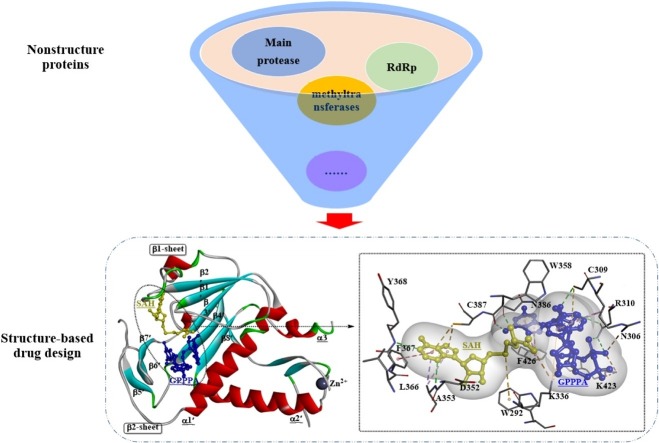
Graphical abstract
Keywords: Coronavirus, Molecular targets, Viral protease, RNA-dependent RNA polymerase, Inhibitors
Highlights
-
•
The pandemic of SARS-CoV-2 has posed significant threats to public health worldwide.
-
•
Target-based drug development is a promising approach against SARS-CoV-2 infection.
-
•
Nonstructural proteins may play critical roles from drug design perspectives.
-
•
Insights into NSPs of different viruses could streamline novel drug development.
Abstract
Outbreaks of severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 have produced high pathogenicity and mortality rates in human populations. However, to meet the increasing demand for treatment of these pathogenic coronaviruses, accelerating novel antiviral drug development as much as possible has become a public concern. Target-based drug development may be a promising approach to achieve this goal. In this review, the relevant features of potential molecular targets in human coronaviruses (HCoVs) are highlighted, including the viral protease, RNA-dependent RNA polymerase, and methyltransferases. Additionally, recent advances in the development of antivirals based on these targets are summarized. This review is expected to provide new insights and potential strategies for the development of novel antiviral drugs to treat SARS-CoV-2 infection.
1. Introduction
The outbreaks of severe acute respiratory syndrome (SARS) in 2003 [1], Middle East respiratory syndrome (MERS) in 2012 [2], and novel coronavirus pneumonia in China in 2020 [3] have posed significant threats to public health worldwide. Given the high case-fatality ratio, strong transmissibility, and shortage of efficient antivirals, human coronaviruses (HCoVs), as well as zoonotic coronaviruses such as SARS-CoV, MERS-CoV and SARS-CoV-2, have become a source of increasing concern. Compared with the inherently high mutation rates of other RNA viruses, the mutation rate of HCoVs may be somewhat low due to their genome-encoded exonuclease (e.g., RNA 3’-to-5’ exoribonuclease and endoribonuclease) associated with RNA processing that improves the low fidelity of RNA replication [4,5]. It has been determined that several virulence-enhancing mutations usually resulted in substantially increased viral polymerase activity [6] and the high mutation rates usually lead to increased adaptability [7].
The pathogenic mechanism of HCoVs is not completely understood; therefore, additional detailed investigations of the pathogenic mechanism, particularly potential targets, might be helpful in the identification of novel antivirals through target-based drug discovery. Coronaviruses, which are composed of four genera (α, β, γ, and δ), are reported to be the largest type of positive-strand RNA viruses (26–32 kb) [8]. To date, seven HCoVs (HCoV-229E, NL63, MERS-CoV, SARS-CoV, HCoVOC43, HCoV-HKU1 and SARS-CoV-2) have been identified, among which SARS-CoV, MERS-CoV and SARS-CoV-2 are the major causes of severe pneumonia in humans. Similarly, these viruses are composed of four major types of structural proteins on the viral membrane [9], including the spike protein (S), envelope protein (E), membrane protein (M), and nucleocapsid (N) proteins. The spike protein is responsible for viral entry into host cells; therefore, it is considered a major therapeutic target for antiviral drug development [10], and the M, E and N proteins play critical roles in viral assembly and RNA synthesis.
Unfortunately, no proven coronavirus-specific therapies are currently recommended for HCoVs, although various antiviral drugs (or vaccines) have been approved (or are in development) for the treatment of specific viral infections. The combined use of different antiviral drugs may be a potential therapeutic strategy. Therefore, the development of efficient antivirals with either virus-specific or pan-CoV activities for the treatment of CoV infections is urgently needed. In this review, we highlight the molecular targets associated with novel drug discovery, strategies for the discovery of novel antiviral drugs, and the identification of successful agents that control CoV infection. Although a large number of studies have been published on the development of novel antiviral compounds against HCoVs, this review is expected to be a resource for the development of efficient antivirals.
2. Potential molecular targets for pancoronavirus antiviral drug development and identified inhibitors
Although the physiology-based approach is a traditional and proven drug discovery paradigm for the discovery of novel drugs, emerging computer-aided drug development methods may become a promising alternative to accelerate the discovery of novel drugs [[11], [12], [13]]. These in silico strategies (e.g., virtual screening and structure-based drug design) very effectively identify novel active scaffolds for a validated target. Therefore, the selection of one (single-target agents) or more (multitarget agents) drug targets is a key starting point for a drug discovery project. In this section, potential molecular targets associated with the development of pancoronavirus antiviral drugs are highlighted; these targets are highly conserved and essential to viral replication or viral pathogenesis among the identified HCoVs.
At least two-thirds of the SARS-CoV genome is occupied by two large open reading frames (ORFs; ORF1a and ORF1b) that together constitute the replicase gene. Polyprotein 1a (PP1a; NSP 1-11) is encoded by ORF1a, while polyprotein 1ab (PP1ab; NSP1-16) is encoded by the combined transcription of ORF1a and ORF1b via a ribosomal frameshift that overreads the stop codon of ORF1a. PP1a and PP1ab are usually further hydrolyzed, releasing the nonstructural proteins (nsp1 to nsp16) that generally interact to form a replication and transcription complex associated with viral RNA synthesis. For example, the polyproteins of SARS-CoV are cleaved into 16 mature nonstructural proteins (16 for SARS-CoV-2 [14]), including NSP 5 (main protease), NSP 12 (an RNA-dependent RNA polymerase), and NSP 14 (3’-to-5’ exoribonuclease) [15,16]. The four main structural proteins (S, E, M, and N) and eight accessory proteins are encoded by the 3’-proximal genes. These proteins are usually not essential for viral replication; however, they may play a role in the pathogenesis of coronavirus infection, particularly in the virus-host interaction [17], for example by modulating interferon signaling pathways [18,19]. Due to their critical roles in the viral life cycle, these important proteins are viewed as potential targets for the development of novel antiviral drugs.
2.1. 3C-like protease (3CLP) or 3C protease (3CP)
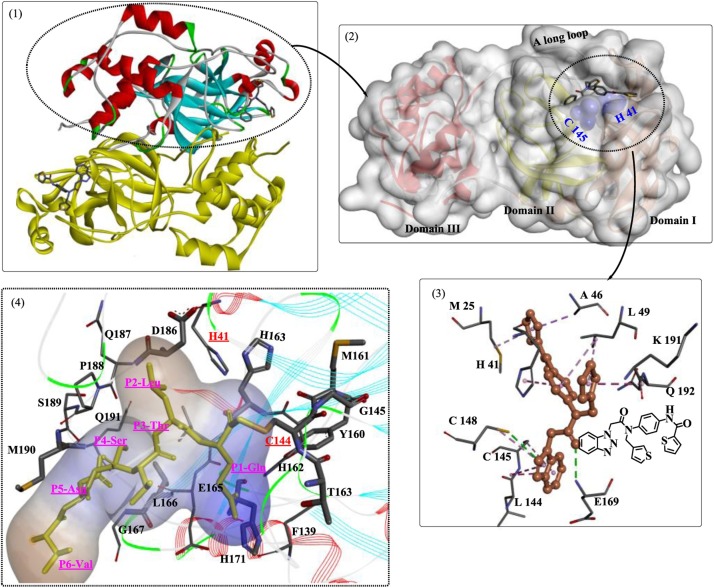
The coronavirus 3CLP (or the main protease, Mpros) is a cysteine protease characterized by chymotrypsin-like folds, a catalytic triad with a nucleophilic cysteine residue, and a preference for a Glu or Gln residue in substrate proteins [20]. It is a key enzyme within the polyproteins, and after autoprocessing [21], active 3CLP cleaves 11 sites (SARS-CoV) in the polyproteins, releasing functional NSPs and structural proteins, such as the RNA-dependent RNA polymerase, helicase, and ribonucleases. Recently, the crystal structure of free SARS-CoV-2 3CLP was solved at room temperature [22], which revealed that the native 3CLP is a homodimer (Fig. 1 ). The dimerization process is essential for its proteolytic activity. Each monomer contains three domains (Domain I, Domain II, and Domain III), and the active site (Cys145-His41 catalytic dyad) of each monomer is located between Domain I and Domain II, constituting the chymotrypsin-like folding scaffold at the interface. Caused by the multiple domains and special location of the active site, the discovery or design of novel inhibitors capable of entering the site readily and forming strong interactions with the target 3CLP might be difficult. Domain III is associated with the dimerization of 3CLP and is connected to the chymotrypsin-like fold by a long loop.
Fig. 1.
Crystal structure of a typical 3C-like protease complexed with the inhibitor and the interaction mode (the structures in 1-3 are drawn based on the crystal structure PDB ID: 4YOI; the structures in 1 and 2 show a solvent accessible surface colored in silver; in the structure presented in 3, the green dashed lines indicate the H-bound, and the pink dashed line indicates the hydrophobic interactions; structures in 4 are drawn based on the crystal structure PDB ID: 1P9U, a hydrophobic surface is colored in blue).
2.1.1. 3CLP inhibitors
To date, many research groups worldwide have focused their drug discovery efforts on the inhibition of 3CLP. The identified inhibitors are divided into two main types: peptidyl compounds and nonpeptide inhibitors. The development of broad-spectrum antivirals based on the inhibition of 3CLP (or 3CP) involves two main fields of research: inhibiting protease activity [23] and targeting the regular dimerization of 3CLP [24]. In addition to the direct screening of potent inhibitors, structure-based drug design is another promising alternative. In this rational strategy, compounds are usually designed to target the dimer interface around the active site [[25], [26], [27]], including (1) the inhibition of binding sites in the active site (S1 site) and (2) the inhibition of the normal dimerization of 3CLP. Due to the highly conserved structure of the substrate-binding pockets among all CoV 3CLPs, potent inhibitors targeting this site are presumed to display wide-spectrum anti-CoV activity.
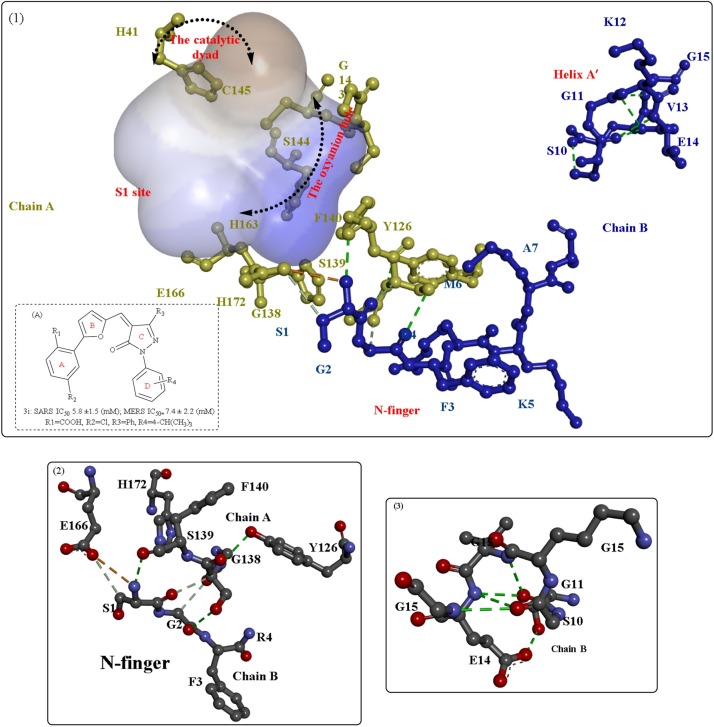
The inhibition of the binding sites is a field of great interest in antiviral drug development, and several important studies have provided insights into the detailed interaction modes of the binding sites [[28], [29], [30]]. Anand et al. [25] synthesized a substrate analog (hexapeptidyl chloromethyl ketone) to investigate the exact binding mode of substrates. The crystal structure showed six critical binding sites occupied by the corresponding residues (Fig. 2 ). In particular, a covalent bond formed between the Sγ atom of Cys145 and the methylene group of CMK (S1 site), and various novel irreversible inhibitors have been developed based on this information [31,32]. The S1 site is located in close proximity to the catalytic dyad, which is stabilized by the oxyanion hole formed by Gly143, Ser144 and His163 (Fig. 2(1)) [33]. In a study by Kumar et al. [34], several compounds with potent inhibitory activity against 3CLP were designed and identified (Fig. 2(1, A)), and these compounds were believed to destabilize the oxyanion hole in 3CLP. The carboxyl group present at P1 of the inhibitors was critical for the destruction of the formed oxyanion hole. Moreover, the conformation of the S1 site or the whole-substrate-binding site was further maintained by the counterpart N-finger (Fig. 2(2)) [35].
Fig. 2.
Schematic of critical residues associated with the rational design of 3CLP inhibitors (PDB ID: 1UK4).
Interference with the correct dimerization of 3CLP, mainly including inhibition of the functions of helix A′ and Domain III, is also a reasonable strategy for the development of antiviral inhibitors. The H-bonds formed between the two A’ helices (Fig. 2(3)) also play important roles in the stabilization of the dimer interface; therefore, these helices are potentially useful antiviral targets. Using this method, several critical residues [28,[36], [37], [38]] were determined to be directly involved in dimerization or dimer stability through hydrogen bonding, such as Gly11, Ser123, Ser 139, Glu169, Arg198 and Gln299. Thus, inhibitors could be designed to target these residues.
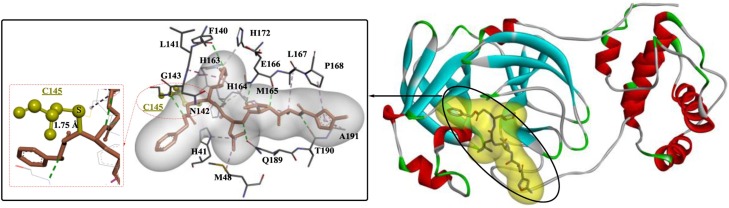
Crystal structures of the main protease of SARS-CoV-2 were elucidated recently and applied in the development of novel antiviral drugs. In one study [39], a wide-spectrum inhibitor (N3) was identified as a potent irreversible inhibitor of the SARS-CoV-2 main protease (Fig. 3 ), with kobs/[I] = 11,300 ± 880 M−1s−1. The structural analysis clearly revealed that the Sγ atom of C145-A formed a covalent bond (1.8 Å) with the Cβ atom of the vinyl group. In addition, the residues Phe140, Asn142, Glu166, His163, His172, and Ser1 of protomer B also play critical roles in S1 subsite formation; His41, Met49, Tyr54, Met165, and Asp187 are associated with S2 subsite formation; and Met165, Leu167, Phe185, Gln192 and Gln189 are involved in S4 subsite formation through H-bond interactions. Then, structure-based drug design, virtual screening and high-throughput screening were used to discover potential inhibitors based on the elucidated crystal structure. The six obtained compounds exerted good inhibitory effects on the main protease of SARS-CoV-2, with IC50 values ranging from 0.67 to 21.4 μM.
Fig. 3.
The crystal structures of the main protease from SARS-CoV-2 and its interactions with the broad-spectrum inhibitor (N3) (PDB ID: 6LU7).
In a similar study [40], a novel inhibitor was designed with an enhanced half-life in the plasma through the incorporation of the P3-P2 amide bond into a pyridone ring. In addition, effective inhibitors of the main protein from SARS-CoV, GC373 and GC376 also exert strong inhibitory effects on SARS-CoV-2, with IC50 values in the nanomolar range (0.40 ± 0.05 μM (GC373) and 0.19 ± 0.04 μM (GC376)) [41]. Furthermore, a structural analysis of the complex showed that the nucleophilic residue Cys145 formed a covalent bond with these inhibitors. Recently, a study of the structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease was reported [42]. The two lead compounds (11a and 11b) obtained by the authors both displayed potent enzyme inhibitory activity (IC50: 0.053 ± 0.005 μM (11a) and 0.040 ± 0.002 μM (11b)), anti–SARS-CoV-2 infection activity (EC50: 0. 53 ± 0.01 μM (11a) and 0.72 ± 0.09 μM (11b)) and good pharmacokinetic properties in vivo. The structure of the complex showed that the aldehyde groups of compounds 11a and 11b were all covalently bound to cysteine 145 of 3CLP, inhibiting its catalytic activity. Panda et al. applied the rational structure-based drug design and immunoinformatics approach to successfully discover efficient drugs against SARS-CoV-2 [43]. The resulting antiviral polymerase inhibitor, PC786, showed multitarget inhibitory effects on the spike glycoprotein, main protease and the receptor binding domain (RBD)/angiotensin-converting enzyme 2 (ACE2) complex of SARS-CoV-2.
2.2. Papain-like protease
Together with 3CLP, papain-like proteases (PLPs) are a critical type of protease that play a vital role in the hydrolysis of viral polyproteins and promote the release of essential nonstructural proteins [44,45]. PLP is also a multifunctional enzyme with deISGylating and deubiquitinating (DUB) activities; moreover, it blocks the interferon regulatory factor 3 (IRF3) pathway [46]. In addition, it functions as a viral ubiquitin-specific protease that is capable of rapidly hydrolyzing the isopeptide bonds of proteins, which are posttranslationally modified by cellular ubiquitin-like (Ubl) molecules, such as ubiquitin (Ub) and interferon-stimulating gene 15 (ISG15) [[47], [48], [49]]. Ub and ISG15 are two important cellular regulators and signaling intermediates that are usually covalently attached to host cell proteins via the formation of an isopeptide bond. Therefore, the development of novel drugs targeting PLP might have great advantages, as these drugs would be capable of not only inhibiting viral replication but also inhibiting the upregulation of collagen expression in infected cells [50].
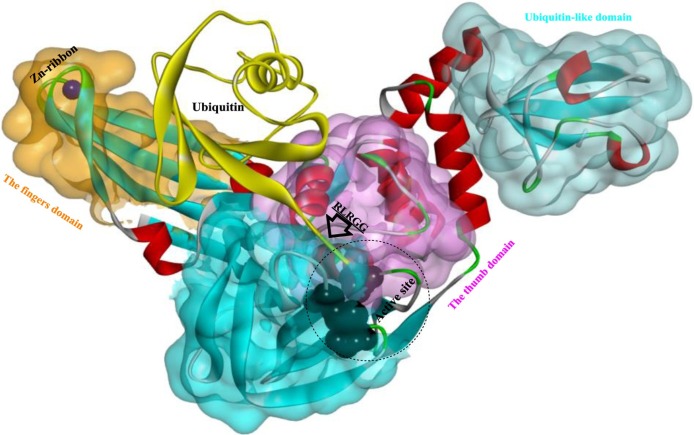
In CoVs, particularly SARS-CoV and MERS-CoV, PLP is composed of four main structural domains (Fig. 4 ): the ubiquitin-like (Ubl), palm, thumb and fingers domains [47,51]. The Ubl domain is highly conserved among CoVs and located directly adjacent to the N terminus of PLP. The peptidase catalytic core is located at the interface of the thumb and palm domains, and the catalytic triad was determined to be Cys112-His273-Asp287. The five C-terminal residues of Ub (RLRGG) efficiently bind to the narrow active site channel between the thumb and palm domains (Fig. 5 ) [52]. In the finger domain, a zinc ion is stabilized by four cysteine residues, and correct binding of zinc is indispensable for protease activity [53]. Remarkably, although their structures are closely related, the PLPs from MERS-CoV and SARS-CoV show significant differences in their capabilities to recognize and hydrolyze the same substrates, such as UB and ISG15 and to cleave the isopeptide bonds of K48- and K63-linked polyubiquitin chains [54].
Fig. 4.
Schematic of the PLP from MERS-CoV in a complex with human ubiquitin (PDB ID: 4RUR).
Fig. 5.
The interaction mode of Ub (particularly the C-terminal five residues (RLRGG)) with the PLP (PDB ID: 4RUR).
Two distinct Ub-binding subsites (the proximal SUb1 and the distal SUb2 sites) have been identified around the active site (S1'), and SARS-PLP preferentially recognizes and then releases diUbLys48 during the cleavage of polyUb chains through a ‘didistributive’ cleavage mechanism [55,56]. In contrast, MERS-PLP shows broad substrate specificity for the cleavage of polyUb chains in a ‘monodistributive’ manner. Among other DUBs, diUb recognition is commonly achieved across S1- S1’ [57,58]. However, S2Ub binding was recently shown to be absolutely vital for SARS-PLP activity, particularly in the specific recognition of K48-linked polyubiquitin [59]. The PLP of MERS-CoV was also reported to display stronger deubiquitinating activity but lower proteolytic activity than the PLP of SARS-CoV [60]. Based on these results, the development of broad-spectrum antiviral drugs against CoV PLPs would be a challenging task, considering the different substrate specificities of the PLPs from different CoVs.
2.2.1. PLP inhibitors
To date, various potent inhibitors of PLPs have been discovered and designed [61], such as thiopurine compounds, natural products (e.g., tanshinones), zinc ions (Zn2+), zinc conjugate inhibitors, and naphthalene inhibitors.
-
I
Natural products
Due to the multifunctional nature of PLP, the development of novel antiviral drugs could be focused on the discovery of inhibitors that may either efficiently inhibit protease activity [62] or inhibit deubiquitinating activities [63]. In a study by Park et al. [62], tanshinones (a natural product) from Salvia miltiorrhiza ethanol extracts were determined to be selective, slow-binding inhibitors of SARS-CoV cysteine proteases (3CLP and PLP) that were more potent than peptide-derived and small-molecule viral cysteine protease inhibitors. Although potent activities were observed, the inhibitory effects of these tanshinones strongly depend on the chemical structure, and the PLP inhibitory activity of these compounds is usually more significant than the 3CLP inhibitory activity. Moreover, they were determined to be selective inhibitors without detectable inhibitory effects on other proteases, such as chymotrypsin, papain, and the HIV protease.
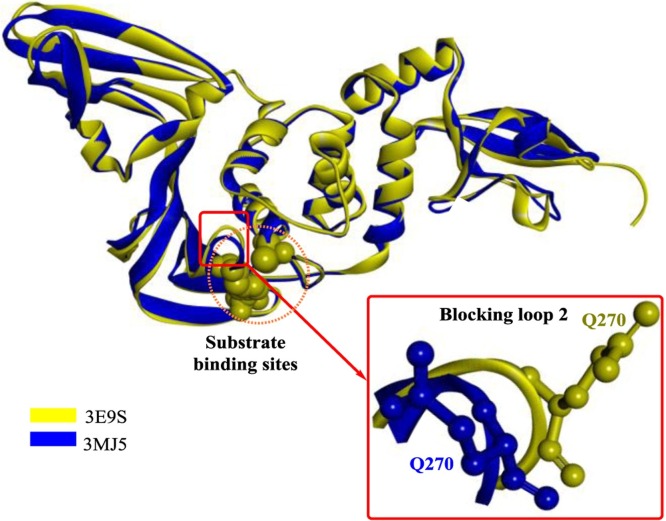
In another study [64], polyphenolic natural products isolated from Broussonetia papyrifera were determined to be potent coronavirus protease inhibitors (3CLP and PLP). Six diarylheptanoid compounds from Alnus japonica with inhibitory activities against SARS-PLP were discovered [65], among which hirsutenone (2) showed the most potent inhibitory activity, with an IC50 of 4.1 μM. Furthermore, the catechol and α,β-unsaturated carbonyl moieties might play a critical role in PLP inhibition. In one study [66], high-throughput screening (HTS) was applied to screen novel inhibitors of both SARS-PLP and MERS-PLP, and a dual noncovalent inhibitor (compound 4) showing potent inhibitory activity against both PLPs was discovered. This compound is the first MERS-PLP inhibitor reported, and it functions as a mixed-type inhibitor of SARS-PLP and a competitive inhibitor of MERS-PLP. Unfortunately, the crystal structures of compound 4 and the MERS-PLP complex were not provided; therefore, the detailed binding mode and interactions were not obtained experimentally. However, an analysis of mutants revealed that Asn109 is a critical residue required for intermediate stabilization through the formation of H-bonds. The structural alignment of SARS-PLP and MERS-PLP showed that the configuration of Gln270 (Fig. 6 ) is quite different from the configuration of the other amino acids, and thus it might function in an unknown way. Based on the results of this study, blocking loop2 (Fig. 6) was further confirmed to play a crucial role in the differences in substrate specificities that lead to the different mechanisms of action of compound 4.
Fig. 6.
The structural alignment of SARS-PLP and MERS-PLP.
Although many inhibitors derived from natural products have been identified and determined to be promising lead compounds for the rational development of more potent anticoronavirus agents [67,68], no detailed studies on their mechanisms of action have been published. Therefore, researchers should focus on performing more in-depth crystallographic analyses of this inhibitory process.
-
II
Naphthalene inhibitors
-
decimal(III)
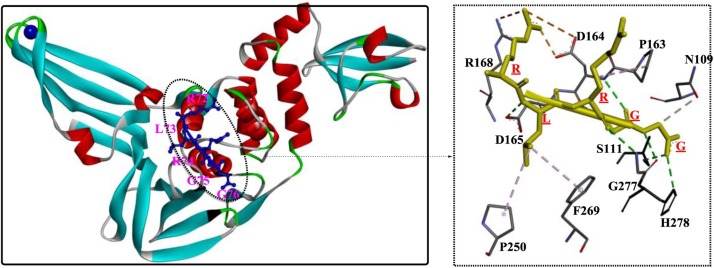
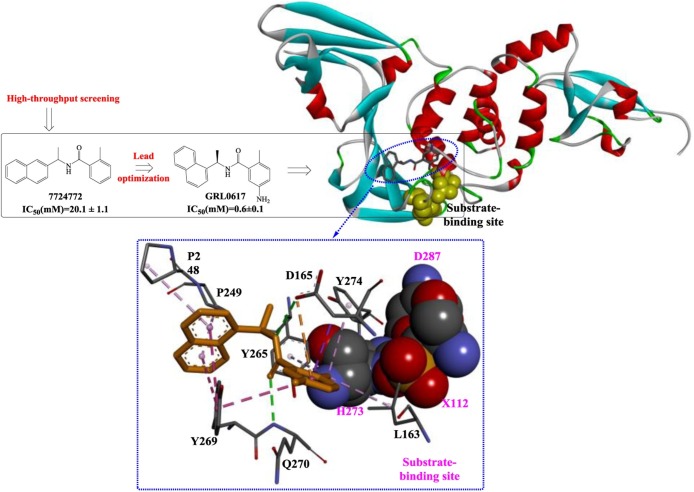
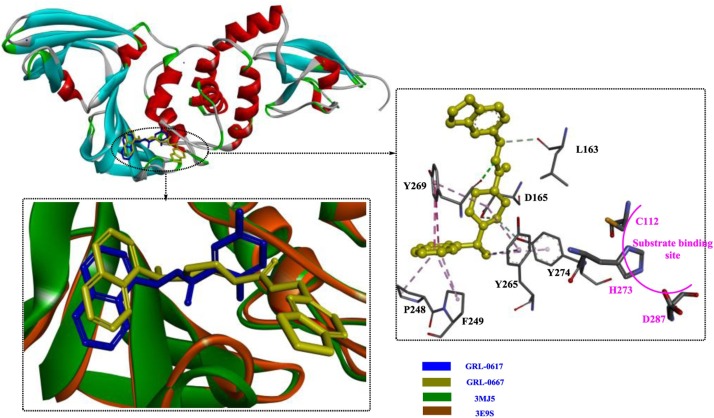
In a study by Ratia et al [69], a first-generation lead compound, 7724772, was discovered in an HTS; this compound functioned as noncovalent competitive inhibitor of the PLP of SARS-CoV. Then, chemical optimization was performed and confirmed by conducting a structure-activity analysis, which showed that the potency dramatically increased to 0.6 ± 0.1 μM (IC50). The crystal structures showed that GRL0617 binds the S4-S3 subsites located around the active site (Fig. 7 ), forming two H-bonds (among Gln270, Asp165 and GRL0617) and a series of hydrophobic interactions to stabilize this conformation. Once bound, GRL0617 further induces the closure of blocking loop 2 and the movement of the side chain of Leu163, blocking access to the catalytic triad and inhibiting deubiquitination activity. In addition, Trp107 and Asn110 were identified as critical residues that support the stabilization of the oxyanion hole of SARS-PLP. These observations are very important for the rational design of novel PLP inhibitors.
-
decimal(IV)
In subsequent studies [70], more potent related derivatives of GRL0617 were identified, in which compound GRL-0667 exerted the strongest inhibitory effect, with an IC50 of 0.32 μM. Based on an analysis of the crystal structure (Fig. 8 ), the binding modes of GRL0617 and GRL-0667 in the hydrophobic pocket are quite different. In particular, for GRL0617, blocking loop 2 shifts away from the inhibitor, resulting in the disruption of the H-bond between Gln270 and the inhibitor. Instead, an H-bond between Tyr269, which is a conserved amino acid residue among CoVs, and GRL-0667 forms. In contrast, the naphthyl rings of both GRL0617 and GRL-0667 are aligned in a similar manner.
-
decimal(V)
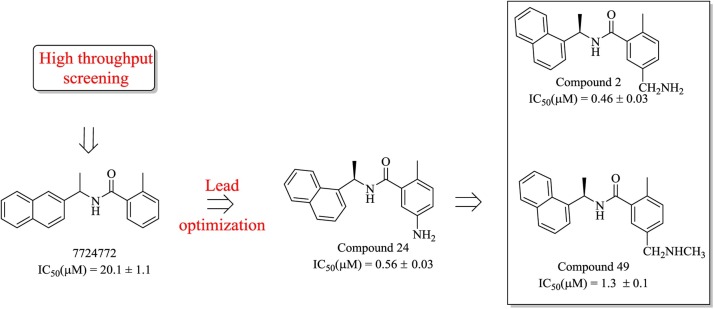
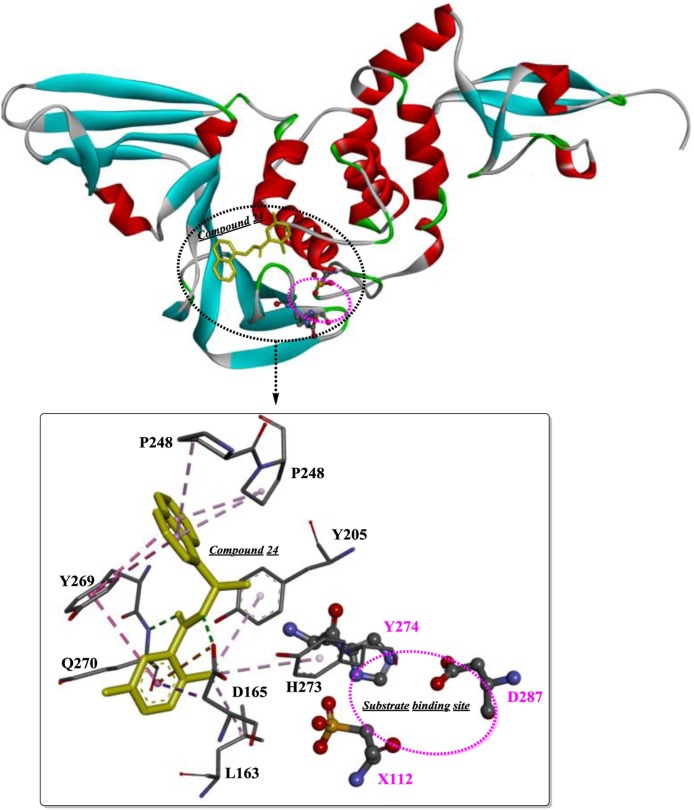
Compound 7724772 was further optimized by Ghosh et al [71] based on a structure-activity analysis, leading to the discovery of compound 24 and compound 2 (Fig. 9 ). As shown in Fig. 10 , the 5-amino-2-methylphenyl ring is located exactly at the entrance of the substrate-binding sites, and the methyl branch forms several weak hydrophobic interactions (shown as light pink dotted lines). Moreover, the two H-bonds formed between the carboxamide group and Asp165 and Gln270 effectively stabilize the binding of compound 24. Therefore, the extension of the methyl branch further into the binding pocket would theoretically strengthen these interactions. The resulting compound 2 with 5-methylamine substituents on the benzamide ring exhibits a slight improvement in inhibitory potency, which was estimated to be caused by the additional H-bonds formed between 5-methylamine and Gln270 and/or Tyr269. Moreover, based on quantitative structure-activity relationship (QSAR) studies, a 3D-QSAR model with the CoMSIA descriptors was provided as y = 0.795 + 0.815x (R2 = 0.9811, y indicates the predicted inhibition, x indicates the actual inhibition). This approach is believed to be a reliable guide for the rational design or lead optimization of novel PLP inhibitors through structure-based drug design.
-
VI
Second-generation naphthalene inhibitors
Fig. 7.
Schematic of the development of GRL0617 and its interaction mode with the PLP from SARS-CoV (PDB ID: 3E9S).
Fig. 8.
The interaction modes of GRL-0617 and GRL-0667 with the target PLP from SARS-CoVs (PDB ID: 3MJ5).
Fig. 9.
The process used to optimize compound 7724772, leading to the discovery of compounds 2 and 49.
Fig. 10.
The binding mode and the interaction of component 24 with PLP (PDB ID: 3E9S).
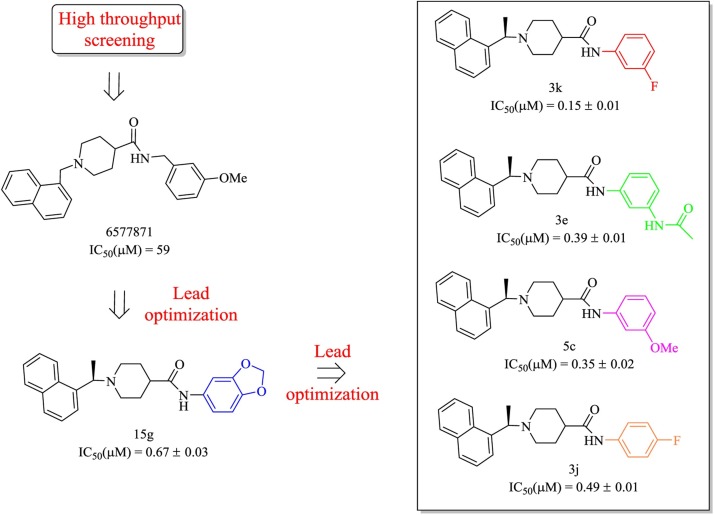
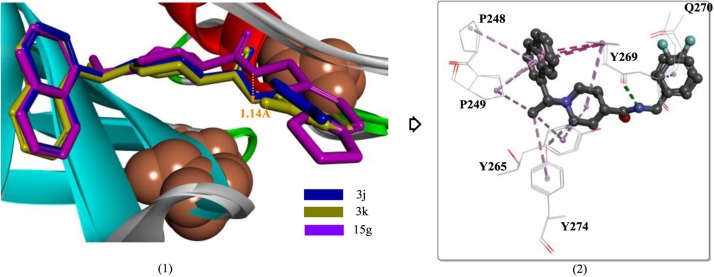
In previous studies, GRL-0667 and compound 24 were successfully designed as potent inhibitors of SARS-CoV PLP. Among the chemical structures, the methyl group at the stereogenic center plays a critical role in the inhibitory potency. Although these two compounds were proven to be highly efficient inhibitors with low toxicity in SARS-CoV-infected Vero E6 cells, they were unable to satisfy the clinical requirements, such as inhibitory potency and physicochemical properties. Further chemical optimization of GRL-0667 led to the discovery of several more promising inhibitors, including the most potent inhibitor, compound 3k (Fig. 11 ) [72]. In particular, compounds 3e and 5c, although they were less potent, both exhibit significantly improved metabolic stability and are proposed to be better candidates for further drug development.
Fig. 11.
The process used to optimize compound 6577871 for the development of the second-generation naphthalene inhibitors.
Based on the corresponding crystal structures, the binding modes of compounds 15 g, 3k and 3 j are quite similar, except for the position of the carboxamide nitrogen of compound 15 g, which exhibits a 1.14-Å difference compared to compound 3k (Fig. 12 (1)). Although the interaction of compound 3k with PLP is almost the same as that of compound 3 j and PLP, the stronger inhibitory activity of compound 3k is presumed to be caused by the stronger interactions (e.g., van der Waals interactions) between m-fluorobenzene and the side chains of Tyr269 and Gln270. This explanation is plausible because the orientation of m-fluorobenzene in compound 3k is exactly facing the region formed by Tyr269 and Gln270 (Fig. 12(2)), which are two key residues associated with inhibitor binding.
-
IV
Thiopurine compounds
Fig. 12.
The binding modes of compounds 15 g, 3k and 3 j in the active site of SARS-CoV PLP (PDB ID:4OW0).
Thiopurine compounds are purine analogs with widespread applications in various therapies, such as cancer treatment. In particular, 6-mercaptopurine (6 M P) and 6-thioguanine (6 TG) were reported to display a broad spectrum of activities, including efficacy in the treatment of children with acute lymphoblastic or myeloblastic leukemia or SARS-CoV infection [73]. In addition, 6 M P and 6 TG also target MERS-CoV PLP synergistically as competitive inhibitors [74]. According to a docking study, 6 M P is capable of binding the catalytic triad of MERS-CoV PLP and is stabilized by three H-bonds with Asn109, His278 and Gly277. Therefore, as promising lead compounds, further optimization of 6 M P and 6 TG to improve their potency will be very important in the development of potent inhibitors of PLPs.
-
V
Other important inhibitors
HTS is a traditional strategy for the discovery and screening of novel hits in the initial phase of drug discovery. As an HTS method, a yeast-based assay was recently developed for rapid and efficient screening of PLP inhibitors [75]. In this method, the expressed PLP would lead to a remarkably slow growth phenotype in Saccharomyces cerevisiae, and the induced slow growth phenotype would probably last for over 60 h. Inhibitors capable of relieving the inhibition caused by PLP and restoring cell growth would be clearly identified with this assay. With this method, compound NSC158362 was identified to specifically and potently inhibit SARS-CoV replication in cell culture without producing cytotoxicity. However, NSC158362 does not inhibit PLP, exhibiting no effect on protease activity or deubiquitination activity, which is believed to be a new antiviral mechanism. Compound NSC158011 was determined to inhibit PLP in a cell-based assay but was unable to inhibit viral replication.
2.3. RNA-dependent RNA polymerase (RdRp)
2.3.1. Analyses of the structure and function of RdRp
The RNA polymerase of RNA viruses represents an attractive target for the development of novel antiviral drugs that inhibit RNA synthesis. In coronaviruses, NSP 12, an RNA-dependent RNA polymerase, is the most conserved protein and functions as a central enzyme associated with viral replication/transcription [76,77]. However, due to the lack of sufficient amounts of highly purified proteins, few crystal structures of CoV NSP 12 are available, and only two cryo-EM (cryo-electron microscopy) structures (PDB ID: 6NUS and 6NUR) [78] have been determined and deposited in the RCSB PDB (https://www.rcsb.org/). Previously, the SARS-CoV RdRp NSP 12 was reported to be a nonprocessive primer-dependent RNA polymerase [79]; however, NSP 7 and NSP 8, two essential cofactors, activate and confer processivity to NSP 12 [80]. The NSP 7-NSP 8 dimer was recently shown to be located too far from the NSP 12 active site (PDB ID: 6M71 and 6YYT) to function as a primase for NSP 12 [81,82]. Recently, Wang et al. [83] provided cryo-EM structures of the SARS-CoV-2 RNA polymerase in complexes with RNA during the RNA translocation process (PDB ID: 7BZF and 7C2K), which showed that the NSP 12/NSP 7/NSP 8 complex would undergo structural rearrangements to accommodate nucleic acid binding.
Currently, the complex (NSP 12/NSP 7/NSP 8) with RNA synthesis and processing activities represents the minimal elements required for nucleotide polymerization, and polymerase activity is strictly dependent on Mn2+ [76]. In addition to the NSP 12/NSP 7/NSP 8 complex, other NSPs associated with RNA processing activity have also been identified, including a 3’-5’ exonuclease in the N-terminal domain of NSP 14 and an endoribonuclease in NSP 15 [77]. However, another study showed that the SARS-CoV-2 NSP 7-NSP 8 complex might have different functions, for example, it may function as a processivity factor, primase or nsp12 activating factor [84]. The crystal structures of endoribonuclease NendoU from SARS-CoV-2 have been already determined [85], and as a hexamer, the structure is homologous to those from SARS- and MERS-CoVs. It is also suggested that inhibitors of SARS-CoV Nsp15 are probably capable of inhibiting the protein from SARS-CoV-2.
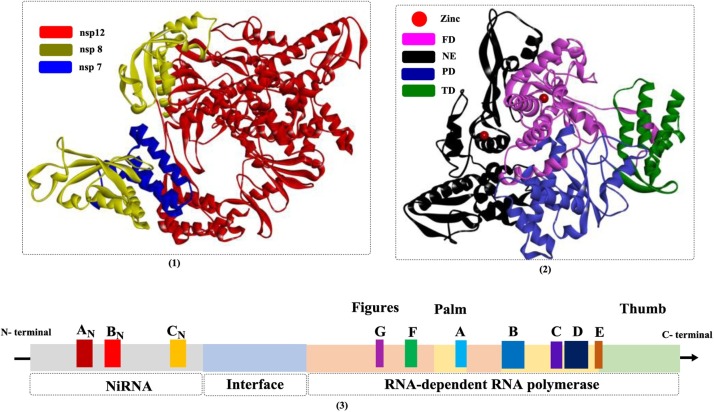
In a previous study [78], the structure of NSP 12 (Fig. 13 ) was clearly shown to be composed of a nidovirus-unique N-terminal extension (NE) and a C-terminal polymerase domain. The NE domain (referred to as nidovirus RdRp-associated nucleotidyltransferase (NiRAN)) is regarded as a nidoviral signature domain, and the region has been further divided into three conserved subdomains (AN, BN, and CN) based on a sequence analysis [86]. The functions of the highly flexible nidovirus-unique extension have not been investigated thoroughly, but this NE is estimated to have nucleotidylation activity essential for the replication of nidoviruses. Several conserved residues (e.g., Lys94, Arg124, Phe166) have been proven to play critical roles in influencing UTP/GTP binding; therefore, inhibiting the activities of these residues would be another promising strategy for the development of anti-CoV drugs. The core polymerase complex (NSP 12/NSP 7/NSP 8, 7BW4) from SARS-CoV-2 has been determined and closely resembles its counterpart in SARS-CoV, exhibiting similar conserved motifs and an activation mechanism catalyzed by cofactors [87]. However, biochemical studies revealed that it showed a lower enzymatic activity and reduced thermostability compared with the SARS-CoV counterpart.
Fig. 13.
The crystal structures of NSP 12-NSP 7-NSP 8 complex (1 and 2) and the diagram of the conserved motifs of SARS-CoV NSP 12 proteins (PDB ID: 6NUR; FD: fingers domain; NE: N-terminal extension; PD: palm domain; TD: thumb domain).
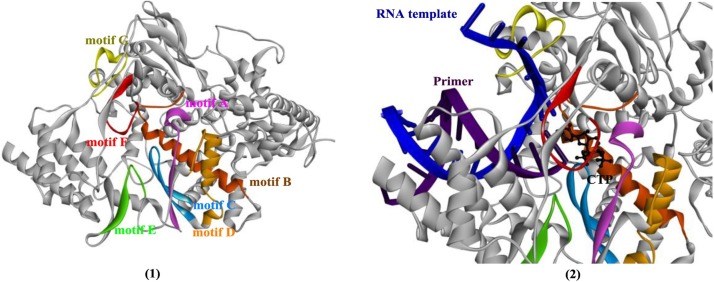
The polymerase domain is usually described as a “cupped right hand” and is specifically composed of a finger domain, a palm domain, and a thumb domain. In addition, CoV NSP 12 also contains two metal (zinc)-binding sites. Each domain is usually responsible for different activities, such as the binding of templates and NTPs, initiation, and elongation [88]. Sequence analysis of the CoV NSP 12 family reveals that the residues involved in metal binding, substrate transport, polymerase active sites and the NE are all highly conserved. The substrate (template and nucleotide)-binding and catalytic sites are composed of seven conserved motif regions (A–G, shown in Fig. 14 ), in which motifs A–E are responsible for RNA polymerization, motif F is capable of NTP binding, and motif G is used for template binding (Fig. 14(2)). The polymerase active site (Ser-Asp-Asp) is conserved in all nidoviruses and located in motif C.
Fig. 14.
The crystal structures of NSP 12 and the seven conserved motifs (1, PDB ID: 6NUR), and the analysis of RNA template, primer and the CTP binding modes by protein structure alignment of 3H5Y (PDB ID).
Thr680 and Asn691 in motif B and Asp623 in motif A facilitate NTP binding through H-bonds. In addition, Val557 in motif F might also contribute to base pairing and modulate polymerase fidelity [89]. Several important conserved residues in NSP 8 (Pro183, Arg190 and Lys58) have also been reported to be essential for the survival of viruses, and three residues in NSP 7 (Lys7, His36, and Asn37) play a role in the RNA binding of the polymerase complex [80]. All the critical residues mentioned above could be targeted for the development of inhibitors of viral RNA replication.
2.3.2. Inhibitors of RdRp
-
I
Remdesivir (RDV; GS-5734)
Remdesivir is a phosphoramidate (1’-cyano-substituted adenosine nucleotide analog) prodrug developed for the treatment of Ebola virus (EC50 = 86 nM) [90] that shows broad-spectrum antiviral activities against Ebola virus, SARS-CoV, and MERS-CoV [91,92]. RDV was designed to transport nucleoside monophosphates into host cells for the efficient formation of active triphosphates. The active triphosphates delivered by RDV show great advantages, specifically potent RdRp-inhibitory effects, high selectivity, and a long intracellular half-life. Once incorporated at position I, RDV will compete with natural NTPs, arresting RNA synthesis at position I + 3 and thereby delaying RNA chain termination [93]. Another study [94] showed that inhibitor (remdesivir-MP) binding at position I did not affect the ensuing nucleotide incorporation event at position I + 1, with an ∼6-fold inhibition observed at this position. However, chain termination was identified predominantly at position I + 5, which might not be overcome by higher substrate concentrations. Moreover, RDV is likely protected from excision performed by the proofreading activity of the 3’-5’ exonuclease (NSP 14) [95].
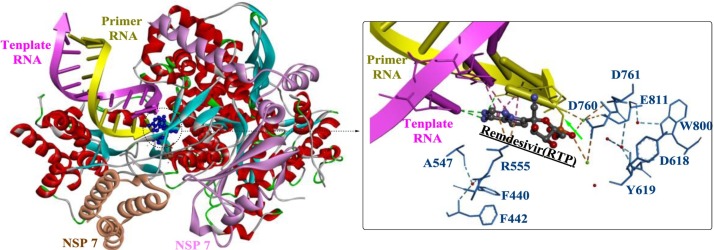
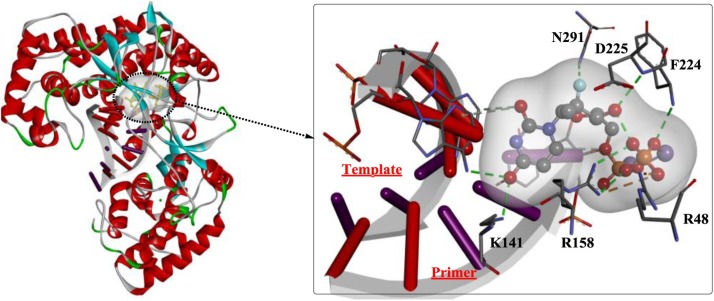
An analysis of the structure-activity relationship revealed that the 1’-CN group and C-linked nucleobase of RDV might play critical roles in the potent anti-Ebola virus (EBOV) potency and selectivity of RDV [96]. Moreover, recent studies provided a structural basis for the mechanism by which remdesivir inhibits the RdRp from SARS-CoV-2 [97,98]. As shown in Fig. 15 , the partial double-stranded RNA template inserts into the central channel of the RdRp, and remdesivir forms a covalent bound with the primer strand at the first replicated base pair (indicate by the green arrow), terminating further chain elongation.
-
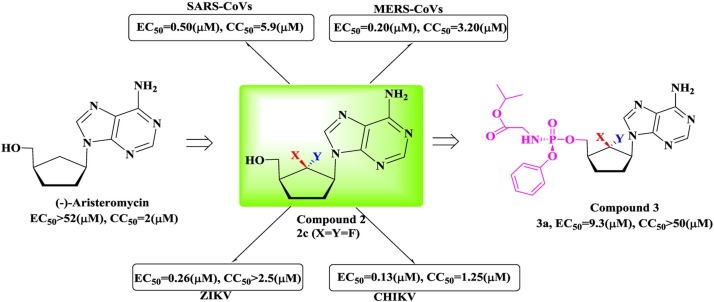
II
6′-Fluorinated-aristeromycin analogs
Fig. 15.
The crystal structure of the remdesivir- and RNA-bound RdRp complex (PDB ID: 7BV2).
The 6’-fluorinated aristeromycins are a series of multifunctional antiviral compounds that inhibit both the RdRp of the virus and the S-adenosyl-l-homocysteine (SAH) hydrolase of host cells [99]. Inhibition of SAH hydrolase indirectly blocks methyltransferases (MTases) via feedback inhibition, which disturbs the normal capping of viral RNA catalyzed by MTases [100,101]. (-)-Aristeromycin is a natural carbocyclic nucleoside that is a type I SAH hydrolase inhibitor, and it displays potent antiviral activity but severe cytotoxicity. Therefore, it was used as a lead compound and optimized to produce novel dual-function drugs (Fig. 16 ). Given the substantial effect of the fluorine introduced at the 6’-position of carbocyclic nucleosides, several 6′-fluorinated-aristeromycin analogs have been designed and synthesized with fluorine introduced at the 6’-position. In addition, 6’-fluorinated purine and pyrimidine nucleosides and a phosphoramidate prodrug (3) were also synthesized for drug development. The results obtained for these molecules showed that all the adenosine derivatives potently inhibit the activity of SAH hydrolase; however, none of the pyrimidine analogs inhibit SAH hydrolase. The most potent inhibitor was determined to be 6’-β-fluoroaristeromycin (2a, IC50 = 0.37 μM), the activity of which was 3.6-fold higher than that of (-)-aristeromycin (IC50 = 1.32 μM). The difference in the inhibitory effect between compounds 2a and 2b reveals that the stereochemistry at the 6′-position may substantially affect the inhibitory activity. Although none of the phosphoramidate prodrugs inhibited SAH hydrolase, the adenosine prodrug an exhibited certain antiviral activities (EC50 = 6.8 μM for SARS-CoV). Thus, compound 3a would probably inhibit the RdRps of RNA viruses.
-
III
Ribavirin
Fig. 16.
The strategy for the design of 6′-fluorinated aristeromycins analog (compound 2) as broad-spectrum antiviral agent.
Ribavirin (1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide) is another nucleoside analog with broad-spectrum antiviral activity that displays inhibitory activity against various RNA viruses [[102], [103], [104]], such as the influenza virus, HCV, RSV, HSV, and CoVs. Although it has been approved for use in the treatment of SARS-CoV-2 [105], ribavirin has been shown to display limited efficacy at very high doses in previous studies [104]. Given this limited efficacy, the combination of ribavirin and other antivirals, such as interferon-α2b [106], telaprevir and boceprevir [107] or favipiravir, is usually effective. Moreover, the incorporated ribavirin 5′-monophosphate is easily excised from RNA due to the effect of the 3’-5’ exoribonuclease (ExoN) [108]. Therefore, in the development of efficient RdRp inhibitors, a promising strategy is to simultaneously inhibit RdRp and ExoN activities, particularly with nucleoside analogs.
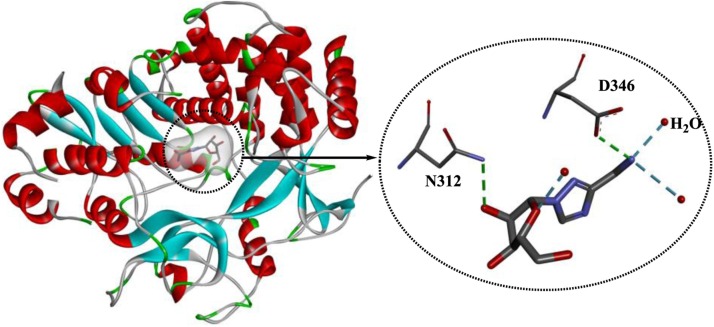
Although ribavirin was identified in 1970 [109], its mechanism of action is still not completely understood, and the mechanisms of action of ribavirin appear to be quite different for various viral pathogens. First, as a nucleoside analog, ribavirin may play an important role in inhibiting viral replication by directly inhibiting the activity of HCV RdRps and disturbing the cellular enzyme IMP dehydrogenase [110]. Second, ribavirin functions as an immunomodulatory agent, promoting interferon signaling [111] and the type 1 cytokine-mediated immune response [112]. Third, ribavirin also functions as a mutagen for HCV and hepatitis E virus, leading to mismatches and subsequent nucleotide substitutions in the viral genome [113,114]. In addition, when used to treat Lassa virus (LASV) infection, ribavirin protects infected cells from dying, probably by modulating the inflammatory response [115]. Various crystal structures of ribavirin in complex with RdRps from different viral pathogens have been released. The crystal structure 3SFU (PDB ID, Fig. 17 ) shows the complex of murine norovirus-1 RdRp with ribavirin in the absence of RNA [116], which illustrates the key interactions involved in recognition of this nucleoside analog within the active site of RdRp. Ribavirin is mainly stabilized by the H-bonds formed between the drug and Asp346/Asn312/H2O. In addition, hydrophobic interactions are also observed between Ser303, Thr309 and Gly345 and ribavirin. Due to the absence of RNA, the interactions identified are associated with the recognition of the nucleoside analogs that block the active site of the viral RdRp, which is expected to be helpful for the design of nucleoside-based inhibitors.
Fig. 17.
The crystal structure of murine norovirus-1 RdRp in complex with ribavirin (PDB ID: 3SFU).
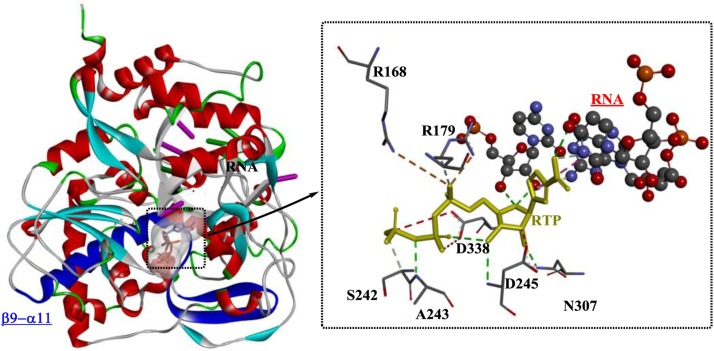
Based on the crystal structures of the RdRp of foot-and-mouth disease virus (FMDV) in complex with different substrates and template-primer molecules, key interactions associated with viral RNA replication have been determined, which provide insights into the mechanism underlying the low fidelity of viral RdRps [117]. As shown in Fig. 18 , RTP (ribavirin triphosphate) is located adjacent to the 3′ terminus of the primer. The position of the RTP base is further stabilized by H-bonds with Ala243, Asp245, Asp338 and Asn307 (within the loop β9–α11) and hydrophilic interactions with Arg168 and Arg179. Clearly, RTP functions as an adenylate analog that base pairs to the acceptor base U4 of the template strand. This action may be caused by the flexible loop β9–α11, which is believed to be capable of adapting its conformation to different template bases to allow the entry of various nucleotides during the RNA elongation process.
Fig. 18.
The crystal structure of RdRp from foot-and-mouth disease virus in complex with RTP and template-primer molecules (PDB ID: 2E9R).
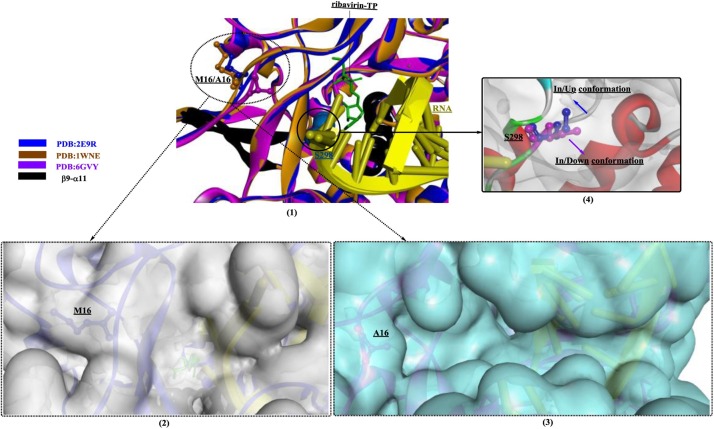
Domingo et al. [118] investigated the functions of the nuclear localization signal (NLS) in the N-terminal region of the RdRp of FMDV. An M16A mutant of FMDV RdRp showed approximately normal polymerase activity; however, the RTP incorporation rate was obviously increased. Therefore, although the mutant virus was capable of replication, it would become more sensitive to ribavirin due to the decreased replication fidelity of RdRp. Through structural comparisons of the mutant RdRp (PDB ID: 6GVY) and the wild-type enzyme (PDB ID: 2E9R and 1WNE), the M16A mutation was shown to lead to the formation of a wider pocket/channel, contributing to the efficient entry and displacement of different nucleotides (e.g., RMP) at position T-2 (Fig. 19 (1–3)). This change is believed to be associated with the altered replication fidelity of RdRps. Interestingly, the conformation of S298 (“in/up” or “in/down”, (Fig. 19(4)) is closely related to the status of RTP incorporation, which may be a structural signature of the lethal mutagenesis of FMDVs.
-
IV
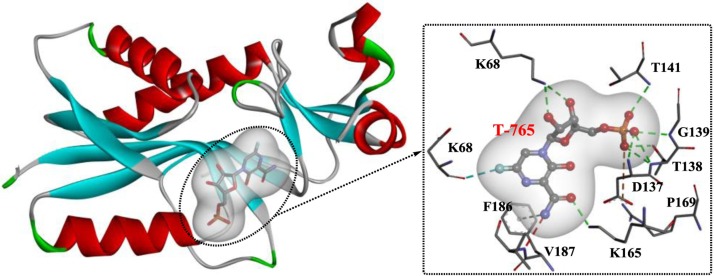
Favipiravir (T-705)
Fig. 19.
Structural comparisons of the mutant RdRp (PDB ID: 6GVY) and the wild-type enzyme (PDB ID: 2E9R, 1WNE) in complex with ribavirin-TP.
Favipiravir (T-705) is a selective and potent inhibitor of the RdRp of RNA viruses that was discovered by Toyama Chemical Co., Ltd., for the treatment of influenza virus [119] through the chemical modification of a pyrazine analog. Favipiravir is a broad-spectrum inhibitor that is effective against most influenza viruses (including drug-resistant influenza [120]) and some other RNA viruses [[121], [122], [123], [124]]. However, due to the risk of teratogenicity and embryotoxicity, it is approved for only restricted clinical use against NAI-resistant pandemic influenza viruses in Japan [125].
Similar to other nucleoside drugs, favipiravir is transported intracellularly and is first converted into its ribose-5′-monophosphate form (T-705-RMP), which is catalyzed by hypoxanthine guanine phosphoribosyltransferase (HGPRT) [126]. Then, T-705-RMP is further processed into the final ribofuranosyl 5′- triphosphate metabolite (T-705-RTP), which is recognized by RNA polymerase as a substrate that competes with GTP, leading to the inhibition of viral RNA synthesis and induction of lethal mutagenesis within the viral genome [127]. A recent study showed that the T-705-modified single-stranded RNA associated with virus nucleoprotein might result in nucleotide splaying [128]. However, the mechanism underlying the activation of T-705 remains unknown, and further rational design or structural optimization of T-705 may be focused on the target of HGPRT. Based on the crystal structure of T-705-RMP in complex with human HGPRT (Fig. 20 ) [126], the fluorine at the 6′-position is required for T-705-RMP antiviral activity and acts by interacting with Lys68 of HGPRT through an H-bond. In addition, the amide at the 3′-position is also necessary because it binds to Lys165 and Phe186 through H-bonds and Pi interactions.
-
V
Sofosbuvir
Fig. 20.
Interaction mode of favipiravir in the active site of HGPRT (PDB ID: 4KN6).
Sofosbuvir (GS-7977) is a potent HCV-specific uridine nucleotide analog inhibitor of the HCV polymerase NS5B [[129], [130], [131], [132]]. However, according to recent studies, it may also exert an inhibitory effect on HCoVs, such as SARS-CoV-2 [133,134]. Sofosbuvir is part of the first class of direct-acting antivirals approved for the treatment of HCV infection in either IFNα-containing regimens or particularly IFNα-free regimens (e.g., velpatasvir/sofosbuvir [135,136], ledipasvir/sofosbuvir [137], and simeprevir/sofosbuvir [138]). Once activated by human histidine triad nucleotide binding protein 1 (hHint1) [139], the released nucleoside monophosphate undergoes further phosphorylation, forming the final uridine triphosphate analog as a chain terminator of RdRp [140]. The final product shows a pangenotypic antiviral effect with high sustained virological response (SVR) rates, minor side effects, a high barrier to resistance, and a low potential for drug-drug interactions (DDIs).
Although crystal structures showing the interaction of sofosbuvir with the RdRps of hCoVs have not been reported, other antiviral targets have been investigated. Appleby et al. explored the RNA replication mechanism catalyzed by hepatitis C virus polymerase and key interactions of inhibitors (sofosbuvir) in the active site of NS5B, the RdRp (Fig. 21 ) [141]. The authors found that the highly conserved residues in the active site are capable of stabilizing the primer for in-line attack on the accepted nucleotide; furthermore, a β loop and a C-terminal membrane-anchoring linker are responsible for occluding the active site cavity in the apo state. During elongation, this linker would retract during primed initiation assembly to enforce replication of the HCV genome from the 3’ terminus and vacate the active site cavity.
Fig. 21.
Interaction mode of sofosbuvir in the active site of hHint1 (PDB ID: 4WTG).
2.4. Methyltransferases (MTases)
Similar to the cellular messenger RNAs of higher eukaryotes, coronavirus viral genomic RNA also possesses a cap moiety at the 5’ end, which would protect it from degradation by 5’–3’ exoribonucleases, improve its stability and ensure its efficient translation [101,142,143]. The 5’ guanosine cap is usually formed by methylation at the N-7 and 2’-O positions mediated by specific MTases, such as guanine-N7-methyltransferase (N7-MTase, NSP 14) [144] and 2’-O-methyltransferase (2’-OMTase, NSP 16) [145]. N-7 methylation is essential for RNA translation, and S-adenosyl-l-methionine (SAM) is usually the methyl group donor of both N7-MTase and 2’-OMTase. Based on the sequence alignment, NSP 10 and NSP 16 are highly conserved among lineage B of the β coronaviruses [146].
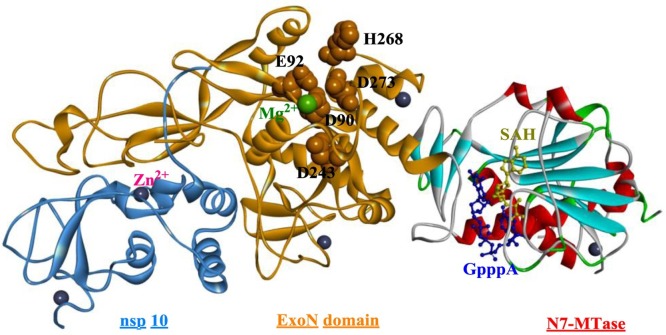
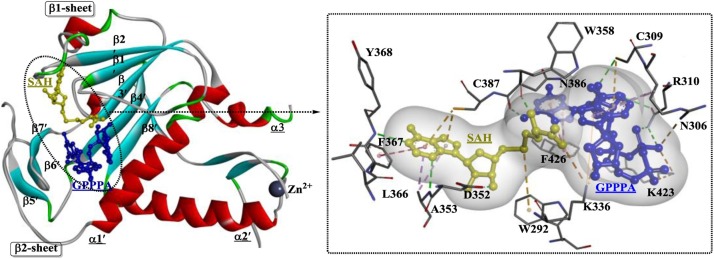
Coronavirus NSP 14 was previously shown to display 3’-to-5’ exoribonuclease activity [147], but detailed studies have shown that NSP 14 is a bifunctional enzyme and that the N7-MTase domain is located in the C-terminus of NSP 14 [148]. Moreover, the exoribonuclease domain is closely associated with the N7-MTase activity. The structural evidence for N7-MTase activity is relatively limited, and crystal structures of the SARS-CoV NSP 14-NSP 10 complex have been generated (Fig. 22 ) [149]. NSP 10 interacts with the ExoN domain to stabilize NSP 14, which is capable of stimulating NSP 14 activity. NSP 14 is a DEDDh-type exoribonuclease, and the DEEDh motif (Asp90, Glu92, Asp243, His268, and Asp273) is responsible for nucleotide excision. Another characteristic that distinguishes NSP 14 from other proofreading exonucleases is the presence of two zinc fingers, which are essential for the normal function of NSP 14. The N7-MTase domain contains a special MTase fold composed of five β-strands, including the parallel β2′, β1′, β3′, and β4′ strands and the antiparallel β8′ strand (Fig. 23 ). Substrates are positioned in a highly constricted pocket surrounded by sheets β1 and β2 and helix α1′. Among the residues associated with the binding of SAH and GPPPA (guanosine-P3-adenosine-5′, 5′-triphosphate), Phe426 and Asn386 play a relatively critical role in determining N7-MTase activity.
Fig. 22.
The crystal structure of the NSP 14–NSP 10 complex from SARS-CoV (PDB ID: 5C8S).
Fig. 23.
The substrate-binding site of the N7-MTase domain of NSP 14 from (SARS)-CoV (PDB ID: 5C8S).
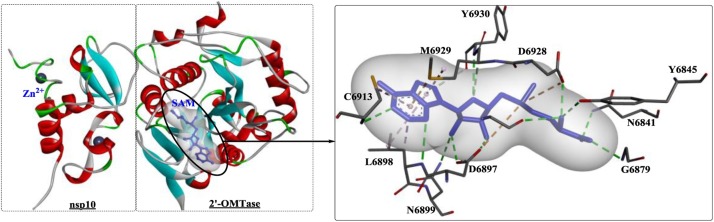
NSP 16 is an m7GpppA-specific, SAM-dependent (nucleoside-2′-O-)-methyltransferase that is active only in the presence of NSP 10 [150]. The 2′-OMTase activity of coronavirus RNA is essential during the viral life cycle and is mainly involved in preventing host recognition and dampening the immune response. To date, various crystal structures of the 2′-OMTase of hCoVs have been identified, such as the crystal structures of complexes of the protein (PDB ID: 3R24, 2XYV, 6YZ1, and 6WVN) with different substrates. In addition, the structures of the 2′-OMTase complex with NSP 10 and different ligands have been determined and deposited in the RCSB PDB (https://www.rcsb.org/) under PDB IDs: 6W4H, 6WKQ, 6WQ3, 6WKS, 6W61, and PDBj (https://bmrbdep.pdbj.org/):7C2I, among others. The crystal structure of the SARS-CoV-2 2′-O-methyltransferase complex with SAM and NSP 10 was deposited under PDB IDs: 6W4H [146] and 6WKS [151]. Similarly, in the complex (Fig. 24 ), two zinc fingers were observed on one side of a central antiparallel pair of β-strands of NSP 10. The structure of NSP 16 is comprised of a Rossmann-like β-sheet fold including eleven α-helices, seven β-strands, and loops, particularly the highly conserved KDKE motif (Lys46, Asp130, Lys170, and Glu203) required for methyl transfer (KDKE). The SAM binding site is coordinated by the critical residues Lys46, Asp130, Lys170, and Glu203. The binding of SAM promotes the normal assembly of the enzymatically active NSP 10/NSP 16 complex by converting 7mGpppG RNA into 7mGpppG2′Om RNA, and the subsequent release of SAH further accelerates NSP 10/NSP 16 dissociation, thereby stimulating reaction turnover. In the complex of 6WKS [151], large conformational changes were discovered as the enzyme transitioned from a binary to a ternary form during substrate binding.
Fig. 24.
The structure of 2′-omethyltransferase complex with SAM from SARS-CoV (PDB ID: 6W4H; SAM: S-adenosylmethionine).
Due to their critical roles in virus replication/transcription, MTases are therefore viewed as promising targets for the development of novel antivirals, which has led to the discovery of various promising inhibitors [[152], [153], [154]]. However, the development of antiviral drugs that target coronavirus MTases has been slow. In a previous study [155], a yeast-based system was developed for the discovery of coronavirus N7-MTase inhibitors by screening 3000 compounds, and three natural product extracts (PF35468, PA48202 and PA48523) with potent inhibitory effects on coronavirus N7-MTase were identified. In this study, the yeast-based system was composed of three yeast strains with yeast, human, or coronavirus N7-MTase activity and was proven to be an effective tool for the discovery of specific inhibitors of coronavirus N7-MTase.
In a study by Wang et al. [156], a peptide (TP29, YGRKKRRQRRRGSGYGGASVCIYCRSRVEHPDVDGLCKLRGKF) was designed based on the sequence of the interaction domain of mouse hepatitis virus (MHV) NSP 10. Based on the results of biochemical assays, TP29 suppressed the 2′-O-MTase activity of different coronaviruses and inhibited viral replication. In particular, TP29 also increased type I interferon production to impair viral virulence and pathogenesis. In addition, sinefungin (a GTP-analog ribavirin triphosphate, IC50 =0.34 mM) and SAH (S-adenosyl-homocysteine, IC50 = 0.63 mM) inhibited 2′-O MTase [157]. In another study [158], a high-throughput screening strategy based on homogenous time-resolved fluorescence (HTRF®) was developed for the identification of viral cap-methyltransferase inhibitors based on the NSP 14 protein. With this method, 2000 molecules were tested, which led to the discovery of 20 hits with inhibitory effects on SARS-CoV NSP 14. Among these hits, compound 1426 and compound 1498 showed the most potent inhibitory effects, with IC50 values ranging from 0.019 μM to 0.74 μM.
2.5. Other important NSPs that have been identified
-
I
SARS-CoV-2 NSP 9 replicase
During the infection of human cells, NSP 9 from SARS-CoV is essential for replication and virulence [159], and it is probably widespread in numerous coronaviruses, including SARS-Cov-2. Recently, the structure of NSP 9 from SARS-CoV-2 was identified and solved (6W9Q and 6WXD) [160], showing 97 % sequence identity with NSP 9 from SARS-CoV. Active CoV NSP 9 usually functions by forming a homodimer through interactions across a hydrophobic interface and is composed of a conserved α-helical GxxxG interaction motif that is directly associated with viral replication [161,162]. Although the detailed interactions with the cofactors of the RNA polymerase have not been determined, the disruption of the dimer interface is presumed to be important for the development of drugs targeting viral replication during infection. In a study by Chandel et al. [163], structure-based drug repurposing of FDA-approved drugs against the NSP 9 replicase and S proteins of SARS-CoV-2 led to the identification of conivaptan and tegobuvir. These findings are important for the treatment of SARS-CoV-2 infections, but further in vitro and in vivo validation and evaluations are indispensable.
-
II
NSP 1 of SARS-CoV-2
NSP 1 is a major virulence factor of SARS-CoVs that suppresses host gene expression by interacting with the ribosome [164,165]. NSP 1 from SARS-CoV-2 was recently shown to be capable of binding to the 40S ribosomal subunit, leading to the inhibition of messenger RNA translation and the subsequent blockade of host protein production [166]. A further structural analysis confirmed that the C-terminus of NSP 1 rigidly binds inside the mRNA entry channel (6ZLW and 6ZN5). In a very recent study [167], a bipartite mechanism of action was suggested for NSP 1. First, the inhabitation of the host ribosome is achieved by a direct interaction of its C-terminal domain with the 40S ribosomal subunit. Then, this inhibition is relieved via a direct interaction of its N-terminal domain with the 5′ untranslated region of SARS-CoV-2 mRNA. This finding is critical for the treatment of SARS-CoV-2, indicating that NSP 1 and the 5′ untranslated region might be promising targets for anti- SARS-CoV-2 therapeutics. In a study by Menezes et al. [168], virtual screening was used to discover potential inhibitors of NSP 1 from the DrugBank database. Although these results have not been validated in vitro and in vivo, the three molecules (tirilazad, phthalocyanine, and Zk-806,450) obtained from this screen might be potential antiviral drugs.
-
III
NSP 3 of SARS-CoV-2
The virus macro domain in NSP 3 is a widespread protein module with multiple functions, such as binding ADP-ribose [169] and the removal of mono (ADP-ribose) from protein [170]. Moreover, based on accumulating evidence, the macro domain might play a critical role in modulating host ADP-ribosylation. Recently, crystal structures of the SARS-CoV-2 macro domain were determined (7C33, 7CZ4 [171] and 6WEY [172]), and the macro domain displays mono-ADP-ribose (ADPR) cleavage enzyme activity by functioning as a poly-ADPR-binding module. Therefore, the macro domain might be a potential therapeutic target. In a very recent study [173], an optimized high-throughput assay combined with further hit prioritization was used to identify potential macro domain inhibitors. The obtained compound cefatrizine (cyclic adenosine monophosphate) was shown to display potent antiviral activity (EC50 = 14 μM). Moreover, through an analysis of the structure of cAMP bound to the SARS-CoV-2 macro domain, scaffolds with a central phosphate (or diphosphate) or sulfate group might be capable of binding to the ADPR-binding cleft.
3. Conclusions and perspectives
Currently, an approved therapy is unavailable for the treatment of coronavirus infection; therefore, the development of novel, broad-spectrum, low-toxicity inhibitors of emerging and endemic CoVs is urgently needed. The identification of natural products with potent antiviral activities against specific CoVs is a routine but usually time-consuming task. Therefore, an efficient HTS strategy is very important, such as the use of a genetically engineered human CoV OC43 strain expressing Renilla luciferase [174]. With the application of an efficient HTS method, repurposing of approved drug libraries has become a promising strategy for the discovery of approved drugs with potent but unknown biological activities.
In addition, with the available structural information on drug-binding sites, structure-based drug discovery could be exploited for rational development of novel antiviral drugs. Therefore, the crystal structures of potential molecular targets are indispensable for identifying key molecular interactions. For hCoVs, the NSPs such as the protease (e.g., the main protease), RNA-dependent RNA polymerase, and methyltransferases, are usually required for the productive stage of the viral life cycle, and are considered potential anti-CoV targets. Drugs targeting these molecules, particularly their most conserved structural domains, might provide options for the development of broad-spectrum antivirals to treat coronavirus infection. However, general cytotoxicity, which is a potential side effect of interfering with normal metabolic pathways, should be noted. The development of drugs based on specific molecular targets will probably provide promising therapeutic solutions to combat CoV infections in the future.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
The authors are supported by the Shandong Provincial Natural Science Foundation, China (No. ZR2019BB062), the Research Fund for Academician Lin He New Medicine (grant no. JYHL2019MS14), Supporting Fund for Teachers' research of Jining Medical University (JYFC2019KJ012), the Doctoral Scientific Research Foundation of Jining Medical University (JY14QD10), National Natural Science Foundation cultivation project of Jining medical university (grant no. JYP201704), the Supporting Fund for Teachers' research of Jining Medical University (grant no. JYFC2018KJ031). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from the one mentioned above.
References
- 1.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguière A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 2.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81(22):12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25(1):35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rolling T., Koerner I., Zimmermann P., Holz K., Haller O., Staeheli P., Kochs G. Adaptive mutations resulting in enhanced polymerase activity contribute to high virulence of influenza A virus in mice. J. Virol. 2009;83(13):6673–6680. doi: 10.1128/JVI.00212-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzsimmons W.J., Woods R.J., McCrone J.T., Woodman A., Arnold J.J., Yennawar M., Evans R., Cameron C.E., Lauring A.S. A speed-fidelity trade-off determines the mutation rate and virulence of an RNA virus. PLoS Biol. 2018;16(6):e2006459. doi: 10.1371/journal.pbio.2006459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neuman B.W., Buchmeier M.J. Supramolecular architecture of the coronavirus particle. Adv. Virus Res. 2016;96:1–27. doi: 10.1016/bs.aivir.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du L., Yang Y., Zhou Y., Lu L., Li F., Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin. Ther. Targets. 2017;21(2):131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talevi A. Computer-aided drug design: an overview. Methods Mol. Biol. 2018;1762:1–19. doi: 10.1007/978-1-4939-7756-7_1. [DOI] [PubMed] [Google Scholar]
- 12.Liu X.H., Zhao J.F., Wang T., Wu M.B. Design, identification, antifungal evaluation and molecular modeling of chlorotetaine derivatives as new anti-fungal agents. Nat. Prod. Res. 2020;34(12):1712–1720. doi: 10.1080/14786419.2018.1528582. [DOI] [PubMed] [Google Scholar]
- 13.Shin W.J., Seong B.L. Novel antiviral drug discovery strategies to tackle drug-resistant mutants of influenza virus strains. Expert Opin. Drug Discov. 2019;14(2):153–168. doi: 10.1080/17460441.2019.1560261. [DOI] [PubMed] [Google Scholar]
- 14.Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39(3):198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.te Velthuis A.J., van den Worm S.H., Snijder E.J. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40(4):1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets. Antiviral Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu D.X., Fung T.S., Chong K.K., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beidas M., Chehadeh W. Effect of human coronavirus OC43 structural and accessory proteins on the transcriptional activation of antiviral response elements. Intervirology. 2018;61(1):30–35. doi: 10.1159/000490566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niemeyer D., Zillinger T., Muth D., Zielecki F., Horvath G., Suliman T., Barchet W., Weber F., Drosten C., Müller M.A. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 2013;87(22):12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y., Lovell S., Tiew K.C., Mandadapu S.R., Alliston K.R., Battaile K.P., Groutas W.C., Chang K.O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86(21):11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muramatsu T., Kim Y.T., Nishii W., Terada T., Shirouzu M., Yokoyama S. Autoprocessing mechanism of severe acute respiratory syndrome coronavirus 3C-like protease (SARS-CoV 3CLpro) from its polyproteins. FEBS J. 2013;280(9):2002–2013. doi: 10.1111/febs.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kneller D.W., Phillips G., O’Neill H.M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L., Kovalevsky A. Structural plasticity of SARS-CoV-2 3CL M(pro) active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020;11(1):3202. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang P.H. Characterization and inhibition of SARS-coronavirus main protease. Curr. Top. Med. Chem. 2006;6(4):361–376. doi: 10.2174/156802606776287090. [DOI] [PubMed] [Google Scholar]
- 24.Hu T., Zhang Y., Li L., Wang K., Chen S., Chen J., Ding J., Jiang H., Shen X. Two adjacent mutations on the dimer interface of SARS coronavirus 3C-like protease cause different conformational changes in crystal structure. Virology. 2009;388(2):324–334. doi: 10.1016/j.virol.2009.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 26.St John S.E., Therkelsen M.D., Nyalapatla P.R., Osswald H.L., Ghosh A.K., Mesecar A.D. X-ray structure and inhibition of the feline infectious peritonitis virus 3C-like protease: structural implications for drug design. Bioorg. Med. Chem. Lett. 2015;25(22):5072–5077. doi: 10.1016/j.bmcl.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.St John S.E., Tomar S., Stauffer S.R., Mesecar A.D. Targeting zoonotic viruses: structure-based inhibition of the 3C-like protease from bat coronavirus HKU4--the likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS) Bioorg. Med. Chem. 2015;23(17):6036–6048. doi: 10.1016/j.bmc.2015.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho B.L., Cheng S.C., Shi L., Wang T.Y., Ho K.I., Chou C.Y. Critical assessment of the important residues involved in the dimerization and catalysis of MERS coronavirus main protease. PLoS One. 2015;10(12):e0144865. doi: 10.1371/journal.pone.0144865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.C., Liao M., Bartlam M., Rao Z. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82(5):2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F., Chen C., Tan W., Yang K., Yang H. Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design. Sci. Rep. 2016;6:22677. doi: 10.1038/srep22677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar V., Shin J.S., Shie J.J., Ku K.B., Kim C., Go Y.Y., Huang K.F., Kim M., Liang P.H. Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CL(Pro) inhibitors. Antiviral Res. 2017;141:101–106. doi: 10.1016/j.antiviral.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abuhammad A., Al-Aqtash R.A., Anson B.J., Mesecar A.D., Taha M.O. Computational modeling of the bat HKU4 coronavirus 3CL(pro) inhibitors as a tool for the development of antivirals against the emerging Middle East respiratory syndrome (MERS) coronavirus. J. Mol. Recognit. 2017;30(11) doi: 10.1002/jmr.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C.G., Cheng S.C., Chen S.C., Li J.Y., Fang Y.H., Chen Y.H., Chou C.Y. Mechanism for controlling the monomer-dimer conversion of SARS coronavirus main protease. Acta Crystallogr. D Biol. Crystallogr. 2013;69(Pt 5):747–755. doi: 10.1107/S0907444913001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar V., Tan K.P., Wang Y.M., Lin S.W., Liang P.H. Identification, synthesis and evaluation of SARS-CoV and MERS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. 2016;24(13):3035–3042. doi: 10.1016/j.bmc.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi J., Sivaraman J., Song J. Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J. Virol. 2008;82(9):4620–4629. doi: 10.1128/JVI.02680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S., Zhang J., Hu T., Chen K., Jiang H., Shen X. Residues on the dimer interface of SARS coronavirus 3C-like protease: dimer stability characterization and enzyme catalytic activity analysis. J. Biochem. 2008;143(4):525–536. doi: 10.1093/jb/mvm246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S., Hu T., Zhang J., Chen J., Chen K., Ding J., Jiang H., Shen X. Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease: crystal structure with molecular dynamics simulations. J. Biol. Chem. 2008;283(1):554–564. doi: 10.1074/jbc.M705240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia B., Kang X. Activation and maturation of SARS-CoV main protease. Protein Cell. 2011;2(4):282–290. doi: 10.1007/s13238-011-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vuong W., Khan M.B., Fischer C., Arutyunova E., Lamer T., Shields J., Saffran H.A., McKay R.T., van Belkum M.J., Joyce M.A., Young H.S., Tyrrell D.L., Vederas J.C., Lemieux M.J. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020;11(1):4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panda P.K., Arul M.N., Patel P., Verma S.K., Luo W., Rubahn H.G., Mishra Y.K., Suar M., Ahuja R. Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci. Adv. 2020;6(28):eabb8097. doi: 10.1126/sciadv.abb8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Báez-Santos Y.M., Mielech A.M., Deng X., Baker S., Mesecar A.D. Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J. Virol. 2014;88(21):12511–12527. doi: 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mielech A.M., Kilianski A., Baez-Santos Y.M., Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450-451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 2006;103(15):5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79(24):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey-Elkin B.A., Knaap R.C., Johnson G.G., Dalebout T.J., Ninaber D.K., van Kasteren P.B., Bredenbeek P.J., Snijder E.J., Kikkert M., Mark B.L. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014;289(50):34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C.Y., Lu C.Y., Li S.W., Lai C.C., Hua C.H., Huang S.H., Lin Y.J., Hour M.J., Lin C.W. SARS coronavirus papain-like protease up-regulates the collagen expression through non-Samd TGF-β1 signaling. Virus Res. 2017;235:58–66. doi: 10.1016/j.virusres.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clasman J.R., Báez-Santos Y.M., Mettelman R.C., O’Brien A., Baker S.C., Mesecar A.D. X-ray structure and enzymatic activity profile of a core papain-like protease of MERS coronavirus with utility for structure-based drug design. Sci. Rep. 2017;7:40292. doi: 10.1038/srep40292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lei J., Hilgenfeld R. Structural and mutational analysis of the interaction between the Middle-East respiratory syndrome coronavirus (MERS-CoV) papain-like protease and human ubiquitin. Virol. Sin. 2016;31(4):288–299. doi: 10.1007/s12250-016-3742-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79(24):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Báez-Santos Y.M., Mielech A.M., Deng X., Baker S., Mesecar A.D. Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J. Virol. 2014;88(21):12511–12527. doi: 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Békés M., Rut W., Kasperkiewicz P., Mulder M.P., Ovaa H., Drag M., Lima C.D., Huang T.T. SARS hCoV papain-like protease is a unique Lys48 linkage-specific di-distributive deubiquitinating enzyme. Biochem. J. 2015;468(2):215–226. doi: 10.1042/BJ20141170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ratia K., Kilianski A., Baez-Santos Y.M., Baker S.C., Mesecar A. Structural Basis for the Ubiquitin-Linkage Specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 2014;10(5):e1004113. doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sato Y., Goto E., Shibata Y., Kubota Y., Yamagata A., Goto-Ito S., Kubota K., Inoue J., Takekawa M., Tokunaga F., Fukai S. Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat. Struct. Mol. Biol. 2015;22(3):222–229. doi: 10.1038/nsmb.2970. [DOI] [PubMed] [Google Scholar]
- 58.Keusekotten K., Elliott P.R., Glockner L., Fiil B.K., Damgaard R.B., Kulathu Y., Wauer T., Hospenthal M.K., Gyrd-Hansen M., Krappmann D., Hofmann K., Komander D. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153(6):1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Békés M., van der Heden van Noort G.J., Ekkebus R., Ovaa H., Huang T.T., Lima C.D. Recognition of Lys48-linked di-ubiquitin and deubiquitinating activities of the SARS coronavirus papain-like protease. Mol. Cell. 2016;62(4):572–585. doi: 10.1016/j.molcel.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin M.H., Chuang S.J., Chen C.C., Cheng S.C., Cheng K.W., Lin C.H., Sun C.Y., Chou C.Y. Structural and functional characterization of MERS coronavirus papain-like protease. J. Biomed. Sci. 2014;21(1):54. doi: 10.1186/1423-0127-21-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Báez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park J.Y., Kim J.H., Kim Y.M., Jeong H.J., Kim D.W., Park K.H., Kwon H.J., Park S.J., Lee W.S., Ryu Y.B. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg. Med. Chem. 2012;20(19):5928–5935. doi: 10.1016/j.bmc.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kemp M. Recent advances in the discovery of deubiquitinating enzyme inhibitors. Prog. Med. Chem. 2016;55:149–192. doi: 10.1016/bs.pmch.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park J.Y., Yuk H.J., Ryu H.W., Lim S.H., Kim K.S., Park K.H., Ryu Y.B., Lee W.S. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzyme Inhib. Med. Chem. 2017;32(1):504–515. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park J.Y., Jeong H.J., Kim J.H., Kim Y.M., Park S.J., Kim D., Park K.H., Lee W.S., Ryu Y.B. Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biol. Pharm. Bull. 2012;35(11):2036–2042. doi: 10.1248/bpb.b12-00623. [DOI] [PubMed] [Google Scholar]
- 66.Lee H., Lei H., Santarsiero B.D., Gatuz J.L., Cao S., Rice A.J., Patel K., Szypulinski M.Z., Ojeda I., Ghosh A.K., Johnson M.E. Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. ACS Chem. Biol. 2015;10(6):1456–1465. doi: 10.1021/cb500917m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cho J.K., Curtis-Long M.J., Lee K.H., Kim D.W., Ryu H.W., Yuk H.J., Park K.H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013;21(11):3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park J.Y., Ko J.A., Kim D.W., Kim Y.M., Kwon H.J., Jeong H.J., Kim C.Y., Park K.H., Lee W.S., Ryu Y.B. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzyme Inhib. Med. Chem. 2016;31(1):23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- 69.Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W., Prabhakar B.S., Johnson M.E., Baker S.C., Ghosh A.K., Mesecar A.D. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 2008;105(42):16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghosh A.K., Takayama J., Rao K.V., Ratia K., Chaudhuri R., Mulhearn D.C., Lee H., Nichols D.B., Baliji S., Baker S.C., Johnson M.E., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation. J. Med. Chem. 2010;53(13):4968–4979. doi: 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghosh A.K., Takayama J., Aubin Y., Ratia K., Chaudhuri R., Baez Y., Sleeman K., Coughlin M., Nichols D.B., Mulhearn D.C., Prabhakar B.S., Baker S.C., Johnson M.E., Mesecar A.D. Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors for the severe acute respiratory syndrome-coronavirus papain-like protease. J. Med. Chem. 2009;52(16):5228–5240. doi: 10.1021/jm900611t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pui C.H., Evans W.E. Treatment of acute lymphoblastic leukemia. N. Engl. J. Med. 2006;354(2):166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 73.Chou C.Y., Chien C.H., Han Y.S., Prebanda M.T., Hsieh H.P., Turk B., Chang G.G., Chen X. Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem. Pharmacol. 2008;75(8):1601–1609. doi: 10.1016/j.bcp.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheng K.W., Cheng S.C., Chen W.Y., Lin M.H., Chuang S.J., Cheng I.H., Sun C.Y., Chou C.Y. Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antiviral Res. 2015;115:9–16. doi: 10.1016/j.antiviral.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frieman M., Basu D., Matthews K., Taylor J., Jones G., Pickles R., Baric R., Engel D.A. Yeast based small molecule screen for inhibitors of SARS-CoV. PLoS One. 2011;6(12):e28479. doi: 10.1371/journal.pone.0028479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahn D.G., Choi J.K., Taylor D.R., Oh J.W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012;157(11):2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sevajol M., Subissi L., Decroly E., Canard B., Imbert I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10(1):2342–2350. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.te Velthuis A.J., Arnold J.J., Cameron C.E., van den Worm S.H., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38(1):203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111(37):E3900–9. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584(7819):154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 83.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang X., Huang Y., Gao H., Liu F., Ge J., Sun Q., Yang X., Xu W., Liu Z., Yang H., Lou Z., Jiang B., Guddat L.W., Gong P., Rao Z. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182(2):417–428. doi: 10.1016/j.cell.2020.05.034. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Konkolova E., Klima M., Nencka R., Boura E. Structural analysis of the putative SARS-CoV-2 primase complex. J. Struct. Biol. 2020;211(2):107548. doi: 10.1016/j.jsb.2020.107548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29(7):1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43(17):8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peng Q., Peng R., Yuan B., Zhao J., Wang M., Wang X., Wang Q., Sun Y., Fan Z., Qi J., Gao G.F., Shi Y. Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020;31(11):107774. doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.te Velthuis A.J. Common and unique features of viral RNA-dependent polymerases. Cell. Mol. Life Sci. 2014;71(22):4403–4420. doi: 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sexton N.R., Smith E.C., Blanc H., Vignuzzi M., Peersen O.B., Denison M.R. Homology-based identification of a mutation in the coronavirus RNA-dependent RNA polymerase that confers resistance to multiple mutagens. J. Virol. 2016;90(16):7415–7428. doi: 10.1128/JVI.00080-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., Warren T.K., Mackman R.L. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of ebola and emerging viruses. J. Med. Chem. 2017;60(5):1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 91.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brown A.J., Won J.J., Graham R.L., Dinnon K.H., 3rd, Sims A.C., Feng J.Y., Cihlar T., Denison M.R., Baric R.S., Sheahan T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 2019;169 doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11(4) doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9(2) doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., Warren T.K., Mackman R.L. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of ebola and emerging viruses. J. Med. Chem. 2017;60(5):1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 97.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang L., Zhou R. Structural basis of the potential binding mechanism of remdesivir to SARS-CoV-2 RNA-dependent RNA polymerase. J. Phys. Chem. B. 2020;124(32):6955–6962. doi: 10.1021/acs.jpcb.0c04198. [DOI] [PubMed] [Google Scholar]
- 99.Yoon J.S., Kim G., Jarhad D.B., Kim H.R., Shin Y.S., Qu S., Sahu P.K., Kim H.O., Lee H.W., Wang S.B., Kong Y.J., Chang T.S., Ogando N.S., Kovacikova K., Snijder E.J., Posthuma C.C., van Hemert M.J., Jeong L.S. Design, synthesis, and anti-RNA virus activity of 6’-fluorinated-aristeromycin analogues. J. Med. Chem. 2019;62(13):6346–6362. doi: 10.1021/acs.jmedchem.9b00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Coutard B., Barral K., Lichière J., Selisko B., Martin B., Aouadi W., Lombardia M.O., Debart F., Vasseur J.J., Guillemot J.C., Canard B., Decroly E. Zika virus methyltransferase: structure and functions for drug design perspectives. J. Virol. 2017;91(5) doi: 10.1128/JVI.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2011;10(1):51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burrows F.S., Carlos L.M., Benzimra M., Marriott D.J., Havryk A.P., Plit M.L., Malouf M.A., Glanville A.R. Oral ribavirin for respiratory syncytial virus infection after lung transplantation: efficacy and cost-efficiency. J. Heart Lung Transplant. 2015;34(7):958–962. doi: 10.1016/j.healun.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 103.Castells L., Rimola A., Manzardo C., Valdivieso A., Montero J.L., Barcena R., Abradelo M., Xiol X., Aguilera V., Salcedo M., Rodriguez M., Bernal C., Suarez F., Antela A., Olivares S., Del Campo S., Laguno M., Fernandez J.R., de la Rosa G., Aguero F., Perez I., Gonzalez-Garcia J., Esteban-Mur J.I., Miro J.M. Pegylated interferon plus ribavirin in HIV-infected patients with recurrent hepatitis C after liver transplantation: a prospective cohort study. J. Hepatol. 2015;62(1):92–100. doi: 10.1016/j.jhep.2014.07.034. [DOI] [PubMed] [Google Scholar]
- 104.Falzarano D., de Wit E., Martellaro C., Callison J., Munster V.J., Feldmann H. Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci. Rep. 2013;3(1):1686. doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Khalili J.S., Zhu H., Mak N.S.A., Yan Y., Zhu Y. Novel coronavirus treatment with ribavirin: groundwork for an evaluation concerning COVID-19. J. Med. Virol. 2020;92(7):740–746. doi: 10.1002/jmv.25798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Falzarano D., de Wit E., Rasmussen A.L., Feldmann F., Okumura A., Scott D.P., Brining D., Bushmaker T., Martellaro C., Baseler L., Benecke A.G., Katze M.G., Munster V.J., Feldmann H. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV–infected rhesus macaques. Nat. Med. 2013;19(10):1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Poordad F., McCone J., Jr., Bacon B.R., Bruno S., Manns M.P., Sulkowski M.S., Jacobson I.M., Reddy K.R., Goodman Z.D., Boparai N., DiNubile M.J., Sniukiene V., Brass C.A., Albrecht J.K., Bronowicki J.P. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 2011;364(13):1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ferron F., Subissi L., Silveira De Morais A.T., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2018;115(2):E162–e171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sidwell R.W., Huffman J.H., Khare G.P., Allen L.B., Witkowski J.T., Robins R.K. Broad-spectrum antiviral activity of Virazole: 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science. 1972;177(4050):705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- 110.Hofmann W.P., Herrmann E., Sarrazin C., Zeuzem S. Ribavirin mode of action in chronic hepatitis C: from clinical use back to molecular mechanisms. Liver Int. 2008;28(10):1332–1343. doi: 10.1111/j.1478-3231.2008.01896.x. [DOI] [PubMed] [Google Scholar]
- 111.Feld J.J., Lutchman G.A., Heller T., Hara K., Pfeiffer J.K., Leff R.D., Meek C., Rivera M., Ko M., Koh C., Rotman Y., Ghany M.G., Haynes–Williams V., Neumann A.U., Liang T.J., Hoofnagle J.H. Ribavirin improves early responses to peginterferon through improved interferon signaling. Gastroenterology. 2010;139(1):154–162. doi: 10.1053/j.gastro.2010.03.037. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tam R.C., Pai B., Bard J., Lim C., Averett D.R., Phan U.T., Milovanovic T. Ribavirin polarizes human T cell responses towards a Type 1 cytokine profile. J. Hepatol. 1999;30(3):376–382. doi: 10.1016/s0168-8278(99)80093-2. [DOI] [PubMed] [Google Scholar]
- 113.Dietz J., Schelhorn S.E., Fitting D., Mihm U., Susser S., Welker M.W., Fuller C., Daumer M., Teuber G., Wedemeyer H., Berg T., Lengauer T., Zeuzem S., Herrmann E., Sarrazin C. Deep sequencing reveals mutagenic effects of ribavirin during monotherapy of hepatitis C virus genotype 1-infected patients. J. Virol. 2013;87(11):6172–6181. doi: 10.1128/JVI.02778-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Todt D., Walter S., Brown R.J., Steinmann E. Mutagenic effects of ribavirin on hepatitis e virus-viral extinction versus selection of fitness-enhancing mutations. Viruses. 2016;8(10) doi: 10.3390/v8100283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carrillo-Bustamante P., Nguyen T.H.T., Oestereich L., Gunther S., Guedj J., Graw F. Determining Ribavirin’s mechanism of action against Lassa virus infection. Sci. Rep. 2017;7(1):11693. doi: 10.1038/s41598-017-10198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alam I., Lee J.-H., Cho K.J., Han K.R., Yang J.M., Chung M.S., Kim K.H. Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase in complex with 2-thiouridine or ribavirin. Virology. 2012;426(2):143–151. doi: 10.1016/j.virol.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 117.Ferrer-Orta C., Arias A., Pérez-Luque R., Escarmís C., Domingo E., Verdaguer N. Sequential structures provide insights into the fidelity of RNA replication. Proc. Natl. Acad. Sci. U. S. A. 2007;104(22):9463–9468. doi: 10.1073/pnas.0700518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.de la Higuera I., Ferrer-Orta C., Moreno E., de Ávila A.I., Soria M.E., Singh K., Caridi F., Sobrino F., Sarafianos S.G., Perales C., Verdaguer N., Domingo E. Contribution of a multifunctional polymerase region of foot-and-Mouth disease virus to lethal mutagenesis. J. Virol. 2018;92(20):e01119–18. doi: 10.1128/JVI.01119-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Furuta Y., Takahashi K., Fukuda Y., Kuno M., Kamiyama T., Kozaki K., Nomura N., Egawa H., Minami S., Watanabe Y., Narita H., Shiraki K. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 2002;46(4):977–981. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sleeman K., Mishin V.P., Deyde V.M., Furuta Y., Klimov A.I., Gubareva L.V. In vitro antiviral activity of favipiravir (T-705) against drug-resistant influenza and 2009 a(H1N1) viruses. Antimicrob. Agents Chemother. 2010;54(6):2517–2524. doi: 10.1128/AAC.01739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017;93(7):449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Oestereich L., Lüdtke A., Wurr S., Rieger T., Muñoz-Fontela C., Günther S. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res. 2014;105:17–21. doi: 10.1016/j.antiviral.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 123.Zhu W., Zhang Z., He S., Wong G., Banadyga L., Qiu X. Successful treatment of Marburg virus with orally administrated T-705 (Favipiravir) in a mouse model. Antiviral Res. 2018;151:39–49. doi: 10.1016/j.antiviral.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Delang L., Abdelnabi R., Neyts J. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res. 2018;153:85–94. doi: 10.1016/j.antiviral.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 125.Nagata T., Lefor A.K., Hasegawa M., Ishii M. Favipiravir: a new medication for the Ebola virus disease pandemic. Disaster Med. Public Health Prep. 2015;9(1):79–81. doi: 10.1017/dmp.2014.151. [DOI] [PubMed] [Google Scholar]
- 126.Naesens L., Guddat L.W., Keough D.T., van Kuilenburg A.B., Meijer J., Vande Voorde J., Balzarini J. Role of human hypoxanthine guanine phosphoribosyltransferase in activation of the antiviral agent T-705 (favipiravir) Mol. Pharmacol. 2013;84(4):615–629. doi: 10.1124/mol.113.087247. [DOI] [PubMed] [Google Scholar]
- 127.Baranovich T., Wong S.S., Armstrong J., Marjuki H., Webby R.J., Webster R.G., Govorkova E.A. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J. Virol. 2013;87(7):3741–3751. doi: 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Omotuyi O.I., Nash O., Safronetz D., Ojo A.A., Ogunwa T.H., Adelakun N.S. T-705-modified ssRNA in complex with Lassa virus nucleoprotein exhibits nucleotide splaying and increased water influx into the RNA-binding pocket. Chem. Biol. Drug Des. 2019;93(4):544–555. doi: 10.1111/cbdd.13451. [DOI] [PubMed] [Google Scholar]
- 129.Asselah T. Sofosbuvir for the treatment of hepatitis C virus. Expert Opin. Pharmacother. 2014;15(1):121–130. doi: 10.1517/14656566.2014.857656. [DOI] [PubMed] [Google Scholar]
- 130.Feld J.J., Jacobson I.M., Hezode C., Asselah T., Ruane P.J., Gruener N., Abergel A., Mangia A., Lai C.L., Chan H.L., Mazzotta F., Moreno C., Yoshida E., Shafran S.D., Towner W.J., Tran T.T., McNally J., Osinusi A., Svarovskaia E., Zhu Y., Brainard D.M., McHutchison J.G., Agarwal K., Zeuzem S. Sofosbuvir and Velpatasvir for HCV Genotype 1, 2, 4, 5, and 6 Infection. N. Engl. J. Med. 2015;373(27):2599–2607. doi: 10.1056/NEJMoa1512610. [DOI] [PubMed] [Google Scholar]
- 131.Lawitz E., Gane E.J. Sofosbuvir for previously untreated chronic hepatitis C infection. N. Engl. J. Med. 2013;369(7):678–679. doi: 10.1056/NEJMc1307641. [DOI] [PubMed] [Google Scholar]
- 132.Naggie S., Cooper C., Saag M., Workowski K., Ruane P., Towner W.J., Marks K., Luetkemeyer A., Baden R.P., Sax P.E., Gane E., Santana-Bagur J., Stamm L.M., Yang J.C., German P., Dvory-Sobol H., Ni L., Pang P.S., McHutchison J.G., Stedman C.A., Morales-Ramirez J.O., Brau N., Jayaweera D., Colson A.E., Tebas P., Wong D.K., Dieterich D., Sulkowski M. Ledipasvir and Sofosbuvir for HCV in patients coinfected with HIV-1. N. Engl. J. Med. 2015;373(8):705–713. doi: 10.1056/NEJMoa1501315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wyles D., Bräu N., Kottilil S., Daar E.S., Ruane P., Workowski K., Luetkemeyer A., Adeyemi O., Kim A.Y., Doehle B., Huang K.C., Mogalian E., Osinusi A., McNally J., Brainard D.M., McHutchison J.G., Naggie S., Sulkowski M., Investigators ft.A.-. Sofosbuvir and velpatasvir for the treatment of hepatitis C virus in patients coinfected with human immunodeficiency virus type 1: an open-label, phase 3 study. Clin. Infect. Dis. 2017;65(1):6–12. doi: 10.1093/cid/cix260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Feld J.J., Jacobson I.M., Hézode C., Asselah T., Ruane P.J., Gruener N., Abergel A., Mangia A., Lai C.-L., Chan H.L.Y., Mazzotta F., Moreno C., Yoshida E., Shafran S.D., Towner W.J., Tran T.T., McNally J., Osinusi A., Svarovskaia E., Zhu Y., Brainard D.M., McHutchison J.G., Agarwal K., Zeuzem S. Sofosbuvir and Velpatasvir for HCV Genotype 1, 2, 4, 5, and 6 Infection. New England J. Med. Surg. Collat. Branches Sci. 2015;373(27):2599–2607. doi: 10.1056/NEJMoa1512610. [DOI] [PubMed] [Google Scholar]
- 137.Lawitz E., Poordad F.F., Pang P.S., Hyland R.H., Ding X., Mo H., Symonds W.T., McHutchison J.G., Membreno F.E. Sofosbuvir and ledipasvir fixed-dose combination with and without ribavirin in treatment-naive and previously treated patients with genotype 1 hepatitis C virus infection (LONESTAR): an open-label, randomised, phase 2 trial. Lancet. 2014;383(9916):515–523. doi: 10.1016/S0140-6736(13)62121-2. [DOI] [PubMed] [Google Scholar]
- 138.Saxena V., Nyberg L., Pauly M., Dasgupta A., Nyberg A., Piasecki B., Winston B., Redd J., Ready J., Terrault N.A. Safety and efficacy of simeprevir/sofosbuvir in hepatitis C-Infected patients with compensated and decompensated cirrhosis. Hepatology. 2015;62(3):715–725. doi: 10.1002/hep.27922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shah R., Maize K.M., Zhou X., Finzel B.C., Wagner C.R. Caught before released: structural mapping of the reaction trajectory for the sofosbuvir activating enzyme, human histidine triad nucleotide binding protein 1 (hHint1) Biochemistry. 2017;56(28):3559–3570. doi: 10.1021/acs.biochem.7b00148. [DOI] [PubMed] [Google Scholar]
- 140.Sofia M.J., Bao D., Chang W., Du J., Nagarathnam D., Rachakonda S., Reddy P.G., Ross B.S., Wang P., Zhang H.R., Bansal S., Espiritu C., Keilman M., Lam A.M., Steuer H.M., Niu C., Otto M.J., Furman P.A. Discovery of a β-d-2’-deoxy-2’-α-fluoro-2’-β-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J. Med. Chem. 2010;53(19):7202–7218. doi: 10.1021/jm100863x. [DOI] [PubMed] [Google Scholar]
- 141.Appleby T.C., Perry J.K., Murakami E., Barauskas O., Feng J., Cho A., Fox D., 3rd, Wetmore D.R., McGrath M.E., Ray A.S., Sofia M.J., Swaminathan S., Edwards T.E. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347(6223):771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 142.García-Sastre A. 2 methylate or not 2 methylate: viral evasion of the type I interferon response. Nat. Immunol. 2011;12(2):114–115. doi: 10.1038/ni0211-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Case J.B., Ashbrook A.W., Dermody T.S., Denison M.R. Mutagenesis of S-adenosyl-l-methionine-binding residues in coronavirus nsp14 N7-Methyltransferase demonstrates differing requirements for genome translation and resistance to innate immunity. J. Virol. 2016;90(16):7248–7256. doi: 10.1128/JVI.00542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen Y., Tao J., Sun Y., Wu A., Su C., Gao G., Cai H., Qiu S., Wu Y., Ahola T., Guo D. Structure-function analysis of severe acute respiratory syndrome coronavirus RNA cap guanine-N7-methyltransferase. J. Virol. 2013;87(11):6296–6305. doi: 10.1128/JVI.00061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Menachery V.D., Yount B.L., Jr., Josset L., Gralinski L.E., Scobey T., Agnihothram S., Katze M.G., Baric R.S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2’-o-methyltransferase activity. J. Virol. 2014;88(8):4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Wiersum G., Kim Y., Jedrzejczak R., Maltseva N.I., Endres M., Jaroszewski L., Godzik A., Joachimiak A., Satchell K.J.F. The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. bioRxiv. 2020 [Google Scholar]
- 147.Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3’-&5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103(13):5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 2009;106(9):3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U. S. A. 2015;112(30):9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lugari A., Betzi S., Decroly E., Bonnaud E., Hermant A., Guillemot J.C., Debarnot C., Borg J.P., Bouvet M., Canard B., Morelli X., Lécine P. Molecular mapping of the RNA Cap 2’-O-methyltransferase activation interface between severe acute respiratory syndrome coronavirus nsp10 and nsp16. J. Biol. Chem. 2010;285(43):33230–33241. doi: 10.1074/jbc.M110.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Viswanathan T., Arya S., Chan S.H., Qi S., Dai N., Misra A., Park J.G., Oladunni F., Kovalskyy D., Hromas R.A., Martinez-Sobrido L., Gupta Y.K. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020;11(1):3718. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lim S.V., Rahman M.B., Tejo B.A. Structure-based and ligand-based virtual screening of novel methyltransferase inhibitors of the dengue virus. BMC Bioinformatics. 2011;12(Suppl. 13):S24. doi: 10.1186/1471-2105-12-S13-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Podvinec M., Lim S.P., Schmidt T., Scarsi M., Wen D., Sonntag L.S., Sanschagrin P., Shenkin P.S., Schwede T. Novel inhibitors of dengue virus methyltransferase: discovery by in vitro-driven virtual screening on a desktop computer grid. J. Med. Chem. 2010;53(4):1483–1495. doi: 10.1021/jm900776m. [DOI] [PubMed] [Google Scholar]
- 154.Stephen P., Baz M., Boivin G., Lin S.X. Structural insight into NS5 of zika virus leading to the discovery of MTase inhibitors. J. Am. Chem. Soc. 2016;138(50):16212–16215. doi: 10.1021/jacs.6b10399. [DOI] [PubMed] [Google Scholar]
- 155.Sun Y., Wang Z., Tao J., Wang Y., Wu A., Yang Z., Wang K., Shi L., Chen Y., Guo D. Yeast-based assays for the high-throughput screening of inhibitors of coronavirus RNA cap guanine-N7-methyltransferase. Antiviral Res. 2014;104:156–164. doi: 10.1016/j.antiviral.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang Y., Sun Y., Wu A., Xu S., Pan R., Zeng C., Jin X., Ge X., Shi Z., Ahola T., Chen Y., Guo D. Coronavirus nsp10/nsp16 methyltransferase can be targeted by nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J. Virol. 2015;89(16):8416–8427. doi: 10.1128/JVI.00948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Selisko B., Peyrane F.F., Canard B., Alvarez K., Decroly E. Biochemical characterization of the (nucleoside-2’O)-methyltransferase activity of dengue virus protein NS5 using purified capped RNA oligonucleotides (7Me)GpppAC(n) and GpppAC(n) J. Gen. Virol. 2010;91(Pt 1):112–121. doi: 10.1099/vir.0.015511-0. [DOI] [PubMed] [Google Scholar]
- 158.Aouadi W., Eydoux C., Coutard B., Martin B., Debart F., Vasseur J.J., Contreras J.M., Morice C., Quérat G., Jung M.L., Canard B., Guillemot J.C., Decroly E. Toward the identification of viral cap-methyltransferase inhibitors by fluorescence screening assay. Antiviral Res. 2017;144:330–339. doi: 10.1016/j.antiviral.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Frieman M., Yount B., Agnihothram S., Page C., Donaldson E., Roberts A., Vogel L., Woodruff B., Scorpio D., Subbarao K., Baric R.S. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J. Virol. 2012;86(2):884–897. doi: 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Littler D.R., Gully B.S., Colson R.N., Rossjohn J. Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. iScience. 2020;23(7) doi: 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zeng Z., Deng F., Shi K., Ye G., Wang G., Fang L., Xiao S., Fu Z., Peng G. Dimerization of coronavirus nsp9 with diverse modes enhances its nucleic acid binding affinity. J. Virol. 2018;92(17) doi: 10.1128/JVI.00692-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hu T., Chen C., Li H., Dou Y., Zhou M., Lu D., Zong Q., Li Y., Yang C., Zhong Z., Singh N., Hu H., Zhang R., Yang H., Su D. Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9. Protein Sci. 2017;26(5):1037–1048. doi: 10.1002/pro.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chandel V., Sharma P.P., Raj S., Choudhari R., Rathi B., Kumar D. Structure-based drug repurposing for targeting Nsp9 replicase and spike proteins of severe acute respiratory syndrome coronavirus 2. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1811773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J. Virol. 2011;85(1):638–643. doi: 10.1128/JVI.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lei L., Ying S., Baojun L., Yi Y., Xiang H., Wenli S., Zounan S., Deyin G., Qingyu Z., Jingmei L., Guohui C. Attenuation of mouse hepatitis virus by deletion of the LLRKxGxKG region of Nsp1. PLoS One. 2013;8(4):e61166. doi: 10.1371/journal.pone.0061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Straub J.H., Stürzel C.M., Fröhlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369(6508):1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shi M., Wang L., Fontana P., Vora S., Zhang Y., Fu T.M., Lieberman J., Wu H. SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism. bioRxiv. 2020 [Google Scholar]
- 168.de Lima Menezes G., da Silva R.A. Identification of potential drugs against SARS-CoV-2 non-structural protein 1 (nsp1) J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1792992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Makrynitsa G.I., Ntonti D., Marousis K.D., Birkou M., Matsoukas M.T., Asami S., Bentrop D., Papageorgiou N., Canard B., Coutard B., Spyroulias G.A. Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose. J. Struct. Biol. 2019;206(1):119–127. doi: 10.1016/j.jsb.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cho C.C., Lin M.H., Chuang C.Y., Hsu C.H. Macro domain from middle east respiratory syndrome coronavirus (MERS-CoV) is an efficient ADP-ribose binding module: CRYSTAL STRUCTURE AND BIOCHEMICAL STUDIES. J. Biol. Chem. 2016;291(10):4894–4902. doi: 10.1074/jbc.M115.700542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lin M.H., Chang S.C., Chiu Y.C., Jiang B.C., Wu T.H., Hsu C.H. Structural, biophysical, and biochemical elucidation of the SARS-CoV-2 nonstructural protein 3 macro domain. ACS Infect. Dis. 2020 doi: 10.1021/acsinfecdis.0c00441. [DOI] [PubMed] [Google Scholar]
- 172.Frick D.N., Virdi R.S., Vuksanovic N., Dahal N., Silvaggi N.R. Molecular basis for ADP-Ribose binding to the Mac1 domain of SARS-CoV-2 nsp3. Biochemistry. 2020;59(28):2608–2615. doi: 10.1021/acs.biochem.0c00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Virdi R.S., Bavisotto R.V., Hopper N.C., Vuksanovic N., Melkonian T.R., Silvaggi N.R., Frick D.N. Discovery of drug-like ligands for the Mac1 domain of SARS-CoV-2 Nsp3. SLAS Discov. 2020 doi: 10.1177/2472555220960428. 2472555220960428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N., Deng Y., Wang H., Ye F., Cen S., Tan W. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J. Virol. 2019;93(12) doi: 10.1128/JVI.00023-19. [DOI] [PMC free article] [PubMed] [Google Scholar]