Abstract
Cell plasticity is important in development and tissue remodeling. Cells can sense physical and chemical cues from their local microenvironment and transduce the signals into the nucleus to regulate the epigenetic state and gene expression, resulting in a change in cell phenotype. In this review, we highlight the role of mechanical cues in regulating stem cell differentiation and cell reprogramming through the modulation of histone modifications. The effects of various mechanical cues, including matrix stiffness, mechanical stretch, and shear stress, on cell fate during tissue regeneration and remodeling will be discussed. Taken together, recent work demonstrates that the alterations in histone modifications by mechanical stimuli can facilitate epigenetic changes during cell phenotypic switching, which has potential applications in the development of biomaterials and bioreactors for cell engineering.
Keywords: Matrix stiffness, mechanical stretch, Shear stress, Histone modification, Cell plasticity
Introduction
Cells exhibit remarkable plasticity to mediate tissue regeneration or restoration of homeostasis after injury. Cellular plasticity includes the interconversion of different cells by reprogramming, stem cell differentiation, transdifferentiation, dedifferentiation, and phenotypic transition of differentiated cells [1]. These changes in cell fate can be regulated by a variety of microenvironmental factors or induced by forced expression of transcriptional factors [2]. In addition to biochemical cues, mechanical stimuli such as substrate stiffness, mechanical stretch and fluid shear stress not only elicit immediate responses in cells but also induce long-term changes in epigenetic state and cell phenotype through biophysical and biochemical modulation of the nucleus and chromatin (Figure 1) [3–5]. Highly ordered chromatin structures, consisting of densely packed heterochromatin and less condensed euchromatin regions, can be regulated by several epigenetic modifications on the amino acid residues of histones, including acetylation, methylation, phosphorylation, ADP-ribosylation, sumoylation and ubiquitination [6,7]. The acetylation of histones generally facilitates chromatin decondensation and gene activation [8]. Methylation of histones at lysine or arginine may induce either a more open or closed chromatin structure, depending on the site of methylation [9]. In addition, histone phosphorylation regulates chromosome condensation and gene activation [10]. Other histone modifications such as ADP-ribosylation and sumoylation can activate and repress gene activation, respectively, and histone ubiquitination is typically involved in chromatin dynamics, transcriptional regulation and DNA repair [11–13]. Furthermore, DNA methylation results in transcription silencing [14]. Altogether, these epigenetic modifications on histones and DNA can alter the on-and-off state of genes, as summarized in Box 1. In this review, we will focus on how mechanical factors such as matrix stiffness, mechanical stretch and shear stress modulate histone modifications to regulate cell plasticity.
Figure 1. Mechanical cues regulate histone and DNA modifications to modulate cell fate.
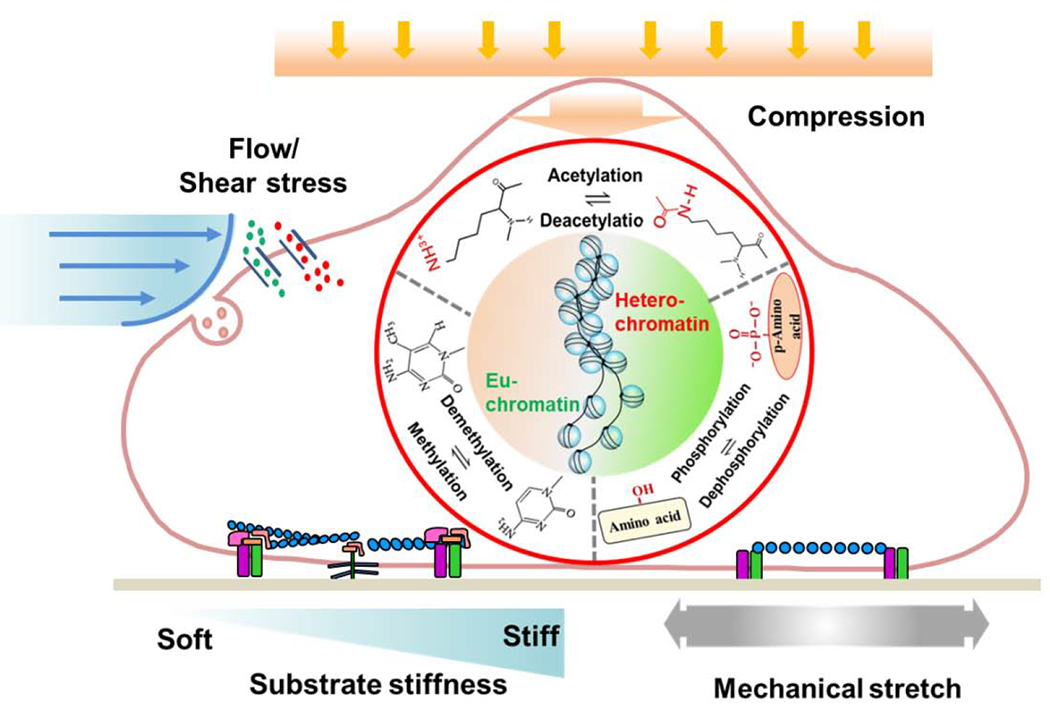
Various mechanical stimuli, including substrate stiffness, mechanical stretch, shear stress and compression, modulate histone and DNA modifications, which regulates chromatin reorganization and cell plasticity.
Box 1. Histone and DNA Modifications.
Acetylation transfers acetyl groups from acetyl coenzyme-A to lysine residues of histones by acetyltransferase, which is typically catalyzed by the activity of enzymes including histone acetyltransferase (HAT) and histone deacetylase (HDAC). Positive charges on the histones are removed by acetylation, and hence this reduces the binding between the N-terminal of histones and the negatively charged phosphate groups of DNA. As a result, the condensed chromatin is transformed into a relaxed structure, making it more accessible to the transcriptional machinery.
Methylation adds methyl (CH3) groups to lysine and arginine residues of histones to promote a chromatin structure that is either open (euchromatin) or more compacted. DNA methylation involves the covalent addition of a methyl group to cytosine or adenine. These methyl groups bind into the major grooves of the DNA, inhibit gene transcription, and maintain DNA integrity and stability.
Phosphorylation of histones occurs on serine, threonine, and tyrosine amino acids and are involved in chromosome condensation and gene activation.
ADP-ribosylation adds one or more ADP-ribose moieties to histone proteins, facilitating gene transcription and DNA repair.
Sumoylation covalently links a small ubiquitin-like modifier (SUMO) moiety to a lysine residue on histones, which is often associated with transcriptional repression.
Ubiquitination binds ubiquitin to a lysine residue on histones and regulates chromatin dynamics, transcriptional regulation and DNA repair.
Regulation by Stiffness of Cell Adhesion Substrates
In tissues and organs, the stiffness of the extracellular matrix (ECM) changes dynamically. Cells, either local or recruited to the site, can sense and respond to the matrix stiffness stimuli by altering their cell behavior, as is evident during cancer cell migration [15], macrophage polarization [16] and stem cell differentiation [17]. Recent studies have shown that ECM stiffness plays an important role in stem cell differentiation and cell reprogramming through the modulation of histone modifications and DNA methylation (Table 1) [18].
Table 1.
Summary of the effects of matrix stiffness on histone modifications and cell phenotype
| Stiffness Tested | Epigenetic Modification | Cell Type | Function | Ref. |
|---|---|---|---|---|
| 0.4-80 kPa | H3K9ac, H3K4me3, H3K27me3 | liver stem cells | Soft substrates increased H3K9ac and decreased H3K4me3, H3K27me3 | [19] |
| 1.5-15 kPa | H4K16ac | MSCs | Soft substrates enhanced MSC reprogramming into iPSCs | [20] |
| 1kPa-plastic | HAT-1, HDAC-1 | Fibroblast | Increase HAT-1, decrease DAC-1 | [22] |
| 0.4-25.6 kPa | H3K27ac | HSCs | Stiff matrices increased H3K27AC | [23] |
| 5.5-32.7 kPa | Histone acetylation | hMSCs | Histone acetylation increased on stiff gels and sustained for days after softening | [24] |
| 0.5-100 kPa | Histone deacetylation | MEFs | HDAC4 expression increased | [28] |
| 0.4-25.6 kPa | H3K4me3 | DLD-1 | Reduction of H3K4me3 on softer matrices | [25] |
| 1-50 kPa | H3K9me2/3 | Fibroblast | Stiff matrices increased global H3K9me2/3 | [26] |
| 60-400 kPa | Histone H3 phosphorylation | MSCs | Soft matrix enhanced Histone H3 phosphorylation | [27] |
| 1kPa-plastic | DNA methylation | Fibroblast | Significant decrease in DNA methylation on soft matrices | [22] |
| 1-20 kPa | DNA demethylation | A549 | DNA demethylation under stiff matrix conditions | [25] |
| 2.16-16.75 kPa | DNA methylation | HUASMCs | Substrate stiffening downregulated DNMT1 and DNA methylation | [28] |
MSCs: Mesenchymal stem cells; iPSCs: Induced pluripotent stem cells; HSCs: Hepatic stellate cells; hMSCs: Human mesenchymal stem cells; MCF10A: Mammary epithelial cells; A549: Human alveolar epithelial adenocarcinoma cells; DLD-1: Colorectal adenocarcinoma cells; MEFs: Mouse embryonic fibroblasts; HUASMCs: Human umbilical artery SMCs; B16-F1: Murine melanoma cells.
Soft matrices (less than 2 kPa) promote global histone acetylation to facilitate cell lineage transitions. For example, during the differentiation of resident liver stem cells into hepatocytes, soft substrates increased the acetylation of histone H3 at lysine 9 (H3K9Ac, known to be an active chromatin modification) and decreased the trimethylation of histone H3 at lysine 4 (H3K4me3) and lysine 27 (H3K27me3); these early epigenetic modifications at the HNF4α promoter enhanced differentiation [19]. The reprogramming of mesenchymal stem cells (MSCs) into induced pluripotent stem cells (iPSCs) occurred faster and more efficiently on a soft matrix, as a result of inducing cytoskeletal and nuclear changes that enhanced histone acetylation, more specifically H4K16Ac [20]. Similar results were found during the reprogramming of fibroblasts into iPSCs, which was attributed to possible suppression of stress fiber formation in cells cultured on soft substrates [21]. One possible mechanism is that soft matrices can promote global acetylation by increasing the expression of histone acetyltransferase 1 (HAT1) while, at the same time, decreasing the expression of histone deacetylase 1 (HDAC-1), as has been shown during the conversion of dermal fibroblasts into insulin-producing cells [22]. However, it has also been reported that human MSCs cultured on stiff surfaces (i.e. higher than 25 kPa) had an elevated expression of HAT1 and a lower expression of HDACs 1, 2, and 3, thereby inducing the nuclear localization of Yes-associated protein (YAP), a mechanosensitive transcription factor. In hepatic stellate cells, stiff surfaces activated p300, a histone acetyltransferase, and p300-dependent acetylation via H3K27ac [23]. Whether this discrepancy of HAT1/HDAC expression and histone acetylation represents a cell type-dependent response to stiffness requires further investigation.
Another interesting question is whether stiffness-induced histone modifications is sustained or easily reversible by dynamic changes in stiffness. It appeared that stiff surface-induced HAT1 expression in MSCs remained high to maintain histone acetylation levels for days. In addition, following one day of culture, softening the substrate from 32.7 kPa to 5.5 kPa resulted in a gradual decrease in histone acetylation, but the overall level was still higher than soft surfaces after five days [24]. It is likely that both the starting stiffness and the length of exposure affect histone modifications and thus, cell phenotype changes, and it is worthwhile to determine the minimum time period required to induce long-lasting histone modifications under different scenarios.
Besides histone acetylation, histone methylation at specific sites may also be regulated by substrate stiffness. For example, colorectal adenocarcinoma cells on a 0.4 kPa soft matrix had much lower H3K4me3 levels compared to those on a stiff surface [25], and human fibroblasts on stiff matrices (> 50 kPa) had higher levels of H3K9me2/3 [26]. In addition, soft matrices (60 kPa) significantly enhanced the phosphorylation of H3 histone to enhance the mesoderm differentiation of human embryonic stem cells, in comparison to stiff matrices (400 kPa) [27]. Despite the evidence that stiffness modulates histone acetylation and methylation, currently, there is still limited information on how stiffness regulates histone ubiquitination, ADP-ribosylation and sumoylation.
Apart from histone modifications, there is also evidence to suggest that DNA methylation is regulated by matrix stiffness. For example, both umbilical artery smooth muscle cells (SMCs) and alveolar epithelial adenocarcinoma cells on a matrix around 20kPa had lower DNA methylation levels than a soft matrix [25,28]. In contrast, fibroblasts on a soft matrix (1 kPa) had significantly lower DNA methylation levels than those on a stiff plastic surface [22]. Further investigation into the effects of stiffness on DNA methylation will aid to reveal whether stiffness regulation of DNA methylation is cell-type dependent.
Effects of Mechanical Strain
Mechanical strain caused by the deformation of the ECM, either a static or repetitive stretch/compression, can be sensed by cells in various tissues, such as fibroblasts in ligaments [29], neuroepithelial cells in the lung [30], muscle cells in the heart and blood vessel, and resident stem cells in the dental pulp [31]. This biophysical factor has also been shown to influence cell differentiation and reprogramming by regulating the epigenetic state (Table 2).
Table 2.
Summary of the effects of mechanical stretch on histone modifications and cell phenotype
| Stretch Parameters | Epigenetic modification | Cell type | Function | Ref. |
|---|---|---|---|---|
| static uniaxial 20% strain | Decrease HDAC activity | MSCs | Promote histone acetylation | [32] |
| static biaxial 10-15% strain | HDAC11, AcH3K14 | OPC | Promoted OPC differentiation | [33] |
| static uniaxial 10-33% strain | Histone H3P | Epithelial cell | Promoted cell proliferation | [34] |
| 0.5Hz, biaxial 10% strain | HDAC1, H3Ac | BMSC | Promoted BMSC differentiation into osteoblasts | [35] |
| 1Hz, biaxial 10% strain | miR-365 downregulated HDAC4 | rBMSC | Promoted chondrogenesis, upregulated ANCN and Sox9 expression | [36] |
| 0.5Hz, biaxial 10% strain | miR-365 reduced HDAC4 | Human Chondrocyte | Cartilage degeneration, downregulated ACAN | [37] |
| 0.5Hz, biaxial 5% strain | Histone 3 dephosphorylation | hASC | Impaired adipogenic differentiation of hASC | [38] |
| 0.5Hz, biaxial 10% strain | Dnmt3b, DNA demethylation | mBMSC | Promoted osteoblastogenesis | [39] |
| 1Hz, biaxial 2.5-15% strain | DNA demethylation | hAT-MSC | GNAS CpG demethylation | [40] |
| 0.5Hz, biaxial 10% strain | KDM4B, H3K9me3 | MSC | Promoted MSC differentiation into fibroblasts | [41] |
| 0.1Hz, biaxial 10% strain | Switched H3K9me2,3 to H3K27me3 | EPC | Repressed EPC differentiation and downregulated gene expression | [42] |
MSC: Mesenchymal stem cells; OPC: Oligodendrocyte progenitor cells; hAT-MSC: Human adipose tissue multipotential stromal cell; BMSC: Bone marrow-derived mesenchymal stem cells/Bone marrow stromal cell; EPC: Epidermal progenitor cells; hAC: Human articular chondrocyte; hASC: Human adipose-derived stem cells.
An early study demonstrated that elongated MSCs had lower HDAC activity and that a static compression or stretch perpendicular to the long axis of the cells further deceased HDAC activity [32]. However, in oligodendrocyte progenitor cells (OPC), biaxial static stretch caused an increase in HDAC11 expression and the global deacetylation of H3K14Ac, an active promoter state, which promoted oligodendrogenesis [33]. It remains to be determined whether uniaxial and biaxial stretch have differential effects and whether these are cell type-dependent responses. In addition, there is evidence that uniaxial static stretch can regulate histone phosphorylation. For instance, histone 3 phosphorylation (H3P) can relax chromatin structure, thereby promoting cell division and cell differentiation, and static strain (10-33% elongation) induced H3P by activating the Piezo1 ion channel [34].
Cyclic stretch also decreased HDAC1 level in bone marrow MSCs (BMSCs) to modulate the histone acetylation of JAG1 promoter, thereby inducing the differentiation of BMSCs into osteoblasts [35]. It appears that microRNAs may mediate biaxial stretch-regulation of HDAC expression. For example, cyclic strain induced miR-365 to suppress HDAC4 gene expression in rat MSCs to facilitate chondrogenic differentiation [36,37]. In contrast to static stretch, biaxial cyclic stretching of human adipose-derived stem cells (hASC) resulted in decreased H3P, repressed FABP4 expression and impaired adipogenic differentiation [38]. These opposing results to those of static stretch may be due to the differences in the stretch mode (static and cyclic), axial direction (uniaxial and biaxial) and even the magnitude of strain, and therefore, should be further studied.
Cyclic stretch has also been reported to regulate the methylation of histones and DNA to enhance cell differentiation. During osteogenic differentiation of both mouse BMSCs and human adipose tissue multipotential stromal cell (hAT-MSC), biaxial cyclic stretch reduced DNA methyltransferase 3b (DNMT3B) expression and enhanced DNA demethylation on CpG islands to promote osteogenesis [39,40]. Additionally, biaxial cyclic stretching of BMSCs downregulated H3K9me3 to promote BMSC differentiation into fibroblasts [41]. In epidermal progenitor cells (EPCs), biaxial cyclic stretch induced the enrichment of Emd, a mechanosensory complex of emerin, at the outer nuclear membrane, leading to a switch from a permanent repression signal (H3K9me3) to a temporary repression signal (H3K27me3) occupancy at constitutive heterochromatin, which directly affected the precocious lineage commitment [42].
Shear Stress Effects
To date, the effects of shear stress stimulation have been mainly studied on endothelial cells (ECs) of the blood vessel as hemodynamic forces play an important role in the homeostasis and remodeling of vascular wall. Different modes of shear stress have been shown to regulate histone modification and cell plasticity (Table 3).
Table 3.
Examples of shear stress-regulated histone modifications in cell phenotype
| Stimulation | Histone/Gene | Cell type | Function | Ref. |
|---|---|---|---|---|
| Steady interstitial flow | HDAC1 phosphorylation | ECs | Increased morphogenesis, MMP14, and angiogenesis | [43] |
| 5 dyne/cm2 steady shear stress | H3K4me3, H3K27ac activation | HUVECs | Increased thrombomodulin expression | [44] |
| Steady shear flow | H2B acetylation | hESCs | Decondensation consolidated the primed state of hESCs | [45] |
| 12±4 dyne/cm2 pulsatile shear stress | H3K27 acetylation | HUVECs | Ca2+-dependent eNOS activation, EC homeostasis | [47] |
| 20 dyne/cm2 steady laminar shear stress | Decrease EZH2 | ECs | Deceased cell cycle and promoted quiescence | [48] |
| 12 dyne/cm2 steady laminar flow | Reduces H3K27me3 | HUVECs | Conferred an anti-inflammatory response | [49] |
| 15-30 dyne/cm2 steady shear stress | Increase DNMT1 | HUVECs | Exhibited DNA hypermethylation | [50] |
ECs: Endothelial cells; ESCs: Embryonic stem cells; HUVECs: Human umbilical vein cells.
During angiogenesis, low shear stress (less than 5 dyne/cm2) increased matrix metalloproteinase-14 (MMP14) expression through the phosphorylation of HDAC1 in ECs, and induced thrombomodulin expression by increasing the levels of H3K4me3 and H3K27ac, which enabled EC growth and migration for angiogenesis [43,44]. In addition, steady shear flow is able to induce H2B acetylation in hESC by CFL2/F-actin cytoskeletal reorganization to consolidate the primed state of cells [45].
High shear stress (HSS) (≥10 dyne/cm2), either steady or pulsatile, has profound effects on vascular remodeling [46]. Pulsatile shear stress (PS) is thought to maintain cell homeostasis and protect blood vessels from atherosclerosis. For instance, PS enhanced H3K27ac through KLF4, which contributed to Ca2+-dependent activation of endothelial nitric oxide synthase (eNOS) and EC homeostasis [47]. In addition, HSS can regulate the expression of the polycomb methyltransferase EZH2. Steady laminar shear stress at 20 dynes/cm2 decreased the expression of EZH2 and a repressive epigenetic mark H3K27me3, leading to a suppression of endothelial inflammatory factors [48,49].
HSS also can regulate cell phenotype through DNA methylation. DNMT1 has been shown to regulate arterial vascular remodeling and arteriogenic capacity. In a femoral artery ligation model, collateral artery segments exposed to increased steady shear stress exhibited increased DNMT1 expression and a global DNA hypermethylation, accompanied by a DNMT1-dependent reduction in proatherogenic monocyte adhesion [50].
Conclusions & Future Directions
Dynamic changes in the tissue microenvironment provide mechanical cues that continuously stimulate local cell populations via mechanotransduction pathways and can alter their fate and function through the modulation of the epigenetic state. Recent work improved our understanding of the epigenetic mechanisms in response to mechanical stimuli. This emerging area of physical genomics will have broad impact on the development of novel materials and new approaches for tissue repair and disease therapies.
In this review, substrate stiffness, mechanical strain and shear stress were selected to exemplify the mechanical regulation of epigenetic state. It is worth noting that many other mechanical stimuli, although not widely studied, also have effects on epigenetic modifications. For example, cyclic hydrostatic pressure activates HDAC4 in osteoarthritis chondrocytes [51]; persistent pulmonary hypertension decreases H4K12ac and increases H3K9me3, which downregulates eNOS in pulmonary artery ECs [52]; and when a T cell passes through a constriction space, the resulting nuclear deformation enhances H3K4me3 [53]. Although there are cumulative findings on the mechanical regulation of epigenetic state, further studies are needed to integrate the information and put together different pieces of the puzzle.
First, how do these mechanical factors, alone or in combination, regulate global and site-specific epigenetic changes? While in-depth studies on certain genes and pathways will provide mechanistic insight, genome-wide analysis such as chromatin accessibility and DNA methylation will help us profile epigenetic changes and have the whole picture of chromatin remodeling. Secondly, are the effects of mechanical factors on histone/DNA modifications dependent on cell type or cell-state? The expression of receptors and signaling molecules, especially those involved in mechanotransduction, may vary in different cells, which can result in variations in epigenetic modifications. In addition, the different epigenetic landscape in euchromatin and heterochromatin in different cell types will also affect the remodeling of chromatin. Single cell analysis is needed to elucidate the heterogeneous responses to mechanical factors. Thirdly, are there any common mechanotransduction mechanisms that account for the various epigenetic changes? For example, the mechanical structure of cells such as focal adhesions, cytoskeleton and nuclear matrix, some of the transcriptional factors, and biophysical properties of the nucleus such as shape, size and deformation may have similar responses to various mechanical stimuli in different cell types. Furthermore, it is important to elucidate how mechanical factors regulate the translocation of transcriptional factors and chromatin modifying enzymes between cytosol and nucleus. For example, there is evidence that force can stretch the nuclear pores to facilitate the transport of YAP into nucleus [54]. These mechanotransduction mechanisms may control a variety of epigenetic modifications and cell functions [55–57], and the understanding of these universal mechanisms will help formulate an overarching theory in physical genomics.
The advancement in biotechnology offers many start-of-art tools to dissect mechanical regulation of epigenetic state. High-throughput genomic analysis such as single cell sequencing and Transposase-Accessible Chromatin using sequencing (ATAC-seq) can be applied to better understand the mechanism of mechanical factor-induced epigenetic and transcriptome changes in various biological processes, including cell reprogramming where conversion efficiencies are relatively low. Additionally, live cell imaging in combination with various biosensor and transgenic cell lines enables real-time tracking of the temporal and spatial modifications of histones and DNA. For example, fluorescence resonance energy transfer (FRET) with specific sensors can detect histone modifications and chromatin reorganization at single cell level [58]. Furthermore, the application of such tools can provide new insights into the mechanisms that regulate the cytosolic to nuclear shuttling of chromatin modifying enzymes and mechanoresponsive transcription factors to facilitate mechanosignaling [56,59,60].
Besides stem cell differentiation and cell reprogramming, mechanical regulation of phenotypic changes of various cells has important implications in tissue remodeling and disease development. For example, a stiff matrix primes macrophage towards a pro-inflammatory phenotype with impaired phagocytosis, while a soft matrix primes cells towards an anti-inflammatory, highly phagocytic phenotype [61]. In addition, an increase in the substrate elastic modulus over the range of 10–200 kPa promotes stronger activation of naive CD4 T cells [62]. However, how epigenetic changes contribute to these processes awaits further study. Moreover, there are also some studies on the physical regulation of cancer cells, although conflicting results have been reported for different cancer cell types. For instance, in breast cancer cells, stiff matrices increase AcH3 [63], but in melanoma cells, stiffening the matrix promotes H3K9 methylation [64]. Unraveling the biophysical regulation of immune cells, cancer cells and many other at epigenetic and phenotypic levels will provide novel insights into the mechanisms of tissue remodeling and disease pathogenesis.
Biomaterials in the form of scaffolds and micro/nano particles can be engineered to provide mechanical cues and regulate the epigenetic state and function of cells. In addition to elastic materials, viscoelastic materials can simulate many soft tissues that exhibit stress relaxation. Even in cases where cells are cultured on substrates that have the same elastic modulus, cell spreading, proliferation, and differentiation are significantly different between viscoelastic and elastic matrices. It is still unclear whether cellular responses to viscoelastic and elastic substrates will induce different histone modifications and cell signaling pathways [65,66]. Furthermore, scaffolds with tunable mechanical properties can induce a change in substrate stiffness that is either permanent or dynamic, providing the possibility to control and study reversible or irreversible epigenetic changes. Moreover, many other biophysical factors such as topography, three-dimensional structures, and porosity can regulate the epigenetic state and cell function [67]. All of these issues require further investigation, and will aid to broaden our understanding of the role of mechanical cues in cell plasticity and the development of novel materials for tissue regeneration and disease therapies.
Highlights.
Mechanical cues modulate the epigenetic state to regulate cell function.
Matrix stiffness regulates histone modifications during cell phenotype transitions.
Mechanical stretch and shear stress regulate cell plasticity through histone modifications.
Acknowledgements
This work is supported in part by grants from National Institute of Health (HL121450 and R56DE029157) and UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
No conflicts of interest to declare.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References and recommended reading
* of special interest
** of outstanding interest
- 1.Varga J, Greten FR: Cell plasticity in epithelial homeostasis and tumorigenesis. Nat Cell Biol 2017, 19:1133–1141. [DOI] [PubMed] [Google Scholar]
- 2.Ofenbauer A, Tursun B: Strategies for in vivo reprogramming. Curr Opin Cell Biol 2019, 61:9–15. [DOI] [PubMed] [Google Scholar]
- 3.Miroshnikova YA, Nava MM, Wickström SA: Emerging roles of mechanical forces in chromatin regulation. J Cell Sci 2017, 130:2243–2250. [DOI] [PubMed] [Google Scholar]
- ** 4.Sun J, Chen J, Mohagheghian E, Wang N: Force-induced gene up-regulation does not follow the weak power law but depends on H3K9 demethylation. Sci Adv 2020, 6:eaay9095. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated that the rapid up-regulation of endogenous mechanoresponsive genes depends on H3K9me3 demethylation.
- 5.Uhler C, Shivashankar GV: Chromosome Intermingling: Mechanical Hotspots for Genome Regulation. Trends Cell Biol 2017, 27:810–819. [DOI] [PubMed] [Google Scholar]
- 6.Allshire RC, Madhani HD: Ten principles of heterochromatin formation and function. Nat Rev Mol Cell Biol 2018, 19:229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janssen A, Colmenares SU, Karpen GH: Heterochromatin: Guardian of the Genome. Annu Rev Cell Dev Biol 2018, 34:265–288. [DOI] [PubMed] [Google Scholar]
- 8.Narita T, Weinert BT, Choudhary C: Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol 2019, 20:156–174. [DOI] [PubMed] [Google Scholar]
- 9.Wu X, Zhang Y: TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat Rev Genet 2017, 18:517–534. [DOI] [PubMed] [Google Scholar]
- 10.Martire S, Gogate AA, Whitmill A, Tafessu A, Nguyen J, Teng YC, Tastemel M, Banaszynski LA: Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nat Genet 2019, 51:941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liszczak G, Diehl KL, Dann GP, Muir TW: Acetylation blocks DNA damage-induced chromatin ADP-ribosylation. Nat Chem Biol 2018, 14:837–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryu H, Su D, Wilson-Eisele NR, Zhao D, López-Giráldez F, Hochstrasser M: The Ulp2 SUMO protease promotes transcription elongation through regulation of histone sumoylation. EMBO J 2019, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worden EJ, Hoffmann NA, Hicks CW, Wolberger C: Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L. Cell 2019, 176:1490–1501.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schübeler D: Function and information content of DNA methylation. Nature 2015, 517:321–326. [DOI] [PubMed] [Google Scholar]
- 15.Gkretsi V, Stylianopoulos T: Cell Adhesion and Matrix Stiffness: Coordinating Cancer Cell Invasion and Metastasis. Front Oncol 2018, 8:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He XT, Wu RX, Xu XY, Wang J, Yin Y, Chen FM: Macrophage involvement affects matrix stiffness-related influences on cell osteogenesis under three-dimensional culture conditions. Acta Biomciter 2018, 71:132–147. [DOI] [PubMed] [Google Scholar]
- 17.Engler AJ, Sen S, Sweeney HL, Discher DE: Matrix elasticity directs stem cell lineage specification. Cell 2006, 126:677–89. [DOI] [PubMed] [Google Scholar]
- 18.Godini R, Lafta HY, Fallahi H: Epigenetic modifications in the embryonic and induced pluripotent stem cells. Gene Expr Patterns 2018, 29:1–9. [DOI] [PubMed] [Google Scholar]
- 19.Cozzolino AM, Noce V, Battistelli C, Marchetti A, Grassi G, Cicchini C, Tripodi M, Amicone L: Modulating the Substrate Stiffness to Manipulate Differentiation of Resident Liver Stem Cells and to Improve the Differentiation State of Hepatocytes. Stem Cells Int 2016, 2016:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerardo H, Lima A, Carvalho J, Ramos JRD, Couceiro S, Travasso RDM, Pires das Neves R, Grãos M: Soft culture substrates favor stem-like cellular phenotype and facilitate reprogramming of human mesenchymal stem/stromal cells (hMSCs) through mechanotransduction. Sci Rep 2019, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higuchi S, Watanabe TM, Kawauchi K, Ichimura T, Fujita H: Culturing of mouse and human cells on soft substrates promote the expression of stem cell markers. J Biosci Bioeng 2014, 117:749–755. [DOI] [PubMed] [Google Scholar]
- 22.Pennarossa G, Santoro R, Manzoni EFM, Pesce M, Gandolfi F, Brevini TAL: Epigenetic Erasing and Pancreatic Differentiation of Dermal Fibroblasts into Insulin-Producing Cells are Boosted by the Use of Low-Stiffness Substrate. Stem Cell Rev Reports 2018, 14:398–411. [DOI] [PubMed] [Google Scholar]
- 23.Dou C, Liu Z, Tu K, Zhang H, Chen C, Yaqoob U, Wang Y, Wen J, van Deursen J, Sicard D, et al. : P300 Acetyltransferase Mediates Stiffness-Induced Activation of Hepatic Stellate Cells Into Tumor-Promoting Myofibroblasts. Gastroenterology 2018, 154:2209–2221.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * 24.Killaars AR, Grim JC, Walker CJ, Hushka EA, Brown TE, Anseth KS: Extended exposure to stiff microenvironments leads to persistent chromatin remodeling in human mesenchymal stem cells In Transactions of the Annual Meeting of the Society for Biomaterials and the Annual International Biomatericds Symposium. Society for Biomaterials; 2019:670. [Google Scholar]; This study utilized tunable hydrogels to demonstrate that epigenetic remodeling can be persistent and might retain cell memory.
- 25.Qu J, Zhu L, Zhou Z, Chen P, Liu S, Locy ML, Thannickal VJ, Zhou Y: Reversing mechanoinductive DSP expression by CRISPR/dCas9-mediated epigenome editing. Am J Respir Crit Care Med 2018, 198:599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ligresti G, Caporarello N, Meridew JA, Jones DL, Tan Q, Choi KM, Haak AJ, Aravamudhan A, Roden AC, Prakash YS, et al. : CBX5/G9a/H3K9me-mediated gene repression is essential to fibroblast activation during lung fibrosis. JCI Insight 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Przybyla L, Lakins JN, Weaver VM: Tissue Mechanics Orchestrate Wnt-Dependent Human Embryonic Stem Cell Differentiation. Cell Stem Cell 2016, 19:462–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Tang CB, Kilian KA: Matrix Mechanics Influence Fibroblast-Myofibroblast Transition by Directing the Localization of Histone Deacetylase 4. Cell Mol Bioeng 2017, 10:405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gracey E, Burssens A, Cambré I, Schett G, Lories R, McInnes IB, Asahara H, Elewaut D: Tendon and ligament mechanical loading in the pathogenesis of inflammatory arthritis. Nat Rev Rheumatol 2020, 16:193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nonomura K, Woo S-H, Chang RB, Gillich A, Qiu Z, Francisco AG, Ranade SS, Liberies SD, Patapoutian A: Piezo2 senses airway stretch and mediates lung inflation-induced apnoea. Nature 2017, 541:176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marrelli M, Codispoti B, Shelton RM, Scheven BA, Cooper PR, Tatullo M, Paduano F: Dental pulp stem cell mechanoresponsiveness: Effects of mechanical stimuli on dental pulp stem cell behavior. Front Physiol 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Chu JS, Kurpinski K, Li X, Bautista DM, Yang L, Sung K-LP, Li S: Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophys J 2011, 100:1902–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagielska A, Lowe AL, Makhija E, Wroblewska L, Guck J, Franklin RJM, Shivashankar GV, Van Vliet KJ: Mechanical Strain Promotes Oligodendrocyte Differentiation by Global Changes of Gene Expression. Front Cell Neurosci 2017, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ** 34.Gudipaty SA, Lindblom J, Loftus PD, Redd MJ, Edes K, Davey CF, Krishnegowda V, Rosenblatt J: Mechanical stretch triggers rapid epithelial cell division through Piezol. Nature 2017, 543:118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study defined a role for mechanical tension in regulating constant epithelial cell numbers, essential for maintaining a tight barrier and preventing cancers from forming.
- 35.Wang J, Wang CD, Zhang N, Tong WX, Zhang YF, Shan SZ, Zhang XL, Li QF: Mechanical stimulation orchestrates the osteogenic differentiation of human bone marrow stromal cells by regulating HDAC1. Cell Death Dis 2016, 7:e2221–e2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan Y, Yang X, Wei L, Chen Q: MiR-365: a mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4. FASEB J 2011, 25:4457–4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng Q, Li X, Xiao L, Shao S, Jiang H, Zhang X, Sun L, Xu H: MicroRNA-365 functions as a mechanosensitive microRNA to inhibit end plate chondrocyte degeneration by targeting histone deacetylase 4. Bone 2019, 128:115052. [DOI] [PubMed] [Google Scholar]
- 38.Paul NE, Denecke B, Kim B-S, Dreser A, Bernhagen J, Pallua N: The effect of mechanical stress on the proliferation, adipogenic differentiation and gene expression of human adipose-derived stem cells. J Tissue Eng Regen Med 2018, 12:276–284. [DOI] [PubMed] [Google Scholar]
- 39.Wang C, Shan S, Wang C, Wang J, Li J, Hu G, Dai K, Li Q, Zhang X: Mechanical stimulation promote the osteogenic differentiation of bone marrow stromal cells through epigenetic regulation of Sonic Hedgehog. Exp Cell Res 2017, 352:346–356. [DOI] [PubMed] [Google Scholar]
- 40.Vlaikou AM, Kouroupis D, Sgourou A, Markopoulos GS, Bagli E, Markou M, Papadopoulou Z, Fotsis T, Nakos G, Lekka MEE, et al. : Mechanical stress affects methylation pattern of GNAS isoforms and osteogenic differentiation of hAT-MSCs. Biochim Biophys Acta - Mol Cell Res 2017, 1864:1371–1381. [DOI] [PubMed] [Google Scholar]
- 41.Zhao B, Wang J, Ren C, Hu M, Wu H, Chen L, Liu X, Wang L, Xu F, Zheng X, et al. : Mechanical stretching promotes the differentiation of BMSC into pelvic floor ligament fibroblasts. Oncotarget 2018, 0. [Google Scholar]
- ** 42.Le HQ, Ghatak S, Yeung C-YC, Tellkamp F, Günschmann C, Dieterich C, Yeroslaviz A, Habermann B, Pombo A, Niessen CM, et al. : Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol 2016, 18:864–875. [DOI] [PubMed] [Google Scholar]; This study identified a force-driven transcriptional rheostat that controls chromatin architecture to fine-tune gene expression.
- 43.Bazou D, Ng MR, Song JW, Chin SM, Maimon N, Munn LL: Flow-induced HDAC1 phosphorylation and nuclear export in angiogenic sprouting. Sci Rep 2016, 6:34046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * 44.Peghaire C, Dufton NP, Lang M, Salles-Crawley II, Ahnström J, Kalna V, Raimondi C, Pericleous C, Inuabasi L, Kiseleva R, et al. : The transcription factor ERG regulates a low shear stress-induced anti-thrombotic pathway in the microvasculature. Nat Commun 2019, 10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified an endogenous, vascular bed-specific anticoagulant pathway in microvasculature in response to low shear stress.
- 45.Wang J, Wu Y, Zhang X, Zhang F, Lü D, Shangguan B, Gao Y, Long M: Flow-enhanced priming of hESCs through H2B acetylation and chromatin decondensation. Stem Cell Res Ther 2019, 10:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiu JJ, Chien S: Effects of disturbed flow on vascular endothelium: Pathophysiological basis and clinical perspectives. Physiol Rev 2011, 91:327–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He M, Huang TS, Li S, Hong HC, Chen Z, Martin M, Zhou X, Huang HY, Su SH, Zhang J, et al. : Atheroprotective Flow Upregulates ITPR3 (Inositol l,4,5-Trisphosphate Receptor 3) in Vascular Endothelium via KLF4 (Krüppel-Like Factor 4)-Mediated Histone Modifications. Arterioscler Thromb Vasc Biol 2019, 39:902–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maleszewska M, Vanchin B, Harmsen MC, Krenning G: The decrease in histone methyltransferase EZH2 in response to fluid shear stress alters endothelial gene expression and promotes quiescence. Angiogenesis 2016, 19:9–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu S, Xu Y, Yin M, Zhang S, Liu P, Koroleva M, Si S, Little PJ, Pelisek J, Jin ZG: Flow-dependent epigenetic regulation of IGFBP5 expression by H3K27me3 contributes to endothelial anti-inflammatory effects. Theranostics 2018, 8:3007–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heuslein JL, Gorick CM, Song J, Price RJ: DNA methyltransferase 1-dependent DNA hypermethylation constrains arteriogenesis by augmenting shear stress set point. J Am Heart Assoc 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheleschi S, De Palma A, Pecorelli A, Pascarelli NA, Valacchi G, Belmonte G, Carta S, Galeazzi M, Fioravanti A: Hydrostatic pressure regulates MicroRNA expression levels in osteoarthritic chondrocyte cultures via the Wnt/β-catenin pathway. Int J Mol Sci 2017, 18:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ke X, Johnson H, Jing X, Michalkiewicz T, Huang YW, Lane RH, Konduri GG: Persistent pulmonary hypertension alters the epigenetic characteristics of endothelial nitric oxide synthase gene in pulmonary artery endothelial cells in a fetal lamb model. Physiol Genomics 2018, 50:828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang P, Dreger M, Madrazo E, Williams CJ, Samaniego R, Hodson NW, Monroy F, Baena E, Sánchez-Mateos P, Hurlstone A, et al. : WDR5 modulates cell motility and morphology and controls nuclear changes induced by a 3D environment. Proc Natl Acad Sci U S A 2018, 115:8581–8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico-Lastres P, Le Roux A-L, et al. : Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 2017, 171:1397–1410.e14. [DOI] [PubMed] [Google Scholar]
- ** 55.Alisafaei F, Jokhun DS, Shivashankar GV, Shenoy VB: Regulation of nuclear architecture, mechanics, and nucleocytoplasmic shuttling of epigenetic factors by cell geometric constraints. Proc Natl Acad Sci U S A 2019, 116:13200–13209. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study utilized a chemomechanical model and micropatterning experiments to further elucidate how chromosome configurations are altered in response to changes in nuclear mechanical properties following cues from the microenvironment.
- 56.Jain N, Iyer KV, Kumar A, Shivashankar G V: Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci 2013, 110:11349–11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roy B, Venkatachalapathy S, Ratna P, Wang Y, Jokhun DS, Nagarajan M, Shivashankar G V: Laterally confined growth of cells induces nuclear reprogramming in the absence of exogenous biochemical factors. Proc Natl Acad Sci 2018, 115:E4741–E4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peng Q, Lu S, Shi Y, Pan Y, Limsakul P, Chernov A V, Qiu J, Chai X, Shi Y, Wang P, et al. : Coordinated histone modifications and chromatin reorganization in a single cell revealed by FRET biosensors. Proc Natl Acad Sci 2018, 115:201811818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Damodaran K, Venkatachalapathy S, Alisafaei F, Radhakrishnan AV, Sharma Jokhun D, Shenoy VB, Shivashankar GV.: Compressive force induces reversible chromatin condensation and cell geometry-dependent transcriptional response. Mol Biol Cell 2018, 29:3039–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kassianidou E, Kalita J, Lim RYH: The role of nucleocytoplasmic transport in mechanotransduction. Exp Cell Res 2019, 377:86–93. [DOI] [PubMed] [Google Scholar]
- 61.Sridharan R, Cavanagh B, Cameron AR, Kelly DJ, O’Brien FJ: Material stiffness influences the polarization state, function and migration mode of macrophages. Acta Biomater 2019, 89:47–59. [DOI] [PubMed] [Google Scholar]
- 62.Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC: Mechanosensing in T lymphocyte activation. Biophys J 2012, 102:L5–L7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stowers RS, Shcherbina A, Israeli J, Gruber JJ, Chang J, Nam S, Rabiee A, Teruel MN, Snyder MP, Kundaje A, et al. : Matrix stiffness induces a tumorigenic phenotype in mammary epithelium through changes in chromatin accessibility. Nat Biomed Eng 2019, 3:1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan Y, Tajik A, Chen J, Jia Q, Chowdhury F, Wang L, Chen J, Zhang S, Hong Y, Yi H, et al. : Matrix softness regulates plasticity of tumour-repopulating cells via H3K9 demethylation and Sox2 expression. Nat Commun 2014, 5:4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaudhuri O, Gu L, Darnell M, Klumpers D, Bencherif SA, Weaver JC, Huebsch N, Mooney DJ: Substrate stress relaxation regulates cell spreading. Nat Commun 2015, 6:6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee H, Lippens E, Duda GN, et al. : Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater 2016, 15:326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Downing TL, Soto J, Morez C, Houssin T, Fritz A, Yuan F, Chu J, Patel S, Schaffer DV, Li S: Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater 2013, 12:1154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]


