-connecting nationality group and age group with underlying conditions.
Table 1. General characteristics of the intensive care unit patients colonized by carbapenem–resistant Enterobacteriaceae.
| Demographic variables | No (%) of patients colonized in | P value | ||
|---|---|---|---|---|
| Total no of patients (%) | Adan hospital (N = 25) | Mubarak hospital (N = 33) | ||
| Age group in years | ||||
| 18–25 | 3 (5.2) | 1 (4) | 2 (6.1) | 0.8 |
| 26–45 | 12 (20.7) | 4 (16) | 8 (24.2) | 0.6 |
| 46–65 | 21 (36.2) | 9 (36) | 12 (36.4) | 0.8 |
| 66–80 | 20 (34.5) | 10 (40) | 10 (30.3) | 0.6 |
| >80 | 2 (3.4) | 1 (4) | 1 (3) | 0.5 |
| Sex | ||||
| Male | 33 (56.9) | 14 (56) | 19 (57.6) | 0.9 |
| Female | 25 (43.1) | 11 (44) | 14 (42.4) | 0.9 |
| Nationality | ||||
| Kuwaiti | 39 (67.2) | 19 (76) | 20 (60.6) | 0.3 |
| Non-Kuwaitis | 19 (32.7) | 6 (24) | 13 (39.4) | 0.6 |
| Underlying conditions | ||||
| Diabetes mellitus | 4 (6.9) | 3 (12) | 1 (3) | 0.4 |
| Hypertension | 19 (32.7) | 8 (32) | 11 (33.3) | 0.9 |
| Ischemic heart disease | 10 (17.2) | 2 (8) | 8 (24.2) | 0.2 |
| Cerebrovascular accident | 5 (8.6) | 2 (8) | 3 (9.1) | 0.7 |
| COPD*** | 10 (17.2) | 0 (0) | 10 (30.3) | 0.007 |
| Previous hospitalization | ||||
| Yes | 6 (10.3) | 2 (8) | 4 (12.1) | 0.9 |
| No | 52 (89.6) | 23 (92) | 29 (87.9) | 0.9 |
| Previous/current antibiotic use | ||||
| Carbapenems | 22 (37.9) | 10 (40) | 12 (36.3) | 0.9 |
| Cephalosporins | 10 (17.2) | 5 (20) | 5 (15.2) | 0.9 |
| Other β-lactams | 2 (3.4) | 0 (0) | 2 (6) | 0.6 |
| Aminoglycoside | 7 (12.1) | 3 (12) | 4 (12.1) | 0.7 |
| Fluoroquinolone | 8 (13.8) | 3 (12) | 5 (15.2) | 0.9 |
| Macrolides | 6 (10.3) | 3 (12) | 3 (9.1) | 0.9 |
| Polymyxin | 3 (5.2) | 1 (4) | 2 (6.1) | 0.8 |
*P<0.001
**P<0.0001
***COPD = chronic obstructive pulmonary disease. Difference between Adan and Mubarak hospitals was statistically significant P = 0.007.
Fig 1. Underlying comorbidity of patients colonized with CRE and their nationality.
Fig 2. Underlying comorbidity of patients colonized with CRE and their age group.
Prevalence of rectal colonization by CRE
Out of the 58 patients colonized by CRE in the two ICUs, 12 were colonized on the day of admission and 46 acquired theirs after 48 hours of admission, giving prevalence rates of 2% and 7.8% community-acquired and hospital-acquired CRE, respectively; this difference is statistically significant (P<0.00009). Analysis per hospital revealed 2.2% (7/320) community-acquired and 5.6% (18/320) hospital-acquired rates (P<0.041), respectively in Adan Hospital, and 1.9% (5/270) and 10.4% (28/270), respectively (P<0.00007) in Mubarak Al Kabeer Hospital. Of these colonized patients, 14 (24.1%) died during the study period: 7 each from Adan and Mubarak.
CRE and antimicrobial susceptibility
The breakdown of the species of the CRE isolates is shown in Table 2. The most prevalent was K. pneumoniae, representing 29 (50%) of all isolates, followed by E. coli, 11 (19%), Enterobacter spp. 5 (8.6%), Serratia spp. 5 (8.6%), and others, 6 (13.7%).
Table 2. Distribution of carbapenem-resistant Enterobacteriaceae isolates in the two hospitals.
| Bacteria | Number (%) of CRE isolates | |||
|---|---|---|---|---|
| Adan hospital (N = 25) | Mubarak hospital (N = 33) | |||
| Community acquired (N = 7) | Hospital acquired (N = 18) | Community acquired (N = 5) | Hospital acquired (N = 28) | |
| Escherichia coli | 0 | 1 (4) | 1 (20) | 9 (32.1) |
| Klebsiella pneumoniae | 5 (71.4) | 11 (64) | 2 (40) | 11 (39.3) |
| Klebsiella oxytoca | 0 | 0 (0) | 0 | 2 (7.1) |
| Enterobacter cloacae | 0 | 1 (4) | 1 (20) | 2 (7.1) |
| Enterobacter gergoviae | 0 | 0 (0) | 0 | 1 (3.6) |
| Morganella morganii | 1 (14.3) | 1 (4) | 0 | 0 (0) |
| Pantoea agglomerans | 0 | 1 (4) | 0 | 0 (0) |
| Proteus mirabilis | 0 | 0 (0) | 0 | 1 (3.6) |
| Roseomonas gilardii | 1 (14.3) | 1 (4) | 0 | 0 (0) |
| Salmonella enterica | 0 | 1 (4) | 0 | 0 (0) |
| Shigella boydii | 0 | 0 (0) | 0 | 1 (3.6) |
| Serratia odoriferae | 0 | 1 (4) | 0 | 0 (0) |
| Serratia marcescens | 0 | 0 (0) | 1 (20) | 0 (0) |
| Serratia plymuthica | 0 | 1 (4) | 0 | 1 (3.6) |
| Serratia rubidaea | 0 | 1 (4) | 0 | 0 (0) |
The MIC range, MIC50, MIC90, and percentage of resistance of the CRE isolates are given in Table 3. All the K. pneumoniae isolates from Adan hospital were resistant to cefoxitin, ciprofloxacin, ertapenem, imipenem and meropenem. About 75% were resistant to tigecycline with MIC50 and MIC90 of 2 μg/ml and 8 μg/ml, respectively while 44% were resistant to colistin with MIC50 and MIC90 of 1 μg/ml and 8 μg/ml, respectively. In Mubarak Al Kabeer Hospital, K. pneumoniae exhibited high resistance to the antibiotics with MIC90s >256 μg/ml for amikacin and cefoxitin, and > 32μg/ml for ciprofloxacin, ertapenem, imipenem, and meropenem.
Table 3. Minimum inhibitory concentrations (MICs) of the antimicrobial agents tested against carbapenem-resistant Enterobacteriaceae (CRE) isolates.
| Bacteria/antibiotics (breakpoint) | Susceptibility of the CRE isolates per hospital | |||||||
|---|---|---|---|---|---|---|---|---|
| Adan hospital | Mubarak hospital | |||||||
| MIC ranges | MIC50 | MIC90 | % resistant | MIC ranges | MIC50 | MIC90 | % resistant | |
| Klebsiella pneumoniae | ||||||||
| Amikacin (16) | 3->256 | >256 | >256 | 94 | >256 | >256 | >256 | 100 |
| Cefoxitin (8) | 64->256 | >256 | >256 | 100 | >256 | >256 | >256 | 100 |
| Ciprofloxacin (1) | >32 | >32 | >32 | 100 | >32 | >32 | >32 | 100 |
| Colistin (2) | 1–16 | 1 | 8 | 44 | 0.5–8 | 1 | 3 | 17 |
| Ertapenem (0.5) | >32 | >32 | >32 | 100 | >32 | >32 | >32 | 100 |
| Imipenem (1) | >32 | >32 | >32 | 100 | 4->32 | >32 | >32 | 100 |
| Meropenem (1) | >32 | >32 | >32 | 100 | >32 | >32 | >32 | 100 |
| Tigecycline (2) | 1->256 | 2 | 8 | 75 | 2–16 | 8 | 16 | 83 |
| Escherichia coli* | ||||||||
| Amikacin (16) | - | - | - | - | 1.5->256 | 24 | >256 | 56 |
| Cefoxitin (8) | - | - | - | - | 12->256 | >256 | >256 | 100 |
| Ciprofloxacin (1) | - | - | - | - | >32 | >32 | >32 | 100 |
| Colistin (2) | - | - | - | - | 0.5–12 | 1 | 2 | 11 |
| Ertapenem (0.5) | - | - | - | - | >32 | >32 | >32 | 100 |
| Imipenem (1) | - | - | - | - | >32 | >32 | >32 | 100 |
| Meropenem (1) | - | - | - | - | >32 | >32 | >32 | 100 |
| Tigecycline (2) | - | - | - | - | 0.25->256 | 12 | >256 | 56 |
*Only one isolate was encountered in Adan Hospital and was resistant to all antibiotics; bp = breakpoints
Comparatively, resistance to tigecycline was 83%. By contrast, only 17% were resistant to colistin with MIC50 and MIC90 of 1 μg/ml and 3 μg/ml, respectively. All, but one, of the E. coli isolates were from Mubarak Al Kabeer Hospital and they were fully resistant to cefoxitin, ciprofloxacin, ertapenem, imipenem and meropenem with MIC90s of >256, >32, >32, >32 μg/ml, respectively. Resistance rates against amikacin and tigecycline were 56% and 56%, respectively. They were moderately resistant (11%) to colistin, with MIC50 and MIC90 of 1 μg/ml, and 2 μg/ml, respectively.
Carbapenemase genes
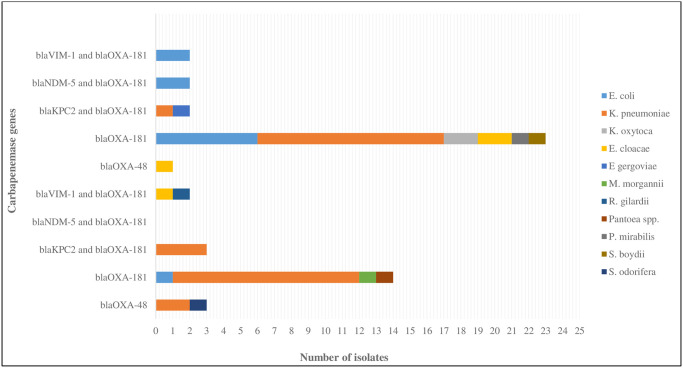
A total of 56 (96.6%) out of the 58 CRE isolates harbored one or 2 carbapenemase-mediating genes as shown in Fig 3. Interestingly, 50 (89.3%) of the 56 carbapenemase-producing Enterobacteriaceae (CPE) isolates harbored blaOXA-181 gene either singly or in combination with other genes. Other genes detected were blaVIM-1 (4 isolates), blaNDM-5 (3), and blaKPC-2 (5). A total of 44 (78.6%) of the 56 isolates harbored single genes and 12 (21.4%) harbored 2 genes. As can be demonstrated in this Fig 3, 38 (86.4%) of those harboring single genes were positive for blaOXA-181, 5 (11.4%) blaOXA-48, and 1 (2.3%) blaKPC-2. By hospital, blaOXA-181 gene alone was harbored by 15 (60%) of 25, and 23 (69.7%) of 33 isolates from Adan and Mubarak Al Kabeer Hospitals, respectively. Of the 12 (21.4%) isolates that carried 2 genes, 4 (33.3%) were K. pneumoniae which harbored blaKPC-2, and blaOXA-181 (3 from Adan Hospital and 1 from Mubarak Al Kabeer Hospital). Two E. coli from Mubarak Al Kabeer Hospital and 1 Salmonella spp. from Adan Hospital carried both blaNDM-5, and blaOXA-181 genes. One each of E. cloacae and Roseomonas gilardii, and 2 E. coli harbored both blaVIM-1, and blaOXA-181 and one E. gergoviae carried blaKPC-2, and blaOXA-181.
Fig 3. Prevalence of different carbapenemase genes harbored by CRE isolates from Adan and Mubarak Al Kabeer Hospitals.
The DNA sequences of blaNDM-5 and blaOXA-181 have been submitted to the GenBank database (US National Library of Medicine, National Center for Biotechnology Information, NCBI) under the accession numbers MK256964.1 (NDM) and MF774789.1 (OXA).
Plasmid study
Analysis of plasmid content revealed that the 3 K. pneumoniae and 2 E. coli isolates contained multiple small and large plasmids, ranging between 80-200kb. Southern hybridization studies using blaOXA-181 probe localized the blaOXA-181 gene on the large 200kb size plasmid in the 3 K. pneumoniae isolates but the blaNDM-5 was located on the small 80kb size plasmid in the 2 E. coli isolates.
Clonal relatedness of isolates
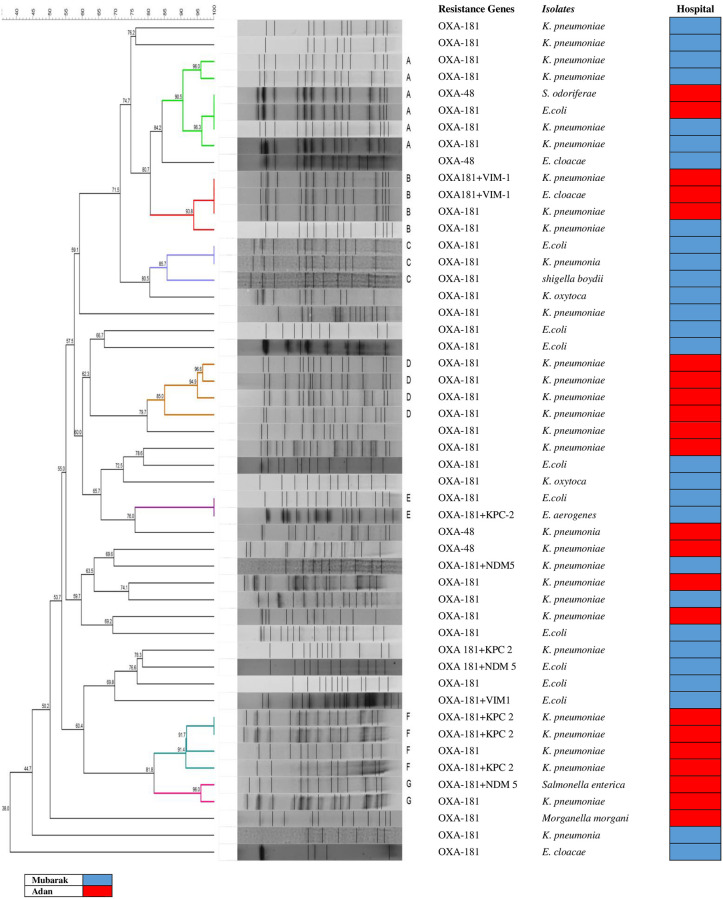
Clonal relationships of 50 carbapenemase-producing Enterobacteriaceae (CPE) isolates that carried either blaOXA-181 or blaOXA-48 genes from the 2 ICUs were determined by PFGE and demonstrated in Fig 4. The XbaI PFGE profile-based dendrograms showed that the overall level of similarity was 38%. However, the 85% criterion resolved 7 pulsotypes (designated A, B, C, D, E, F, and G). There was no single predominant pulsotype.
Fig 4. Dendrogram based on UPGMA cluster analysis of Dice similarity values of PFGE fingerprint patterns of 50 carbapenemase-producing Enterobacteriaceae (CPE) isolates in Kuwait.
Band profiles of each strain are shown corresponding with the lines of the dendrogram. Seven pulsotypes (designated A, B, C, D, E, F, and G) diverging 85% criterion. The gene, isolate identification, and hospital are represented in the columns.
Further investigation revealed that certain isolates were collected from certain areas or units in the ICU e.g. in Mubarak Al Kabeer Hospital ICU, there were 7 subpulsotypes i.e. A1, A2, A5, A6, C1, C2, and E1 which were collected from unit D in the Medical ICU. While in Adan Hospital ICU, there were 13 different subpulsotypes i.e. A3, A4, B1, B3, D1, D2, D3, D4, F1, F2, F3, G1, and G2 which were isolated from the both Surgical and Medical ICUs.
Discussion and conclusion
The prevalence of rectal colonization by CRE among patients has been described previously with rates varying from 0.5 to 12.2% in different areas of the world [26–31]. To our knowledge, this is the first study to report the prevalence of CRE gut colonization among ICU patients in Kuwait and, indeed, in the entire Gulf Cooperation Council (GCC) countries. We take cognizance of the fact that all colonized patients with CRE cannot be detected by depending on cultures collected according to the clinical condition of the patients. Detection of asymptomatic carriers in patients is exceedingly important because these patients may act as reservoir for transmission of CRE [15]. The main reservoir for CRE is the gut and fecal samples are usually used for its screening. But the isolation of CRE from stool specimens is often difficult due to the fact that these strains compose a small proportion of the gut flora and absence of efficient or standardized screening method for their detection makes the process more difficult [32, 33]. In addition, it has been shown that surveillance of low level resistance, such as IMP and OXA-48 producers, is difficult and, more often than not, the numbers are underestimated [34, 35]. Experience has shown that colonized patients with CRE are often transferred between wards within the same hospital or between different hospitals.
In this study, the overall prevalence of CRE among the 2 ICU patients was 9.8% which is lower than those of 12.2% reported in long-term care hospitals and acute-care hospitals in Japan [29] and 37.9% in Iran [36]. Similarly, higher figures of 12% have been reported in post-acute-care facilities wards in Israel [26] and 20.4% in the hematology-oncology wards in India [37]. In contract, our prevalence rate is higher than the reported figures of 6.6% in China [28] and 5.4% in USA although these studies were carried out in the ICU, medical, surgical, and acute rehabilitation units [27]. It is pertinent to point out that a direct comparison with these studies should be interpreted with caution as the settings and patients were different from ours. However, in studies, similar to ours, reported on hospitalized patients in Spain and France, the prevalence rates were 2.9% and 5.3%, respectively [38, 39]. Similarly, a prevalence of 3.8%, much lower than ours, was also reported in a large refractory nosocomial outbreak of KPC-producing E. coli in the Central Manchester University Hospital, UK [31] as well as in another study conducted in a large teaching hospital in Northern Italy, where the percentage of CRE rectal colonization dropped significantly from 0.2–0.05% after introduction of infection control interventions over a period of 30 months [30]. It is important to note that only a few of these studies were conducted on ICU patients. In concordance with the low community-acquired colonization rates encountered in our study, a study in Spain did report a much lower community-acquired colonization of rate of 0.4% [38].
An interesting characteristic feature found in our study was the preponderance of colonized Kuwaitis over non-Kuwaitis in each hospital, a finding that attained statistically significant difference (P<0.0001). This paradox has an important epidemiological implication because the population of Kuwait consists of about 35% indigenes versus 65% non-indigenes with strict segregation and yet more colonized patients were found among the Kuwaitis. Furthermore, the majority of these Kuwaitis had not traveled outside the country in the preceding year. It is conceivable that the CRE were present in the community and hospital environment in low numbers perhaps acquired silently from their foreign helpers who might have brought them from their respective countries over time.
The commonest CRE isolates in our study was K. pneumoniae followed by E. coli as in previous reports [37, 38] but a study from Japan had previously reported a reversal in prominence between the two bacteria [29]. It is noteworthy that 2 of the CRE colonized patients were asymptomatically positive for Salmonella enterica and S. boydii, a finding that has important infection control implications, especially in the ICU setting.
Not surprisingly, all our CRE isolates were resistant to the carbapenems, cephalosporins, and ciprofloxacin. This is concordant with the previous reports from Kuwait [6, 7]. Resistance to colistin and tigecycline appears to be on the ascendancy when compared to previous experience in Kuwait [8]. This is a dangerous trend as these antibiotics are the drugs of last resort for treating patients infected by multi-drug resistant (MDR) Gram-negative bacteria. The activities of the antibiotics tested against E. coli were equally poor although there were some differences in the susceptibility levels to carbapenems, aminoglycoside, colistin, and tigecycline. We foresee a situation where treatment options will be severely restricted in our hospitals should our patients become infected with these CRE strains.
By far the most important highlight of this study is the high prevalence (67.9%) of the CPE mediated by blaOXA-181 gene. This finding is new to Kuwait and very uncommon in the surrounding Gulf countries. Evidence from this study showed that this finding may represent rapid and widespread dissemination of blaOXA-181 in our country. Our assertion is based on the fact that most cases of infections and colonization have been in areas outside this region usually in patients who had recently traveled to Southeast Asia and the Asian Pacific region as these are the areas suspected to be the origin of this gene and an important reservoir of this carbapenemase gene. Although OXA-181-producing Enterobacteriaceae have also been reported in several other non-Asian countries [13], only a few anecdotal reports of clinical cases in Abu Dhabi, Oman and Lebanon have been reported from other countries in the Middle East [25, 40, 41]. It is pertinent to recognize that about 68% of the patients colonized in this study were indigenous Kuwaitis who had no history of previous travels to these regions. More intriguing is the fact that the 17% who were Indians did not admit traveling home in the last one year preceding admission to the ICU. Our speculation, at this time, is that the gene might have been imported by the returning expatriate Asian workers who form a very large population of domestic and middle class work force in the country. It is conceivable that this gene is present at low levels within this community and has found its way into the indigenous population.
Other interesting findings include the 2 E. coli and 1 S. boydii isolates that harbored blaNDM-5 and blaOXA-181. This is the first of such findings in Kuwait. The first report of a combination of these two genes was previously encountered in a patient, with urinary tract infection managed at a Singapore hospital in 2013, who had been transferred from Bangladesh [11]. Similarly, OXA-181 and NDM-5-producing CRE was reported in 2015 in a K. pneumoniae isolate from a South Korean patient transferred from a tertiary hospital in Abu Dhabi, UAE [42]. Recently, a cluster of NDM-5 and chromosomally mediated OXA-181-producing pan-resistant K. pneumoniae from 4 patients in Abu Dhabi with possible connections to the strains subsequently isolated in South Korea, was reported by Sonnevend et al., 2017 [25]. However, finding this combination in E. coli, like ours, has only been reported once before in Egypt [43] and Saudi Arabia [44]. It should be noted that in our study this combination was also found in one S. boydii isolated from a patient with diabetes mellitus and ischemic heart disease in Adan hospital representing the first of such report in this bacterium particularly in this country and the region. Furthermore, blaOXA-181 gene in association with other carbapenemase genes e.g. blaNDM-1 and blaVIM-1 has been reported in K. pneumoniae [42]. But the occurrence of this co-existence in E. coli, Enterobacter cloaceae and Roseomonas gilardii is alarming because of the belief that worldwide spread of this enzyme is a mirror image of that of NDM-1 [13].
As a result of the widespread colonization by OXA-181-producing CRE the isolates in the 2 hospital ICUs were investigated for any evidence of horizontal intra or inter–hospital spread using the PFGE. A relatively high percentage of the isolates (38%) were related. This suggests some form of spread within the ICUs of each hospital. Two isolates from both hospitals were also similar, a finding that can be explained, in part, by the occasional movements of patients between hospitals in Kuwait with possible transmission between the 2 hospital ICUs that remains a cause for grave concern in Kuwait. There is a significant correlation of isolates with identical pulsotypes between the date of collection and the hospital.
In our study, the gene encoding OXA-181 was carried on 200kb plasmid which is similar to the previous study reported by Castanheira et al. where blaOXA-181 gene was carried on plasmids of 200-250kb molecular sizes [45] but unlike the study of Potron et al which showed that the blaOXA-181 was located on a 7.6-kb ColE-type plasmid in K. pneumoniae isolate [46]. The NDM-5 plasmid, in our study, was located on a small 80kb plasmid which is smaller (110kb) than that detected in E. coli in Poland [47].
Limitations of this study include the following: only two centers were included and plasmid studies were limited to a few isolates. In addition, we did not study all carbapenemase resistance genes and other mechanisms of resistance especially for those 2 isolates which were carbapenem-resistant but carbapenemase genes negative.
In conclusion, the prevalence of rectal colonization by CRE in these 2 ICUs was higher than expected. Detection of blaOXA-181 and blaNDM-5 is new to the milieu of genes so far described in isolates from Kuwait. The emergence of blaOXA-181-mediated CRE is a cause for concern as there is the possibility of rapid horizontal spread among hospital patients. It is highly recommended that active surveillance cultures, strict patients isolation, and improved epidemiologic and hygienic situation should be carried out in our ICUs even when CRE prevalence remains relatively low in order to avoid an endemic trend.
Supporting information
(TIF)
Acknowledgments
We thank Dr. Ahlam Jeragh of Adan hospital and all the nurses in the 2 ICUs who collected the rectal swabs.
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
This work was supported by Kuwait University, Research Grant no. YM07/17. The funder had no rule in study design, data collection and analysis, decision to publish or preparing of the manuscript.
References
- 1.Pitout JD, Laupland KB. Extended-spectrum β-lactamase producing Enterobacteriaceae: an emerging public-health problem. Lancet Infect Dis. 2008; 8: 159–166. 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- 2.Bogaerts P, Rodriguez-Villalobos H, Laurent C, Deplano A, Struelens MJ, Glupczynski T. Emergence of extended–spectrum AmpC-expressing Escherichia coli isolates in Belgian hospitals. J Antimicrob Chemother. 2009; 63: 1073–1075. 10.1093/jac/dkp046 [DOI] [PubMed] [Google Scholar]
- 3.Meyer E, Schwab F, Schroeren-Boersch B, Gastmeier P. Dramatic increase of third- generation cephalosporin-resistant E. coli in German intensive care units: secular trends in antibiotic drug use and bacterial resistance, 2001 to 2008. Crit Care. 2010; 14: R113 10.1186/cc9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng E, Earnest A, Lye DC, Ling ML, Ding Y, Hsu L. The excess financial burden of multidrug resistance in severe gram-negative infections in Singaporean hospitals. Ann Acad Med Singapore. 2012; 41: 189–193. [PubMed] [Google Scholar]
- 5.Jamal W, Rotimi VO, Albert MJ, Khodakhast F, Udo EE, Poirel L. Emergence of nosocomial New Delhi metallo-β-lactamase-1 (NDM-1)-producing Klebsiella pneumoniae in patients admitted to a tertiary care hospital in Kuwait. Int J Antimicrob Agents. 2012; 39: 183–184. 10.1016/j.ijantimicag.2011.10.002 [DOI] [PubMed] [Google Scholar]
- 6.Jamal W, Rotimi VO, Albert MJ, Khadakhast F, Nordmann P, Poirel L. High Prevalence of VIM-4 and NDM-1 metallo-β-lactamase among carbapenem–resistant Enterobacteriaceae. J Med Microbiol. 2013; 62: 1239–1244. 10.1099/jmm.0.059915-0 [DOI] [PubMed] [Google Scholar]
- 7.Jamal WY, Albert MJ, Khodakhast F, Poirel L, Rotimi VO. Emergence of new sequence type OXA-48 carbapenemase-producing Enterobacteriaceae in Kuwait. Microb Drug Resist. 2015; 21: 329–334. 10.1089/mdr.2014.0123 [DOI] [PubMed] [Google Scholar]
- 8.Jamal WY, Albert MJ, Rotimi VO. High prevalence of New Delhi metallo-β-lactamase-1 (NDM-1) producers among carbapenem-resistant Enterobacteriaceae in Kuwait. PLoS One. 2016; 11: e0152638 10.1371/journal.pone.0152638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rojas LJ, Hujer AM, Rudin SD, Wright MS, Domitrovic TN, Marshall SH, et al. NDM-5 and OXA-181 Beta-lactamases, a significant threat continues to spread in the Americas. Antimicrob Agents Chemother. 2017; 61: e00454–17. 10.1128/AAC.00454-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. Early dissemination of NMD-1-and OXA-181–producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob Agents Chemother. 2011; 55: 1274–1278. 10.1128/AAC.01497-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balm MN, La MV, Krishnan P, Jureen R, Lin R, Teo JWP. Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin Microbiol Infect. 2013; 19: E421—E423. 10.1111/1469-0691.12247 [DOI] [PubMed] [Google Scholar]
- 12.Hall JM, Corea E, Sanjeewani HAD, Inglis TJ. Molecular mechanisms of β-lactam resistance in carbapenemase-producing Klebsiella pneumoniae from Sri Lanka. J Med Microbiol. 2014; 63:1087–1092. 10.1099/jmm.0.076760-0 [DOI] [PubMed] [Google Scholar]
- 13.Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012; 67:1597–1606. 10.1093/jac/dks121 [DOI] [PubMed] [Google Scholar]
- 14.Lerner A, Alder A, Abu-Hanna J, Meitus I, Navon-Venezia S, Carmeli Y. Environmental contamination by carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 2013; 51: 177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben-David D, Maor Y, Keller N, Regev-Yochay G, Tal I, Shachar D, et al. Potential of active surveillance in the control of a hospital–wide outbreak of carbapenem-resistant Klebsiella pneumoniae infection. Infect Control Hosp Epidemiol. 2010; 6: 620–626. 10.1086/652528 [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Lee JY, Kim SI, Song W, Kim JS, Jung S, et al. Rates of fecal transmission of extended-spectrum β-lactamase-producing and carbapenem-resistant Enterobacteriaceae among patients in intensive care units in Korea. Ann Lab Med. 2014; 34: 20–25. 10.3343/alm.2014.34.1.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CLSI, 2018. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 28th informational supplement. CLSI publication 2018; M100- S26, Wayne, Pennsylvania, USA.
- 18.Giani T, Tascini C, Arena F, Ciullo I, Conte V, Leonildi A, et al. Rapid detection of intestinal carriage of Klebsiella pneumoniae producing KPC carbapenemase during an outbreak. J Hosp Infect. 2012; 81: 119–122. 10.1016/j.jhin.2012.04.004 [DOI] [PubMed] [Google Scholar]
- 19.EUCAST. Breakpoints tables for interpretation of MICs and zone diameters 2016. www.eucast.org (accessed December 18, 2016).
- 20.Toleman MA, Biedenbach D, Bennett D, Jones RN, Walsh TR. Genetic characterization of a novel metallo-beta-lactamase gene, blaIMP-13, harboured by a novel Tn5051-type transposon disseminating carbapenemase in Europe: report from SENTRY worldwide antimicrobial surveillance programme. J Antimicrob Chemother. 2003; 52: 583–590. 10.1093/jac/dkg410 [DOI] [PubMed] [Google Scholar]
- 21.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemases genes. Diag Microbiol Infect Dis. 2011; 70: 119–123. [DOI] [PubMed] [Google Scholar]
- 22.Poirel L, Heritier C, Tolun V, Nordmann P, 2004, Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother, 48; 15–22. 10.1128/aac.48.1.15-22.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995; 33: 2233–2239. 10.1128/JCM.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kado CI, Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981, 145: 1365–1373. 10.1128/JB.145.3.1365-1373.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonnevend A, Ghazawi A, Hashmey R, Haidermota A, Girgis S, Alfaresi M, et al. Multihospital occurrence of pan-resistant Klebsiella pneumoniae sequence type 147 with an ISEcpi1-directed blaOXA-181 insertion sequence in the mgrB gene in the United Arab Emirates. Antimicrob Agents Chemother. 2017; 61: pii: e00418–17. 10.1128/AAC.00418-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-David D, Masarwa S, Navon-Venezia S, Mishali H, Fridental I, Robinovitch B, et al. Carbapenem-resistant Klebsiella pneumoniae in post-acute–care facilities in Israel. Infect Control Hosp Epidemiol. 2011; 32: 845–853. 10.1086/661279 [DOI] [PubMed] [Google Scholar]
- 27.Swaminathan M, Sharma S, Poliansky Blash S, Patel G, Banach DB, Phillips M, et al. Prevalence and risk factors for acquisition of carbapenem-resistant Enterobacteriaceae in the setting of endemicity. Infect Control Hosp Epidemiol. 2013: 34: 809–817. 10.1086/671270 [DOI] [PubMed] [Google Scholar]
- 28.Zhao ZC, Xu XH, Liu MB, Wu J, Lin J, Li B. Fecal carriage of carbapenem-resistant Enterobacteriaceae in Chinese university hospital. Am J Infect Control. 2014; 42: 61–64. 10.1016/j.ajic.2014.01.024 [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto N, Asada R, Kawahara R, Hagiya H, Akeda Y, Shanmugakani RK, et al. Prevalence of, and risk factors for, carriage of carbapenem-resistant Enterobacteriaceae among hospitalized patients in Japan. J Hosp Infect. 2017; 97: 212–217. 10.1016/j.jhin.2017.07.015 [DOI] [PubMed] [Google Scholar]
- 30.Viale P, Tumietto F, Giannella M, Bartoletti M, Tedeschi S, Ambretti S, et al. Impact of a hospital–wide multifaceted programme for reducing carbapenem-resistant Enterobacteriaceae infections in a large teaching hospital in northern Italy. Clin Microbiol Infect. 2015; 21: 242–247. 10.1016/j.cmi.2014.10.020 [DOI] [PubMed] [Google Scholar]
- 31.Decraene V, Phan HTT, George R, Wyllie DH, Akinremi O, Aiken Z, et al. A large, refractory nosocomial outbreak of Klebsiella pneumoniae carbapenemase–producing Escherichia coli demonstrates carbapenemase gene outbreaks involving sink sites require novel approaches to infection control. Antimicrob Agents Chemother. 2018; 62: e01689–18. 10.1128/AAC.01689-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanner WD, Atkinson RM, Goel RK, Porucznik CA, Benson LS, VanDerslice JA. Effect of meropenem concentration on the detection of low numbers of carbapenem–resistant Enterobacteriaceae. Antimicrob Agents Chemother. 2015; 60: 712–713. 10.1128/AAC.01904-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viau R, Frank KM, Jacobs MR, Wilson B, Kaye K, Donskey CJ, et al. Intestinal carriage of carbapenemase–producing organisms: current status of surveillance methods. Clin Microbiol Rev. 2016; 1: 29: 1–27. 10.1128/CMR.00108-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girlich D, Bouihat N, Poirel L, Benouda A, Nordman P. High rate of faecal carriage of extended-spectrum β–lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a university hospital in Morocco. Clin Microbiol Infect. 2014; 20: 350–354. 10.1111/1469-0691.12325 [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto N, Kawahara R, Akeda Y, Shanmugakani RK, Yoshida H, Hagiya H, et al. Development of selective media for IMP-type carbapenemase-producing Enterobacteriaceae in stool specimens. BMC Infect Dis. 2017; 17: 229 10.1186/s12879-017-2312-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solgi H, Badmasti F, Aminzadeh Z, Giske CG, Pourahmad M, Vaziri F, et al. Molecular characterization of intestinal carriage of carbapenem-resistant Enterobacteriaceae among inpatients at two Iranian university hospitals: first report of co-production of blaNDM-7 and blaOXA-48. Eur J Clin Microbiol Infect Dis. 2017; 36: 2127–2135. 10.1007/s10096-017-3035-3 [DOI] [PubMed] [Google Scholar]
- 37.Jaiswal SR, Gupta S, Kumar RS, Sherawat A, Rajoreya A, Dash SK, et al. Gut colonization with carbapenem-resistant Enterobacteriaceae adversely impacts the outcome in patients with hematological malignancies: results of a prospective surveillance study. Mediterr J Hematol Infect Dis. 2018; 10: e2018025 10.4084/MJHID.2018.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gijon D, Curiao T, Baquero F, Coque TM, Canton R. Fecal carriage of carbapenemase-producing Enterobacteriaceae: a hidden reservoir in hospitalized and nonhospitalized patients. J Clin Microbiol. 2012; 50: 1558–1563. 10.1128/JCM.00020-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vidal-Navarro L, Pfeiffer C, Bouzinges N, Sotto A, Lavigne JP. Faecal carriage of multi-drug resistant Gram-negative bacilli during a non-outbreak situation in a French university hospital. J Antimicrob Chemother. 2010, 65: 2455–2458. 10.1093/jac/dkq333 [DOI] [PubMed] [Google Scholar]
- 40.Dortet L, Poirel L, Al Yaqoubi F, Nordmann P. NDM-1, OXA-48 and OXA-181 carbapenemase-producing Enterobacteriaceae in Sultanate of Oman. Clin Microbiol Infect. 2012; 18: E144—E148. 10.1111/j.1469-0691.2012.03796.x [DOI] [PubMed] [Google Scholar]
- 41.Bitar I, Dagher C, Salloum T, Araj G, Tokajian S. First report of Escherichia coli from Lebanon carrying an OXA-181 carbapenemase resistance determinant. J Glob Antimicrob Resist. 2018; 12: 113–114. 10.1016/j.jgar.2018.01.002 [DOI] [PubMed] [Google Scholar]
- 42.Cho SY, Huh HJ, Baek JY, Chung NY, Ryu JG, Ki C-S, et al. Klebsiella pneumoniae co-producing NDM-5 and OXA-181carbapenemases, South Korea. Emerg Infect Dis. 2015; 21: 1088–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gamal D, Fernandez-Martinez M, El-Defrawy I, Ocampo-Sosa AA, Martinez-Martinez L. First identification of NDM-5 associated with OXA-181 in Escherichia coli from Egypt. Emerg Microb Infect. 2016; 5: e.30 10.1038/emi.2016.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abd El Ghany M, Sharaf H, Al-agamy MH, Shibl A, Hill-Cawthorne GA, Hong P-Y. Genomic characterization of NDM-1 and 5, and OXA-181 carbapenemases in uropathogenic Escherichia coli isolates from Riyadh, Saudi Arabia. PLoS One. 2018; 13: e0201613 10.1371/journal.pone.0201613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. Early dissemination of NDM-1 and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance program, 2006–2007. Antimicrob Agents Chemother. 2011; 55: 1274–1278. 10.1128/AAC.01497-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashi F, Poirel L. Characterization of OXA-181, a carbapenem-hydrolyzing class D β-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2011; 55: 4896–4899. 10.1128/AAC.00481-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baraniak A, Izdebski R, Fiett J, Gawryszewska I, Bojarska K, Herda M, et al. NDM-producing Enterobacteriaceae in Poland, 2012–14: inter-regional outbreak of Klebsiella pneumoniae ST11 and sporadic cases. J Antimicrob Chemother. 2016; 71: 85–91. 10.1093/jac/dkv282 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.