Abstract
Defining patient-to-patient similarity is essential for the development of precision medicine in clinical care and research. Conceptually, the identification of similar patient cohorts appears straightforward; however, universally accepted definitions remain elusive. Simultaneously, an explosion of vendors and published algorithms have emerged and all provide varied levels of functionality in identifying patient similarity categories. To provide clarity and a common framework for patient similarity, a workshop at the American Medical Informatics Association 2019 Annual Meeting was convened. This workshop included invited discussants from academics, the biotechnology industry, the FDA, and private practice oncology groups. Drawing from a broad range of backgrounds, workshop participants were able to coalesce around 4 major patient similarity classes: (1) feature, (2) outcome, (3) exposure, and (4) mixed-class. This perspective expands into these 4 subtypes more critically and offers the medical informatics community a means of communicating their work on this important topic.
Keywords: patient matching; precision medicine; patients like me; personalized medicine, similar patients
INTRODUCTION
The premise of precision medicine is deceptively simple: similar patients with similar features have similar outcomes. While traditional clinical trial design creates strong evidence in regard to the activity of a singular intervention, it does not provide the basis for personalizing medical care for a specific patient.1 Finding similar patients furthers the pursuit of precision medicine by identifying key traits and features of patients that may identify their clinical course.2,3 Patient matching provides the opportunity to improve patient care and clinical research by identifying and potentially controlling for key covariates that may help predict a patient’s outcome.4–6
Previously identified key challenges in the patient similarity space included data heterogeneity and data-sharing algorithm selection.7 Significant progress has been made in these areas, especially within the field of oncology. Efforts by the cancer community, such as those of Minimal Common Oncology Data Element (mCODE) and Global Alliance for Genomics and Health (GA4GH), continue to develop and refine standards for parsing ever-evolving patient features. Further, data elements like tumor genomics and PD-L1 positivity have rapidly evolved to become commonplace in research and clinical care. In the domain of data sharing, consortia efforts, such as the Oncology Research Information Exchange Network and the American Association for Cancer Research Genie project, have amassed large volumes of clinico-genomic patient data.8,9 Continued publication of results of high enrollment, multi-arm treatment trials, such as the tumor-agnostic National Cancer Institute Molecular Analysis for Therapy Choice and the lung cancer Alchemist trials, have been anxiously awaited to evaluate the utility of their patient-matching criteria. Naturally, multi-dimensional patient-matching algorithms have proliferated in part due to the variety of use cases, specific features, and outcome variables available.
While the science of patient matching has vastly improved, our ability to communicate about the type of patient similarity we use has become a significant challenge.10,11 Advances in multi-dimensional patient matching have been slow to develop due in part to the heterogeneous interpretations that exist within similarity matching.12 A continued lack of consensus regarding terminology, methods, and data types has resulted in poor consistency of resultant findings of patient-matching studies.13 While the science of patient matching has vastly improved, our ability to communicate about the type of patient similarity we endeavor to accomplish has become a significant challenge.10,11 Indeed, heterogeneous interpretations exist within similarity matching.12 A continued lack of consensus regarding terminology, methods, and data types has resulted in poor consistency of resultant findings of patient-matching studies.13
DEFINING THE PROBLEM: AMBIGUOUS NOMENCLATURE IN THE PATIENT SIMILARITY SPACE
Identification of common language and methods among the many efforts of quantifying and improving patient similarity is vital to improve precision patient care.14 In addition to a need for improved standardization of medical terminology and categorization, there is further need for an accepted framework for synthesizing data elements that create a generalizable “computable phenotype” as a basis for matching similar patients.15 Defining similar patients, therefore, may require disease- or task-specific methods.
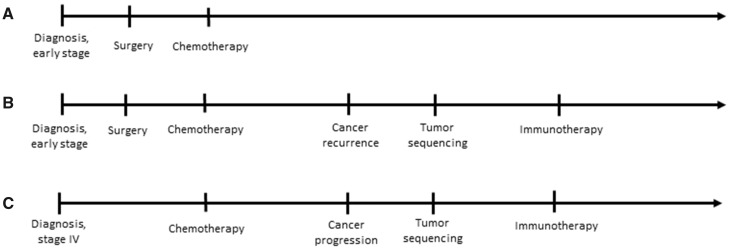
Shifts in the core features of a patient’s disease over time adds additional complexity to defining similarity. Features such as genomic similarity are distinct from features such as similar response to therapy. An example of temporal complexity can be seen in the treatment of cancer where 2 patients diagnosed with early-stage disease may be quite similar early in their disease trajectories, but if 1 of those patients develops recurrent disease, that patient may subsequently be much more similar to a third patient who had advanced disease at diagnosis (Figure 1). Standardization of language and methodology when discussing patient similarity is vital to the progression of its study and implementation.
Figure 1.
Defining patient similarity. These diagrams represent the clinical courses of 3 hypothetical patients with non-small cell lung cancer. Patient A corresponds to a patient who was diagnosed with early stage disease, who underwent surgery and adjuvant chemotherapy and, so far, has not developed recurrent disease. Patient B had a trajectory that began similarly but developed cancer recurrence, leading their oncologist to order tumor genomic sequencing and prescribe immunotherapy. Patient C had metastatic disease at diagnosis which was treated initially with chemotherapy and, subsequently, with immunotherapy. Any definition of similarity among these patients must necessarily be time-dependent; early in the cancer trajectory, patients A and B are most similar, but later in the trajectory, patients B and C are most similar.
PATIENT SIMILARITY WORKSHOP DETAILS
To define a common framework for relating patient similarity, a workshop was convened at the American Medical Informatics Association (AMIA) 2019 Annual Meeting entitled: “What defines a patient like mine? A collaborative effort to provide clarity into the computational nomenclature of patient similarity, their requisite data categories, and associated algorithms.” Open to all registrants of the meeting, attendees participated in a series of focused presentations by expert discussants with academic, industry, and regulatory viewpoints. This perspective builds on the consensus recommendations presented by discussants and among attendees.
CONSENSUS RECOMMENDATIONS
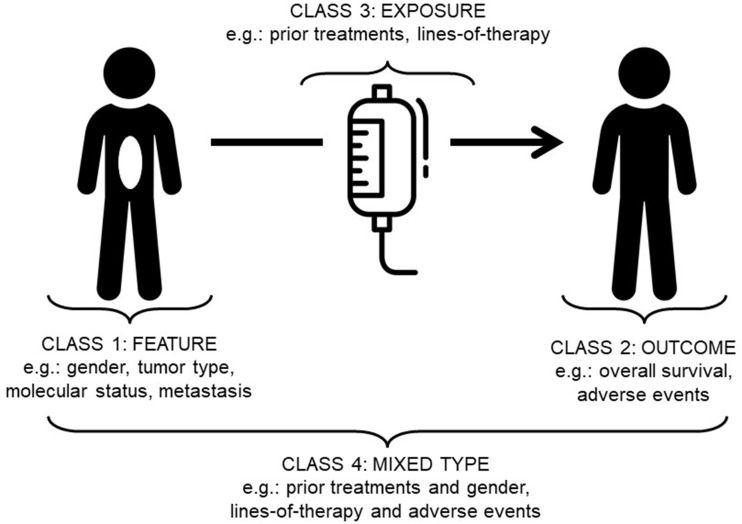
Patient similarity can be divided into 4 classes: 1) feature; 2) outcome; 3) exposure; and 4) mixed-class (Figure 2; Table 1). Each class has particular characteristics of temporality (snapshot versus change over time), and whether the feature describes an object or an action. By object, we refer to features that are properties of physical objects (ie, people or tumors), also commonly thought of as baseline characteristics or attributes. By actions we refer to processes performed (ie, various treatment modalities).
Figure 2.
Patient similarity categories. Classes of patient similarity proposed in this perspective. Drawing from a broad range of backgrounds, workshop participants were able to coalesce around 4 major patient similarity categories: (1) Feature, (2) Outcome, (3) Exposure, and (4) Mixed-Class.
Table 1.
Classes of patient similarity
| Similarity Class | Temporality | Object or Action | Examples |
|---|---|---|---|
| Feature | Snapshot | Object | Disease type/status, past medical history, treatments received |
| Outcome | Snapshot | Object | Adverse event, treatment efficacy |
| Exposure | Change over time | Action | Prior lines of therapy define a cohort for study and reflect disease status |
| Mixed-class | Snapshot/change over time | Object/Action | Molecularly and disease-matched patients who exhibit a similar outcome to therapy |
Class 1: feature similarity
Feature similarity can be considered as the state of a physical object or short period of a “snapshot.” This would include the mutational status of the tumor, the state of the disease, cancer stage, as well as more complex features, such as past medical history, previous therapies, and allergies. A common example of feature-based similarity in the biomedical informatics domain is the use of diagnostic billing codes to define groups of patients. Despite their demonstrated utility, abstracted features are nevertheless problematic due to their imprecision.16,17 Historically, feature similarity has been well-studied and implemented in clinical practice. However, developing high-dimensional feature similarity quickly results in inaccurate or minimal similarities between patients, particularly when dimensionality exceeds the number of patients in a study.18,19 Methods to identify features with the greatest predictive value of a given outcome are necessary to improve the utility of this class of patient similarity measures.
Class 2: outcome similarity
Outcome similarity focuses on finding matches in temporal-based endpoints. These metrics try to answer the question, “How did the patient do?” Outcome measures used to match similar patients can also be considered a “snapshot” of a patient’s health. These outcome measures can be process measures of other related interventions, toxicities related to disease or treatment, or classic therapeutic benchmarking outcome measures, in addition to others. Using these metrics, it may be possible to find similar patients for a control group, therapeutic benchmarking, and granular dynamic “features” of a patient reflecting “outcomes” of disease control. Outcome similarity metrics may ultimately be used to develop quintessential real-world evidence (RWE). As RWE is not without its limitations, developing granular understanding of its contribution to data from existing clinical trials can help clinical trialists select patient populations, help companies prioritize research efforts, or reduce uncertainty for patients and practitioners surrounding treatment decisions.20 Pulling these outcome measures from systematically mapped sources of structured data reduces variability and enhances RWE as a modeling tool. Data in this space are inherently challenging to analyze and are highly subject to selection bias and confounding.
Class 3: exposure similarity
Exposure similarity identifies patients based on the presence or absence of therapeutic interventions or other exposures which affect their health status. These exogenously applied “actions” may include drugs, devices, surgical and radiation therapy, and environmental exposures. Feature similarity addresses patient and disease characteristics as baseline objects, and outcome similarity treats these objects as endpoints. In contrast, exposure similarity defines changes over time, adding a temporal dimension to patient similarity. In an observational cohort study of a therapeutic intervention, exposure similarity is used to define 1 or more groups for comparison. In clinical trials, exposure to prior lines of therapy are used as inclusion criteria in order to enhance the precision of likely disease activity status and response to therapy. These prior lines of therapy are often described in the indications for approved drugs and biologics. Use of RWE as an external comparator for a single-arm trial places special emphasis on temporal issues as well as exposure and feature similarity. Because the groups are not necessarily ascertained in the same temporal period with the same background availability of therapeutic exposures and with the same level of granularity regarding the details of the therapeutic interventions, secular trends in therapeutic patterns, differing availability of therapeutics, or differential ascertainment of the details of exposure may impact outcomes.
Class 4: mixed-class similarity
When considering the 3 previous classes of patient similarity, the last significant class of similarity is the interaction of these classes, or a mixed-class similarity. For example, the interaction of comorbidity status and diuretic therapy exposure in a patient creates a mixed metric more complex and indicative of true patient similarity.21 In the case of 3 different cancer patients outlined in Figure 1, the interaction of baseline feature, exposure, and outcome provided vastly different similarity possibilities temporally. In modern clinical medicine, attempting to derive general phenotypes for patient matching may be extremely challenging; in effect, suffering from a “curse of dimensionality” would imply no 2 patients are similar in any meaningful way given the near infinite data necessary to accurately portray a patient.22 Mixed-class similarity represents a challenge computationally that has yet to be well-addressed. It is likely that computable similarity efforts that are task- and setting-dependent will improve its applicability.
OPPORTUNITIES FOR IMPROVEMENT
Ultimately, multiple sources of data derived from the previously discussed classes of patient similarity must be integrated to adequately construct patient cohorts that are similar in phenotype and genotype. Previous studies have demonstrated a preference for study of molecular measures of patient similarity; however, multi-class phenotype calculation is also necessary.5,23–25 One approach to harmonizing the collection and sharing of data is the creation of networks between stakeholders in order to agree on key parameters, such as patient consent and data dictionaries.26 Recognizing that patients’ diseases are heterogeneous and molecularly evolve following treatment, may require sequential clinical and molecular analysis to accurately assign patients to the most similar patient cohort. Approaches based on sequence alignment may provide promising solutions for matching patients while considering important temporal information.27–29 The application of machine learning (ML) to analyze observational cohorts also has the potential to improve clinical decision making but will require very large populations followed prospectively throughout the clinical course for each patient.30 Patient similarity will also be key to a type of ML called reinforcement learning (RL). In contrast to traditional supervised learning methods that usually rely on single-episode training, RL tackles clinical questions with sequential decision-making problems using sampled, evaluative, and delayed feedback.31
Identifying common health variables is a vital element of biomedical research. Currently utilized general ontologies for medical concepts (eg, SNOMED, ICD) provide mechanisms for structuring the often-unstructured data contained in health records.32–35 These coding systems have improved the structure of the medical record but lack the ability to define key clinical characteristics for many aspects of clinical care. Newer frameworks, such as mCODE, are specifically designed to capture such key concepts and may serve to further standardize the language of medical data and provide a platform to improve the computation of patient similarity.36,37
CONCLUSION
In many respects, it is easier to sequence a whole cancer genome in 2020 than to readily and reproducibly define a group of “similar” patients. Similarity classes create a framework for defining groups of patients who are likely to have similar defining traits, outcomes, and/or temporal experiences. This aids clinicians in their treatment decisions and patients in anchoring themselves to a defined wellness or illness group. While every patient is unique and every journey is different, practically, treatments are targeted toward a group of patients with similar characteristics for whom we reasonably would expect a similar response. This is the same reason nomenclature has moved from personalized medicine to precision medicine. This objective approach to similarity has major advantages.38 First, people want to develop kinship with patients facing similar medical issues as themselves—as demonstrated through the development of cancer biomarker-defined patient advocacy groups (eg, ROS1ders, EGFR Resisters).39,40 These groups demonstrate how patient similarity can provide a community for patients while also serving as a launchpad for further research. Second, reproducible similarity metrics are also used in drug development as industry and regulatory bodies approach drug approvals in defined patient cohorts, with biomarkers and prior treatment-specific indications granted by the FDA.
Taken together, this perspective represents a nascent effort to bring together a variety of stakeholders in patient similarity to define common nomenclature. Communities that centralize stakeholders, such as AMIA, must continue to unify future clinical and research efforts in this space. We believe these aforementioned classes will provide a clear and useful basis for communicating work surrounding patient similarity.
AUTHOR CONTRIBUTIONS
All authors contributed to the manuscript preparation and have approved the final version of the manuscript and agree to be accountable for all aspects of the work.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the organizers and attendees of the AMIA 2019 annual meeting for providing a fruitful forum for discussion. This article reflects the views of the authors and should not be construed to represent FDA’s views or policies.
CONFLICT OF INTEREST STATEMENT
WSD: employment (M2Gen); intellectual property and royalties (Moffitt Cancer Center). JLW: consulting fees (Westat); advisory board (IBM Genomics Health); owns stock (HemOnc.org; no monetary value). No other authors declared conflicts of interest.
REFERENCES
- 1. Pallmann P, Bedding AW, Choodari-Oskooei B, et al. Adaptive designs in clinical trials: why use them, and how to run and report them. BMC Med 2018; 16 (1): 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sharafoddini A, Dubin JA, Lee J. Patient similarity in prediction models based on health data: a scoping review. JMIR Med Inform 2017; 5 (1): e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Henriques J, Carvalho P, Paredes S, et al. Prediction of heart failure decompensation events by trend analysis of telemonitoring data. IEEE J Biomed Health Inform 2015; 19 (5): 1757–69. [DOI] [PubMed] [Google Scholar]
- 4. Sackett DL, Rosenberg WM, Gray JA, Haynes RB, Richardson WS. Evidence based medicine: what it is and what it isn't. 1996. Clin Orthop Relat Res 2007; 455: 3–5. [PubMed] [Google Scholar]
- 5. Pai S, Bader GD. Patient similarity networks for precision medicine. J Mol Biol 2018; 430 (18): 2924–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seligson ND, Hobbs ALV, Leonard JM, Mills EL, Evans AG, Goorha S. Evaluating the impact of the addition of cladribine to standard acute myeloid leukemia induction therapy. Ann Pharmacother 2018; 52 (5): 439–45. [DOI] [PubMed] [Google Scholar]
- 7. Johnson T, Liebner D, Chen JL. Opportunities for patient matching algorithms to improve patient care in oncology. JCO Clin Cancer Inform 2017; 1 (1): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Oncology Research Information Exchange Network (ORIEN). https://www.oriencancer.org/ Accessed June 9, 2020
- 9.American Association for Cancer Research Genomics Evidence Neoplasia Information Exchange. Secondary American Association for Cancer Research Genomics Evidence Neoplasia Information Exchange. https://www.aacr.org/professionals/research/aacr-project-genie/ Accessed June 15, 2020.
- 10. Feinstein AR, Rubinstein JF, Ramshaw WA. Estimating prognosis with the aid of a conversational-mode computer program. Ann Intern Med 1972; 76 (6): 911–21. [DOI] [PubMed] [Google Scholar]
- 11. Concato J, Horwitz RI. Beyond randomised versus observational studies. Lancet 2004; 363 (9422): 1660–1. [DOI] [PubMed] [Google Scholar]
- 12. Campbell-Scherer D. Multimorbidity: a challenge for evidence-based medicine. Evid Based Med 2010; 15 (6): 165–6. [DOI] [PubMed] [Google Scholar]
- 13. Just BH, Marc D, Munns M, Sandefer R. Why patient matching is a challenge: research on master patient index (MPI) data discrepancies in key identifying fields. Perspect Health Inf Manag 2016; 13: 1e. [PMC free article] [PubMed] [Google Scholar]
- 14. Kuhn KA, Knoll A, Mewes HW, et al. Informatics and medicine–from molecules to populations. Methods Inf Med 2008; 47 (04): 283–95. [PubMed] [Google Scholar]
- 15. Mo H, Thompson WK, Rasmussen LV, et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. J Am Med Inform Assoc 2015; 22 (6): 1220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010; 26 (9): 1205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Warner JL, Denny JC, Kreda DA, Alterovitz G. Seeing the forest through the trees: uncovering phenomic complexity through interactive network visualization. J Am Med Inform Assoc 2015; 22 (2): 324–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen Q, Nian H, Zhu Y, Talbot HK, Griffin MR, Harrell FE. Too many covariates and too few cases? A comparative study. Stat Med 2016; 35 (25): 4546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lussier YA, Stadler WM, Chen JL. Advantages of genomic complexity: bioinformatics opportunities in microRNA cancer signatures. J Am Med Inform Assoc 2012; 19 (2): 156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Webster J, Smith BD. The case for real-world evidence in the future of clinical research on chronic myeloid leukemia. Clin Ther 2019; 41 (2): 336–49. [DOI] [PubMed] [Google Scholar]
- 21. Burnier M, Bakris G, Williams B. Redefining diuretics use in hypertension: why select a thiazide-like diuretic? J Hypertens 2019; 37 (8): 1574–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bellman R. Dynamic Programming. Princeton, NJ: Princeton University Press; 1957. [Google Scholar]
- 23. Parimbelli E, Marini S, Sacchi L, Bellazzi R. Patient similarity for precision medicine: a systematic review. J Biomed Inform 2018; 83: 87–96. [DOI] [PubMed] [Google Scholar]
- 24. König J, Kranz B, König S, et al. Phenotypic spectrum of children with nephronophthisis and related ciliopathies. CJASN 2017; 12 (12): 1974–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen X, Garcelon N, Neuraz A, et al. Phenotypic similarity for rare disease: ciliopathy diagnoses and subtyping. J Biomed Inform 2019; 100: 103308. [DOI] [PubMed] [Google Scholar]
- 26. Dalton WS, Sullivan D, Ecsedy J, Caligiuri MA. Patient enrichment for precision-based cancer clinical trials: using prospective cohort surveillance as an approach to improve clinical trials. Clin Pharmacol Ther 2018; 104 (1): 23–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang M, Shah ND, Yao L. Evaluating global and local sequence alignment methods for comparing patient medical records. BMC Med Inform Decis Mak 2019; 19 (S6): 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Temporal sequence alignment in electronic health records for computable patient representation. In: 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); December 3–6, 2018; Madrid, Spain.
- 29.KELSA: A knowledge-enriched local sequence alignment algorithm for comparing patient medical records health intelligence. In: workshop at the 34th AAAI Conference on Artificial Intelligence; February 7–12, 2020; New York.
- 30. Gottesman O, Johansson F, Komorowski M, et al. Guidelines for reinforcement learning in healthcare. Nat Med 2019; 25 (1): 16–8. [DOI] [PubMed] [Google Scholar]
- 31. Zhang Z. Reinforcement learning in clinical medicine: a method to optimize dynamic treatment regime over time. Ann Transl Med 2019; 7 (14): 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee D, de Keizer N, Lau F, Cornet R. Literature review of SNOMED CT use. J Am Med Inform Assoc 2014; 21 (e1): e11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen F, Peng S, Fan Y, et al. HPO2Vec+: leveraging heterogeneous knowledge resources to enrich node embeddings for the human phenotype ontology. J Biomed Inform 2019; 96: 103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shen F, Liu H. Incorporating knowledge-driven insights into a collaborative filtering model to facilitate the differential diagnosis of rare diseases. AMIA Annu Symp Proc 2018; 2018: 1505–14. [PMC free article] [PubMed] [Google Scholar]
- 35. Major P, Kostrewski BJ, Anderson J. Analysis of the semantic structures of medical reference languages: part 2. Analysis of the semantic power of MeSH, ICD and SNOMED. Med Inform (Lond) 1978; 3 (4): 269–81. [DOI] [PubMed] [Google Scholar]
- 36. Rubinstein WS. CancerLinQ: cutting the Gordian knot of interoperability. JOP 2019; 15 (1): 3–6. [DOI] [PubMed] [Google Scholar]
- 37. Bodenreider O, Cornet R, Vreeman DJ. Recent developments in clinical terminologies - SNOMED CT, LOINC, and RxNorm. Yearb Med Inform 2018; 27 (01): 129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cooper R. What is wrong with the DSM? Hist Psychiatry 2004; 15 (1): 5–25. [Google Scholar]
- 39.EGFR Resisters. http://egfrcancer.org Accessed June 9, 2020
- 40.ROS1ders. http://ros1cancer.com Accessed June 9, 2020