Extracellular vesicles (EVs) are membrane-derived lipid bilayers secreted by bacteria and eukaryotic cells. Bacterial membrane vesicles were discovered over 60 years ago and have been extensively studied in Gram-negative bacteria. During their production, EVs are loaded with proteins, nucleic acids, and various compounds that are subsequently released into the environment. Depending on the packaged cargo, EVs have a broad spectrum of action and are involved in pathogenesis, antibiotic resistance, nutrient uptake, and nucleic acid transfer.
KEYWORDS: EVs, extracellular vesicles, OMV, Gram-positive bacteria, membrane vesicles
ABSTRACT
Extracellular vesicles (EVs) are membrane-derived lipid bilayers secreted by bacteria and eukaryotic cells. Bacterial membrane vesicles were discovered over 60 years ago and have been extensively studied in Gram-negative bacteria. During their production, EVs are loaded with proteins, nucleic acids, and various compounds that are subsequently released into the environment. Depending on the packaged cargo, EVs have a broad spectrum of action and are involved in pathogenesis, antibiotic resistance, nutrient uptake, and nucleic acid transfer. Due to differences in cell wall structure, EVs in Gram-positive bacteria have been disregarded for decades, and our understanding of their biogenesis and host cell interaction is incomplete. Recently, studies on bacteria such as Staphylococcus aureus, Streptococcus spp., Bacillus subtilis, and Mycobacterium spp. have demonstrated EV production in Gram-positive bacteria and shown the great importance EVs have in Gram-positive bacterial physiology and disease progression. Here, we review the latest findings on the biogenesis and functions of EVs from Gram-positive bacteria and identify key areas for future research.
INTRODUCTION
Living and surviving in an environment requires strong adaptation and communication with surrounding partners. The secretion of cytosolic compounds plays a key role in inter- and intraspecies interactions, one of which can lead to the destruction of external invaders. In bacteria, nearly 35% of proteins are secreted and are involved in various functions ranging from nutrient acquisition to the killing of host cells through toxins (1). In general, secreted compounds follow one of the two well-known secretion machineries (i.e., the general secretion system [Sec] [2] or the twin-arginine transport [Tat] pathway [3]) and need the presence of a particular signaling domain to be transported outside the cell. The Sec system relies on the use of ATP and proton motive force (PMF) to translocate unfolded chaperone-associated proteins containing an N-terminal signal peptide sequence. The Tat system, composed of the TatA and TatC subunits (at least), directly transports folded proteins with an N-terminal ST-R-R-x-FLK consensus sequence, by use of PMF (3). Despite the existence of these two secretion systems, approximately one-quarter of secreted proteins have no known export signals. This raises important questions regarding the existence of alternative delivery and secretion systems (1).
Extracellular vesicles (EVs) are membrane-derived lipid bilayers that are found in all three domains of life. EVs are capable of packaging cytosolic compounds, including proteins and nucleic acids, and are generated in a process known as vesicle biogenesis or vesiculogenesis (4). Unjustly neglected for a long time, EVs have gained growing attention in recent decades and have been extensively studied primarily in eukaryotic cells and Gram-negative bacteria (4–7). In eukaryotes, EVs are called microvesicles, exosomes, or apoptotic bodies (depending on their size and origin in the cell) and can deliver cargos to cells in close proximity or cells located throughout the body (4). They are involved in the maintenance of normal physiology but can also be associated with disease (8–10). In Gram-negative bacteria, EVs are pinched off from the outer membrane and are therefore known as outer membrane vesicles (OMVs) (6). OMVs, which are spherical, usually trap periplasmic components and are implicated in many bacterial processes, such as biofilm formation, antibiotic resistance, and nucleic acid transfer (6). During infection, host cells absorb OMVs through a variety of different routes, including receptor-mediated endocytosis and membrane fusing (11). Host cell responses to OMV uptake have been characterized in different cell types and range from the initiation of a proinflammatory response in immune cells to an immunomodulatory effect in nonimmune cells (12).
Although it was long thought to be impossible (due to cell wall architecture), the production of EVs in Gram-positive bacteria was confirmed at the end of the last decade, thus demonstrating the universality of membrane vesicles (13). However, our knowledge of the biogenesis and composition of EVs in Gram-positive bacteria and their interaction with host cells lags behind that of EVs in other cell types. This review outlines recent advances in our understanding of EVs in Gram-positive bacteria, from genetic determinants leading to vesiculogenesis to interactions with host cells, while highlighting major questions that remain to be answered in this rapidly developing field of research.
VESICULOGENESIS
Genetic regulation of vesiculogenesis in Gram-positive bacteria.
EV biogenesis in Gram-positive bacteria remains an elusive mechanism. Experiments with heat-inactivated bacteria show that they are incapable of EV synthesis, demonstrating that vesiculogenesis relies on cells that are metabolically active (14–16). In Gram-negative bacteria, comparison of OMV production in wild-type bacteria within isogenic mutant lineages has greatly enriched our knowledge of the genes involved in vesiculogenesis (17). OMV formation is complex and not solely dependent on one gene. One study in Escherichia coli reported that nearly 150 genes were involved in OMV biogenesis (4, 17–21).
Since the analysis of EVs in Gram-positive bacteria is a relatively recent development (compared to OMV study in Gram-negative bacteria), very few studies have identified genetic determinants responsible for vesiculation (15, 22–25). In Listeria monocytogenes, EVs from a ΔsigB mutant were less abundant and appeared deformed relative to those in the wild-type strain (22). The two-component system CovRS in Streptococcus pyogenes and the Pst/SenX3-RegX3 signal transduction system in Mycobacterium tuberculosis have also been shown to regulate vesiculogenesis (15, 25). These global regulators drive the expression of numerous genes; consequently, their involvement suggests that EV production is not dependent on a small number of genes and that instead, a complex global gene network is involved. Widely conserved regulators (e.g., sigB) could be common genetic determinants of EV production in Gram-positive bacteria; however, this does not exclude the existence of intraspecies-specific genes that also contribute to vesiculogenesis. For instance, inactivation of the virR gene in M. tuberculosis leads to hypervesiculation via an unknown mechanism (23). Conversely, in S. aureus, deletion of the psmα genes but not the psmβ gene decreased the number of EVs recovered in the supernatant (24) (Fig. 1). Phenol-soluble modulins (PSMs) are a family of amphipathic alpha-helical peptides with surfactant-like activity. PSMs are divided into two groups; alpha-type PSMs (including δ-toxin), and beta-type PSMs. Even though PSMs have been observed in other staphylococcal species (26, 27), only S. aureus carries psmα genes with a described role in EV production (24). Our understanding of the genetic factors that contribute to vesiculogenesis in Gram-positive bacteria is in its infancy. Additional studies using global approaches (including the analysis of transposon mutant libraries and transposon insertion sequencing [TN-seq]) will likely identify species-specific factors that control vesiculogenesis and may lead to the discovery of common genetic determinants involved in vesiculogenesis in Gram-positive bacteria.
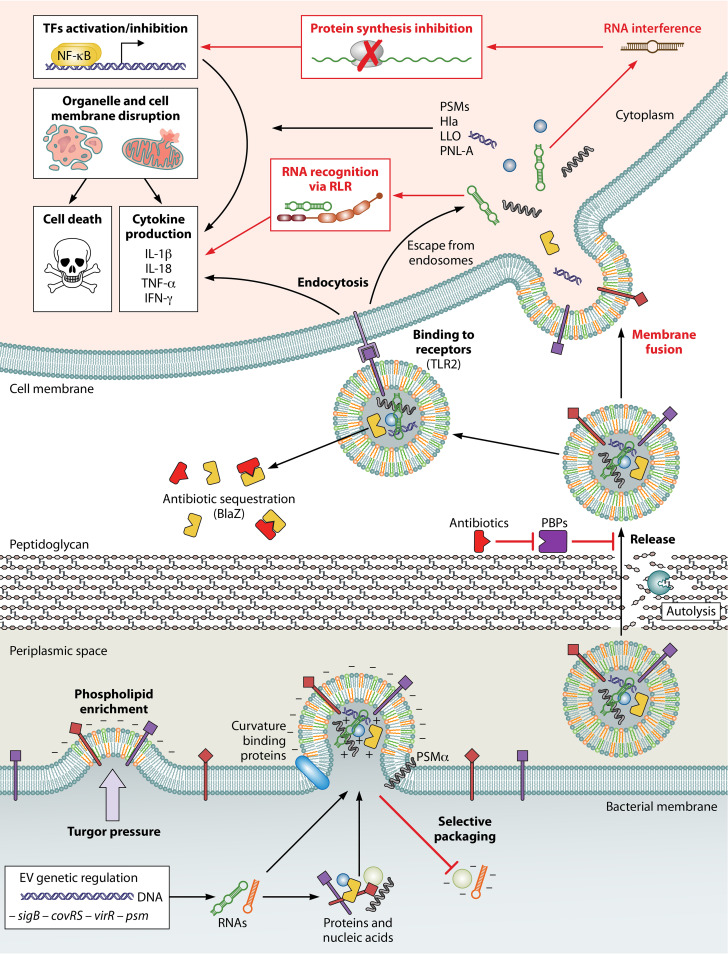
FIG 1.
Biogenesis and functions of Gram-positive EVs. Key steps are shown in bold. Red font indicates the hypothetical involvement of proteins or RNAs that have yet to be demonstrated. Turgor pressure promotes membrane curvature, aided by the overall negative charge due to phospholipid enrichment. A selective sorting mechanism occurs during vesiculogenesis, driven by electrostatic interactions within EVs. The involvement of curvature-recognizing proteins is also possible. Specific proteins, such as the αPSMs in S. aureus, facilitate the liberation of EVs from the plasma membrane. Autolysins and antibiotics that inhibit penicillin-binding proteins (PBPs) loosen the peptidoglycan and thus promote the release of EVs. The cargo can be discharged directly into the environment. This process can facilitate increased antibiotic resistance if, for example, antibiotic resistance proteins (e.g., β-lactamase [BlaZ]) have been preincorporated into EVs. Once EVs come into contact with host cells, surface-associated molecules, such as lipopolysaccharides, can interact with Toll-like receptor 2 (TLR-2) and trigger EV endocytosis. Direct membrane fusion, as described for Gram-negative OMVs, is also a potential delivery mechanism for Gram-positive EVs. Cargo released inside the cell can have a wide variety of effects, such as activation or inhibition of transcription factors (TFs), disruption of host cell membranes (Hla, PSMs, listeriolysin O [LLO], or pneumolysin A [PNL-A]), production and release of cytokines, and cell death. Finally, as described for Gram-negative bacteria, the presence of RNA in Gram-positive bacterial EVs could interfere with host cell protein synthesis and/or gene regulation by a base-pairing mechanism. Alternatively, prokaryote-specific RNA modifications may trigger immune system activation. (Created with BioRender.com.)
Release of EVs.
The nature of the peptidoglycan cell wall was long thought to rule out the possibility of EV production in Gram-positive bacteria. However, in recent years, several studies have isolated and observed 20- to 400-nm spherical lipid-bilayer structures in the supernatants of Gram-positive bacteria, demonstrating that EVs are widely spread throughout the prokaryote domain (13, 28–30).
The first step in the release of EVs is budding of the cytoplasmic membrane. Reduction of the turgor pressure in S. aureus hindered the release of EVs, demonstrating the importance of a hypotonic environment in vesiculogenesis (Fig. 1) (31). Lipidomic studies showed that EVs and cytoplasmic membranes are broadly composed of similar lipids, indicating the membrane origin of EVs (29, 32). However, in several bacteria, differences in fatty acids and phospholipid content between vesicles and membranes have been observed. In S. pyogenes, thin-layer chromatography-based quantification of membrane and EV lipid species identified anionic phosphatidylglycerol as being enriched and cardiolipin as being depleted in EVs (15). Similarly, vesicles from L. monocytogenes were enriched in phosphatidylethanolamine, sphingolipids, and triacylglycerols (16). The dissimilarities in phospholipid content between membranes and vesicles suggest that vesicle budding may be promoted at particular lipid-enriched domains within cytoplasmic membranes. Supporting this assumption are numerous studies in Gram-negative bacteria showing asymmetrical distribution of phospholipids, which led to the proposal by Roier et al. that the major force triggering vesiculation was the accumulation of phospholipids in the outer leaflet of the outer membrane, which leads to rapid extension and an outward bulging of the membrane (20). A similar mechanism involving lipid distribution is possible within cytoplasmic membranes of Gram-positive bacteria; however, studies have shown that additional factors can contribute to the release of EVs from membranes. For instance, reduction in lipoprotein content in S. aureus improved membrane fluidity, which was implicated in increased EV discharge (33). In addition, due to their surfactant-like properties and amphipathic helical structure, the αPSMs are thought to disrupt membranes, facilitating the formation of EVs and their subsequent release into the periplasmic space (24). Given that αPSMs are unique to S. aureus, it is possible that specific mechanisms exist in different bacterial species to promote the formation and release of EVs from the plasma membrane (Fig. 1).
Passage through the cell wall is the final step in EV release. The cell wall of Gram-positive bacteria primarily consists of peptidoglycan, a polymer made up of two alternating sugar residues (N-acetylglucosamine and N-acetylmuramic acid) cross-linked by peptide bonds. The mycobacterial cell wall is more complex than that found in other Gram-positive bacteria. It is made of mycolic acids and arabinogalactan anchored in a unique peptidoglycan composed of N-glycolyl and N-acetyl-muramic acids (34). Cell wall homeostasis relies on a complex balance of synthesis and degradation, orchestrated by proteins such as transpeptidases (also called penicillin-binding proteins [PBPs]) and autolysins (34, 35). Mass spectrometry proteomic analyses of EVs from S. aureus, Bacillus anthracis, L. monocytogenes, S. pyogenes, Propionibacterium acnes, Filifactor alocis, and mycobacterial species (including M. tuberculosis) detected the presence of PBPs and autolysins inside EVs, suggesting that cell wall modification plays a critical role in vesicle release (13–15, 22, 29, 36, 37). One study of group B streptococcus (GBS) did not find cell wall-associated proteins in EVs, possibly because a targeted proteomic analysis was used, rather than a global approach (32). The precise mechanism leading to EV trafficking through the cell wall remains poorly understood, but some evidence in S. aureus has demonstrated that the degree of peptidoglycan cross-linking can play a role in EV release (24). Indeed, sublethal or lethal exposure to β-lactam antibiotics reduced peptidoglycan cross-linking and massively increased the number of EVs recovered from S. aureus supernatants, relative to untreated cells (24, 38). Importantly, a strain defective in PBP production exhibited increased EV production, whereas an autolysin mutant (with a mutation in the sle1 gene), and to a lesser extent an atl1 mutant, exhibited reduced EV size and release, respectively, consistent with roles for sle1 and atl1 in cross-linking and peptidoglycan degradation (24). Likewise, wall teichoic acids negatively influenced S. aureus EV production (24).
In Gram-negative bacteria, a process called explosive cell lysis has also been shown to participate in OMV production (39). Increased expression of prophage-derived endolysins under DNA-damaging stress conditions provokes thinning of the peptidoglycan, resulting in cell lysis. Membranes from dead cells can round up and form OMVs (7). A similar mechanism has been observed in Gram-positive bacteria, where prophage-derived endolysins facilitate EV release under SOS response-inducing conditions (e.g., exposure to ciprofloxacin) (38, 40). Unlike Gram-negative bacteria, Bacillus subtilis preserves its morphology during this process. Bacterial membrane is pressed out of holes in the peptidoglycan, and EVs are subsequently created (40). EVs have been isolated within different growth phases (e.g., exponential and stationary phase) and during sporulation in B. subtilis, showing that EV production is a continuous process (41). Studies in Gram-negative bacteria revealed that vesicles can be formed at the division septa between the two daughter cells. (42). It is thought that OMVs are easily released during cell division, because this region is transiently composed of a thinner peptidoglycan layer (42). A similar process has yet to be proven for Gram-positive bacteria.
Hence, membrane fluidity and cell wall integrity (in particular, peptidoglycan cross-linking) appear to be critical determinants of vesicle release in Gram-positive bacteria (Fig. 1).
COMPOSITION OF EV CARGOS
Extracellular-vesicle content.
EVs can contain numerous different compounds such as cytosolic proteins, secreted proteins, and nucleic acids (DNA and RNA). The first report describing the composition of EVs in a Gram-positive bacterium was published in 2009 by Lee and colleagues (13). Ninety proteins were identified in S. aureus EVs, the majority of which were cytoplasmic proteins (56.7%), followed by extracellular proteins (23.3%) (13). A subsequent study further characterized the protein content in S. aureus EVs showing more than 200 proteins associated with EVs, 160 of which were cytoplasmic proteins (43). Recently, the same group confirmed these results in a variety of S. aureus strains, isolated from humans and animals, and demonstrated the existence of a “core S. aureus EV proteome” consisting of 119 proteins (44). Others groups have found similar results in different bacteria, such as M. tuberculosis, Streptococcus pneumoniae, and Bacillus anthracis (29, 45–47). Proteinase protection assays showed that proteins could either be localized in the EV lumen (and thus be protected from proteinase activity) or be anchored in the EV membrane (33, 48), demonstrating a complex protein organization within vesicles. Given the origin of EVs, it seems likely that most EV membrane-associated proteins originate from the cytoplasmic membrane, while proteins found in the lumen are cytoplasmic proteins packaged during vesiculogenesis. It is still unknown to what extent cytoplasmic proteins are “selected” and loaded into EVs. Functional category analyses of EV proteins indicates that the most common type of protein contained in EVs are metabolism associated (13, 29, 36, 43, 46). The function of these non-virulence-related proteins in EVs is still debated; however, several studies have speculated that they may play a role in distribution of nitrogen sources within bacterial communities (49, 50).
Genetic material has also been reported to be associated with EVs, either in the lumen or associated with the EV membrane (51, 52). Most research investigating the nucleic acid content of vesicles has been performed in Gram-negative bacteria, and the presence of DNA (chromosomal, plasmid, or phage origin) and RNA (including mRNA, rRNA, sRNA, and tRNA) has been demonstrated (53–59). To the best of our knowledge, only two studies have characterized the genetic material in Gram-positive-bacterial EVs (15, 32). Vesicles from group B streptococcus contained DNA at a concentration of 33 ng/μg of EV protein, and the virulence factor gene cfb (a pore-forming toxin-encoding gene) was successfully amplified by PCR from EVs (32). Using deep RNA sequencing, Resch et al. (15) observed that the majority of vesicular RNA sequences corresponded to rRNAs and tRNAs in S. pyogenes; however, the overall RNA profile was not the same as that found inside the parent cells (see below). This suggests that (as is the case for proteins) some type of selection process exists that controls which RNA species are loaded inside EVs. Characterization of nucleic acid composition in Gram-positive EVs has only begun, and additional studies in other species (e.g., S. aureus and M. tuberculosis) using global approaches (next-generation sequencing [NGS] and transcriptome sequencing [RNA-seq]) are needed to ascertain if any commonalities exist with regard to EV nucleic acid content and function.
A potential sorting mechanism.
To our knowledge, all studies conducted to date to define EV content noted differences in EV protein and nucleic acid composition relative to the bacterial cell, suggesting that an unknown sorting mechanism exists to package EV cargos (15, 16, 33, 43–47). SDS-PAGE analysis showed similarities and differences between EVs, whole cells, membranes, and supernatants in streptococcus species (15, 47). Notably, mass spectrometry analysis indicated that 169 proteins were enriched in S. pyogenes EVs, and 3 membrane proteins were entirely unique to EVs (15). Similarly, in B. subtilis, 30 proteins were found to be associated only with EVs and not the bacterial cell (45). Studies in S. aureus, M. tuberculosis, and L. monocytogenes showed that EVs were enriched for proteins related to metabolism, such as translation and energy production (16, 43, 46). It is noteworthy that in several pathogenic bacteria, certain virulence factor proteins are found exclusively in vesicles or are more abundant than in the bacterium, suggesting a key role for EVs in virulence (16, 24, 29, 43, 46, 47, 60). Moreover, as mentioned above, the discovery by Tartaglia et al. that diverse S. aureus strains possess a core proteome composed of 119 proteins strongly suggests that a conserved sorting mechanism exists to load S. aureus EVs with their protein cargo (44).
The nucleic acid composition of EVs (both RNA and DNA) has also been shown to be different from that of the parent cell. An analysis of RNAs in S. pyogenes EVs showed that 207 RNA species were differentially present relative to bacterial cells, with 120 RNA transcripts being more abundant inside EVs (15). The adcR, nrdG, rexA, and hsdR transcripts were highly abundant in EVs while the sagA and nagB transcripts were depleted (15). In GBS, DNA encoding the cfb gene could be detected within vesicles; however, no amplification was obtained for other virulence factors (such as cylE, pepB, and zooA) or housekeeping genes (32). Although the precise mechanism leading to EV cargo loading/differential packaging is still unknown, several nonexclusive hypotheses can be made from what we know about EVs in eukaryotic cells and Gram-negative bacteria (Fig. 1).
In Gram-negative bacteria that contain both neutral (O lipopolysaccharide [O-LPS]) and negatively charged (A-LPS) O antigen residues in the outer membrane, only A-LPS has been found to be associated with OMVs. This suggests the pre-existence of patches of negatively charged outer membrane enriched in A-LPS (6). In Porphyromonas gingivalis OMVs, virulence factors such as gingipains were highly abundant, but the packaging process responsible for this enrichment was abolished in an A-LPS mutant strain. Hence, outer membrane patches with high A-LPS composition may play a role in sorting proteins as a consequence of their affinities for the overall charge (61). Furthermore, in eukaryotic cells, exosomes are formed in lipid raft-like regions enriched in cholesterol and particular types of glycolipids and phospholipids, such as phosphatidylcholine and sphingomyelin (62). It has been proposed that the selective loading of RNAs inside exosomes was based on the affinity of RNAs for lipid raft-like regions. Specific nucleotide sequences or even RNA hydrophobic modifications could determine their localization in rafted regions (62).
Such passive mechanisms could also exist in Gram-positive bacteria. A recent study by Wang et al. in S. aureus revealed that EVs from an lgt-deficient strain (which are impaired in the first step of lipoprotein synthesis) had a cargo profile different from that of EVs from the parental strain (i.e., a reduction of pore-forming toxins inside EVs) (33). Given that lipoproteins are commonly found within EVs from many Gram-positive bacteria (15, 33, 45–47), it is possible that a passive lipoprotein-dependent loading mechanism helps sort proteins and nucleic acids into EVs based on their charges, similarly to the mechanism described for A-LPS in Gram-negative bacteria. Furthermore, a recent study in S. aureus showed that proteins packed inside EVs were overall positively charged and contained more small residues and fewer aromatic, aliphatic, and hydrophobic amino acids than the whole-cell proteome, supporting the important role of the physicochemical properties (and potentially charge) of cargos (44). The asymmetric distribution of lipids in EVs suggests that EVs could be synthesized in lipid domains, which could potentiate the lipoprotein-dependent effect on cargo loading. The existence of such domains in bacterial membranes has been studied, mostly in B. subtilis, and one of the best examples is functional membrane microdomains (63, 64). These nanoscale regions in membranes are enriched in particular types of lipids from the polyisoprenoid family and enriched in specific proteins, such as flotillins (63–65).
Flotillins were originally found in eukaryotic lipid rafts and promote protein interactions within lipid raft regions. In B. subtilis, flotillins are essential for the kinase signaling cascade leading to sporulation (66). In S. aureus, flotillins are involved in type VII secretion system assembly within functional membrane microdomains (67). Interestingly, flotillins are not found in the core proteome of S. aureus EVs defined by Tartaglia et al., and this suggests that vesiculogenesis may occur in lipid-enriched regions different from functional membrane microdomains (44). Similarly, cardiolipin domains have also been identified in S. pyogenes and B. subtilis, but the depletion of cardiolipin in S. pyogenes EVs suggests a different origin from cardiolipin-enriched domains for vesiculogenesis (15, 63). Finally, we cannot exclude the existence of an active sorting mechanism that is specifically responsible for packing EVs. Studies in B. subtilis and E. coli reported that proteins such as SpoVM could recognize convex or concave topology of membranes (68). The budding of EVs from the bacterial membrane creates a region of curvature, and it is possible that curvature-detecting proteins could sense this and guide compounds to the appropriate location for them to be inserted into EVs (Fig. 1).
INTERACTION OF EVs WITH CELLS
Interactions with eukaryotic hosts.
The function of vesicles in Gram-positive bacteria is dependent, to a large extent, on the cargo packaging during vesiculogenesis. The presence of toxins, siderophores, immune evasion proteins, adhesins, and antibiotic resistance proteins clearly indicates a role for EVs in virulence (16, 43, 46–48). S. aureus EVs contain superantigens that induce the activation of human T cells (e.g., enterotoxin SeQ), lipase, immune evasion proteins (e.g., protein A and SbI), toxins (such as PSMs and the bicomponent pore-forming toxins alpha-toxin, LukSF-PV, and LukAB), β-lactamase-degrading β-lactam antibiotics, and staphopain A (responsible for extracellular matrix degradation promoting tissue invasion) (13, 33, 43, 44, 48). Likewise, L. monocytogenes EVs contain the pore-forming toxin listeriolysin O, which contributes to the escape of this bacterium from host vacuoles (16). The S. pneumoniae cytosolic pore-forming toxin pneumolysin lacks an export signal sequences and is released in host cells via EV secretion only, highlighting the importance of EVs in S. pneumoniae virulence (47).
There is strong evidence suggesting that EV-associated toxins have a greater impact in the progression of certain diseases than the action of soluble secreted forms of the same toxin(s). For example, the development of atopic dermatitis is strongly influenced by EV-associated alpha-toxin from S. aureus (69). This disease is characterized by chronic skin inflammation triggered by keratinocyte necrosis, resulting in the loss of the skin barrier function. Both soluble and EV-associated alpha-toxin induce keratinocyte death, but only the EV-associated form provoked keratinocyte necrosis and eosinophilic infiltration in the dermis of mice—a process specific to atopic dermatitis disease (69). It has also been shown that EVs enriched in extracellular-matrix-degrading enzymes from GBS are involved in physical barrier disruption and host cell death. In mice, treatment of fetal membranes with GBS EVs resulted in collagen fragmentation, leukocytic and macrophage infiltration, and loss of membrane integrity, leading to preterm birth (32). One potential explanation for the increased potency of EV-associated toxins is that once embedded within EVs, toxins can be delivered at higher concentrations, as they are not diluted as a function of distance and are protected from immune system clearance (e.g., antibodies and protease activity). Furthermore, the method of delivery (either soluble or EV associated) could potentially explain the differential effect of associated and soluble forms. While soluble toxins can attack host cells only from the outside, EV-associated toxins can be directly distributed into recipient cells, disrupt host cell organelles, and potentially attack host cell membranes from the inside (Fig. 1).
To date, only three potential modes of delivery (into host cells) have been described for EVs from Gram-positive bacteria. They are (i) dynamin-dependent endocytosis, (ii) membrane fusion, and (iii) clathrin-dependent endocytosis. Pretreatment of THP-1 cells or monocyte-derived macrophages with the inhibitor dynasore (which inhibits dynamin-dependent endocytosis) prevented the internalization of S. aureus EVs and in doing so inhibited the delivery of pore-forming toxins inside host cells (33). The cholesterol-destroying agent MβCD abolished the internalization of EV-associated protein A from S. aureus inside Hep2 cells (human laryngeal carcinoma cells), potentially demonstrating that EVs fuse with cholesterol-rich domains of host cell membranes (70). In P. acnes, clathrin-dependent endocytosis appears to be the major route of internalization of EVs in human epidermal keratinocyte cells, although the receptor involved remains unknown (Fig. 1) (71). While these are the only known delivery routes for Gram-positive EVs, it is likely that additional modes of entry exist, given that multiple routes of internalization have been described for OMVs and the route taken is often dependent on the size of vesicles and the type of cells being infected (12).
Interactions with bacteria.
While the ability of OMVs from Gram-negative bacteria to target other bacterial species has long been known (72, 73), the potential interaction of EVs with other bacteria have only recently been reported (41, 74). EVs labeled with the lipophilic probe R18 from B. subtilis can fuse to other B. subtilis cells (41). Lactobacillus acidophilus EVs merge with Lactobacillus delbrueckii and E. coli membranes. The high content of bacteriocin (antimicrobial peptide) inside L. acidophilus EVs leads to growth inhibition of the target cells (74). Cell wall-degrading enzymes are commonly found in Gram-positive vesicles and could contribute to the antibacterial action of EVs (50).
Bactericidal activity (against other bacterial cells) is not the only role EVs are thought to play in bacterial interactions. Several studies have shown that vesicles incorporate quorum-sensing molecules (e.g., Pseudomonas quinolone signal and N-acylhomoserine lactones) and could therefore be used as an alternative delivery system for long-distance signaling within bacterial populations (7, 75, 76). The delivery of quorum-sensing molecules embedded in EVs could specifically activate the recipient cells, leading to heterogenous gene activation within a bacterial population. However, as intriguing a prospect as this is, the presence of quorum-sensing molecules within Gram-positive EVs has yet to be shown.
As described above, bacterial vesicles can package DNA, and they have already been associated with horizontal gene transfer in several Gram-negative bacteria (51). For instance, Acinetobacter baumannii can acquire resistance to carbapenem antibiotics via OMVs harboring the blaOXA-24 gene (54). It is not yet clear whether vesicle-based horizontal gene transfer could occur across species or even bacterial genera. Nonetheless, since EVs can fuse to bacterial membranes (41, 74), it is highly likely that horizontal gene transfer via EVs can occur in Gram-positive bacteria, at least within the same bacterial species. A similar mechanism could also occur with EV-embedded-RNAs, in which recipient bacteria could transiently use RNAs delivered by EVs.
IMMUNOMODULATION OF EVs
Proinflammatory activity.
Because EVs originate from bacterial membranes, it is not surprising that their presence induces a proinflammatory response, involving the recognition of pathogen-associated molecular patterns (PAMPs) by host pattern recognition receptors (PRRs), leading to cytokine and chemokine production. As each bacterium loads different content into EVs, the extent to which a proinflammatory response is generated can differ. Treatment of THP-1 cells with F. alocis EVs caused increased secretion of CCL1, CCL2, MIP-1, CCL5, CXCL1, CXCL10, ICAM-1, interleukin 1 receptor agonist (IL-1RA), IL-6, IL-8, migration inhibition factor (MIF), and SerpinE, while the secretion of CXCL1, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-6, and IL-8 was increased in HOK-16B cells (oral keratinocyte cell line) relative to controls infected with whole bacterial cells (37). In vivo intra-amniotic injection in mice with GBS EVs increased the mRNA levels of genes encoding IL-1β, tumor necrosis factor alpha (TNF-α), gamma interferon (IFN-γ), KC (homolog of IL-8 in humans), and IL-6 (32). Similarly, S. pneumoniae EVs induced production of IL-6, IL-8, IL-10, and TNF in human monocyte-derived dendritic cells, independently of pneumolysin content (47, 77).
The precise mechanism through which S. aureus activates the immune system response via EVs in human macrophages has recently been described, and that report remains the only study describing the signaling pathway involved in EV-mediated cytokine production thus far (33). The establishment of an inflammatory response relied on two essential steps. First, lipoproteins within the S. aureus EV membrane stimulate Toll-like receptor 2 (TLR-2), resulting in NF-κB-dependent production of pro-IL-1β and pro-IL-18 (the inactive precursor forms of IL-1β and IL-18). Second, EV-associated pore-forming toxins (such as LukAB and alpha-toxin) activate the NLRP3 inflammasome through K+ efflux, which results in caspase-1 activation and the release of IL-1β and IL-18 after cleavage of the precursor forms (33). Consistent with this study, EVs from bacillus Calmette-Guérin (BCG), M. tuberculosis, and Mycobacterium smegmatis produced an intense TLR-2 inflammatory response in mice with the secretion of IL-1β, IL-6, IL-10, IL-12, TNF, CXCL1, and MIP-1α/CCL3, consistent with the high content of TLR-2 ligands in EVs, such as the lipoproteins LprA and LprG and the polar lipids PIM2 and PIM6 (14). While the TLR-2-dependent activation pathway appears to be an important signaling cascade in the proinflammatory effect of EVs, others are likely to be involved. In studies of OMVs, several different pathways leading to proinflammatory cytokine production have been described. TLR-4 and TLR-8 activation, recognition of OMV-associated LPS outside cells, recognition of OMV-associated RNA inside cells, and NOD1 interaction with peptidoglycan (PGN) inside host cells (delivered via OMVs) are among the known OMV proinflammatory activation mechanisms (12, 78). It is probable that some of these mechanisms are shared, and additional unique activation pathways exist, for Gram-positive EVs.
Anti-inflammatory activity.
Anti-inflammatory activity of EVs has mostly been described in nonpathogenic species such as probiotic bacteria. EVs from Lactobacillus paracasei reduced the expression of LPS-induced cytokines (IL-1α, IL-1β, IL-2, and TNF-α) and increased the production of anti-inflammatory cytokines IL-10 and TGF-β in HT29 human colorectal cancer cells (79). Furthermore, in vivo oral administration of L. paracasei EVs protected mice against dextran sulfate sodium-induced colitis, demonstrating a biologically meaningful anti-inflammatory impact of EVs in vivo (79). EVs from additional lactobacilli species have also been shown to damp the proinflammatory secretion of peripheral blood mononuclear cells (80). In addition to these studies on probiotic bacteria, one study demonstrated the anti-inflammatory activity of EVs from a pathogenic bacterium, M. tuberculosis. Athman and colleagues demonstrated that during macrophage infection, mycobacterial EVs were released and could inhibit the activation of CD4+ T cells (81). Lipoglycans on the EVs were transferred to T cells and stimulated expression of the T cell anergy factor GRAIL, which diminished the capacity of the cells to proliferate upon subsequent restimulation (81).
Immune stimulation can also be carried out by nucleic acids packaged inside membrane vesicles. One example of this is the small RNA sRNA52320, found in P. aeruginosa OMVs, which can modulate the immune response (59). sRNA52320, abundant in OMVs, is delivered inside human airway cells and downregulates the expression of several major genes involved in the LPS-stimulated mitogen-activated protein kinase (MAPK) signaling pathway, leading to a reduction in OMV/LPS-induced IL-8 secretion (59). Based on bioinformatic analysis, this sRNA was predicted to interact, by a base pair mechanism, with several human RNAs within the MAPK signaling pathway (59). This elegant system opens the door to a whole new world of interkingdom communication, facilitated by RNA molecules delivered by bacterial vesicles. Although an equivalent mechanism has yet to be described for a Gram-positive bacterium, the discovery that EVs are replete with RNA suggests that similar interactions are possible.
BIOENGINEERED EVs TO FIGHT INFECTIONS
The presence of multiple immunogenic epitopes within EVs leads to the production of antibodies against these epitopes in mice. This has raised the interesting prospect of using EVs as new vaccine vectors. Immunization of mice with EVs from B. anthracis led to increased survival upon challenge, compared to control mice, and generated a higher IgM response (29). The vaccine potential of M. tuberculosis EVs was evaluated in a mouse model; it was shown to be just as effective as BCG vaccination and to enhance the humoral and cellular immune responses of BCG-vaccinated mice (82). Interestingly, no adjuvant was necessary with EVs to induce immunity, and the antibody response generated was exclusively directed toward lipoproteins and toxins (82). The protective activity of S. pneumoniae EVs has been demonstrated in mice, where they were shown to be more immunoreactive than pneumococcal cell extracts (83). It has been suggested that to improve the immunogenicity and safety of EV vaccines, bioengineered EVs could be constructed that contain increased amounts of immunoreactive determinants while toxins and deleterious compounds are removed. This process has already been applied to produce a vaccine platform to protect mice against S. aureus lethal sepsis (24). Immunization with EVs from S. aureus which expressed nontoxic forms of the cytolytic toxins Hla and LukE provided significant protection against lethal sepsis in mice (24). It is noteworthy that vesicle-based vaccines have already been commercialized for Neisseria meningitidis (VA-MENGOC-BC, MenBvac, and MeNZB), and several others are under investigation (84).
Bacteriophage are emerging as one potential treatment option to fight antibiotic-resistant bacteria, and bacteriophage therapy has already shown clinical efficiency in resolving infections (85). Despite their promise, the narrow host range of phages limits their use, and bacteria can rapidly become resistant to phage infection (85). Studies have shown that in B. subtilis, phage-resistant cells treated with phages could be lysed if phage-sensitive cells were present in the bacterial population (86). Phage-resistant cells transiently acquired phage entry receptors from phage-sensitive cells through EV transfer (86). Remarkably, EVs also transferred phage entry receptors to nonhost bacterial species, allowing the attachment of phage (86). EVs harboring phage receptors could be used in conjunction with phage therapy to enhance the host range and prevent the selection of phage-resistant bacteria. In a similar way, EVs could be used to carry antibiotics, facilitating their delivery to previously inaccessible sites. The intracellular lifestyle (either within a vacuole or in the host cell cytoplasm) is a particularly efficient method for bacteria to escape from antibiotics because of reduced accumulation and/or retention of antibiotics inside cells. Intracellular reservoirs of bacteria (such as S. aureus) have been speculated to represent a sustained source of recolonization and/or reinfection of the host. Exosomes filled with linezolid have already been shown to demonstrate bactericidal effects on S. aureus inside host macrophages (87). Loading EVs with antibacterial compounds may increase their rate of delivery to Gram-positive pathogens and could represent a promising new delivery system to fight infection with intracellular bacteria.
CONCLUDING REMARKS
EVs in Gram-positive bacteria play an important role in various processes. Although research has accelerated in recent years, many aspects of EV biology remain unclear. Some compounds inside EVs have an obvious role in virulence and triggering immune system activation (e.g., antibiotic-sequestering proteins and toxins) while others have an undefined role during the interaction with host cells. The presence of nucleic acids (RNA and DNA) in EVs leads to speculation about their roles inside host cells. It is tempting to think that EV-delivered bacterial RNA could interfere with the host transcriptome (as described for viral RNAs), leading to protein synthesis inhibition. In addition, bacterial RNA modifications could be recognized by Rig-1-like receptors (RLR), initiating the immune system response. Further studies are needed to precisely define the role of bacterial nucleic acids once they are delivered inside host cells. The use of new technologies to isolate and study EVs and our knowledge of EVs in Gram-negative and eukaryotic cells will improve the characterization of vesicles and broaden our understanding of this fascinating and rapidly emerging field.
ACKNOWLEDGMENTS
We extend special thanks to R.K.C. lab members for their helpful discussions.
This work was supported in part by grant AI143743 from the National Institute of Allergy and Infectious Diseases.
Biographies

Paul Briaud received his master’s degree in microbiology at the University of Tours, France, in 2016 and then continued with a Ph.D. degree at the Centre International de Recherche en Infectiologie (CIRI) in the team Pathogénie des Staphylocoques, Lyon, France. For 3 years, he worked on Staphylococcus aureus transcriptomic response during polymicrobial interaction with Pseudomonas aeruginosa. His work resulted in a number of publications, and he was awarded first presentation prize at the French Cystic Fibrosis Association. He successfully graduated with a Ph.D. in 2019 from the University Claude Bernard, Lyon, France. In 2020, he joined the lab of Ronan Carroll at Ohio University (Athens, OH) as a postdoctoral fellow to continue his research on the contribution of RNAs to S. aureus pathogenesis. Paul has an interest in extracellular vesicles in S. aureus as a potential carrier to deliver RNAs to the environment.

Ronan K. Carroll is an Associate Professor in the Department of Biological Sciences at Ohio University. He received a B.A. and Ph.D. from Trinity College Dublin in his native Ireland before moving to the United States in 2005. After completing postdoctoral research at the University of Illinois at Chicago (Chicago, IL), The Methodist Hospital Research Institute (Houston, TX), and The University of South Florida (Tampa, FL), he accepted a faculty position at Ohio University in 2014. Research in the Carroll lab at Ohio University is focused on molecular pathogenesis of the Gram-positive pathogen Staphylococcus aureus. In particular, the lab is interested in the variety of ways in which RNA can contribute to pathogenesis. The Carroll lab also investigates PPIase enzymes and protein secretion. Recently the lab has become interested in extracellular vesicles (EVs) due to their ability to secrete RNA but also due to the fact that PPIases may also contribute to EV formation via regulation of αPSM production in S. aureus.
REFERENCES
- 1.Anné J, Economou A, Bernaerts K. 2017. Protein secretion in Gram-positive bacteria: from multiple pathways to biotechnology. Curr Top Microbiol Immunol 404:267–308. doi: 10.1007/82_2016_49. [DOI] [PubMed] [Google Scholar]
- 2.Beckwith J. 2013. The Sec-dependent pathway. Res Microbiol 164:497–504. doi: 10.1016/j.resmic.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanley NR, Palmer T, Berks BC. 2000. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J Biol Chem 275:11591–11596. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- 4.Gill S, Catchpole R, Forterre P. 2019. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol Rev 43:273–303. doi: 10.1093/femsre/fuy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colombo M, Raposo G, Théry C. 2014. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 6.Jan AT. 2017. Outer membrane vesicles (OMVs) of Gram-negative bacteria: a perspective update. Front Microbiol 8:1053. doi: 10.3389/fmicb.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toyofuku M, Nomura N, Eberl L. 2019. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol 17:13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 8.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. 2008. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann A, Muth C, Dabrowski O, Krasemann S, Glatzel M. 2017. Exosomes and the prion protein: more than one truth. Front Neurosci 11:194. doi: 10.3389/fnins.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig N, Jackson EK, Whiteside TL. 2020. Role of exosome-associated adenosine in promoting angiogenesis. Vessel Plus 2020:8. doi: 10.20517/2574-1209.2019.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Donoghue EJ, Krachler AM. 2016. Mechanisms of outer membrane vesicle entry into host cells. Cell Microbiol 18:1508–1517. doi: 10.1111/cmi.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaparakis-Liaskos M, Ferrero RL. 2015. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol 15:375–387. doi: 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- 13.Lee E-Y, Choi D-Y, Kim D-K, Kim J-W, Park JO, Kim S, Kim S-H, Desiderio DM, Kim Y-K, Kim K-P, Gho YS. 2009. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9:5425–5436. doi: 10.1002/pmic.200900338. [DOI] [PubMed] [Google Scholar]
- 14.Prados-Rosales R, Baena A, Martinez LR, Luque-Garcia J, Kalscheuer R, Veeraraghavan U, Camara C, Nosanchuk JD, Besra GS, Chen B, Jimenez J, Glatman-Freedman A, Jacobs WR, Porcelli SA, Casadevall A. 2011. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Invest 121:1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Resch U, Tsatsaronis JA, Le Rhun A, Stübiger G, Rohde M, Kasvandik S, Holzmeister S, Tinnefeld P, Wai SN, Charpentier E. 2016. A two-component regulatory system impacts extracellular membrane-derived vesicle production in group A Streptococcus. mBio 7:e00207-16. doi: 10.1128/mBio.00207-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coelho C, Brown L, Maryam M, Vij R, Smith DFQ, Burnet MC, Kyle JE, Heyman HM, Ramirez J, Prados-Rosales R, Lauvau G, Nakayasu ES, Brady NR, Hamacher-Brady A, Coppens I, Casadevall A. 2019. Listeria monocytogenes virulence factors, including listeriolysin O, are secreted in biologically active extracellular vesicles. J Biol Chem 294:1202–1217. doi: 10.1074/jbc.RA118.006472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker JL, Chen L, Rosenthal JA, Putnam D, DeLisa MP. 2014. Microbial biosynthesis of designer outer membrane vesicles. Curr Opin Biotechnol 29:76–84. doi: 10.1016/j.copbio.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernadac A, Gavioli M, Lazzaroni J-C, Raina S, Lloubès R. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol 180:4872–4878. doi: 10.1128/JB.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulp AJ, Sun B, Ai T, Manning AJ, Orench-Rivera N, Schmid AK, Kuehn MJ. 2015. Genome-wide assessment of outer membrane vesicle production in Escherichia coli. PLoS One 10:e0139200. doi: 10.1371/journal.pone.0139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, Klug L, Gadermaier B, Weinzerl K, Prassl R, Lass A, Daum G, Reidl J, Feldman MF, Schild S. 2016. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun 7:10515. doi: 10.1038/ncomms10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nevermann J, Silva A, Otero C, Oyarzún DP, Barrera B, Gil F, Calderón IL, Fuentes JA. 2019. Identification of genes involved in biogenesis of outer membrane vesicles (OMVs) in Salmonella enterica serovar Typhi. Front Microbiol 10:104. doi: 10.3389/fmicb.2019.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, Choi C-W, Lee T, Kim SI, Lee J-C, Shin J-H. 2013. Transcription factor σB plays an important role in the production of extracellular membrane-derived vesicles in Listeria monocytogenes. PLoS One 8:e73196. doi: 10.1371/journal.pone.0073196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rath P, Huang C, Wang T, Wang T, Li H, Prados-Rosales R, Elemento O, Casadevall A, Nathan CF. 2013. Genetic regulation of vesiculogenesis and immunomodulation in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 110:E4790–4797. doi: 10.1073/pnas.1320118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Thompson CD, Weidenmaier C, Lee JC. 2018. Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nat Commun 9:1379. doi: 10.1038/s41467-018-03847-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White DW, Elliott SR, Odean E, Bemis LT, Tischler AD. 2018. Mycobacterium tuberculosis Pst/SenX3-RegX3 regulates membrane vesicle production independently of ESX-5 activity. mBio 9:e00778-18. doi: 10.1128/mBio.00778-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Da F, Joo H-S, Cheung GYC, Villaruz AE, Rohde H, Luo X, Otto M. 2017. Phenol-soluble modulin toxins of Staphylococcus haemolyticus. Front Cell Infect Microbiol 7:206. doi: 10.3389/fcimb.2017.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin L, Da F, Fisher EL, Tan DCS, Nguyen TH, Fu C-L, Tan VY, McCausland JW, Sturdevant DE, Joo H-S, Queck SY, Cheung GYC, Otto M. 2017. Toxin mediates sepsis caused by methicillin-resistant Staphylococcus epidermidis. PLoS Pathog 13:e1006153. doi: 10.1371/journal.ppat.1006153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorward DW, Garon CF. 1990. DNA is packaged within membrane-derived vesicles of Gram-negative but not Gram-positive bacteria. Appl Environ Microbiol 56:1960–1962. doi: 10.1128/AEM.56.6.1960-1962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivera J, Cordero RJB, Nakouzi AS, Frases S, Nicola A, Casadevall A. 2010. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci U S A 107:19002–19007. doi: 10.1073/pnas.1008843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prados-Rosales R, Brown L, Casadevall A, Montalvo-Quirós S, Luque-Garcia JL. 2014. Isolation and identification of membrane vesicle-associated proteins in Gram-positive bacteria and mycobacteria. MethodsX 1:124–129. doi: 10.1016/j.mex.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlatterer K, Beck C, Hanzelmann D, Lebtig M, Fehrenbacher B, Schaller M, Ebner P, Nega M, Otto M, Kretschmer D, Peschel A. 2018. The mechanism behind bacterial lipoprotein release: phenol-soluble modulins mediate Toll-like receptor 2 activation via extracellular vesicle release from Staphylococcus aureus. mBio 9:e01851-18. doi: 10.1128/mBio.01851-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Surve MV, Anil A, Kamath KG, Bhutda S, Sthanam LK, Pradhan A, Srivastava R, Basu B, Dutta S, Sen S, Modi D, Banerjee A. 2016. Membrane vesicles of group B Streptococcus disrupt feto-maternal barrier leading to preterm birth. PLoS Pathog 12:e1005816. doi: 10.1371/journal.ppat.1005816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Eagen WJ, Lee JC. 2020. Orchestration of human macrophage NLRP3 inflammasome activation by Staphylococcus aureus extracellular vesicles. Proc Natl Acad Sci U S A 117:3174–3184. doi: 10.1073/pnas.1915829117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abrahams KA, Besra GS. 2018. Mycobacterial cell wall biosynthesis: a multifaceted antibiotic target. Parasitology 145:116–133. doi: 10.1017/S0031182016002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeon J, Mok HJ, Choi Y, Park SC, Jo H, Her J, Han J-K, Kim Y-K, Kim KP, Ban C. 2017. Proteomic analysis of extracellular vesicles derived from Propionibacterium acnes. Prot Clin Appl 11:1600040. doi: 10.1002/prca.201600040. [DOI] [PubMed] [Google Scholar]
- 37.Kim HY, Lim Y, An S-J, Choi B-K. 2020. Characterization and immunostimulatory activity of extracellular vesicles from Filifactor alocis. Mol Oral Microbiol 35:1–9. doi: 10.1111/omi.12272. [DOI] [PubMed] [Google Scholar]
- 38.Andreoni F, Toyofuku M, Menzi C, Kalawong R, Mairpady Shambat S, François P, Zinkernagel AS, Eberl L. 2018. Antibiotics stimulate formation of vesicles in Staphylococcus aureus in both phage-dependent and -independent fashions and via different routes. Antimicrob Agents Chemother 63:e01439-18. doi: 10.1128/AAC.01439-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Cárcamo-Oyarce G, Gloag ES, Shimoni R, Omasits U, Ito S, Yap X, Monahan LG, Cavaliere R, Ahrens CH, Charles IG, Nomura N, Eberl L, Whitchurch CB. 2016. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 7:11220. doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toyofuku M, Cárcamo-Oyarce G, Yamamoto T, Eisenstein F, Hsiao C-C, Kurosawa M, Gademann K, Pilhofer M, Nomura N, Eberl L. 2017. Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat Commun 8:481. doi: 10.1038/s41467-017-00492-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim Y, Edwards N, Fenselau C. 2016. Extracellular vesicle proteomes reflect developmental phases of Bacillus subtilis. Clin Proteom 13:6. doi: 10.1186/s12014-016-9107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deatherage BL, Cookson BT. 2012. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun 80:1948–1957. doi: 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tartaglia NR, Breyne K, Meyer E, Cauty C, Jardin J, Chrétien D, Dupont A, Demeyere K, Berkova N, Azevedo V, Guédon E, Le Loir Y. 2018. Staphylococcus aureus extracellular vesicles elicit an immunostimulatory response in vivo on the murine mammary gland. Front Cell Infect Microbiol 8:277. doi: 10.3389/fcimb.2018.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tartaglia NR, Nicolas A, Rodovalho V. d R, da Luz BSR, Briard-Bion V, Krupova Z, Thierry A, Coste F, Burel A, Martin P, Jardin J, Azevedo V, Le Loir Y, Guédon E. 2020. Extracellular vesicles produced by human and animal Staphylococcus aureus strains share a highly conserved core proteome. Sci Rep 10:8467. doi: 10.1038/s41598-020-64952-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown L, Kessler A, Cabezas-Sanchez P, Luque-Garcia JL, Casadevall A. 2014. Extracellular vesicles produced by the Gram-positive bacterium Bacillus subtilis are disrupted by the lipopeptide surfactin. Mol Microbiol 93:183–198. doi: 10.1111/mmi.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J, Kim S-H, Choi D-S, Lee JS, Kim D-K, Go G, Park S-M, Kim SH, Shin JH, Chang CL, Gho YS. 2015. Proteomic analysis of extracellular vesicles derived from Mycobacterium tuberculosis. Proteomics 15:3331–3337. doi: 10.1002/pmic.201500037. [DOI] [PubMed] [Google Scholar]
- 47.Codemo M, Muschiol S, Iovino F, Nannapaneni P, Plant L, Wai SN, Henriques-Normark B. 2018. Immunomodulatory effects of pneumococcal extracellular vesicles on cellular and humoral host defenses. mBio 9:e00559-18. doi: 10.1128/mBio.00559-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J, Lee E-Y, Kim S-H, Kim D-K, Park K-S, Kim KP, Kim Y-K, Roh T-Y, Gho YS. 2013. Staphylococcus aureus extracellular vesicles carry biologically active β-lactamase. Antimicrob Agents Chemother 57:2589–2595. doi: 10.1128/AAC.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Defourny KAY, Smid EJ, Abee T. 2018. Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front Microbiol 9:1502. doi: 10.3389/fmicb.2018.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Domingues S, Nielsen KM. 2017. Membrane vesicles and horizontal gene transfer in prokaryotes. Curr Opin Microbiol 38:16–21. doi: 10.1016/j.mib.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 52.Tsatsaronis JA, Franch-Arroyo S, Resch U, Charpentier E. 2018. Extracellular vesicle RNA: a universal mediator of microbial communication? Trends Microbiol 26:401–410. doi: 10.1016/j.tim.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 53.Kolling GL, Matthews KR. 1999. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol 65:1843–1848. doi: 10.1128/AEM.65.5.1843-1848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rumbo C, Fernández-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, Mosquera A, Chaves F, Bou G. 2011. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother 55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaudin M, Krupovic M, Marguet E, Gauliard E, Cvirkaite‐Krupovic V, Cam EL, Oberto J, Forterre P. 2014. Extracellular membrane vesicles harbouring viral genomes. Environ Microbiol 16:1167–1175. doi: 10.1111/1462-2920.12235. [DOI] [PubMed] [Google Scholar]
- 56.Ho M-H, Chen C-H, Goodwin JS, Wang B-Y, Xie H. 2015. Functional advantages of Porphyromonas gingivalis vesicles. PLoS One 10:e0123448. doi: 10.1371/journal.pone.0123448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blenkiron C, Simonov D, Muthukaruppan A, Tsai P, Dauros P, Green S, Hong J, Print CG, Swift S, Phillips AR. 2016. Uropathogenic Escherichia coli releases extracellular vesicles that are associated with RNA. PLoS One 11:e0160440. doi: 10.1371/journal.pone.0160440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi J-W, Kim S-C, Hong S-H, Lee H-J. 2017. Secretable small RNAs via outer membrane vesicles in periodontal pathogens. J Dent Res 96:458–466. doi: 10.1177/0022034516685071. [DOI] [PubMed] [Google Scholar]
- 59.Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA, Mielcarz DW, Demers EG, Dolben EL, Hammond JH, Hogan DA, Stanton BA. 2016. A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog 12:e1005672. doi: 10.1371/journal.ppat.1005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thay B, Wai SN, Oscarsson J. 2013. Staphylococcus aureus α-toxin-dependent induction of host cell death by membrane-derived vesicles. PLoS One 8:e54661. doi: 10.1371/journal.pone.0054661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, Feldman MF. 2011. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem 286:1269–1276. doi: 10.1074/jbc.M110.185744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Janas T, Janas MM, Sapoń K, Janas T. 2015. Mechanisms of RNA loading into exosomes. FEBS Lett 589:1391–1398. doi: 10.1016/j.febslet.2015.04.036. [DOI] [PubMed] [Google Scholar]
- 63.Strahl H, Errington J. 2017. Bacterial membranes: structure, domains, and function. Annu Rev Microbiol 71:519–538. doi: 10.1146/annurev-micro-102215-095630. [DOI] [PubMed] [Google Scholar]
- 64.Lopez D, Koch G. 2017. Exploring functional membrane microdomains in bacteria: an overview. Curr Opin Microbiol 36:76–84. doi: 10.1016/j.mib.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bramkamp M, Lopez D. 2015. Exploring the existence of lipid rafts in bacteria. Microbiol Mol Biol Rev 79:81–100. doi: 10.1128/MMBR.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Donovan C, Bramkamp M. 2009. Characterization and subcellular localization of a bacterial flotillin homologue. Microbiology (Reading) 155:1786–1799. doi: 10.1099/mic.0.025312-0. [DOI] [PubMed] [Google Scholar]
- 67.Mielich-Süss B, Wagner RM, Mietrach N, Hertlein T, Marincola G, Ohlsen K, Geibel S, Lopez D. 2017. Flotillin scaffold activity contributes to type VII secretion system assembly in Staphylococcus aureus. PLoS Pathog 13:e1006728. doi: 10.1371/journal.ppat.1006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barák I, Muchová K. 2013. The role of lipid domains in bacterial cell processes. Int J Mol Sci 14:4050–4065. doi: 10.3390/ijms14024050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hong S-W, Choi E-B, Min T-K, Kim J-H, Kim M-H, Jeon SG, Lee B-J, Gho YS, Jee Y-K, Pyun B-Y, Kim Y-K. 2014. An important role of α-hemolysin in extracellular vesicles on the development of atopic dermatitis induced by Staphylococcus aureus. PLoS One 9:e100499. doi: 10.1371/journal.pone.0100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gurung M, Moon DC, Choi CW, Lee JH, Bae YC, Kim J, Lee YC, Seol SY, Cho DT, Kim SI, Lee JC. 2011. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS One 6:e27958. doi: 10.1371/journal.pone.0027958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choi E-J, Lee HG, Bae I-H, Kim W, Park J, Lee TR, Cho E-G. 2018. Propionibacterium acnes-derived extracellular vesicles promote acne-like phenotypes in human epidermis. J Invest Dermatol 138:1371–1379. doi: 10.1016/j.jid.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 72.Li Z, Clarke AJ, Beveridge TJ. 1998. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J Bacteriol 180:5478–5483. doi: 10.1128/JB.180.20.5478-5483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim JH, Lee J, Park J, Gho YS. 2015. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin Cell Dev Biol 40:97–104. doi: 10.1016/j.semcdb.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 74.Dean SN, Rimmer MA, Turner KB, Phillips DA, Caruana JC, Hervey WJI, Leary DH, Walper SA. 2020. Lactobacillus acidophilus membrane vesicles as a vehicle of bacteriocin delivery. Front Microbiol 11:710. doi: 10.3389/fmicb.2020.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toyofuku M, Morinaga K, Hashimoto Y, Uhl J, Shimamura H, Inaba H, Schmitt-Kopplin P, Eberl L, Nomura N. 2017. Membrane vesicle-mediated bacterial communication. ISME J 11:1504–1509. doi: 10.1038/ismej.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote 7057. Nature 437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 77.Mehanny M, Koch M, Lehr C-M, Fuhrmann G. 2020. Streptococcal extracellular membrane vesicles are rapidly internalized by immune cells and alter their cytokine release. Front Immunol 11:80. doi: 10.3389/fimmu.2020.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han E-C, Choi S-Y, Lee Y, Park J-W, Hong S-H, Lee H-J. 2019. Extracellular RNAs in periodontopathogenic outer membrane vesicles promote TNF-α production in human macrophages and cross the blood-brain barrier in mice. FASEB J 33:13412–13422. doi: 10.1096/fj.201901575R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi JH, Moon CM, Shin T-S, Kim EK, McDowell A, Jo M-K, Joo YH, Kim S-E, Jung H-K, Shim K-N, Jung S-A, Kim Y-K. 2020. Lactobacillus paracasei-derived extracellular vesicles attenuate the intestinal inflammatory response by augmenting the endoplasmic reticulum stress pathway. Exp Mol Med 52:423–437. doi: 10.1038/s12276-019-0359-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mata Forsberg M, Björkander S, Pang Y, Lundqvist L, Ndi M, Ott M, Escribá IB, Jaeger M-C, Roos S, Sverremark-Ekström E. 2019. Extracellular membrane vesicles from lactobacilli dampen IFN-γ responses in a monocyte-dependent manner. Sci Rep 9:17109. doi: 10.1038/s41598-019-53576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Athman JJ, Sande OJ, Groft SG, Reba SM, Nagy N, Wearsch PA, Richardson ET, Rojas R, Boom WH, Shukla S, Harding CV. 2017. Mycobacterium tuberculosis membrane vesicles inhibit T cell activation. J Immunol 198:2028–2037. doi: 10.4049/jimmunol.1601199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prados-Rosales R, Carreño LJ, Batista-Gonzalez A, Baena A, Venkataswamy MM, Xu J, Yu X, Wallstrom G, Magee DM, LaBaer J, Achkar JM, Jacobs WR, Chan J, Porcelli SA, Casadevall A. 2014. Mycobacterial membrane vesicles administered systemically in mice induce a protective immune response to surface compartments of Mycobacterium tuberculosis. mBio 5:e01921-14. doi: 10.1128/mBio.01921-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olaya-Abril A, Prados-Rosales R, McConnell MJ, Martín-Peña R, González-Reyes JA, Jiménez-Munguía I, Gómez-Gascón L, Fernández J, Luque-García JL, García-Lidón C, Estévez H, Pachón J, Obando I, Casadevall A, Pirofski L, Rodríguez-Ortega MJ. 2014. Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J Proteomics 106:46–60. doi: 10.1016/j.jprot.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 84.Piccini G, Torelli A, Gianchecchi E, Piccirella S, Montomoli E. 2016. Fighting Neisseria meningitidis: past and current vaccination strategies. Expert Rev Vaccines 15:1393–1407. doi: 10.1080/14760584.2016.1187068. [DOI] [PubMed] [Google Scholar]
- 85.Kortright KE, Chan BK, Koff JL, Turner PE. 2019. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25:219–232. doi: 10.1016/j.chom.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 86.Tzipilevich E, Habusha M, Ben-Yehuda S. 2017. Acquisition of phage sensitivity by bacteria through exchange of phage receptors. Cell 168:186–199.E12. doi: 10.1016/j.cell.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 87.Yang X, Shi G, Guo J, Wang C, He Y. 2018. Exosome-encapsulated antibiotic against intracellular infections of methicillin-resistant Staphylococcus aureus. Int J Nanomedicine (Lond) 13:8095–8104. doi: 10.2147/IJN.S179380. [DOI] [PMC free article] [PubMed] [Google Scholar]