Supplemental Digital Content is available in the text.
Keywords: artificial intelligence, critical care, digital twin, directed acyclic graph, organ failure
Objectives:
To develop and verify a digital twin model of critically ill patient using the causal artificial intelligence approach to predict the response to specific treatment during the first 24 hours of sepsis.
Design:
Directed acyclic graphs were used to define explicitly the causal relationship among organ systems and specific treatments used. A hybrid approach of agent-based modeling, discrete-event simulation, and Bayesian network was used to simulate treatment effect across multiple stages and interactions of major organ systems (cardiovascular, neurologic, renal, respiratory, gastrointestinal, inflammatory, and hematology). Organ systems were visualized using relevant clinical markers. The application was iteratively revised and debugged by clinical experts and engineers. Agreement statistics was used to test the performance of the model by comparing the observed patient response versus the expected response (primary and secondary) predicted by digital twin.
Setting:
Medical ICU of a large quaternary- care academic medical center in the United States.
Patients or Subjects:
Adult (> 18 year yr old), medical ICU patients were included in the study.
Interventions:
No additional interventions were made beyond the standard of care for this study.
Measurements and Main Results:
During the verification phase, model performance was prospectively tested on 145 observations in a convenience sample of 29 patients. Median age was 60 years (54–66 d) with a median Sequential Organ Failure Assessment score of 9.5 (interquartile range, 5.0–14.0). The most common source of sepsis was pneumonia, followed by hepatobiliary. The observations were made during the first 24 hours of the ICU admission with one-step interventions, comparing the output in the digital twin with the real patient response. The agreement between the observed versus and the expected response ranged from fair (kappa coefficient of 0.41) for primary response to good (kappa coefficient of 0.65) for secondary response to the intervention. The most common error detected was coding error in 50 observations (35%), followed by expert rule error in 29 observations (20%) and timing error in seven observations (5%).
Conclusions:
We confirmed the feasibility of development and prospective testing of causal artificial intelligence model to predict the response to treatment in early stages of critical illness. The availability of qualitative and quantitative data and a relatively short turnaround time makes the ICU an ideal environment for development and testing of digital twin patient models. An accurate digital twin model will allow the effect of an intervention to be tested in a virtual environment prior to use on real patients.
Early diagnosis and treatment are synonymous with improved outcomes in sepsis and other life-threatening conditions (1–3). It is likely that poor outcomes in sepsis and related common ICU syndromes are at least in part due to suboptimal early management (4). Indeed, a failure to recognize and treat deteriorating patients accounts for an estimated 11% of total number of deaths in hospitalized patients (5). The complex nature of sepsis and critical illness, in general, calls for alternative approaches to assist in timely diagnosis and management.
The availability of electronic health record (EHR) data has resulted in a proliferation of artificial intelligence (AI) applications to be used in critical care medicine. A group of computer scientists and clinicians from the Imperial College, London, United Kingdom, used an AI approach to develop a decision support model aptly named AI Clinician (6). AI Clinician is a computational model based on reinforcement learning, which is able to suggest dynamically optimal treatments for adult patients with sepsis in the ICU. It was developed and validated in retrospective data from two clinical databases: Medical Information Mart for Intensive Care-III and electronic-ICU research database (7, 8). A similar approach was also used for the continuous prediction of acute kidney injury (AKI) (9). Although this exciting new approach yielded clinically plausible results, its utility as a bedside tool remains yet to be proven of benefit in the clinical practice.
A major drawback of the current “black-box” AI modes is that the complex analytics are provided to the clinician without the disclosure of the data that is used or the analytical algorithm that is applied. This lack of transparency can be hazardous and unreliable in situations of highly complex decision-making and is unlikely to be accepted by the bedside clinicians. Prediction models used in research, administration, and clinical practice aim to use the available data to predict an outcome (e.g., clinical state), which has not yet been observed (10).
Considering the abovementioned drawbacks of associative AI models and utilizing the understanding from prediction models, we developed a digital twin model of critically ill patient with sepsis that is explicitly designed based on causal relationships using the directed acyclic graph (DAG) approach (11). To have a better understanding of DAGs and development of the model, we need a basic understanding of Bayesian networks. Bayesian networks are DAGs whose nodes equate to variables in the Bayesian sense: they can be observable quantities, latent variables, unknown parameters, or hypotheses (12–14). Edges represent conditional dependencies; nodes that are not connected (no path connects one node to another) represent variables that are conditionally independent of each other. Each node is associated with a probability function that takes, as input, a particular set of values for the node’s parent variables and gives (as output) the probability (or probability distribution, if applicable) of the variable represented by the node. Directed acyclic graphical model is a probabilistic graphical model (a type of statistical model) that represents a set of variables and their conditional dependencies—also known as the Bayesian network Model. DAGs are helpful graphical tool providing a visually easy way to represent and understand the key concepts of exposure, outcome, causation, confounding, and bias (15). Here, we present the iterative development, prospective verification, and preliminary performance of a DAG-based causal AI model to predict the response to specific treatment during the first 24 hours of sepsis.
MATERIALS AND METHODS
Model Coding and Design
A hybrid approach that combines agent-based modeling and simulation (ABMS), discrete-event simulation, and Bayesian network model was used to develop model and simulate the patient and the environment across multiple stages. ABMS of sepsis was used to simulate the actions and interactions of major organ systems (cardiovascular, neurologic, renal, respiratory, gastrointestinal, inflammatory, and hematology). Sepsis-3 definition was used to identify the patients included in our study (16). Organ systems (agents) act and interact based on preprogrammed expert rules, with the aim of identifying further insights into their collective behavior. Expert rules were derived using literature supporting the current standard of clinical practice including textbook chapters, critical care board reviews, and original research articles. The execution of the rules and state of these major organ systems is dependent on the time, medication, and medication dose that is administered. Cointerventions (mechanical ventilation and source control) and patient’s baseline state (age, preexisting illnesses, baseline functional status, etc.) among other factors also had an effect on the patient’s change of state, but these were not measured. Organ systems interact among themselves based on the expert rules, to recreate and predict the future state of the patient. Effort was made to reduce information overload and describe the organ systems in terms of the most important and most relevant clinical markers (over 60 clinical markers for the major organ systems described above) (17).
Three fundamental concepts that underlie and distinguish our model and allow it to generate better future predictions are as follows:
Patient’s case timeline: Patient’s timeline was divided in carefully chosen steps and all predictions were made for the patient’s health state in the next step, given the previous state (state of organ systems), baseline health state, and the interventions (including medications). After admission, the patient timeline was focused on “golden hours” (15 min intervals during the first hour, hourly during the first 12 hr) and then scales to days (every 24 hr), until 1 week (7 d) after admission.
Actionable clinical markers: Clinical markers are color coded as yellow, red, and white. Yellow and red characterized a disturbance that needs to be acted upon. A typical clinical marker might have one of the flowing three values: white (normal), yellow (disturbance or abnormal), and red (major disturbance or highly abnormal).
Graduated clinical interventions. Interventions have three states: off, low dose, and high dose.
Definition of success and failure of the simulation: with each intervention made in the digital twin, the model was programmed to have a change in the variable by one level. For example, based on the expert rules used to design the model, for a critically ill patient with hypotension, we expected that administration of vasopressors would improve the mean arterial pressure (MAP) from red (very low) to yellow (low) or from yellow (low) to white (normal) range. If the changes in the digital twin were concordant with the real patient, this was considered as a success. However, if the output was different from what was expected, it would fall under one of the types of error that is explained in the subsequent sections of the article (Supplement Table 1, http://links.lww.com/CCX/A405). For example, “coding error” was defined as any error resulting from dissonance between the rules established in the expert rule book at the time of model design and the output resulting from a faulty coding during model programming. For example, rule states that MAP should increase by one level (from red to yellow or from yellow to white range) after administration of vasopressors. If the output results were noted in a different direction, it would be considered as a coding error.
Model Development
DAGs for this model have been developed based on the preexisting medical knowledge (Fig. 1). DAGs were based on the information in the expert rules. Furthermore, these DAGs are iteratively refined by critical care experts from various backgrounds (18). The final model is derived from the curated expert rules and critical data points, thereby effectively embedding mechanistic understanding in the model. After imputation of appropriate clinical and laboratory data derived from a real patient into this model (virtual patient, Online Supplement Fig. 1, http://links.lww.com/CCX/A406, and Online Supplement Fig. 2, http://links.lww.com/CCX/A407), it predicts clinical output (response to intervention, Online Supplement Fig. 3, http://links.lww.com/CCX/A408) with the help of incorporated AI algorithm and thus generating a “digital twin” (Online supplement Fig. 4, http://links.lww.com/CCX/A409). The functioning of the model and data capturing is also explained in Supplement Video 1 (http://links.lww.com/CCX/A410).
Figure 1.
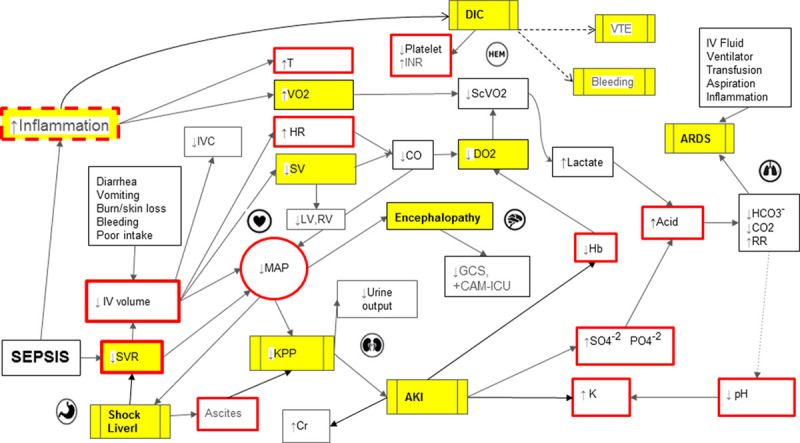
Directed acyclic graph depicting complex pathophysiologic interactions in sepsis-associated multiple organ dysfunction. Yellow boxes represent concepts, red solid border indicates actionable clinical points, and red interrupted border denotes semiactionable clinical points. AKI = acute kidney injury, ARDS = acute respiratory distress syndrome, CAM-ICU = confusion assessment method for ICU, CO = cardiac output, CO2 = serum carbon dioxide, DIC = disseminated intravascular coagulation, DO2 = arterial oxygen delivery, GCS = Glasgow Coma Scale, Hb = serum hemoglobin, HR = heart rate, INR = international normalized ratio, IVC = inferior vena cava, KPP = kidney perfusion pressure, LV = left ventricle, MAP = mean arterial pressure, RR = respiratory rate, RV = right ventricle, ScVO2 = central venous oxygen saturation, SV = stroke volume, SVR = systemic vascular resistance, T = temperature, VO2 = oxygen uptake, VTE = venous thromboembolism.
The application was iteratively revised and debugged while working in synchrony with program developers and engineers. The “rule book” included response to a medical intervention and is compartmentalized based on organ systems. For example, as evident in Figure 1, low MAP will lead to low kidney perfusion pressure, which will ultimately lead to AKI. A Bayesian network approach to modeling was used to model the conditional dependencies of major organ systems: cardiovascular, neurologic, renal, respiratory, gastrointestinal, hematologic, and immune. Conditional dependencies between various organ systems were defined upon the existing medical knowledge. Probabilistic relationships defined interactions and impact on organ systems. Expert rules defined the effects that variables have on each other, and various causes (interventions and interactions) lead to certain effects on organ systems reflected by clinical markers (i.e., increased heart rate, decreased urine output, decreased Glasgow Coma Scale [GCS], and dozens of others). Finally, program coding was done based on the master rule book to show the anticipated response of medical interventions in the virtual patient or the digital twin.
Model Verification
The study was approved by the Mayo Clinic institutional review board; requirement for informed consent was waived for this observational study. As a part of this pilot study, we prospectively validated the performance of the causal AI model by creating a digital twin for medical ICU patients admitted from the emergency department. Treatment response was observed in a convenience sample of 29 adult (> 18) patients with sepsis admitted to the ICU from the emergency department. Patients received standard of care in the ICU without any additional intervention from the research team. Care provided by the bedside clinician was independent of the patient’s inclusion in the study.
Data abstraction: Demographic data and clinical data points needed for the study were abstracted from the EHRs. Patient’s baseline data and clinical data points were used to create a clinical state for the digital twin.
Observations were made to assess, refine, and validate model performance. Time “zero” was the time of admission to the ICU. The model was tested for the type of errors in the design and functioning of the model such as coding error, timing error,), expert rule error, EHR artifact, error due to unaccounted influence by coadministration of a known medication, and unknown error. For detailed definition of errors, please refer to Supplemental Table 1 [http://links.lww.com/CCX/A405]. Primary response to an intervention was defined as a clinically significant change in the digital twin and real patient, which is expected to be directly related to an intervention. The secondary response was an additional or ancillary change in the clinical state of the patient in response to an intervention, which is not a direct (intended) effect but is often seen as an association (i.e., primary response to propofol administration is a drop in GCS, whereas the secondary response is a concomitant drop in MAP). Effect of these interventions was studied (as outcome) in the digital twin and compared with real patient outcome. Various outcomes observed were grouped together based on the organ system such as respiratory (respiratory rate, oxygen saturation, etc.), cardiovascular (MAP, heart rate, arrhythmia, etc.), neurologic (GCS), fluid (including pH, electrolytes, etc.), and immune homeostasis (inflammatory biomarkers such as CRP, WBC count, etc.). Studied interventions were categorized as medications (including antibiotics, vasoactive agents, cardiac drugs, anesthetics and sedatives, etc.), noninvasive or mechanical ventilation, IV fluids, source control (including endoscopic retrograde cholangiopancreatography, abscess drainage, etc.), or blood product transfusion. Model calibration checks were also conducted regularly by making no interventions in the virtual patient simulating the times of no intervention in the real patient. In the absence of an intervention, the expected observation was maintenance of a “steady clinical state” with no major deviation of clinical parameters.
The model was iteratively debugged during this pilot run based on the commonly occurring errors.
Data Analysis
Data were analyzed using the JMP statistical software (Version 14.0; SAS Institute, Cary, NC). Observations were descriptively summarized using frequencies and percentages for categorical data, and median and interquartile ranges (IQRs) for continuous variables. The response of the virtual patient was compared against the real patient response (gold standard) by studying agreement statistics (kappa coefficient) as appropriate.
RESULTS
During the verification phase, model performance was prospectively tested on 145 observations in a convenience sample of 29 patients. Median age was 60 years (54–66) with a median Sequential Organ Failure Assessment score of 9.5 (IQR, 5.0–14.0). The most common source of sepsis was pneumonia, followed by hepatobiliary. The observations were made during the first 24 hours of the ICU admission with one-step interventions, comparing the output in the digital twin with the real patient response.
Demographics, clinical features, type of interventions, and outcomes are described in Table 1.
TABLE 1.
Descriptive Analysis
| Age in years (median and range) | 60.2 (54.2–66.3) |
| Gender (n = 29), n (%) | |
| Male | 13 (44.8) |
| Female | 16 (55.2) |
| Sequential Organ Failure Assessment score (interquartile range) | 9.5 (5.0–14.0) |
| Source of sepsis (n = 29), n (%) | |
| Pneumonia | 21 (72) |
| Hepatobiliary | 4 (14) |
| Urinary tract infection | 2 (7) |
| Enterocolitis | 2 (7) |
| Type of interventions tested (n = 145), n (%) | |
| Medication effect | 73 (50.3) |
| Noninvasive and mechanical ventilation | 26 (17.9) |
| IV fluids | 23 (15.9) |
| No intervention | 9 (6.4) |
| Hemodialysis | 8 (5.5) |
| Blood product transfusion | 4 (2.7) |
| Source control | 2 (1.4) |
| Outcomes studied (n = 145) | |
| Fluid hemostasis | 33 (22.7) |
| Respiratory effects | 28 (19.3) |
| Cardiovascular effects | 24 (16.5) |
| Neurologic effects | 16 (11.0) |
| Metabolic effects (including renal) | 14 (9.6) |
| Model calibration checks | 9 (6.2) |
| Inflammatory (sepsis homeostasis) | 18 (12.4) |
| Transfusion effects | 3 (2.1) |
Out of the 145 observations, any error was detected in 75 (51.7%). The most common error detected was coding error in 50 observations (35%), followed by expert rule error in 29 (20%) and timing error in seven (5%) observations (Table 2). The agreement between the observed versus the expected response ranged from fair (kappa coefficient of 0.41) for primary response to good (kappa coefficient of 0.65) for secondary response to the intervention (Table 3).
TABLE 2.
Model Testing Analysis
| Total number of patients | 29 |
| Total number of interventions tested (n) | 145 |
| Any error detected | 75 (51.7%) |
| Coding error | 50 (34.5%) |
| Expert rule error | 29 (20.0%) |
| Unaccounted error secondary to a known medication | 5 (3.4%) |
| Electronic health record error | 1 (0.7%) |
| Unknown error | 7 (4.8%) |
| Error Secondary to preexisting illness | 0 (0.0%) |
| Timing error | 7 (4.8%) |
Data are presented as number of patients (%).
TABLE 3.
Agreement Statistics
| Kappa Coefficient | se | 95% CI | |
|---|---|---|---|
| Degree of agreement for actual observed patient effect (primary) versus the expected patient effect | 0.41 | 0.08 | 0.25–0.57 |
| Degree of agreement for actual observed patient effect (secondary) versus the expected patient effect | 0.65 | 0.06 | 0.53–0.77 |
DISCUSSION
The results from this pilot study of verification of a causal AI model of early sepsis are encouraging. We proved the feasibility of developing a digital twin and established methodology for prospective evaluation and testing. The development of a functional digital twin of critically ill patients will have wide application from teaching and evaluating cognitive skills of critical care trainees, to bedside decision support and in silico clinical experiments. Digital twin allows clinical or research interventions to be tested first in a virtual environment, minimizing safety concerns to real patients. In addition, visualization of clinical markers and the interplay of underlying expert rules describing known interactions among organ systems will allow researchers to identify important knowledge gaps.
Causal AI has been previously used in developing a model for diabetes mellitus (Archimedes) to study the interaction of about 50 continuously interacting variables (19). The Archimedes model was developed and refined over the period of years due to the chronic nature of the disease pathophysiology. The design objective for the model was to simulate the real-life health state that is realistic with the natural level of detail. In contrast, our model is more dynamic due to the nature and temporal pace of critical illness and rapidly changing clinical state (along with clinically relevant data points). Information-gathering phase followed by refinement could be achieved within a relatively short period of time.
The availability of qualitative and quantitative data in relatively short turnaround time for critically ill patients makes ICU an ideal environment for testing and frequent debugging of the model. This approach allows rapid iterative verification of expert rules in the digital-twin (in silico model) prospectively. Causal associations are explicitly sought and refined using the DAGs that are developed during the early phase of model design. The model verification is done prospectively so that the model learning could be augmented by finer observations made by the clinician at every stage and not based on the retrospective data points captured from the EHRs.
Our model is designed to provide predictive value (how will the patient respond with or without specific treatment) in contrast to purely prognostic value of associative AI models. The models developed from database and repositories are primarily focused on prognostic enrichment. For example, model developed to prognosticate the risk of developing AKI and need for hemodialysis can provide information on which patient will eventually need hemodialysis, but it provides little value for the bedside decision-making (9).
Studying the primary and secondary outcomes in the digital twin during the verification phase of our model helped us to focus on the most essential but limited elements. This was done intentionally to avoid building a model, which is too complicated while maintaining an appropriate level of granularity (20). Correct identification and adjustment of the confounders was an important component that helped with calibration and debugging.
The limitations of our study include the small, convenience sample size compared with a large number of data points that could be extracted from data repositories. Development of AI models that are debugged, verified, and validated prospectively are time-consuming and very resource-intensive. For this study, we only considered one-step interventions. In the ICUs, patients undergo multiple interventions at the same time with largely unknown interactions that remain to be studied.
To address the high error rate and attain a greater level of accuracy, the model will be further refined using a much larger sample size in the next phase. Tuning the model according to the raw data (rather than threshold changes) may further improve the fidelity of the model in the next phases. This analysis is helpful in the initial stages of model development and verification, as this provides the much-needed transparency for the complex analytics to the involved clinicians with appropriate disclosure of the data that are used to refine the analytical algorithm applied in the model. This is one of the major advantages of causal AI model over the currently used “black-box” AI models where the complex analytics are provided to the clinician without the clear revelation of the data that is used or the analytical algorithm (21, 22).
Technology-based innovation adoption is challenging due to a lack of confidence in the model, addition of a supplementary electronic tools to the already existing EHR, and resource utilization (23). We have aimed to mitigate the lack of transparency seen in many “black-box” AI models; this is done with an intent to successfully facilitating technology adoption and addressing cognitive, emotional, and contextual concerns of the clinician who will have potential utility for this tool (24, 25).
CONCLUSIONS
We developed and pilot-tested a digital twin of critically ill patient with sepsis and established methodology for verification and prospective validation of causal AI applications. Future studies will determine the utility of digital twin models for education, research, and bedside decision support.
Supplementary Material
Footnotes
Supported, in part, by Mayo Clinic, Critical Care Independent Multidisciplinary Program.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (http://journals.lww.com/ccejournal).
The authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.Bledsoe BE. The golden hour: Fact or fiction? Emerg Med Serv. 2002; 31:105. [PubMed] [Google Scholar]
- 2.Damluji A, Colantuoni E, Mendez-Tellez PA, et al. Short-term mortality prediction for acute lung injury patients: External validation of the acute respiratory distress syndrome network prediction model. Crit Care Med. 2011; 39:1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinhart K, Daniels R, Kissoon N, et al. Recognizing sepsis as a global health priority - a WHO resolution. N Engl J Med. 2017; 377:414–417 [DOI] [PubMed] [Google Scholar]
- 4.Rivers E, Nguyen B, Havstad S, et al. ; Early Goal-Directed Therapy Collaborative Group. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N Engl J Med. 2001; 345:1368–1377 [DOI] [PubMed] [Google Scholar]
- 5.National Patient Safety Agency: Safer Care for the Acutely Ill Patient: Learning From Serious Incidents: The Fifth Report From the Patient Safety Observatory. Great Britain: National Patient Safety Agency, 2007 [Google Scholar]
- 6.Komorowski M, Celi LA, Badawi O, et al. The Artificial Intelligence Clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med. 2018; 24:1716–1720 [DOI] [PubMed] [Google Scholar]
- 7.Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016; 3:160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollard TJ, Johnson AEW, Raffa JD, et al. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci Data. 2018; 5:180178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomašev N, Glorot X, Rae JW, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. 2019; 572:116–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leisman DE, Harhay MO, Lederer DJ, et al. Development and reporting of prediction models: Guidance for authors from editors of respiratory, sleep, and critical care journals. Crit Care Med. 2020; 48:623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lederer DJ, Bell SC, Branson RD, et al. Control of confounding and reporting of results in causal inference studies. Guidance for authors from editors of respiratory, sleep, and critical care journals. Ann Am Thorac Soc. 2019; 16:22–28 [DOI] [PubMed] [Google Scholar]
- 12.Cooper GF. The computational complexity of probabilistic inference using Bayesian belief networks. Artif Intell. 1990; 42:393–405 [Google Scholar]
- 13.Lal A, Pinevich Y, Gajic O, et al. Artificial intelligence and computer simulation models in critical illness. World J Crit Care Med. 2020; 9:13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neapolitan RE: Learning Bayesian Networks. Upper Saddle River, NJ: Pearson Prentice Hall, 2004 [Google Scholar]
- 15.Williams TC, Bach CC, Matthiesen NB, et al. Directed acyclic graphs: A tool for causal studies in paediatrics. Pediatr Res. 2018; 84:487–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 2016; 315:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickering BW, Gajic O, Ahmed A, et al. Data utilization for medical decision making at the time of patient admission to ICU. Crit Care Med. 2013; 41:1502–1510 [DOI] [PubMed] [Google Scholar]
- 18.Longo DL, Fauci AS, Kasper DL, et al. : Harrison’s Manual of Medicine. New York, NY: McGraw-Hill Medical, 2013 [Google Scholar]
- 19.Eddy DM, Schlessinger L. Archimedes: A trial-validated model of diabetes. Diabetes Care. 2003; 26:3093–3101 [DOI] [PubMed] [Google Scholar]
- 20.Grimm V, Revilla E, Berger U, et al. Pattern-oriented modeling of agent-based complex systems: Lessons from ecology. Science. 2005; 310:987–991 [DOI] [PubMed] [Google Scholar]
- 21.Giannini HM, Ginestra JC, Chivers C, et al. A machine learning algorithm to predict severe sepsis and septic shock: Development, implementation, and impact on clinical practice. Crit Care Med. 2019; 47:1485–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ginestra JC, Giannini HM, Schweickert WD, et al. Clinician perception of a machine learning-based early warning system designed to predict severe sepsis and septic shock. Crit Care Med. 2019; 47:1477–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King AJ, Cooper GF, Hochheiser H, et al. Using machine learning to predict the information seeking behavior of clinicians using an electronic medical record system. AMIA Annu Symp Proc. 2018; 2018:673–682 [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, Kaushal R, Khullar D. Should health care demand interpretable artificial intelligence or accept “black box” medicine? Ann Intern Med. 2020; 172:59–60 [DOI] [PubMed] [Google Scholar]
- 25.Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell. 2019; 1:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


