Abstract
Karrikins and strigolactones are two classes of butenolide molecules that have diverse effects on plant growth. Karrikins are found in smoke and strigolactones are plant hormones, yet both molecules are likely recognized through highly similar signaling mechanisms. Here we review the most recent discoveries of karrikin and strigolactone perception and signal transduction. Two paralogous α/β hydrolases, KAI2 and D14, are respectively karrikin and strigolactone receptors. D14 acts with an F-box protein, MAX2, to target SMXL/D53 family proteins for proteasomal degradation, and genetic data suggest that KAI2 acts similarly. There are striking parallels in the signaling mechanisms of karrikins, strigolactones, and other plant hormones, including auxins, jasmonates, and gibberellins. Recent investigations of host perception in parasitic plants have demonstrated that strigolactone recognition can evolve following gene duplication of KAI2.
Diverse Roles for Karrikins and Strigolactones in Plant Growth
Karrikins (KARs) and strigolactones (SLs) have been studied intensively over the past decade as two new classes of plant growth regulators. KARs and SLs are both butenolide molecules (Box 1), but they have very different sources and effects on growth. KARs, which are found in smoke, activate germination of many species after fire [1–3]. KAR effects are not limited to plants from fire-prone environments, however. KARs enhance Arabidopsis thaliana germination and seedling responses to light [4,5]. KARs also increase seedling vigor and stress tolerance of several crop species [6–8]. SLs were first discovered in cotton root exudates nearly 50 years ago as germination stimulants of parasitic weeds [9]. SLs have since been found to act as rhizosphere signals that promote symbiotic interactions between roots and arbuscular mycorrhizal fungi [10–12], and as hormones that influence shoot branching [11,13], root architecture [14–16], leaf shape [17–19], leaf senescence [20,21], and cambial growth [22].
Box 1. Comparing Karrikins and Strigolactones.
KARs are produced by pyrolysis of cellulose or sugars, and are therefore generated by all wildfires [75] (Figure IA). KARs in smoke are thought to be deposited on the soil surface during a fire, and then absorbed by seeds buried in the soil in the following months after being dissolved by rain [3]. KAR1 is the most abundant KAR in smoke-water solutions, and is usually the most bioactive. Five additional KARs with methyl group substitutions have been identified [1].
SLs are synthesized from carotenoids via a carlactone intermediate by a carotene isomerase (D27), two carotenoid cleavage dioxygenases (CCD7/MAX3 and CCD8/MAX4), and a cytochrome P450 (MAX1) [23–28]. Carlactone is converted to carlactonoic acid by MAX1 in three-step oxidative reactions. Carlactonoic acid is a precursor of SLs as well as methyl carlactonoate, a D14 substrate with SL-like activities [25]. Approximately 20 SLs have been found in plants. All feature a tricyclic lactone (ABC-ring) connected to a butenolide (D-ring) in a 2′R configuration [76]. MAX1 paralogs in rice contribute to SL diversity [23,77]. SLs fall into two classes based upon stereochemistry at the B–C-ring junction that are typified by 5-deoxystrigol (5DS) and 4-deoxyorobanchol (Figure IB). The D-ring and a cleavable ether linkage are essential elements of active SLs. However, stereochemistry at the 2′ carbon atom is also critical for signaling specificity [37,39]. GR24 is a commonly used synthetic SL analog that is typically a racemic mixture of two enantiomers (Figure IC). GR245DS mimics 5DS stereochemistry; however, its enantiomer GR24ent–5DS has an unnatural 2′S configuration. The Arabidopsis SL receptor D14 preferentially recognizes 2′R SLs (blue) versus 2′S SLs, while its paralog KAI2 is responsive to KARs and GR24ent–5DS (red) [37]. True SL responses are thus best verified by testing SL-deficient mutants and optically pure SL enantiomers.
Although KARs and SLs are both butenolide molecules that trigger germination, from the perspective of a seed these signals convey opposite messages. As fire-derived compounds, KARs indicate a low-competition environment in which ‘plants are absent’, whereas SLs found in soil are exuded by roots, therefore signaling ‘plants are present’. Selective germination responses to KARs and SLs are important ecological adaptations for some plants. For example, the fire-following species Brassica tournefortii germinates in response to KARs, but is much less responsive to rac-GR24 [5]. By contrast, root parasitic weeds in the Orobanchaceae family that require a host plant for survival are highly sensitive to SLs and unresponsive to KARs [50].
Figure I. Structures of Karrikins (KARs) and Strigolactones (SLs).
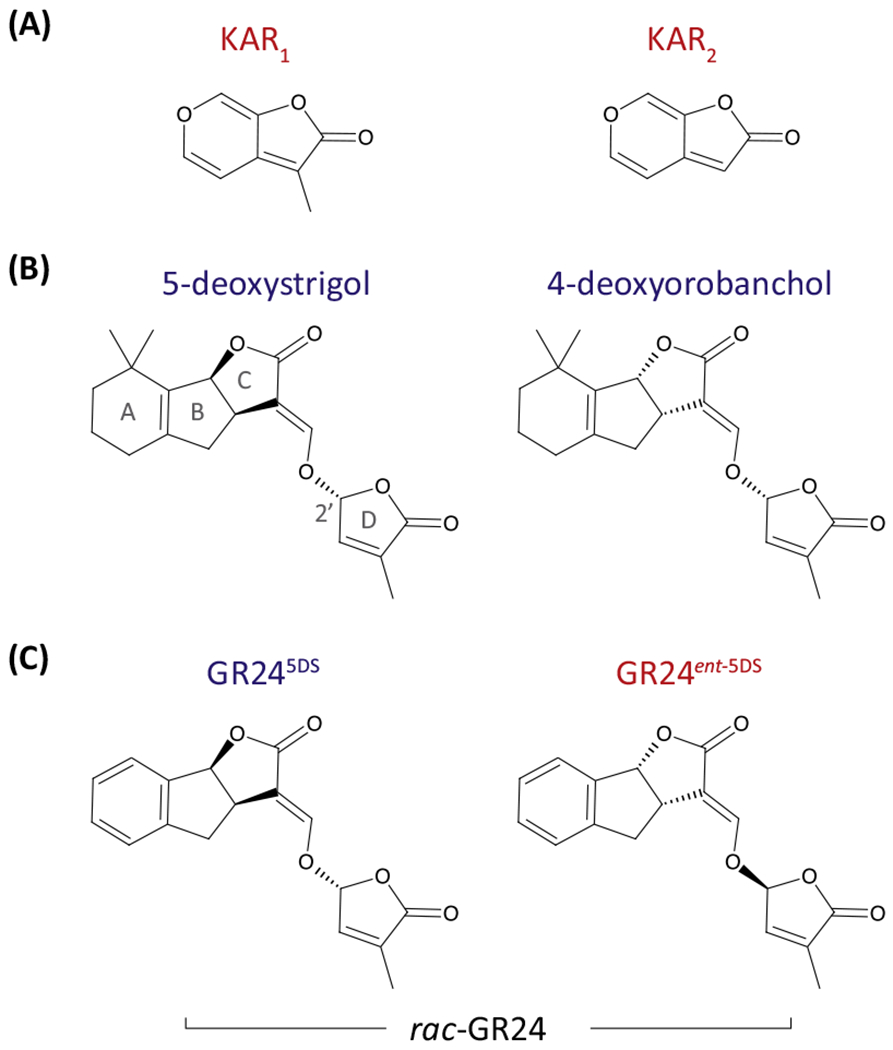
Molecular structures of KARs (A), representatives of the two major classes of natural SLs (B), and a commonly used synthetic SL analog GR24 (C). KAR2 is commonly used in Arabidopsis thaliana experiments because of its higher activity than KAR1 [5]. Note the stereochemistry at the 2′ carbon of the butenolide D-ring in SL structures. rac-GR24 is a mixture of GR245DS and its enantiomer. Compounds that signal primarily or exclusively through KAI2 are labeled in red; those that signal through D14 are labeled in blue. Abbreviations: KAI2, KARRIKIN-INSENSITIVE2; D14, DWARF14.
Substantial progress has been made toward understanding how SLs are synthesized [23–28] and transported [29–31], how KARs and SLs are sensed, and how these signals control different aspects of plant growth. Strikingly, KAR and SL signaling mechanisms involve homologous genetic components. Therefore, this system can provide broader insights into how new signaling mechanisms arise and how signaling specificity is achieved. In this review, we focus on recent breakthroughs in the mechanisms and evolution of KAR and SL signaling.
Genetic Screens Link KAR and SL Signaling
Despite a basic structural similarity, KARs and SLs are not interchangeable signals (Box 1). KARs do not restore branching suppression to SL-deficient mutants, implying that KARs and SLs are recognized by different signaling pathways. Therefore, it was surprising when the first genetic screen for KAR-insensitive mutants identified two loss-of-function alleles of MORE AXILLARY GROWTH2 (MAX2) [32], which is also required for SL responses [11,13]. As KAR and SL signaling pathways converge on MAX2 but cause different growth effects, this implied that KAR and SL are distinguished at the point of signal perception and likely initiate different signal transduction cascades as well.
The basis for SL and KAR discrimination was resolved by characterization of DWARF14 (D14) and KARRIKIN-INSENSITIVE2 (KAI2). The d14 mutant in rice has increased tiller numbers and reduced stature, but unlike SL-deficient mutants is not responsive to the synthetic SL GR24 [33] (Box 1). Thus, its shoot growth is phenotypically similar to rice d3/max2. Loss-of-function mutations in D14 orthologs in Arabidopsis and petunia (DAD2) also cause SL-insensitive branching phenotypes [34,35]. However, Arabidopsis d14 only shares phenotypes with max2 that are also found in SL-deficient mutants [34]; max2 uniquely has increased seed dormancy and reduced seedling growth responses to light [17,32,36], implying that MAX2 also mediates non-SL, D14-independent signaling.
A fortuitous KAR-insensitive mutation and reverse genetic analysis of a close homolog of D14 led to the identification of KAI2 in Arabidopsis. kai2 loss-of-function alleles mimic the seed and seedling phenotypes of max2, but have normal branching [34]. max2 phenotypes are a combination of d14 and kai2 phenotypes, resulting from a loss of both D14- and KAI2-dependent signaling [34,37,38]. KAI2 mediates responses to KARs and SL analogs with a non-natural 2′S configuration (e.g., GR24ent-5DS); by contrast, D14 preferentially mediates responses to SL stereoisomers with a natural 2′R configuration SL stereoisomers (e.g., GR245DS) and is unresponsive to KARs (Box 1) [37,39,40].
Another homolog of KAI2 and AtD14 in Arabidopsis, D14-like 2 (DLK2), intriguingly has one of the strongest known positive transcriptional responses to KAR/SL treatment and is down-regulated in kai2 and max2 [34]. To date, however, no dlk2 mutant phenotypes have been found [34]. It is possible that DLK2 contributes to mesocotyl elongation in dark-grown rice seedlings, as RNAi-mediated suppression of D14L/KAI2 in a d14 background does not fully replicate a d3/max2 mesocotyl phenotype [41].
KAI2 and D14 are Receptors and Enzymes
There is substantial evidence that KAI2 and D14, which are members of the α/β-hydrolase superfamily, encode receptors that require enzymatic activity for signal transduction. We will first discuss D14, which has been more thoroughly characterized. SL binding to D14 has been shown through isothermal calorimetry, scintillation proximity assays, and crystallography of D14 complexes with the synthetic SL GR24 [40,42–44]. D14 slowly hydrolyzes GR24 at a rate of approximately one molecule per 3 min [44]. Although this hydrolytic activity is weak, it is necessary for signal transduction, as mutation of the conserved Ser-His-Asp catalytic triad abolishes D14 function in vivo [35]. The products of GR24 hydrolysis do not rescue the excess branching phenotype of a petunia dad2/d14 mutant, however. This suggests that SL signaling occurs during the act of SL hydrolysis by D14, rather than by another protein perceiving a SL byproduct [35].
Indeed, DAD2/D14 protein undergoes thermal destabilization in differential scanning fluorimetry assays in the presence of GR24, and this requires an active catalytic site [35]. The degree of GR24-induced D14 destabilization varies in different species, but it has been shown repeatedly that SL enhances D14 interactions with MAX2 and its downstream targets D53/SMXL6,7,8 (see later). Mutations that reduce SL binding by D14 also reduce its association with D3/MAX2. D14–D3/MAX2 association is only activated by 2′R SL stereoisomers, with GR245DS having the most potent effect; non-natural 2′S stereoisomers have no activity [40]. There is surprisingly little difference between the surface conformations of apo-D14 (no ligand bound) and D14 bound to GR24, the GR24 hydrolysis intermediate, or the D-ring hydrolysis product. However, SL-induced destabilization of D14 is enhanced by D3/MAX2 binding, suggesting that D14–MAX2 complex formation proceeds through a multistep process that is initiated by SL and stabilized by MAX2 binding [40]. It is unclear if D14 complexes first with MAX2 or its downstream targets following SL perception. Intriguingly, D14 itself is degraded following SL treatment in a MAX2-dependent manner, suggesting a feedback mechanism to dampen SL signaling [45].
KAI2 function is more enigmatic. KAR1 binding to KAI2 has been demonstrated through equilibrium microdialysis, heteronuclear single-quantum coherence (HSQC) NMR, isothermal titration calorimetry, and crystallization of a KAI2–KAR1 complex [42,46]. The catalytic triad is necessary for KAI2 function, suggesting that KAI2 signal transduction is similar to D14 [47–49]. However, the proposed mechanism of KAI2 action – hydrolysis of a Ser–KAR1 intermediate following nucleophilic addition – is expected to regenerate the original KAR1 molecule, in contrast to the destructive release of the SL D-ring by D14 hydrolysis [44,49]. Also, in the reported KAR1–KAI2 complex, the position of KAR1 at the opening of the active site is not close enough to the catalytic Serto enable nucleophilic attack [46,49]. Finally, KAR1 and KAR2 have no thermal destabilization effect on KAI2, and rac-GR24 does not promote KAI2–D3/MAX2 association [40,47]. Nonetheless, almost all of the surface residues identified as important for D14 interactions with D3/MAX2 [40] are strongly conserved in KAI2. Also, KAI2 has hydrolytic activity against GR24ent–5DS and is thermally destabilized by GR24ent–5DS [47]. Therefore, it may be that in vitro experiments with KAR substrates simply do not replicate KAI2 action in vivo. Similar to D14, KAI2 is degraded following KAR perception; however, its degradation is MAX2-independent and does not involve polyubiquitination [48].
Two final observations support that D14 and KAI2 are receptors. First, the loss-of-function allele d14-2/seto5 encodes a Pro169Leu substitution. This highly conserved residue lies on the surface of the D14 protein and is unlikely to influence SL hydrolysis, suggesting that a protein–protein interaction site necessary for signal transduction is disrupted [45]. Likewise, a Phe28Trp substitution that minimally affects GR24 binding by D14 causes a strong decrease in GR24-induced D14–D3/MAX2 association [40]. Second, a class of KAI2 paralogs from parasitic plants confers SL responses to Arabidopsis kai2 seeds, but AtD14 does not [47,50–52]. If the only functions of these parasite KAI2 and AtD14 are to convert SL into an active signaling molecule, then both types of genes should enable SL responses in seed. Instead, these results imply that the parasite KAI2 have a nonenzymatic signal transduction activity that AtD14 lacks (e.g., specific protein–protein interactions), which can activate Arabidopsis germination after SL treatment.
SMXL/D53 Proteins are Targets of MAX2
MAX2 encodes an F-box protein with C-terminal leucine-rich repeats. F-box proteins confer target specificity to the SKP1–CULLIN–F-box (SCF) class of E3 ubiquitin ligase complexes, which conjugate ubiquitin to protein substrates. Polyubiquitination of a protein typically triggers its rapid proteolysis by the 26S proteasome [53]. MAX2/D3 interacts with SKP1 and Cullin orthologs in Arabidopsis and rice, and is therefore likely to act in proteolytic targeting [54,55].
Hormone-triggered proteolysis is a shared feature of auxin, jasmonate, and gibberellin signaling mechanisms. Interactions between F-box proteins and their substrates are activated by hormone perception, leading to degradation of the substrate and activation of downstream responses to the hormone [56–58]. In auxin and jasmonate signaling, the hormone acts as ‘molecular glue’ that directly promotes interactions between the F-box protein and its targets [59,60]. In gibberellin (GA) signaling, however, GA binding causes allosteric activation of the receptor GID1, exposing hydrophobic surfaces that can interact with DELLA proteins [61]. DELLA proteins lack DNA-binding domains, but indirectly regulate transcription. Formation of the GA–GID1–DELLA complex enhances recognition of DELLA by the F-box protein SLEEPY1, leading to degradation of DELLA and growth responses to GA [62]. As GID1 is an α/β-hydrolase protein like KAI2 and D14, and SLEEPY1 and MAX2 are F-box proteins, there is an intriguing parallel between GA and KAR/SL signaling components.
It is therefore likely that max2 phenotypes result from overaccumulation of its protein substrate(s). Indeed, overexpressing a MAX2 transgene that lacks the F-box domain can cause dominant-negative effects that increase shoot branching, presumably due to sequestering its substrates from polyubiquitination by wild-type MAX2 [54]. Identifying MAX2 targets was a long-standing roadblock for the field that has been resolved with the identification of SMAX1 and D53 (Figure 1, Key Figure). A screen for genetic suppressors of max2 phenotypes at seed and seedling stages led to the discovery of SMAX1 in Arabidopsis [63]. Loss-of-function smax1 alleles cause rapid seed germination, reduced hypocotyl elongation and enlarged cotyledons in seedlings, and restore expression of KAR-responsive transcriptional markers in a max2 background; these phenotypes mimic responses to KAR treatment. However, smax1 does not suppress SL-related max2 effects on shoot branching, lateral root density, or leaf senescence. Eight genes including SMAX1 compose a SMAX1-like (SMXL) gene family in Arabidopsis, which raised the possibility that some SMXLs may control SL-related growth [63].
Figure 1.
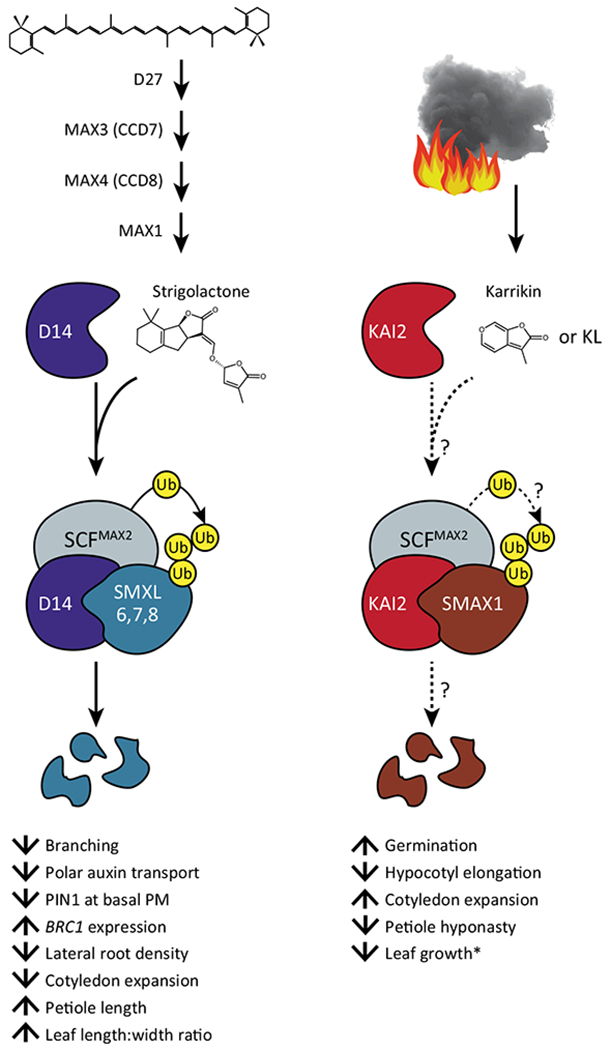
Key Figure
Models of Strigolactone (SL) and Karrikin (KAR) Signaling
SLs are synthesized from all-trans-β-carotene by the sequential action of D27, CCD7, CCD8, and MAX1. SLs are recognized by D14, triggering association of D14 with SCFMAX2 and SMXL6,7,8/D53. SMXL6,7,8/D53 proteins are then targeted for proteasomal degradation, enabling growth responses to SL. Other carotenoid-derived SL-like molecules such as methyl carlactonoate may act similarly through D14. KARs are produced by burning vegetation. Based on genetic evidence and analogy to the SL pathway, we hypothesize that KAR or a putative KAI2 ligand (KL) are recognized by KAI2, triggering formation of a SCFMAX2–KAI2–SMAX1 complex. SMAX1 is then polyubiquitinated and degraded by the 26S proteasome, allowing KAR/KL responses such as increased germination. Dashed lines and question marks indicate that the KAI2-dependent signaling mechanism is an untested hypothesis. Upward arrows indicate an increase in a growth response, downward arrows indicate a decrease. *Leaf growth effects of kai2 and smax1 are influenced by photoperiod. Abbreviations: KAI2, KARRIKIN-INSENSITIVE2; D14, DWARF14; MAX, MORE AXILLARY GROWTH; SCF, SKP1–CULLIN–F-box.
At the same time, two teams characterized a dominant high tillering dwarf mutant in rice, dwarf53 (d53) [64,65]. Like d14 and d3/max2 mutants, d53 is insensitive to GR24 and has increased SL production. Positional cloning identified D53 as a paralog of SMAX1 that is grouped with the SMXL6, SMXL7, and SMXL8 phylogenetic clade. D53 is polyubiquitinated and rapidly degraded after GR24 treatment in a D3/MAX2- and D14-dependent manner. The d53 mutant protein is hypermorphic, however, and is stable in the presence of GR24. D53 physically interacts with D14, and GR24 enhances this interaction, but an active D14 catalytic site is required. Because d53 protein is not polyubiquitinated after GR24 treatment but maintains normal GR24-responsive interactions with D14 [64,65], it is possible that the d53 mutation disrupts D53–D3/MAX2 interactions or a key ubiquitin attachment site. RNAi-mediated suppression of D53 restores tillering control to d3 and d14 mutants, providing genetic evidence that D53 acts downstream of D3 and D14 [64,65].
Further evidence that these genes are epistatic to MAX2 in SL-regulated growth was provided by loss-of-function mutations in Arabidopsis SMXL6, SMXL7, and SMXL8 (collectively called SMXL6,7,8 here), which are dicot orthologs of D53 [38,66]. SMXL6,7,8 have redundant functions in branching control, although SMXL7 makes the strongest contribution. This redundancy explains why previous attempts to identify suppressors of the max2 branching phenotype through forward genetic screens were unsuccessful. Thesmxl6,7,8 triple mutant restores branching suppression to max2, perhaps due to decreased polar auxin transport in the stem and/or strongly increased expression of the transcription factor BRANCHED1 in axillary buds. SMXL6,7,8 also control lateral root density, rosette leaf morphology, and cotyledon expansion [38,66].
Regulation of SMXL6,7,8 by GR24 is similar to D53 regulation in rice; these proteins are polyubiquitinated and degraded in a D14- and MAX2-dependent manner after GR24 treatment, but not by KAR1 [38,66] (Figure 1). Replication of the d53 mutation in SMXL6 and SMXL7 produces dominant isoforms that are resistant to GR24-induced polyubiquitination and proteolysis, as in rice. A four amino acid deletion is sufficient to stabilize SMXL6, causing increased branching, rounded leaf shape, and upregulation of the SL biosynthesis gene MAX4 [66]. This motif is highly conserved in SMAX1, SMXL2, and SMXL6,7,8 homologs in angiosperms but is absent in SMXL3, SMXL4, and SMXL5, which suggests that SMXL3,4,5 may be regulated differently than other SMXL proteins [38].
D14 interactions with D3/MAX2 and D53/SMXL6,7,8 are enhanced by GR24 treatment [35,39,40,55,64–66]. By contrast, D3/MAX2 and D53/SMXL6,7,8 interact in pull-down and co-immunoprecipitation assays independently of GR24 treatment or D14 [64,66]. Relative to the input levels of the tested components these interactions appear to be weak, however, and D14 is clearly important for D53/SMXL6,7,8 proteolysis. Thus, it remains to be determined whether D14-independent interaction with MAX2 is relevant for SMXL6,7,8 regulation in vivo, and how D14 association influences the ubiquitination activity of SCFMAX2 on SMXL6,7,8. It would also be beneficial to test D14 interactions in a SL-deficient mutant background, to determine the extent to which they are SL-dependent versus SL-enhanced.
It must be emphasized that the current model of KAR signaling through KAI2 is a hypothesis based only upon genetic evidence and analogy to the SL signaling mechanism. KAR-induced KAI2-SMAX1 interactions and MAX2-dependent proteolysis of SMAX1 remain to be tested through biochemical experiments. It is also currently unknown how SMXL/D53 proteins regulate growth. SMXL proteins have weak similarity to HSP100/ClpB heat-shock proteins, which function as hexameric complexes that unravel protein aggregates in an ATP-dependent manner [63]. This suggests that SMXL could unfold or remodel protein complexes. However, the motifs for ATP binding and hydrolysis are poorly conserved. An alternative hypothesis favored in the literature is that SMXL proteins function as transcriptional corepressors (Box 2).
Box 2. TOPLESS-Mediated Corepression: A Conserved Hormone Signaling Module?
SMXL/D53 may function as transcriptional repressors in KAR and SL signaling, analogous to Aux/IAA proteins in auxin signaling and JAZ or JAZ–NINJA proteins in jasmonate signaling. Aux/IAA and JAZ/JAZ–NINJA do not directly bind DNA, but repress transcription through interactions with transcription factors and TOPLESS (TPL) and TOPLESS-RELATED (TPR) proteins. Interactions with TPL/TPR are mediated by ETHYLENE RESPONSE FACTOR-associated amphiphilic repression (EAR) motifs. TPL/TPR are members of the Groucho/Tup1 corepressor family first identified in fruit flies and yeast that influence gene expression by recruiting the histone deacetylase HDA19 to DNA-bound complexes [78,79]. Aux/IAA proteins recruit TPL/TPR to ARF transcription factors via an EAR motif; transcriptional repression is relieved by proteasomal degradation of Aux/IAA, which follows the auxin-triggered association of Aux/IAA and TIR1 family F-box proteins [80]. Jasmonate signaling is similar, except that the adapter function of an Aux/IAA can involve two proteins. JAZ proteins bind to transcription factors (e.g., MYC) and are targeted for degradation after jasmonate-Ile perception triggers JAZ association with the F-box protein COI1. Some JAZ proteins have an EAR motif, while others associate with TPL/TPR by binding NINJA, which has an EAR motif [81–83].
Like Aux/IAA and JAZ, SMXL proteins lack DNA-binding motifs. However, a C-terminal EAR motif is conserved in SMXL homologs throughout basal plants and angiosperms [38]. Evidence for EAR motif-dependent TPL/TPR interactions with D53, SMXL6,7,8, and SMAX1 has been shown through mammalian and yeast two-hybrid assays, immunoprecipitation, and bimolecular fluorescence complementation assays [38,64,66]. Furthermore, SMXL6,7,8–GAL4 fusions repress a GAL4–UAS transcriptional reporter in plants in an EAR motif-dependent manner [66].
SL and KAR responses may therefore share a signaling module with auxin and jasmonate pathways, with the exception that SMXL degradation requires a receptor in addition to an F-box protein (Figure I). This is an appealing hypothesis, but genetic tests will be required to determine the functional significance of the EAR motif and TPL interactions in SMXL proteins. Indeed, the transcriptional responses to KAR and SL reported thus far have been modest in terms of differentially expressed gene number and the magnitude of fold changes [4,84].
The conserved N-terminal domain of TPL/TPR proteins (TPD), which mediates interactions with EAR motifs, forms a tetrameric structure [85]. Interestingly, the affinity of TPD for EAR motifs is enhanced by oligomerization of the repressor partner and may be important for efficient transcriptional repression. It will be important to determine if SMXL/D53 oligomerize like their homologs, HSP100/ClpB.
Figure I. A Putative Plant Hormone Signaling Module.
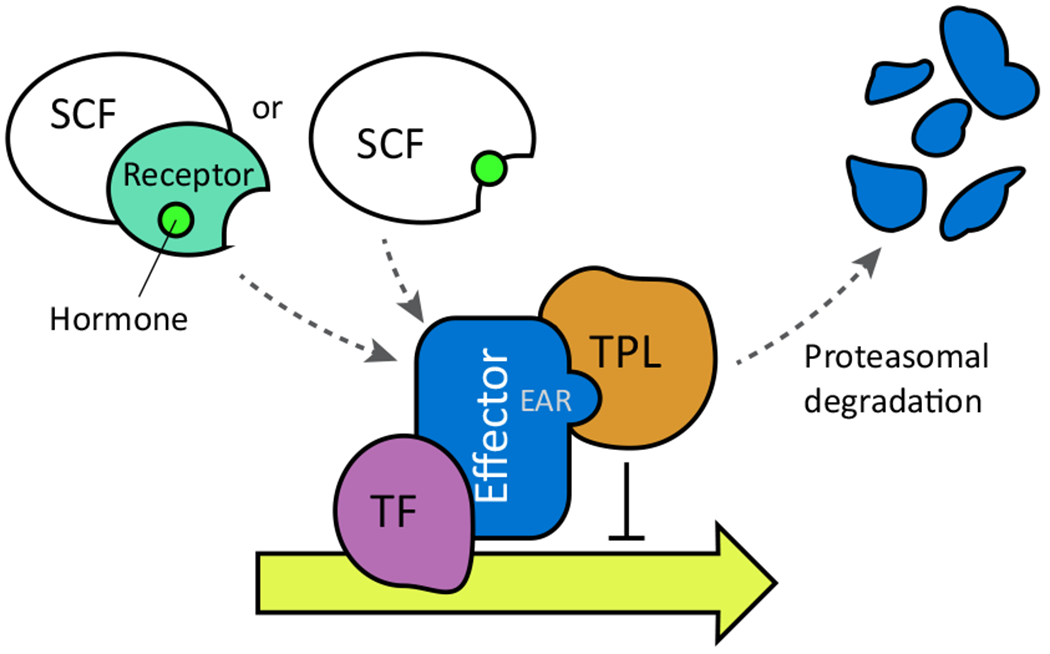
Transcriptional repression is mediated by an effector protein that interacts with a transcription factor (TF) and TPL/TPR proteins via an EAR motif. In some cases, two proteins (e.g., JAZ–NINJA) work together as the effector, although only one may be degraded (e.g., JAZ). Hormone perception by a receptor triggers complex formation with the effector and a SCF E3 ubiquitin ligase complex, leading to polyubiquitination and degradation of the effector by the 26S proteasome. In auxin and jasmonate signaling, the F-box protein itself is a coreceptor with the effector. Abbreviations: TPL, TOPLESS; TPR, TOPLESS-RELATED; EAR, ETHYLENE RESPONSE FACTOR-associated amphiphilic repression; SCF, SKP1–CULLIN–F-box.
KAR and SL Signaling Specificity
The epistatic relationships of SMAX1 and SMXL6,7,8 to MAX2 are limited to distinct subsets of max2 phenotypes, which combine kai2 and d14 defects. So far, the roles of SMXL family members partition by phylogenetic clade [38]. SMAX1 controls growth processes that are associated with KAI2-dependent signaling and KAR responses, whereas the SMXL6,7,8/D53 clade controls growth that is associated with D14-dependent signaling and SL responses (Figure 1). While KAI2 and D14 pathways often regulate growth in different tissues (e.g., seeds versus axillary buds), both contribute to leaf morphology. Rosette leaves of smax1 max2 phenocopy d14, implying that the kai2-related phenotypes of max2 have been suppressed; smxl6,7,8 max2 leaves phenocopy kai2, indicating the SL-insensitive, d14-related phenotypes of max2 have been suppressed [38]. The branching and leaf shape phenotypes of the SL-deficient mutant max3 are also fully suppressed by smxl6,7,8 [66]. Altogether, this suggests that SMAX1 is a specific target of KAI2, and SMXL6,7,8 are specific targets of D14. Epistasis tests of smax1 kai2 and smxl6,7,8 d14 could strengthen this conclusion.
This raises the question of how specific receptor–effector pairing among homologous components could occur, which is presumably necessary to avoid crosstalk between KAR and SL pathways. Spatiotemporal differences in gene regulation is one means for signaling specificity; D14 and SMXL6,7,8 may have overlapping expression patterns that are distinct from the expression of KAI2 and SMAX1. Indeed, SMAX1 is the most highly expressed SMXL family member in Arabidopsis seed, and KAI2 is expressed 100-fold higher than D14 in seed [34,63]. This hypothesis can be tested by promoter-swapping experiments. KAI2pro:D14 does not rescue kai2 or confer germination responses to rac-GR24, nor does D14pro:KAI2 rescue d14 [47]. Therefore, the expression patterns of KAI2 and D14 are not sufficient to account for signaling specificity. Instead, specific receptor–SMXL protein interactions may be the basis for KAR and SL responses. Seven potential specificity determining positions that may influence KAI2 and D14 protein interactions with downstream partners have been proposed [45]. Functional tests of these residues and determination of the affinities of ligand-activated KAI2 and D14 for different SMXL proteins will be important future experiments.
Evolution of SL Signaling in Parasitic Plants
The identification of central components of KAR and SL signaling has enabled studies of how these mechanisms have evolved. One of the most striking evolutionary examples is also at the heart of a major agricultural problem. Obligate parasitic plants in the Orobanchaceae family germinate after detecting SLs exuded from a nearby host root. This is an important adaptation for the parasite, which will die several days after germination if it fails to attach to a host [67]. Several species within this family are weeds that cause $US billions in crop losses each year, and particularly impact smallholder farmers in sub-Saharan Africa. Three recent reports have demonstrated that a clade of KAI2 paralogs in parasitic plants evolved the ability to detect SLs.
KAI2 has undergone extensive gene duplication in the parasitic lineage. Although most angiosperms have only one or two copies of KAI2, the parasites examined in the Orobanchaceae have on average five or six KAI2 copies and as many as 13 [50]. By contrast, D14 is maintained as a single gene copy. KAI2 paralogs are divided into three phylogenetic clades under different rates of selection that also differ in terms of ligand-binding pocket shape and ligand specificity. KAI2c (for conserved) paralogs are under the strongest purifying selection, and are typically present as a single copy in both parasite and non-parasite genomes. A clade under an intermediate rate of selection, KAI2i, has representatives in some, but not all, Lamiid genomes. The most rapidly evolving clade, KAI2d (for divergent), is found only in parasites and comprises the majority of KAI2 paralogs in those genomes. Homology modeling based on an AtKAI2 structure predicts that KAI2d paralogs have substantially larger ligand-binding pockets than KAI2c or KAI2i in parasites, and several residues that shape the ligand-binding pocket are no longer highly conserved [50]. A crystal structure of a receptor in the parasite Striga hermonthica, ShHTL5, validates the prediction of enlarged ligand-binding pockets in KAI2d proteins [52]. KAI2d transgenes confer SL-responsive germination to Arabidopsis kai2 seed. Interestingly, KAI2i genes from S. hermonthica (ShHTL2 and ShHTL3) confer KAR-responsive germination [50,52]. A KAI2c gene from Phelipanche aegyptiaca rescues kai2 seed dormancy but does not respond to KAR or SL [50]. Therefore, KAI2 have evolved new ligand specificities following gene duplication in the Orobanchaceae family (Figure 2).
Figure 2. Evolution of KAI2 in Parasitic Plants.
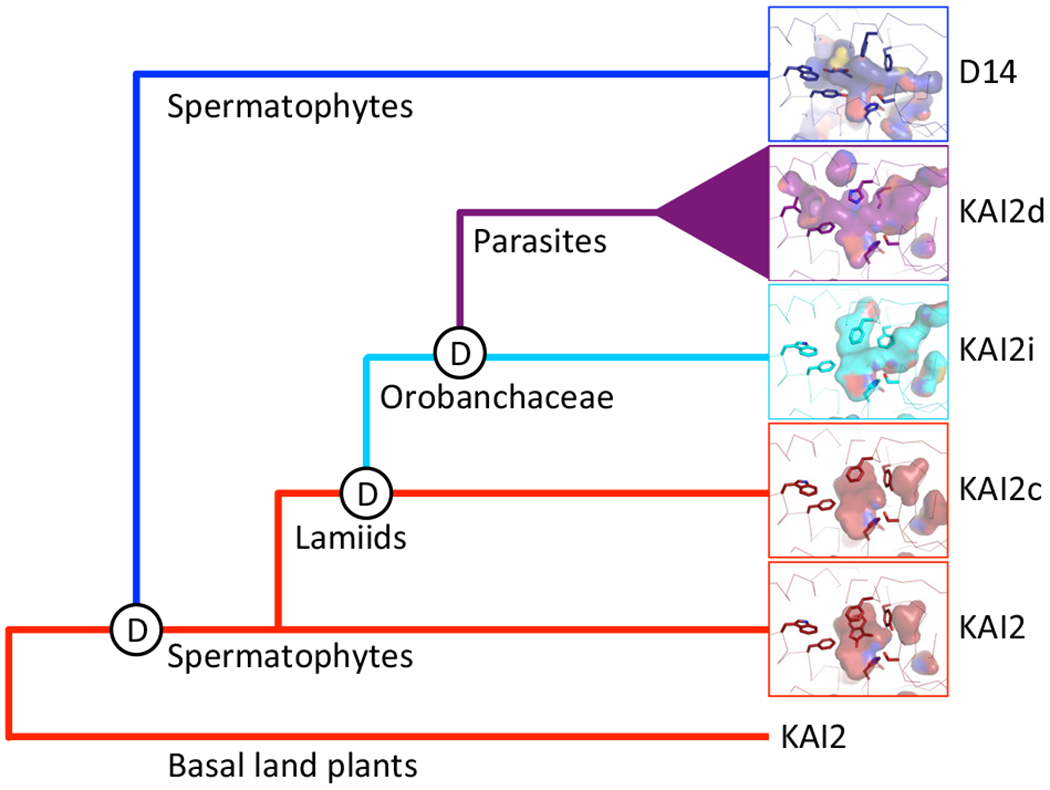
D14 is an ancient paralog of KAI2 that has SL specificity. Duplication of KAI2 in the Lamiid lineage likely gave rise to KAI2c and KAI2i paralogs. Further, extensive duplication of KAI2 in the Orobanchaceae family gave rise to the KAI2d clade found in parasitic plants. The ligand-binding pockets of rice D14 (top, blue) and Arabidopsis KAI2 (bottom, red) are shown with key pocket residues and shading of the hydrophobic cavities. Homology modeling of KAI2 paralogs in parasitic plants indicates merging of two cavities in KAI2i and an increase in ligand-binding pocket size in the KAI2d clade. Similar to D14, KAI2d proteins recognize SLs, suggesting convergent evolution. KAI2i respond to KARs and KAI2c may recognize KL; Arabidopsis KAI2 has both properties, suggesting subfunctionalization may have occurred in the Lamiids. Abbreviations: KAI2, KARRIKIN-INSENSITIVE2; D14, DWARF14; KAR, karrikin; SL, strigolactone; KL, KAI2 ligand.
The diversification of KAI2d paralogs in parasites may enable detection of a variety of SL or SL-like butenolide molecules, which could in turn influence host range [50]. A fluorogenic agonist for SL receptors that has SL-like activity, Yoshimulactone Green (YLG), has enabled tests of SL specificity among KAI2 [51]. YLG is hydrolyzed by Arabidopsis D14 and by ten KAI2/HTL paralogs in S. hermonthica, but not by the basal ShKAI2c/HTL1 protein. Competitive inhibition assays of YLG hydrolysis demonstrate a range of in vitro sensitivities to different SLs among the parasite KAI2. These results support the idea that KAI2d paralogs have evolved specialized SL preferences, but signaling outcomes may be more complicated in vivo; all tested KAI2/HTL have similar IC50 values for 5-deoxystrigol and 4-deoxyorobanchol, yet S. hermonthica seeds have ~100-fold differences in sensitivity to these SLs during germination [51].
A similar chemical probe, YLGW, which has reduced selectivity but increased signal strength, was used to test for ShKAI2/HTL activity in vivo. Two waves of YLGW hydrolysis occur during germination of S. hermonthica seed. Although it cannot be excluded that YLGW hydrolysis can occur in parasite tissues independently of KAI2, this result is exciting as it might reflect an early SL recognition step that primes the seed for a second round of SL detection before committing to germination [51].
KAI2 Function in Basal Land Plants
Phylogenetic analyses indicate that KAI2 homologs are present in basal land plants, but D14 homologs are not [34,68]. Therefore, D14 is likely to have arisen from an ancient duplication of KAI2, prior to the divergence of the spermatophyte lineage. While KAI2 in non-parasitic angiosperms may not be SL receptors, the function of KAI2 in basal land plants is still unknown. One hypothesis is that KAI2 recognizes an undiscovered endogenous signal (KAI2 ligand, KL) that is neither KAR nor SL [69,70], and that SL perception evolved after KAI2 gene duplication in paralogs such as D14 and KAI2d [50]. Cross-species complementation analysis of basal KAI2c, which rescue multiple Atkai2 phenotypes but do not respond to KAR1 or SL, supports this idea [50,70]. However, SL biosynthetic genes, SLs of unknown stereochemistry, and growth responses to rac-GR24 are found in mosses and charophyte green algae [68,71,72]. It is possible that a subset of KAI2 paralogs found in basal plants function as SL receptors but are not readily recognized as such because they evolved independently of D14.
Waters et al. (2015) investigated the function of KAI2 homologs in the basal land plants Selaginella moellendorffii and Marchantia polymorpha [47]. SmKAI2a had hydrolase activity against a generic substrate but not GR24 stereoisomers. SmKAI2b, by contrast, had strong hydrolytic activity against GR245DS. Interestingly, SmKAI2a partially rescued Arabidopsis kai2 phenotypes, particularly at later stages of development, but was unresponsive to KAR or SL. Its ability to rescue was dependent upon a functional catalytic domain. SmKAI2a has effects that are weaker but consistent with KAI2c genes; therefore, it may also recognize KL. In contrast to this, SmKAI2b was not functional in cross-species complementation assays. M. polymorpha KAI2 paralogs were also inactive in Arabidopsis, even though MpKAI2b had strong hydrolase activity against the non-natural SL stereoisomer GR24ent-5DS [47]. The limited functionality of these transgenes may be due to an inability to interact with Arabidopsis signaling components (e.g., MAX2 or SMAX1). Reverse genetic analysis in basal land plants will resolve KAI2 function more directly. Nonetheless, the in vitro activity of SmKAI2b supports the idea that it may be a SL receptor.
KAI2 is Required for Symbiosis with Arbuscular Mycorrhizal Fungi
New roles for MAX2-dependent signaling pathways continue to be discovered. Most recently, D14L/KAI2 in rice was shown to be necessary for the establishment of arbuscular mycorrhizal (AM) symbiosis, which is used by more than 80% of land plants to take up mineral nutrients such as phosphorus [73]. SL biosynthesis and exudation from roots is important for AM colonization, but SL perception in the host by D14 is not [29,73,74]. Intriguingly, loss of D14L/KAI2 blocks early colonization events in AM symbiosis and renders roots insensitive to signals in the exudates of germinated AM spores. It remains to be determined whether D14L/KAI2 directly perceives a precontact signal from AM fungi, or if it regulates the competency of roots for symbiotic interactions [73].
Concluding Remarks and Future Perspectives
Rapid progress has been made toward understanding KAR and SL signaling. The core components of the signaling mechanisms have been identified, and their functions are understood at a basic level. Strikingly, ligand-activated degradation of transcriptional regulators has emerged as a repeated theme of hormone signaling mechanisms in plants. The unprecedented availability of genome sequence information enabled by next-generation sequencing technology now makes it possible to move beyond model system foundations and explore the evolution of signaling pathways in specific ecological contexts. Comparative genetic analyses are likely to be among the most promising strategies for gaining insights into critical residues or forms of regulation for KAR and SL pathways.
There are still many unanswered questions facing the field (see Outstanding Questions). An important technological limitation is the ability to detect SLs easily. The development of in vivo SL reporters, perhaps similar in design to the auxin and jasmonate sensors, will reveal sites of SL synthesis and perception. This tool would enable genetic screens for new mutants in the SL pathway, and new ways to assess crosstalk with other hormone and abiotic signaling pathways.
Outstanding Questions.
How and in what order do SL signaling complexes assemble? High-resolution crystal structures of D14 complexes with MAX2/D3 and SMXL6,7,8/D53 may resolve key events in the SL signaling mechanism as well as the active form of SL.
What are the functions of SMXL proteins? If SMXLs are transcriptional repressors, identifying their DNA-binding partners and direct transcriptional targets will be an important breakthrough.
How is branching regulated by strigolactones? This is a hotly contested subject that will benefit from understanding the roles that SMXL proteins play and how SL signaling rapidly influences PIN1 levels at the plasma membrane.
Does SMAX1 regulation by KAI2 mirror the SL signaling mechanism as hypothesized? Are SMXL3,4,5 targeted for degradation by MAX2, and in response to which signals?
Which SMXL genes control other MAX2-regulated traits such as senescence, cambial growth, and drought tolerance? What roles do SMXL3, SMXL4, and SMXL5 have in plant growth?
What is the molecular basis of KAR and SL signaling specificity? How are different SLs made and perceived? This is an important research area that may foster innovative approaches to combat parasitic weeds. It is now possible to investigate the molecular basis of ligand specificity in D14 and different classes of KAI2 paralogs found in parasitic plants. This will help answer evolutionary questions about how new ligand specificities arise, how SLs are perceived in basal plants, and how fire followers distributed throughout the angiosperms have evolved high sensitivity to karrikins.
What are the origins and functions of the SMXL family in land plants? A coevolutionary analysis of receptor–effector interaction domains may yield insights into the basis of signaling specificity.
What is the endogenous KAI2 ligand? This ligand(s) may be an undiscovered hormone, and its identification may give clues about the original functions of KAI2 in plants.
Trends.
Compelling evidence that KAI2 and D14 are karrikin (KAR) and strigolactone (SL) receptors with hydrolytic activity has been provided through recent genetic, biochemical, and structural studies in several plant species. Ligand hydrolysis promotes protein–protein interactions.
The elusive targets of KAR and SL signaling have been discovered. Two clades within the SMXL/D53 family regulate growth processes that are associated with either KAR or SL responses.
SL receptors in parasitic weeds, which enable host-triggered germination, evolved from KAI2 paralogs. Other KAI2 paralogs in parasites detect KARs specifically or an unidentified endogenous ligand, KL.
KAR and SL signaling mechanisms share features of other plant hormone pathways, including hormone-activated proteolysis of EAR motif-containing proteins that likely interact with transcriptional corepressors.
Acknowledgments
Funding support was provided by the National Science Foundation (IOS-1350561) to D.C.N. and L.F., and the National Institute of General Medical Sciences (NIGMS) National Institutes of Health Award T32GM007103 to N.M. We apologize to those whose work could not be cited owing to space limitations.
References
- 1.Flematti GR et al. (2009) Identification of alkyl substituted 2H-Furo[2,3-c]pyran-2-ones as germination stimulants present in smoke. J. Agric. Food Chem 57, 9475–9480 [DOI] [PubMed] [Google Scholar]
- 2.Flematti GR et al. (2004) A compound from smoke that promotes seed germination. Science 305, 977. [DOI] [PubMed] [Google Scholar]
- 3.Nelson DC et al. (2012) Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu. Rev. Plant Biol 63, 107–130 [DOI] [PubMed] [Google Scholar]
- 4.Nelson DC et al. (2010) Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A 107, 7095–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson DC et al. (2009) Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 149, 863–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Staden J et al. (2006) Post-germination effects of the smoke-derived compound 3-methyl-2H-furo[2,3-c]pyran-2-one, and its potential as a preconditioning agent. Field Crops Res. 98, 98–105 [Google Scholar]
- 7.Kulkarni MG et al. (2006) Stimulation of rice (Oryza sativa L.) seedling vigour by smoke-water and butenolide. J. Agron. Crop Sci 192, 395–398 [Google Scholar]
- 8.Jain N et al. (2006) A butenolide, isolated from smoke, can overcome the detrimental effects of extreme temperatures during tomato seed germination. Plant Growth Regul. 49, 263–267 [Google Scholar]
- 9.Cook CE et al. (1966) Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154, 1189–1190 [DOI] [PubMed] [Google Scholar]
- 10.Akiyama K et al. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827 [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Roldan V et al. (2008) Strigolactone inhibition of shoot branching. Nature 455, 189–194 [DOI] [PubMed] [Google Scholar]
- 12.Kohlen W et al. (2012) The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 196, 535–547 [DOI] [PubMed] [Google Scholar]
- 13.Umehara M et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200 [DOI] [PubMed] [Google Scholar]
- 14.Kapulnik Y et al. (2011) Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233, 209–216 [DOI] [PubMed] [Google Scholar]
- 15.Ruyter-Spira C et al. (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 155, 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen A et al. (2012) Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 158, 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stirnberg P et al. (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129, 1131–1141 [DOI] [PubMed] [Google Scholar]
- 18.Lauressergues D et al. (2015) Strigolactones contribute to shoot elongation and to the formation of leaf margin serrations in Medicago truncatula R108. J. Exp. Bot 66, 1237–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scaffidi A et al. (2013) Carlactone-independent seedling morphogenesis in Arabidopsis. Plant J. 76, 1–9 [DOI] [PubMed] [Google Scholar]
- 20.Ueda H and Kusaba M (2015) Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol. 169, 138–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada Y et al. (2014) Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240, 399–408 [DOI] [PubMed] [Google Scholar]
- 22.Agusti J et al. (2011) Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc. Natl. Acad. Sci. U.S.A 108, 20242–20247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y et al. (2014) Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat. Chem. Biol 10, 1028–1033 [DOI] [PubMed] [Google Scholar]
- 24.Seto Y et al. (2014) Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. U.S.A 111, 1640–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abe S et al. (2014) Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. U.S.A 111, 18084–18089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waters MT et al. (2012) The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol. 159, 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alder A et al. (2012) The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351 [DOI] [PubMed] [Google Scholar]
- 28.Lin H et al. (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21, 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kretzschmar T et al. (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483, 341–344 [DOI] [PubMed] [Google Scholar]
- 30.Sasse J et al. (2015) Asymmetric localizations of the ABC transporter PaPDR1 trace paths of directional strigolactone transport. Curr. Biol 25, 647–655 [DOI] [PubMed] [Google Scholar]
- 31.Fridlender M et al. (2015) Influx and efflux of strigolactones are actively regulated and involve the cell-trafficking system. Mol. Plant 8, 1809–1812 [DOI] [PubMed] [Google Scholar]
- 32.Nelson DC et al. (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A 108, 8897–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arite T et al. (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424 [DOI] [PubMed] [Google Scholar]
- 34.Waters MT et al. (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295 [DOI] [PubMed] [Google Scholar]
- 35.Hamiaux C et al. (2012) DAD2 is an alpha/betahydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol 22, 2032–2036 [DOI] [PubMed] [Google Scholar]
- 36.Shen H et al. (2012) MAX2 affects multiple hormones to promote photomorphogenesis. Mol. Plant 5, 750–762 [DOI] [PubMed] [Google Scholar]
- 37.Scaffidi A et al. (2014) Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 165, 1221–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soundappan I et al. (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27, 3143–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umehara M et al. (2015)Structural requirements of strigolactones for shoot branching inhibition in rice and Arabidopsis. Plant Cell Physiol. 56, 1059–1072 [DOI] [PubMed] [Google Scholar]
- 40.Zhao LH et al. (2015) Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 25, 1219–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kameoka H and Kyozuka J (2015) Downregulation of rice DWARF 14 LIKE suppress mesocotyl elongation via a strigolactone independent pathway in the dark. J. Genet. Genomics 42, 119–124 [DOI] [PubMed] [Google Scholar]
- 42.Kagiyama M et al. (2013) Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18, 147–160 [DOI] [PubMed] [Google Scholar]
- 43.Nakamura H et al. (2013) Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun 4, 2613. [DOI] [PubMed] [Google Scholar]
- 44.Zhao LH et al. (2013) Crystal structures of two phytohormone signal-transducing alpha/beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23, 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chevalier F et al. (2014) Strigolactone promotes degradation of DWARF14, an alpha/beta hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26, 1134–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo Y et al. (2013) Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 110, 8284–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waters MT et al. (2015) A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27, 1925–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waters MT et al. (2015) Substrate-induced degradation of the alpha/beta-fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Mol. Plant 8, 814–817 [DOI] [PubMed] [Google Scholar]
- 49.Waters MT et al. (2014) The karrikin response system of Arabidopsis. Plant J. 79, 623–631 [DOI] [PubMed] [Google Scholar]
- 50.Conn CE et al. (2015) PLANT EVOLUTION. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349, 540–543 [DOI] [PubMed] [Google Scholar]
- 51.Tsuchiya Y et al. (2015) PARASITIC PLANTS. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349, 864–868 [DOI] [PubMed] [Google Scholar]
- 52.Toh S et al. (2015) Structure–function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350, 203–207 [DOI] [PubMed] [Google Scholar]
- 53.Somers DE and Fujiwara S (2009) Thinking outside the F-box: novel ligands for novel receptors. Trends Plant Sci. 14, 206–213 [DOI] [PubMed] [Google Scholar]
- 54.Stirnberg P et al. (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50, 80–94 [DOI] [PubMed] [Google Scholar]
- 55.Zhao J et al. (2014) DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching. Plant Cell Physiol. 55, 1096–1109 [DOI] [PubMed] [Google Scholar]
- 56.Gray WM et al. (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414, 271–276 [DOI] [PubMed] [Google Scholar]
- 57.Thines B et al. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665 [DOI] [PubMed] [Google Scholar]
- 58.Chini A et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 [DOI] [PubMed] [Google Scholar]
- 59.Tan X et al. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 [DOI] [PubMed] [Google Scholar]
- 60.Sheard LB et al. (2010) Jasmonate perception by inositolphosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murase K et al. (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463 [DOI] [PubMed] [Google Scholar]
- 62.Hauvermale AL et al. (2012) Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol. 160, 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stanga JP et al. (2013) SUPPRESSOR OF MORE AXILARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 163, 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang L et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou F et al. (2013) D14–SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang L et al. (2015) Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27, 3128–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoneyama K et al. (2010) Strigolactones as germination stimulants for root parasitic plants. Plant Cell Physiol. 51, 1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delaux PM et al. (2012) Origin of strigolactones in the green lineage. New Phytol. 195, 857–871 [DOI] [PubMed] [Google Scholar]
- 69.Flematti GR et al. (2013) Karrikin and cyanohydrin smoke signals provide clues to new endogenous plant signaling compounds. Mol. Plant 6, 29–37 [DOI] [PubMed] [Google Scholar]
- 70.Conn CE and Nelson DC (2016) Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci 6, 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Proust H et al. (2011)Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138, 1531–1539 [DOI] [PubMed] [Google Scholar]
- 72.Hoffmann B et al. (2014) Strigolactones inhibit caulonema elongation and cell division in the moss Physcomitrella patens. PLoS ONE 9, e99206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gutjahr C et al. (2015) Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350, 1521–1524 [DOI] [PubMed] [Google Scholar]
- 74.Yoshida S et al. (2012) The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytol. 196, 1208–1216 [DOI] [PubMed] [Google Scholar]
- 75.Flematti GR et al. (2011) Production of the seed germination stimulant karrikinolide from combustion of simple carbohydrates. J. Agric. Food Chem 59, 1195–1198 [DOI] [PubMed] [Google Scholar]
- 76.Xie X et al. (2013) Confirming stereochemical structures of strigolactones produced by rice and tobacco. Mol. Plant 6, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Challis RJ et al. (2013)A role for more axillary growth1 (MAX1) in evolutionary diversity in strigolactone signaling upstream of MAX2. Plant Physiol. 161, 1885–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Long JA et al. (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523 [DOI] [PubMed] [Google Scholar]
- 79.Krogan NT et al. (2012) APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Szemenyei H et al. (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386 [DOI] [PubMed] [Google Scholar]
- 81.Pauwels L et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Causier B et al. (2012)The TOPLESS inleraclome: a framework for gene repression in Arabidopsis. Plant Physiol. 158, 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shyu C et al. (2012) JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24, 536–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mashiguchi K et al. (2009) Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci. Biotechnol. Biochem 73, 2460–2465 [DOI] [PubMed] [Google Scholar]
- 85.Ke J et al. (2015) Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci. Adv 1, e1500107. [DOI] [PMC free article] [PubMed] [Google Scholar]


