Abstract
Lysozyme is an important component of the innate immune system, and has roles in peptidoglycan cleavage of gram positive organisms. Myeloid cells highly express the isoform, lysozyme M, and its promoter has been used to direct Cre-recombinase expression to target deletion of floxed genes in myeloid cells. However, generation of the LysM-Cre mouse effectively disrupts the LysM-gene, and mice homozygous for the Cre allele lack the LysM gene product. To test the contribution of LysM in sterile acute lung injury, we generated LysM-Cre mice homozygous for the Cre allele (+/+) or wild-type allele (−/−). These mice were challenged with LPS delivered via oropharygneal aspiration. Mice were monitored and weighed daily, and BAL cell counts, differential, protein, and cytokine levels were assessed at days 2 and 4. LysMCre+/+ and LysMCre−/− had similar weight loss and recovery, and similar inflammatory responses to LPS at days 2 and 4. These findings indicate that that loss of LysM and expression of Cre-recombinase are non-contributory in sterile acute lung injury.
Introduction:
Lysozyme is a glycoside hydrolase that catalyzes the hydrolysis the β-1,4 glycosidic bond between N-acetyl muramic acid and N-acetyl gucosamine[1]. Its main substrates are predominantly found in the cell wall of Gram-positive bacteria, but shorter saccharides and chitin have also been shown to be substrates of this enzyme [2, 3]. Beyond its catalytic activity, non-catalytic antimicrobial roles have also been ascribed to Lysozyme [4], as well as roles in monocyte-mediated tumoricidal activity[5]. Lysozyme M (LysM or Lyz2), one of two isoforms, is abundant in the airway and functions as a critical anti-microbial in the lung. Augmentation of its expression in lung increases antimicrobial killing of P. aeruginosa and group B Streptococcus [6], and LysM deletion mice leads to increased susceptibility to Klebsiella pneumoniae [7].
Lysozyme M is expressed by myeloid cells, and its promoter has been utilized to direct Cre recombinase to target deletion of floxed genes in these cells. LysMcre mice have a nuclear-localized Cre recombinase inserted into the first coding ATG of the lysozyme M gene, abolishing endogenous LyzM gene function, and placing the Cre recombinase under the control of the LyzM promoter [8]. When crossed with reporter mice (ROSA-EYFP) to characterize efficiency and specificity, use of this strain targets monocytes, neutrophils, and macrophages [9]. In addition to myeloid cells, a portion of type II alveolar epithelial cells also express Cre recombinase [10].
Given the growing interest of assessing myeloid-specific genes in acute lung injury, LysMcre mice are frequently employed in these models [11, 12]. However, there is an under appreciation of loss of LysM expression in homozygous LysMcre mice, with occasional lack of reporting of the zygosity for LysMcre alleles or use of appropriate controls to account for LysM contribution. In this study, we tested the contribution of lysozyme M in LPS-induced lung injury. Mice homozygous for the LysMcre allele (and thus, LysM deficient) and littermate wild-type controls were challenged with oropharygeal aspiration of E. coli LPS and assessed at days 2 and 4 for BAL cell count, differential, protein, and cytokines. We observed similar changes in total body weight in response to LPS and a similar neutrophilic predominant inflammatory influx at days 2 and 4. These findings indicate that deletion of LysM and expression of Cre-recombinase does not modulate the inflammatory response to LPS-induced ALI.
Materials and Methods:
Animals.
LysMcre mice (B6.129P2-Lyz2tm1(cre)Ifo/J) were obtained from Jackson Laboratory (Bar Harbor, ME) and crossed with C57BL/6J mice. Heterozygous LysMcre (+/−) mice from these crosses were established as breeders. Offspring were genotyped, and homozygous Cre+/+ mice and Cre−/− mice were used for these studies. Mice were age- and gender-matched, and LPS doses (1.5 μg/g) were administered by oropharygeal aspiration as described [13]. Mice were euthanized at days 2 and 4. BAL and cytospins were performed as described [14]. The University of Washington Office of Animal Welfare approved all animal protocols.
Genotyping:
The following primers were used to detect Cre recombinase or wildtype alleles: Cre 5’ CCC AGA AAT GCC AGA TTA CG , Common 5’ CTT GGG CTG CCA GAA TTT CTC, and Wild type 5’ TTA CAG TCG GCC AGG CTG AC.
Broncho-alveolar lavage:
In euthanized mice, the trachea was cannulated with an angiocatheter. Broncho-alveolar lavage was performed with three serial instillations of lavage buffer (PBS, 0.5% EDTA) (total volume 2.5 ml). BAL cells were pelleted and re-suspended in RPMI + 10% FBS, and approximately 50,000 cells placed on slides using a cytospin. Cells were stained with Differential quik (VWR, Radnor, PA), and a manual differential was performed on 100 cells.
Lung Histolopathology:
Lungs were inflated and fixed with 10% formalin, dehydrated with serial dilutions of ethanol, and embedded in paraffin. Lung sections (5 μm) were stained with hematoxylin and eosin. Lungs were scored on a 5-point scale (0=no abnormality; 1= minimal inflammation involving <10% of lung; 2= mild involving 10-25%; 3=moderate involving 26-50%; 4=moderate-severe involving 51-75%; 5= severe involving >75%).
Quantitative RT-PCR (qRT-PCR):
Total RNA from cells was isolated using RNeasy Mini kit (Qiagen, Valencia, CA). The quantity and quality of RNA were determined using a NanoDrop spectrophotometer (NanoDrop Inc., Wilmington DE). Primers and TaqMan probes (FAM dye-labeled) for Lyz2 (Mm01612741; ThermoFisher Scientific, Waltham, MA) and Hprt were added to cDNA synthesized from total RNA with a High-Capacity cDNA Archive kit (Applied Biosystems, Carlsbad, CA). Product amplification was measured with an ABI HT7900 Fast real-time PCR system (Applied Biosystems). The threshold cycle (Ct) was obtained from duplicate samples and averaged. The ΔCt was the difference between the average Ct for the target gene and the housekeeping gene, Hprt. The ΔΔCt was the average ΔCt for a given sample point minus the average ΔCt of control samples; relative quantification was calculated as 2−ΔΔCt.
Cytokine Measurements:
Cytokines and chemokines concentrations for CXCL1/KC, and CCL2/MCP-1 were measured in broncho-alveolar lavage (BAL) samples using ELISA per manufacture instructions (R&D Systems, Minneapolis, MN) and analyzed using a Synergy 4 plate reader (BioTeck, Winooski, VT).
Statistics:
Results are expressed as means ± SEM. Statistical significance was determined using one-way ANOVA with multiple comparisons. Differences were considered significant if the P-value was <0.05.
Results:
We generated and bred LysMcre mice heterozygous for the Cre allele (+/−). Littermates homozygous for the Cre allele (Cre+/+) or wildtype allele (Cre−/−) were identified by genotyping (Fig 1A). Marked reduction of the LysM gene product was confirmed by qPCR using RNA isolated from broncho-alveolar (BAL) macrophages (Fig 1B).
Figure 1. Loss of LysM expression in Cre+/+ mice.
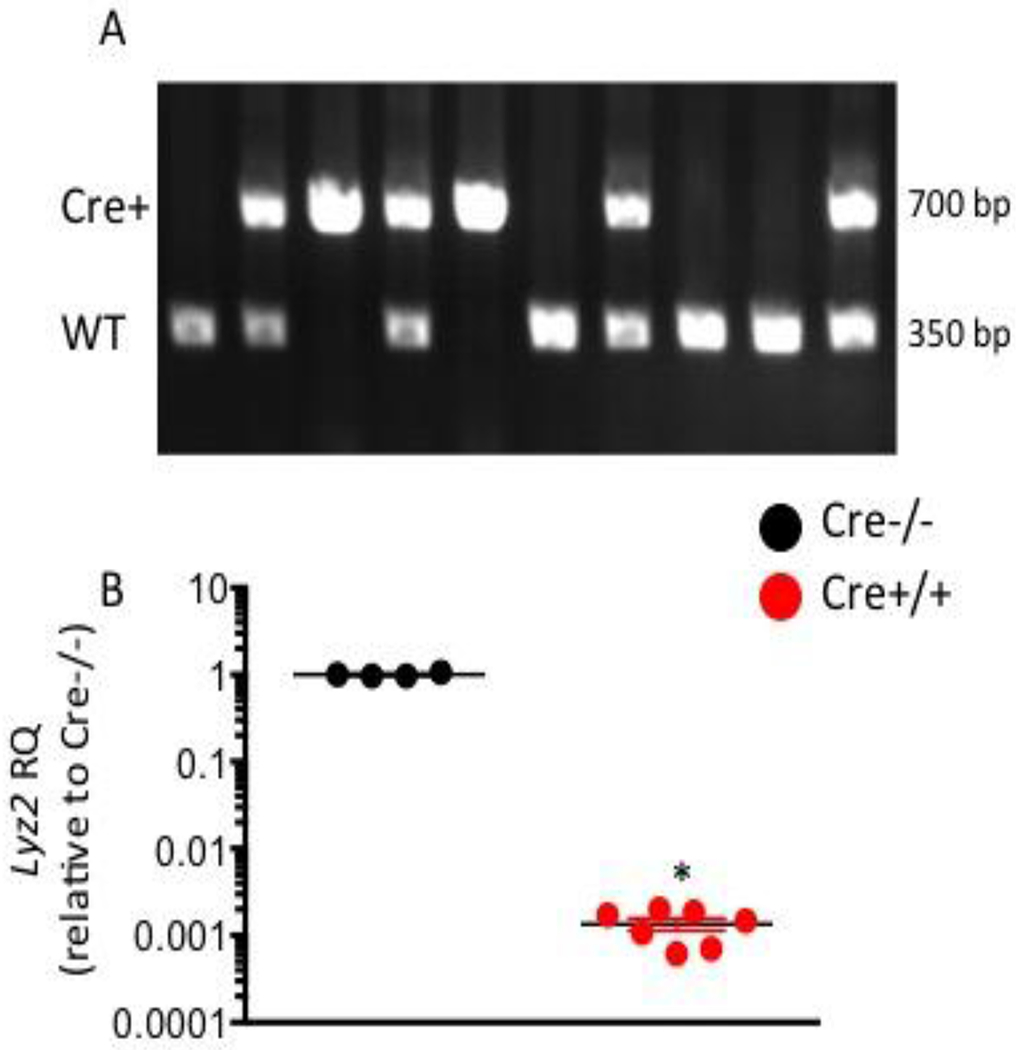
(A) Representative PCR demonstrating amplification of the Cre gene (700 bp band) or Wild-type allele (350 bp band) used to identify Cre+/+ (single 700 bp band) or Cre−/− (single 350 bp band) mice. (B) RT-PCR for LysM (Lyz2) mRNA expression. Results shown as relative quantification (RQ) to the average of Cre−/− samples (black circle). * p value < 0.05
Next, Cre+/+ and Cre−/− mice were challenged with E. coli LPS and assessed daily for changes in their body weight, a surrogate marker for illness severity induced by LPS. The absence of lysozyme M did not alter the initial weight loss, greatest at day 2, or recovery of weights seen at days 3-4 (Fig 2A). Mice were euthanized at days 2 and 4 for broncho-alveolar lavage, cell count and differential. At day 2, both genotypes had similar LPS-induced increases in BAL cell count (Fig 2B) and a neutrophil-predominant influx into the alveolar compartment (Fig 2C). By day 4, BAL cell counts and BAL PMNs were significantly reduced between days 2 and 4, with no difference by genotype (Fig 2B,C). BAL macrophages were also similar in LPS-treated groups (Fig 2D) across genotypes and time points, with a trend for an increase after injury. We assessed lung histopathology to assess degree and localization of inflammatory influx. There was predominant alveolar and perivascular inflammation with similar extent of patchy lung involvement in LPS-injured groups (Fig 2E) with a similar assessment of inflammation based on scoring: 2.9±0.2 vs. 3.1±0.3 D2 Cre+/+ vs. D2 Cre−/− p value=0.53; and 3.0±0.3 vs. 2.8±0.5 D4 Cre+/+ vs. D4 Cre−/− p value=0.64.
Figure 2. LPS-induced ALI.
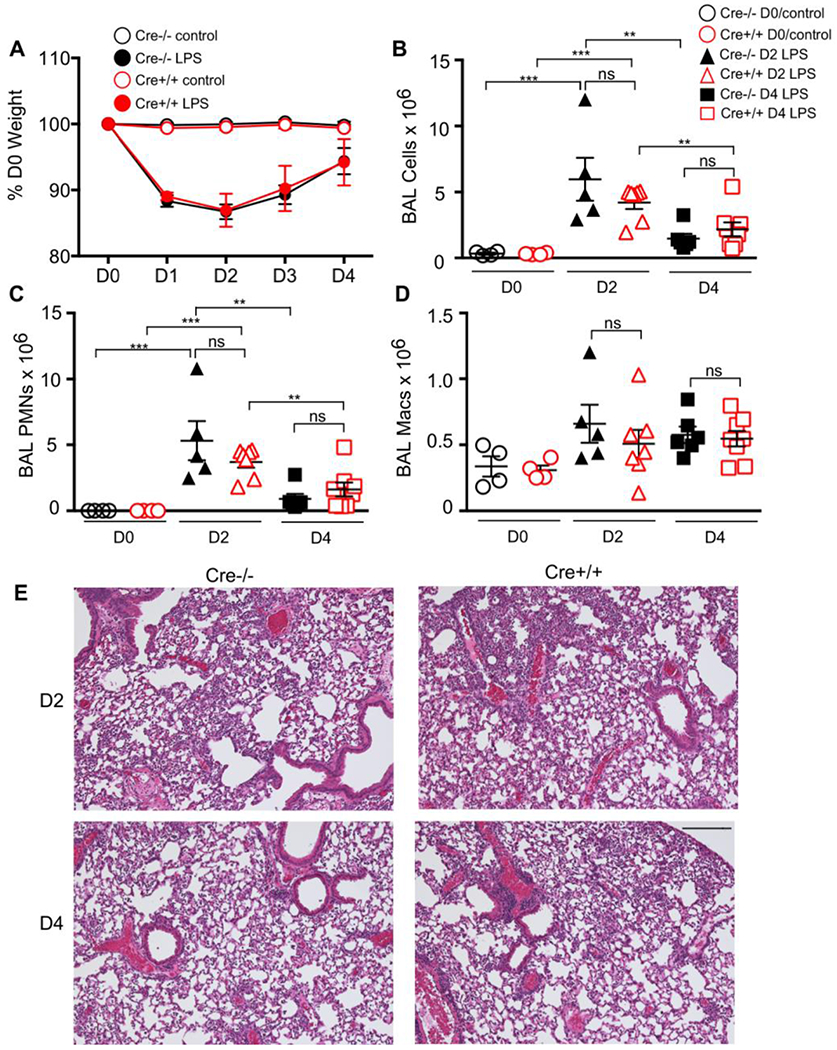
LysMCre+/+ or –LysMCre−/− mice received LPS via oropharygneal aspiration or were unchallenged (control). (A) Mice were weighed daily. There was a similar decrease in weight in both LPS groups, independent of genotype with recovery of weights between days 3-4. Broncho-alveolar lavage was performed on days 2 and 4 for (B) total cell counts, (C) neutrophil (PMN) cell count, (D) alveolar macrophage (Macs) cell count. There was no difference in BAL cell counts, neutrophils, and macrophages between LPS-treated groups matched for time-point. (E) Representative lung histopathology of D2 (top) and D4 (bottom) post-LPS. Scale bar= 200 μm. *** p value < 0.0001; ** p value <0.001, ns=non-significant
BAL protein, a marker of vascular injury was increased at day 2 compared to day 0, with a similar response between LPS-treated genotypes (Fig 2E). We assessed for differences in inflammatory chemokines, CXCL1, and CCL2. At day 2, both chemokines were increased in LPS-treated groups compared to control, with no difference in responses between genotypes. By day 4, the chemokine levels were significantly reduced or below the level of detection in both genotypes.
Discussion:
Use of LysMCre-recombinase mice to target myeloid cell-depletion of floxed genes is commonly employed. However, deletion of LysM gene and its protein product is a confounder when homozygous mice are generated. To develop an understanding of how often the genetics and use of appropriate controls for LysMCre mice are reported in manuscripts, we examined 12 articles identified using PubMed and the search string, “LysM and Cre and lung.” Upon review of these manuscripts, we found that a majority (7/12) did not directly report LysM zygosity and/or use appropriate controls to account for the loss of the lysozyme 2 gene. Based on the ARRIVE guidelines, it is recommended that authors provide information about the experimental animals used in a study including mouse strain, sex, weight, and genetic modification status [15]. In particular, because the targeted insertion of LysM Cre results in the loss of lysozyme 2 gene, it is important to report the zygosity of these mice.
In infectious models, the role of LysM in controlling infection and inflammation is well-established [7]; however, in sterile models of ALI, the contribution of LysM has not been reported. Given the broad use of these mice to model myeloid contribution in lung injury, we sought to test if the LysM gene is contributory to LPS-induced acute lung injury. We generated LysCre+/− mice and studied littermates that were homozygous for the wildtype (Cre−/−) or Cre (Cre+/+) allele. Using commercially available primer probe sets for Lyz2 mRNA, there was a large reduction in mRNA detected. However, there was still detectable signal from LysM-deleted mice possibly a result of detection of its isoform. Importantly, we found that LysM was non-contributory to the inflammatory phenotype of LPS-treated mice as both genotypes had similar inflammatory cell recruitment to the lung, protein leak, and cytokine expression.
Limitations of this study are the assessment of inflammation in a single model of ALI, limiting the generalizability to other models of lung injury. However, recognition of this potential confounder is important in future experimental design in other injury models as is the reporting of zygosity of the LysM allele. Hence, addition of controls using Cre+/+ mice that lack the floxed gene or use of mice with a single copy of the Cre transgene to assure functionality of at least one allele of the LysM gene are necessary.
Figure 3.
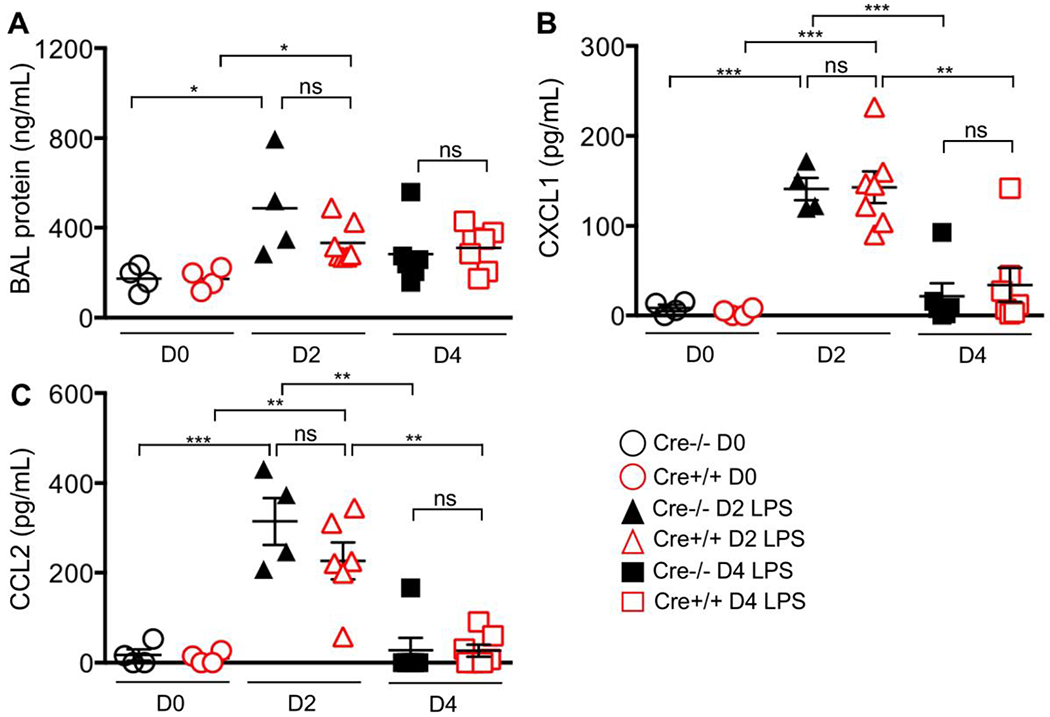
BAL protein and chemokine response. LysMCre+/+ or –LysMCre−/− mice received LPS via oropharygneal aspiration or were unchallenged (control). Broncho-alveolar lavage was performed on days 2 and 4 for (A) total protein, (B) CXCL1, and (C) CCL2 concentration. There was no difference in these measurements between LPS-treated groups matched for time-point. *** p value < 0.0001; ** p value <0.001, *p value <0.05, ns=non-significant
Acknowledgments
This work was supported by R01 HL116514 (AMM), P30-ES-007033-19-6363 (AMM)
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of Interest: None
References:
- 1.Cross M, Mangelsdorf I, Wedel A, Renkawitz R. Mouse lysozyme M gene: isolation, characterization, and expression studies. Proc Natl Acad Sci U S A 1988;85: 6232–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishiguro K, Yoshie N, Sakurai M, Inoue Y. A 1H NMR study of a fragment of partially n-acetylated chitin produced by lysozyme degradation. Carbohydr Res 1992;237: 333–338. [DOI] [PubMed] [Google Scholar]
- 3.Pangburn SH, Trescony PV, Heller J. Lysozyme degradation of partially deacetylated chitin, its films and hydrogels. Biomaterials 1982;3: 105–108. [DOI] [PubMed] [Google Scholar]
- 4.Ibrahim HR, Matsuzaki T, Aoki T. Genetic evidence that antibacterial activity of lysozyme is independent of its catalytic function. FEBS Lett 2001;506: 27–32. [DOI] [PubMed] [Google Scholar]
- 5.LeMarbre P, Rinehart JJ, Kay NE, Vesella R, Jacob HS. Lysozyme enhances monocyte-mediated tumoricidal activity: a potential amplifying mechanism of tumor killing. Blood 1981;58: 994–999. [PubMed] [Google Scholar]
- 6.Akinbi HT, Epaud R, Bhatt H, Weaver TE. Bacterial killing is enhanced by expression of lysozyme in the lungs of transgenic mice. J Immunol 2000;165: 5760–5766. [DOI] [PubMed] [Google Scholar]
- 7.Markart P, Korfhagen TR, Weaver TE, Akinbi HT. Mouse lysozyme M is important in pulmonary host defense against Klebsiella pneumoniae infection. Am J Respir Crit Care Med 2004;169: 454–458. [DOI] [PubMed] [Google Scholar]
- 8.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 1999;8: 265–277. [DOI] [PubMed] [Google Scholar]
- 9.Abram CL, Roberge GL, Hu Y, Lowell CA. Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J Immunol Methods 2014;408: 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCubbrey AL, Allison KC, Lee-Sherick AB, Jakubzick CV, Janssen WJ. Promoter Specificity and Efficacy in Conditional and Inducible Transgenic Targeting of Lung Macrophages. Front Immunol 2017;8: 1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang Z, Chen Z, Li L, Zhou W, Zhu L. Lack of SOCS3 increases LPS-induced murine acute lung injury through modulation of Ly6C(+) macrophages. Respir Res 2017;18: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia H, Ren X, Bolte CS, Ustiyan V, Zhang Y, Shah TA, Kalin TV, Whitsett JA, Kalinichenko VV. Foxm1 regulates resolution of hyperoxic lung injury in newborns. Am J Respir Cell Mol Biol 2015;52: 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy WE, Gong KQ, Bell B, Parks WC, Ziegler SF, Manicone AM. Stat5 Is Required for CD103(+) Dendritic Cell and Alveolar Macrophage Development and Protection from Lung Injury. J Immunol 2017;198: 4813–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manicone AM, Gharib SA, Gong KQ, Eddy WE, Long ME, Frevert CW, Altemeier WA, Parks WC, Houghton AM. Matrix Metalloproteinase-28 Is a Key Contributor to Emphysema Pathogenesis. Am J Pathol 2017;187: 1288–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG, Group NCRRGW. Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol 2010;160: 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]


