Abstract
Understanding the pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and clarifying antiviral immunity in hosts are critical aspects for the development of vaccines and antivirals. Mice are frequently used to generate animal models of infectious diseases due to their convenience and ability to undergo genetic manipulation. However, normal adult mice are not susceptible to SARS-CoV-2. Here, we developed a viral receptor (human angiotensin-converting enzyme 2, hACE2) pulmonary transfection mouse model to establish SARS-CoV-2 infection rapidly in the mouse lung. Based on the model, the virus successfully infected the mouse lung 2 days after transfection. Viral RNA/protein, innate immune cell infiltration, inflammatory cytokine expression, and pathological changes in the infected lungs were observed after infection. Further studies indicated that neutrophils were the first and most abundant leukocytes to infiltrate the infected lungs after viral infection. In addition, using infected CXCL5-knockout mice, chemokine CXCL5 was responsible for neutrophil recruitment. CXCL5 knockout decreased lung inflammation without diminishing viral clearance, suggesting a potential target for controlling pneumonia.
Keywords: SARS-CoV-2, Mouse model, Lung infection, ACE2, Neutrophil, CXCL5
INTRODUCTION
Since December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected more than 37 million people and caused over one million deaths worldwide as of April 2020. SARS-CoV-2 infection causes a respiratory disease (known as coronavirus disease 2019, COVID-19) with symptoms ranging from mild to severe. Furthermore, SARS-CoV-2 infection can lead to pneumonia and acute respiratory distress syndrome (ARDS) in up to 20% of COVID-19 cases (Moore & June, 2020). Clinical findings show that severe cases are associated with dysregulated immune responses and uncontrolled local and systemic inflammation. Patients with severe COVID-19 have higher leukocyte counts and increased neutrophil numbers in circulation, together with lower lymphocyte counts and reduced percentages of monocytes, eosinophils, and basophils (Huang et al., 2020; Qin et al., 2020; Xu et al., 2020b). In addition, most severe cases exhibit elevated serum levels of inflammatory cytokines, including TNF-α, IL-1β, IL-6, IL-2, IL-8, IL-17, G-CSF, GM-CSF, MCP1, and MIP1α (Huang et al., 2020; Qin et al., 2020; Xu et al., 2020b). Thus, understanding the immune-inflammatory activation mechanism of SARS-CoV-2 infection is a crucial step for the prevention and treatment of COVID-19.
Mouse infection models are the most commonly used animal models for infectious diseases due to their convenience and ability to undergo genetic mutagenesis. However, during our preliminary experiments, we found that two commonly used experimental mouse strains, BALB/c and C57BL/6, were not susceptible to SARS-CoV-2 as adults (10–12 weeks old). Similar to the highly homologous SARS virus, SARS-CoV-2 also uses human angiotensin-converting enzyme 2 (hACE2) as its entry receptor for infection (Li et al., 2003; Zhou et al., 2020). Previous studies successfully established a SARS infection model using hACE2 transgenic mice (McCray et al., 2007; Tseng et al., 2007; Yang et al., 2007) and a SARS-CoV-2 infection model using Ad5-hACE2-sensitized mice (Sun et al., 2020). Thus, to study the pulmonary immune/inflammatory response upon SARS-CoV-2 infection in a mouse model, we used an in vivo lung hACE2 gene transfection method to establish a SARS-CoV-2 transient lung infection model in mice. Based on this model, viral infection, innate immune cell infiltration, inflammatory cytokine expression, and pathological changes were observed in the infected lungs. Furthermore, the mouse neutrophil chemokine CXCL5 was found to play an important role in neutrophil infiltration in these lungs.
MATERIALS AND METHODS
Plasmids, cells, and virus
The human ACE2 gene was inserted into the pCMV3-C-GFPSpark plasmid to construct a pCMV-ACE2-GFPSpark plasmid (Sino Biological, China). The human ACE2 gene was transcribed using the CMV promoter. MLE-12 mouse lung epithelial cells (ATCC, CRL-2110) purchased from the BeNa Culture Collection (China) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, USA) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Gibco, USA). Virus strain SARS-CoV-2-KMS1/2020 (GenBank accession No.: MT226610.1) was isolated by the Institute of Medical Biology (Chinese Academy of Medical Sciences) and propagated and titered on Vero cells in DMEM (Gibco, US) supplemented with 2% (vol/vol) fetal calf serum (FCS) (Gibco, USA), 50 U/mL penicillin, and 50 μg/mL streptomycin (Gibco, USA).
Transfection
MLE-12 cells were cultured in 6-well plates or glass bottom cell culture dishes for immunofluorescence. At approximately 80% confluent, the cells were either transfected with the human ACE2 (hACE2) expression plasmid pCMV-ACE2-GFPSpark or the control pCMV3 plasmid using FuGENE HD Transfection Reagent (Promega, USA) according to the manufacturer’s instructions. At 48 h post-transfection, expression of the hACE2 protein was detected by western blotting and immunofluorescence.
Animal experiments
All animal experiments were performed at the Kunming National High-Level Biosafety Research Center for Non-Human Primates and approved by the Ethics Committee of the Institute of Medical Biology, Chinese Academy of Medical Sciences (Permit number, DWSP202002 003-2).
C57BL/6 mice were obtained from the animal housing center of the Institute of Medical Biology, Chinese Academy of Medical Sciences. CXCL5−/− mice (Mei et al., 2010) were obtained from Dr. Scott Worthen at the Children’s Hospital of Philadelphia, USA. The mice were bred and housed in individually ventilated cages at an ABSL-3 laboratory for study under specific pathogen-free conditions. Mice aged 10–12 weeks (males, due to availability of both C57BL/6 and CXCL5−/− mice at the beginning of our study) were used in all experiments. For in vivo transfection of the pCMV-ACE2-GFPSpark plasmid or control pCMV3 plasmid, an atraumatic orotracheal intubation method was used to intratracheally administer plasmids and TurboFect in vivo Transfection Reagent (Thermo Fisher, USA) into the lungs (Das et al., 2013; Guo et al., 2017). Briefly, wild-type (WT) C57BL/6 and CXCL5−/− mice were anesthetized using an intraperitoneal injection of pentobarbital (50 mg per body weight) and orotracheally intubated with a 20-gauge angiocatheter (BD Biosciences, USA) with the guidance of an optical fiber source. Mice were subsequently placed in a vertical position suspended by their upper incisors. A polyethylene catheter was advanced into the trachea and used to instill transfection compounds containing 35 μg of hACE2 expression plasmid (pCMV-ACE2-GFPSpark) or control pCMV3 plasmid. At 48 h post-transfection, mice were anaesthetized and then intranasally inoculated with SARS-CoV-2 (1×105 CCID50) in 25 μL of phosphate-buffered saline (PBS).
Western blotting
MLE-12 cells were transfected with 2 μg of pCMV-ACE2-GFPSpark plasmid or control pCMV3 plasmid per 106 cells using TransIT-2020 Transfection Reagent (Mirus, USA). Protein extracts were prepared at 48 h post-transduction using RIPA lysis buffer. Lung tissue samples were harvested and mechanically homogenized using a TGrinder instrument (Tiangen Biotechnologies, China). Protein extracts were prepared using a tissue protein extraction kit (BC3790; Solarbio, China). Equal amounts of protein were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were stained with rabbit anti-human ACE2 polyclonal antibody (ab15348; Abcam, UK) or rabbit anti-β-actin polyclonal antibody (ab8227; Abcam, UK). Proteins were detected using the SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher, USA).
Immunofluorescence
MLE-12 cells were transfected with 2 μg of pCMV-ACE2-GFPSpark plasmid or control pCMV3 plasmid per 106 cells using TransIT-2020 Transfection Reagent (Mirus, USA). At 48 h post-transfection, the cells were either infected with virus or underwent immunofluorescence staining. After blocking with 5% bovine serum albumin, the cells were incubated overnight at 4 °C with rabbit anti-human ACE2 monoclonal antibody (ab108209; Abcam, UK), mouse anti-GFP monoclonal antibody (ab1218; Abcam, UK), or rabbit anti-SARS-CoV-2 spike monoclonal antibody (40150-R007; Sino Biological, China), depending on the experiment design. The cells were then incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (ab150113; Abcam, UK) and Alexa Fluor 647-conjugated goat anti-rabbit IgG (ab150075; Abcam, UK) for 1 h at room temperature. Nuclei were counterstained with 4',6-diamidino-2-phenylindole (DAPI, Beyotime, China). Images were obtained with a Leica TCS SP8 laser confocal microscope (Leica Microsystems, Germany).
Lung tissue sections in paraffin were subjected to deparaffinization in xylene and rehydration in a graded series of ethanol, followed by rinsing in double-distilled water. The rehydrated tissue sections were blocked for 2 h in 10% FBS. The slides were then incubated with rabbit anti-prosurfactant protein C (proSP-C) polyclonal antibody (ab90716; Abcam, UK) for 2 h at room temperature. After washing three times with PBS plus 0.05% Tween 20 (PBST), the tissue slides were permeabilized with 0.1% Triton-X100 for 15 min and labeled with mouse anti-viral nucleoprotein monoclonal antibody (40143-MM05; Sino Biological, China) or mouse anti-human ACE2 monoclonal antibody (ab89111; Abcam, UK) for 2 h at room temperature. Finally, the ACE2 and pro-SP-C proteins were visualized using Alexa Fluor 488-conjugated goat anti-rabbit IgG (ab150077; Abcam, UK) and Alexa Fluor 647-conjugated goat anti-mice IgG (ab150115; Abcam, UK) for 1 h, respectively. The viral nucleoprotein and proSP-C protein were visualized by Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa Fluor 647-conjugated goat anti-mice IgG for 1 h, respectively. Cell nuclei were stained with DAPI (ab104139; Abcam, USA) and images were captured using the Leica TCS SP8 laser confocal microscope.
Viral RNA and titers
Mice were euthanized at 1, 2, 3, 5, and 7 days post-infection (dpi), and lung tissue samples were harvested and mechanically homogenized using a TGrinder instrument (Tiangen Biotechnologies, China). RNA was isolated from the lung homogenates using TRIzol Reagent (Tiangen Biotechnologies, China) according to the manufacturer’s guidelines. The RNA concentration of each sample was determined by measuring the absorbance at 260 nm using a NanoDrop 2000 (Thermo Fisher, USA). In all, 100 ng of total RNA was reverse transcribed and amplified using a One Step PrimeScriptTM RT-PCR Kit (Takara Biotechnologies, China) on a 7500 Fast Real-Time PCR system (Applied Biosystems, USA). To determine viral loads, primers and a viral nucleocapsid (N) gene probe were used: 5'-GGGGAACTTCTCCTGCTAGAAT-3' (forward), 5'-CAGACATTTTGCTCTCAAGCTG-3' (reverse), and 5'-(FAM)-TTGCTGCTGCTTGACAGATT-(TAMRA)-3' (probe). The N gene of the virus was cloned into the pMD18-T vector, which was used to create a standard curve by 10-fold serial dilution. Viral copy numbers were normalized to the mass of the original tissue samples and calculated based on the standard curve described above.
Cell culture supernatants from the infected cells and lung homogenates from the infected mice were centrifuged at 5 000 r/min for 10 min at 4 °C, after which the supernatants were collected. Viral titers were determined using CCID50 titers by serial titration of viruses in Vero cells. Titers were calculated based on the method of Reed and Muench (Read & Muench, 1938).
Lung bronchoalveolar lavage fluid (BALF) collection and flow cytometry
The mice were sacrificed, and their lungs were lavaged three times with 0.8 mL of cold, sterile PBS. Cells were counted using a hemocytometer. BALF cells were washed in fluorescence-activated cell sorting buffer after lysis with BD FACS Lysing Solution (BD Biosciences, USA). Cell surface staining was conducted using Mouse BD Fc Block (Cat. No. 551163, clone 2.4G2), anti-mouse CD3-PerCP (Cat. No. 551163, clone 145-2C11), and anti-mouse CD64-PE (Cat. No. 553652, clone H129.19) (BD Biosciences, USA). Flow cytometry data were collected on a CytoFLEX system (Beckman Coulter, USA) and analyzed using CytoExpert v2.3.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
RNA was isolated from the lung homogenates using TRIzol Reagent (Tiangen Biotechnologies, China). Total RNA quality and concentration were determined based on the 260/280 absorbance ratio and absorbance at 260 nm, respectively, using a NanoDrop 2000 (Thermo Fisher, USA). qRT-PCR was carried out using a 7500 Fast Real-Time PCR system (Applied Biosystems, USA) with a One Step SYBR®PrimeScriptTM RT-PCR Kit (Takara Biotechnologies, China) according to the manufacturer’s guidelines. The primers used for qRT-PCR are listed in Table S1. Fold changes in the mRNA expression of these genes were calculated using the 2−ΔΔCt method for relative quantification, with GAPDH applied as the endogenous reference gene. All reactions were carried out in triplicate.
Lung histological and immunohistochemical analyses
Mice were euthanized, and their lungs were slowly inflated by intratracheally instilling 1 mL of 4% formaldehyde. The trachea was tightened, and the lungs were fixed in formalin for 48 h at 4 °C and embedded in paraffin. Lung tissue sections (5 mm) were stained with hematoxylin and eosin for histological analysis. Lung histological scores were determined based on a 0–4 point scale of combined scores for alveolar structure, inflammatory cell infiltration, aggregation in the alveolar space and septa, bronchiolitis, and lung edema by a pathologist blind to the study. A score of 0 indicated no damage, 1 indicated mild damage, 2 indicated moderate damage, 3 indicated severe damage, and 4 indicated very severe histological changes; increments of 0.5 were used if the levels of inflammation fell between two integers. Three random areas in each tissue sample were scored, and the mean value was calculated. The histological score was the median of values from three mice. For immunohistochemical studies, sections were stained with rabbit anti-human ACE2 polyclonal antibody (ab15348; Abcam, UK) overnight at 4 °C, then incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (ab6721; Abcam, UK) for 30 min, and counterstained with hematoxylin. Images were obtained using a Slide Converter (3DHISTECH, Hungary).
Neutrophil depletion
Mice were intraperitoneally injected with 500 mg of rat anti-Ly6G [1A8] antibody (ab210204; Abcam, UK) or isotype control antibody to deplete neutrophilsin vivo(Tate et al., 2009). Mice were treated 24 h before infection and every 48 h thereafter. Depletion of neutrophils (>90%) from the blood was confirmed by differential leukocyte counts. The SARS-CoV-2 viral RNA and lung pathologies of neutrophil-depleted mice were assessed.
Statistical analysis
All statistical analyses were performed with GraphPad Prism v8. The data obtained from all experiments are presented as mean±SD, and P<0.05 using Student’st-test indicated statistical significance.
RESULTS
Characterization of in vitro expression of hACE2 in a mouse cell line
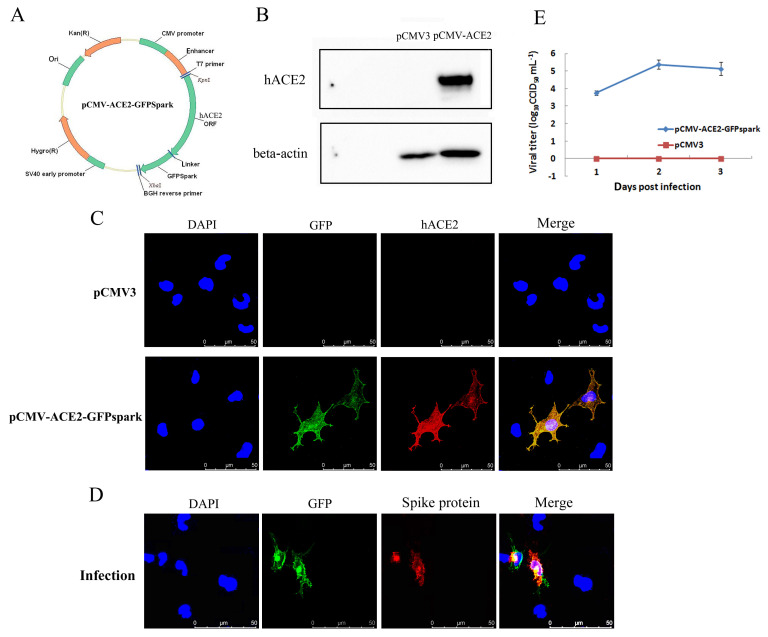
The mammalian expression plasmid pCMV3-C-GFPSpark, which contains the cDNA coding sequence of hACE2, was transfected into mouse lung epithelial cells (MLE-12), with hACE2 expression then verified. Cells were harvested at 48 h post-transfection, and the expression of ACE2 was examined by western blotting and immunofluorescence analysis. As shown in Figure 1, abundant hACE2 expression was detected using a specific anti-human ACE2 antibody, whereas expression of hACE2 was not detected in the control pCMV3-transfected cells (Figure 1B). An immunofluorescence assay also detected expression of the hACE2 protein and GFP tag in hACE2-transfected cells but not in control pCMV3-transfected cells (Figure 1C). Furthermore, infection of hACE2-transfected MLE-12 cells with SARS-CoV-2 was evaluated. Immunofluorescence detected the expression of the viral spike protein only in GFP-expressing (hACE2 expressing) MLE-12 cells at 48 h post-infection (Figure 1D). Viral titer detection revealed that SARS-CoV-2 proliferated to high titers in hACE2-transfected cells at 48 h post-infection, whereas control pCMV3-transfected cells were resistant to SARS-CoV-2 infection (Figure 1E). These data suggest that the expression of human ACE2 rendered mouse lung epithelial cells susceptible to SARS-CoV-2.
Figure 1.
Characterization of in-vitro transfection of hACE2 and SARS-CoV-2 infection in MLE-12 cells
A: MLE-12 cells were transfected with human ACE2 (hACE2) expression plasmid pCMV-ACE2-GFPSpark or pCMV3 plasmid as a control. B: hACE2 protein expression was detected by western blotting using anti-hACE2 antibody. C: hACE2 protein expression was detected by immunofluorescence using anti-hACE2 antibody. D: hACE2-transfected cells were infected with SARS-CoV-2 at an MOI of 5 at 48 h post-transfection, and immunofluorescence detection of viral spike protein and GFP protein was performed at 24 h post-infection by confocal microscopy. E: hACE2-transfected cells and control-transfected cells were infected with SARS-CoV-2 at an MOI of 1 at 48 h post-transfection, and viral titers at different days post-infection (dpi) were determined by a viral CCID50 assay (lower). Error bars indicate standard deviation of triplicate biological samples.
Pulmonary SARS-CoV-2 infection in hACE2-transfected mice
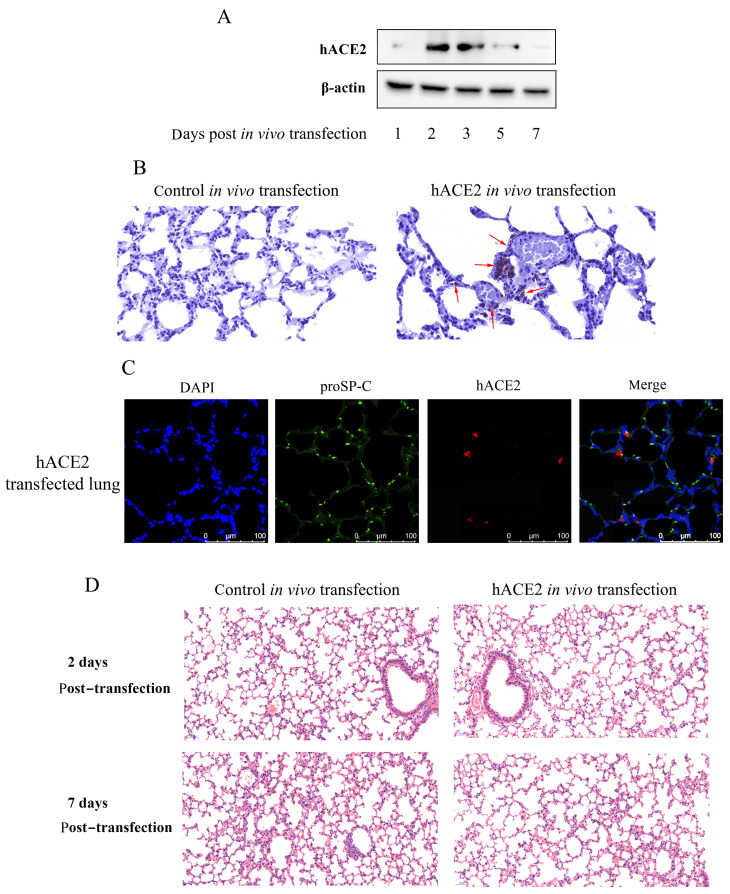
We next transfected the hACE2 expression plasmid into the mouse lung by delivering a plasmid-transfection reagent complex into the lung using the orotracheal intubation method described above. Exogenous hACE2 expression was detected in the hACE2-transfected mouse lungs by western blotting on days 1 to 7 post-transfection, with peak values found on days 2 and 3 (Figure 2A). Furthermore, the expression of hACE2 in lung tissues was determined by immunohistochemical analysis, which indicated that exogenous hACE2 was expressed in alveolar epithelial cells (Figure 2B). To confirm hACE2 expression in alveolar epithelial cells, immunofluorescence detection against prosurfactant protein C (proSP-C), a molecule specifically expressed in alveolar epithelial type II (AEII) cells, and hACE2 protein was conducted in hACE2-transfected lung tissue. As shown in Figure 2C, hACE2 expression was localized in lung AEII cells that expressed proSP-C, thus indicating the expression of hACE2 in lung epithelial cells. No visible inflammatory pathological changes were observed in the lungs of transfected mice at 2 and 7 days post-transfection (Figure 2D). Considering that ACE2 is an interferon stimulated gene (Ziegler et al., 2020), lung immune response after hACE2 transfection was analyzed. As shown in Supplementary Figure 1, no elevated BALF leukocyte numbers or immune/inflammatory cytokine levels were observed in the hACE2-transfected mice compared to the mock-transfected (pCMV3 plasmid) mice. These results indicate that exogenous expression of hACE2 in the mouse lung did not induce a pulmonary immune response.
Figure 2.
In vivo hACE2 pulmonary transfection in mice
Mice underwent pulmonary transfection with hACE2 expression plasmid (35 μg each) or control pCMV3 plasmid via orotracheal intubation, after which lungs were harvested at different days post-transfection. A: Expression of hACE2 in transfected lung tissue via western blotting. B: Lung sections at 2 days post-transfection were stained with anti-hACE2 antibody and hematoxylin (original magnification, 40×). Red arrows indicate obvious expression of hACE2. C: Immunofluorescence detection of proSP-C and hACE2 proteins in lung tissue sections at 2 days post-transfection. D: Lung sections at 2 and 7 days post-transfection were stained with hematoxylin and eosin for histopathological analysis (original magnification, 20×).
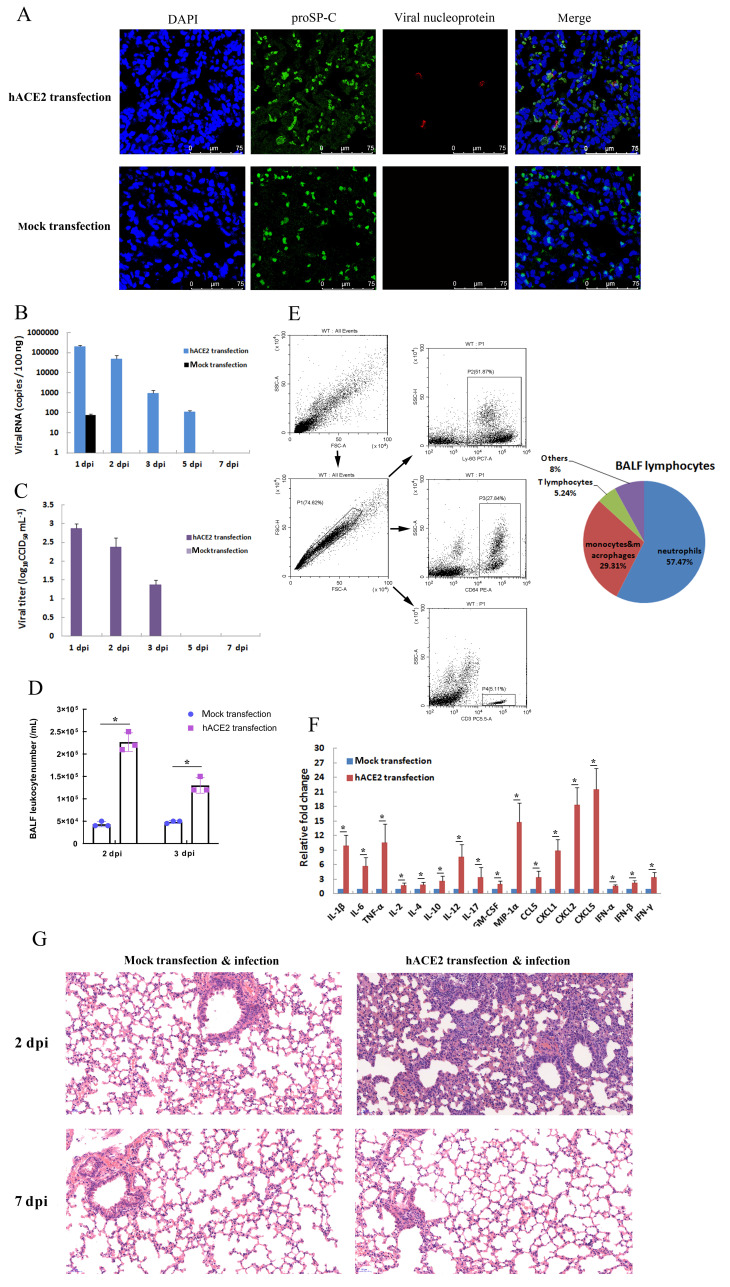
Based on the expression of transfected hACE2 in the lungs, hACE2-transfected mice (hACE2 mice) were intranasally infected with SARS-CoV-2 (1×105 CCID50 each) at 48 h post-transfection, with mice transfected with the pCMV3 plasmid (mock-transfected mice) serving as negative controls. Although body weight loss was not observed in either group of mice at 7 dpi (data not shown), viral RNA, viral protein, and infectious virions were detected in the infected lungs of hACE2 mice but not those of mock-transfected mice at 2 dpi (Figure 3A-C). As shown in Figure 3B, viral RNA in the lungs of hACE2 mice was detected by qRT-PCR at 1 dpi, 2 dpi, 3 dpi, and 5 dpi, and the number of viral RNA copies peaked at 1–2 dpi (approximately 105 copies/100 ng). Viral RNA began to decline after 2 dpi, and the virus was cleared by 7 dpi. Consistent with the viral RNA data, infectious virions (viral titers) in the lungs of hACE2 mice were detected at 1 dpi, 2 dpi, and 3 dpi, with the peak viral load detected from 1–2 dpi (approximately 102.5 CCID50/mL) (Figure 3C).
Figure 3.
Pulmonary SARS-CoV-2 infection in hACE2-transfected mice
Two days (48 h) after in vivo transfection with hACE2 plasmid or control plasmid (mock transfection), mice were intranasally infected with SARS-CoV-2 (1×105 CCID50). A: Lungs were harvested from infected mice at 2 days post-infection (dpi), fixed in formalin, and embedded in paraffin. Lung sections were stained with anti-proSP-C and viral nucleoprotein for immunofluorescence detection. B, C: Lungs were harvested at different dpi, viral RNA was determined based on number of nucleocapsid (N) gene RNA copies detected by qRT-PCR (B), and viral titers were determined using a CCID50 assay (C). Error bars indicate standard deviation of triplicate biological samples. D: Total numbers of leukocytes in BALF at different dpi were determined. E: Percentages of neutrophils, monocytes and macrophages, and lymphocytes in BALF of infected hACE2 mice at 2 dpi were analyzed by flow cytometry using anti-Ly6G, anti-CD64, and anti-CD3 antibodies, respectively (left). Statistical analysis of BALF lymphocyte composition was carried out with data from triplicate biological samples (right). F: mRNA levels of cytokines from lung tissue homogenates of infected mice at 2 dpi were examined by qRT-PCR (normalized to β-actin). Error bars indicate standard deviation of triplicate biological samples. *: P<0.05 based on Student’st-test. G: Lungs were harvested from infected mice at 2 dpi, fixed in formalin, and embedded in paraffin. Lung sections were stained with hematoxylin and eosin for histopathological analysis (original magnification, 20×).
Viral infection in hACE2-transfected mouse lungs can activate pulmonary antiviral immunity, and the innate immune system will respond to viral infection first. To assess activation of the pulmonary innate immune response of hACE2 mice, lung leukocyte infiltration and cytokine expression were examined at different dpi. As shown in Figure 3D, the number of leukocytes in the BALF at the early stage of infection increased significantly (2–3 dpi) in hACE2 mice compared to that in mock-transfected mice, suggesting that leukocytes were recruited to the infected lung. Flow cytometry revealed that the dominant types of leukocytes in the BALF were neutrophils (57.47%), monocytes and macrophages (29.31%), and T lymphocytes (5.24%) (Figure 3E). To determine the effect of SARS-CoV-2 infection on the expression of immune and inflammatory cytokines in the infected lungs, the mRNA levels of genes encoding different cytokines were measured by qRT-PCR. Figure 3F shows that the expression levels of cytokines IL-1β, IL-6, TNF-α, IL-2, IL-4, IL-10, IL-12, IL-17, GM-CSF, MIP-1α, CCL5, CXCL1, CXCL2, CXCL5, IFN-α, IFN-β, and IFN-γ in the lung tissues at 2 dpi were up-regulated in hACE2 mice compared to that in mock-transfected mice. Together, the accumulation of BALF leukocytes and increased expression of lung cytokines indicate that SARS-CoV-2 infection in the hACE2 mouse model activated the pulmonary antiviral innate immune response. Furthermore, histopathological analysis of the infected lungs was performed using hematoxylin and eosin staining of lung tissue. Results revealed the presence of lung inflammation in hACE2 mice but not in mock-transfected mice (Figure 3G). The symptoms included inflammatory cell infiltration in the lung parenchyma and interstitium and massive inflammatory cell aggregation in the alveolar wall and spaces (Figure 3G). Thus, based on the above data, hACE2-transfected mice were susceptible to SARS-CoV-2 infection, which induced a pulmonary innate immune/inflammatory response in these mice.
Comparison of SARS-CoV-2 infection in hACE2-transfected WT mice and CXCL5−/− mice
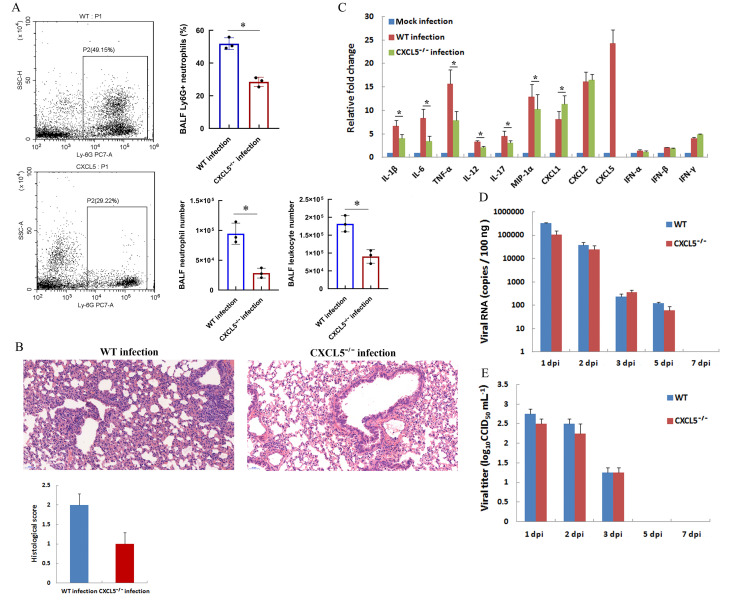
A large proportion of neutrophils (>50% BALF leukocytes) infiltrated the infected lungs of the hACE2 model mice soon after SARS-CoV-2 replication and proliferation (Figure 2F). Upon respiratory infection, infected epithelial and immune cells release inflammatory cytokines to recruit large numbers of granulocytes, macrophages, and monocytes to the lung parenchyma and alveoli. Chemokine production is a critical step for leukocyte accumulation in the lungs. In pathogen-infected mice, keratinocyte-derived chemoattractant (KC, also called CXCL1), macrophage inflammatory protein-2 (MIP-2, also called CXCL2), and lipopolysaccharide-induced CXC chemokine (LIX, also called CXCL5), which act by binding to the CXCR2 receptor, are major chemoattractants for neutrophil recruitment in inflamed lungs (Balamayooran et al., 2010; Puneet et al., 2005). In the SARS-CoV-2-infected hACE2 mouse model, the expression levels of the CXCL1, CXCL2, and CXCL5 chemokines increased (Figure 3F), with CXCL5 showing the highest up-regulation (up to 25-fold). To further determine the effect of CXCL5 on neutrophil recruitment to the infected lungs at the innate immune phase of pulmonary SARS-CoV-2 infection, a SARS-CoV-2 infection CXCL5−/− mouse model was established using the same method applied as for the hACE2 transfection mouse model. As shown in Figure 4A, CXCL5 knockout significantly decreased the percentage and number of BALF neutrophils compared to those in WT-infected mice at 2 dpi (Figure 4A), indicating that CXCL5 is likely responsible for neutrophil recruitment to infected lungs upon SARS-CoV-2 infection. Additionally, the total number BALF leukocytes was reduced compared to that in WT-infected mice due to the reduced neutrophil number in CXCL5−/− infected mice (Figure 4A). Furthermore, lung inflammation was compared in infected WT and CXCL5−/− mice. Histopathological examination revealed that CXCL5−/− mice presented less inflammatory cell aggregation in the bronchial walls and alveolar spaces at 2 dpi than WT mice (Figure 4B). Histological scores indicated that CXCL5−/− mice exhibited less severe lung pathology than WT mice (Figure 4B). The qRT-PCR results showed that the mRNA expression levels of immune and inflammatory cytokines IL-1β, IL-6, TNF-α, IL-12, IL-17, MIP-1α, and CXCL1 in the infected lungs of CXCL5−/− mice were significantly lower than that in the infected lungs of WT mice (Figure 4C). These results suggest that CXCL5 contributes to neutrophil infiltration in infected lungs and plays a role in the induction of lung inflammation at the early innate immune response phase of SARS-CoV-2 pulmonary infection. CXCL5 knockout did not affect viral clearance in the infected mice (Figure 4D, E), suggesting a limited role of CXCL5 in the antiviral immune response of hACE2-infected mice.
Figure 4.
Comparison of SARS-CoV-2 infection in hACE2-transfected WT mice and CXCL5−/− mice
Two days (48 h) after in vivo transfection with hACE2 plasmid, WT and CXCL5-knockout mice (CXCL5−/−) were intranasally infected with SARS-CoV-2 (1×105 CCID50). A: Percentages of neutrophils (Ly6G antibody-positive) from two infected mouse strains at 2 dpi were analyzed by flow cytometry (left) and calculated from triplicate samples (right). Total numbers of leukocytes and neutrophils in BALF of infected mice at 2 dpi were counted (lower right). B: Lungs were harvested from infected mice at 2 dpi, fixed in formalin, and embedded in paraffin. Lung sections were stained with hematoxylin and eosin for histopathological analysis (original magnification, 20×). Lung histological scores were assessed by a veterinary pathologist blind to the study, as described in the Methods and Materials section. C: mRNAs levels of indicated cytokines in lung tissue homogenates of infected mice at 2 dpi were examined by qRT-PCR (normalized to β-actin). Mock-infected mice were transfected with control pCMV3 plasmid. D, E: Lungs were harvested at different dpi, viral RNA was determined based on number of nucleocapsid (n) gene RNA copies detected by qRT-PCR (D), and viral titers were determined using a CCID50 assay (E). Error bars indicate standard deviation of triplicate biological samples. *: P<0.05 based on Student’st-test.
DISCUSSION
Animal models serve as an indispensable platform with which to study the emerging pathogenesis of infectious viruses and evaluate the efficacy of potential drugs and vaccines. To date, a number of animal species, including nonhuman primates (Deng et al., 2020; Lu et al., 2020; Rockx et al., 2020; Shan et al., 2020; Song et al., 2020), mice (Bao et al., 2020; Jiang et al., 2020; Sun et al., 2020; Zhang et al., 2020), ferrets (Shi et al., 2020), tree shrews (Xu et al., 2020a), golden Syrian hamsters (Chan et al., 2020), and cats (Shi et al., 2020), have been proposed as models of SARS-CoV-2 infection in humans. Mice exhibit several advantages over other animals in the development of a SARS-CoV-2 infection model, namely low cost, convenience, and ability to undergo genetic manipulation. However, based on our work and other studies (Bao et al., 2020; Jiang et al., 2020; Sun et al., 2020; Zhang et al., 2020), normal male and female adult mice, at least the BALB/c, C57BL/6, and ICR strains, are not susceptible to SARS-CoV-2 infection. SARS-CoV-2 uses hACE2 as its host cell entry receptor. Based on this, Bao et al. (2020) successfully established a SARS-CoV-2 infection model using hACE2 transgenic mice. Sun et al. (2020) also recently established a SARS-CoV-2 infection model using Ad5-hACE2-sensitized mice. Thus, to establish a SARS-CoV-2 infection model in C57BL/6 mice with different genetic backgrounds, we transfected the hACE2 gene into the mouse lung for transient expression and then infected mice with SARS-CoV-2 at 2 days post-transfection. Data revealed that the transfected mice acquired susceptibility to SARS-CoV-2. Viral RNA, viral protein, and infectious virions were observed in the infected lung tissue, and a pulmonary innate immune response and lung inflammation were also detected after infection. However, there were limitations to the hACE2 transfection mouse model. in vivo transient transfection can induce short-term expression and limited tissue distribution of the exogenous viral receptor protein (Figure 2A, B), thereby preventing severe disease and mortality induced by viral infection and proliferation in mice. Based on our in vivo pulmonary hACE2 transfection mouse model, the virus showed infection and replication in the target cells, with hACE2 expression in the infected lungs, as supported by the presence of viral RNA (Figure 3B), viral nucleocapsid protein (Figure 3A), and infectious virions (Figure 3C) at 2 dpi; in contrast, no viral RNA, nucleocapsid protein, or virion was detected in the infected lungs of non-hACE2-transfected control mice at the same time. Indeed, only a small portion of the lung epithelial cells transfected and expressed the hACE2 protein (Figure 2B), suggesting that the inoculation virus only showed infection and replication in the hACE2-transfected epithelial cells during the initial round. No substantial replication or proliferation of the virus to other pulmonary cells beyond the initial infected cells was observed, as demonstrated in the viral RNA and viral titer detection analyses (Figures 3B, C; 4D, E). Thus, the decrease in viral load in the infected lungs of the hACE2 transfection model mice could be attributed to the limited expression of hACE2 in the transfected lungs. However, considering viral infection and immune/inflammatory responses were observed during the early infection stage (2–3 dpi) in hACE2-transfected mice, this model may be useful for exploring host antiviral innate immune response and inflammation at the early infection stage.
Expression of the hACE2 protein was mainly observed in the lung epithelial cells of transfected mice, which suggests that SARS-CoV-2 infected these cells first for proliferation. Upon infection, pulmonary epithelial cells release inflammatory cytokines to recruit many granulocytes, macrophages, and monocytes to the lung parenchyma and alveoli. These innate immune cells then clear the virus via phagocytosis and dissolution of infected cells, and further release cytokines and inflammatory chemokines to exert additional antiviral innate immune effects and induce inflammation. In our infection model, the lung immune response was activated from the early infection stage (2–3 dpi) based on BALF leukocyte infiltration and BALF cytokine expression (Figure 3D, F). Among BALF leukocytes, neutrophils were the most abundant to infiltrate the infected lung, and they did so at the early stage of infection. ELR (glutamic acid-leucine-arginine)+CXC chemokines are neutrophil chemoattractants, with seven members of this class of chemokine so far identified in humans, including interleukin (IL)-8, neutrophil-activating peptide 2 (NAP-2), growth-related oncogenes (FRO)-α/β/γ, epithelium-derived neutrophil-activating peptide 78 (ENA-78), and granulocyte chemotactic protein 2 (GCP-2). Among these chemokines, IL-8 is considered the most potent neutrophil chemoattractant in humans (Baggiolini et al., 1993; Lukacs et al., 1999). No human IL-8 homologs have been identified in rodents. However, KC (CXCL1), MIP-2 (CXCL2), LIX (CXCL5), and lungkine in mice are important neutrophil chemoattractants during lung inflammation (Balamayooran et al., 2012; Chen et al., 2001; Driscoll et al., 1995; Frevert et al., 1995). Here, the expression of neutrophil chemoattractants CXCL1, CXCL2, and especially CXCL5 increased in the infected lungs after SARS-CoV-2 infection. Further infection studies using CXCL5−/− mice confirmed that the CXCL5 protein was likely responsible for neutrophil recruitment to the infected lungs. These results indicate that pulmonary infection with SARS-CoV-2 induced massive neutrophil infiltration in the lungs of infected mice at the early innate immune stage and that CXCL5 is a key chemokine responsible for neutrophil recruitment. Because mouse CXCL5 is a close homolog of ENA-78 in humans and GCP-2 in rodent species (Puneet et al., 2005), our findings suggest the possible roles of ENA-78 and GCP-2 in human infection with SARS-CoV-2. In addition, lung inflammation was reduced in CXCL5−/−-infected mice, suggesting that reduced neutrophil infiltration in infected lungs contributes to a decrease in lung inflammation. This was confirmed by a neutrophil depletion assay, in which depletion of neutrophils contributed to the alleviation of lung pathology of SARS-CoV-2 infection in hACE2-transfected mice at 2 dpi (Supplementary Figure 2A). Neutrophil depletion did not affect overall viral clearance, although viral elimination was reduced at 2 dpi (Supplementary Figure 2B). It is worth noting that controlling lung inflammation through regulating neutrophil infiltration in SARS-CoV-2-infected lungs could be beneficial for the suppression of pneumonia development without affecting the antiviral immune response.
In short, we established a SARS-CoV-2 infection C57BL/6 mouse model using in vivo pulmonary transfection of an hACE2-expression plasmid. Moreover, infection of hACE2-transfected CXCL5−/− mice with SARS-CoV-2 revealed the role of CXCL5 in regulating neutrophil infiltration in lungs at the early stage of pulmonary infection, further suggesting a potential target for controlling pneumonia in future research.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
L.D.L. and L.G. conceived and designed the experiments. L.G., Y.L., H.L., J.L., H.W.Z., J.L.L., and Z.N.Y. performed the experiments. L.G. and H.J.S. analyzed the data and wrote the manuscript. Y.L. and L.D.L. reviewed and corrected the manuscript. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We thank Jun-Jie Mei and G. Scott Worthen for providing CXCL5−/− mice.
Funding Statement
This work was supported by the National Natural Science Foundation of China (82041017) and Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (2016-I2M-1-014)
Contributor Information
Lei Guo, Email: shried@163.com.
Long-Ding Liu, Email: longdingl@gmail.com.
References
- 1.Baggiolini M, Dewald B, Moser B Lnterleukin-8 and related chemotactic cytokines–CXC and CC chemokines. Advances in Immunology. 1993;55:97–179. doi: 10.1016/S0065-2776(08)60509-X. [DOI] [PubMed] [Google Scholar]
- 2.Balamayooran G, Batra S, Cai SS, Mei JJ, Worthen GS, Penn AL, et al Role of CXCL5 in leukocyte recruitment to the lungs during secondhand smoke exposure. American Journal of Respiratory Cell and Molecular Biology. 2012;47(1):104–111. doi: 10.1165/rcmb.2011-0260OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balamayooran G, Batra S, Fessler MB, Happel KI, Jeyaseelan S Mechanisms of neutrophil accumulation in the lungs against bacteria. American Journal of Respiratory Cell and Molecular Biology. 2010;43(1):5–16. doi: 10.1165/rcmb.2009-0047TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao LL, Deng W, Huang BY, Gao H, Liu JN, Ren LL, et al The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 5.Chan JFW, Zhang AJ, Yuan SF, Poon VKM, Chan CCS, Lee ACY, et al. 2020. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clinical Infectious Diseases: ciaa325.
- 6.Chen SC, Mehrad B, Deng JC, Vassileva G, Manfra DJ, Cook DN, et al Impaired pulmonary host defense in mice lacking expression of the CXC chemokine lungkine. The Journal of Immunology. 2001;166(5):3362–3368. doi: 10.4049/jimmunol.166.5.3362. [DOI] [PubMed] [Google Scholar]
- 7.Das S, MacDonald K, Chang HYS, Mitzner W A simple method of mouse lung intubation. Journal of Visualized Experiments. 2013;(73):e50318. doi: 10.3791/50318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng W, Bao LL, Liu JN, Xiao C, Liu JY, Xue J, et al Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369(6505):818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Driscoll KE, Hassenbein DG, Howard BW, Isfort RJ, Cody D, Tindal MH, et al Cloning, expression, and functional characterization of rat MIP-2: a neutrophil chemoattractant and epithelial cell mitogen. Journal of Leukocyte Biology. 1995;58(3):359–364. doi: 10.1002/jlb.58.3.359. [DOI] [PubMed] [Google Scholar]
- 10.Frevert CW, Huang S, Danaee H, Paulauskis JD, Kobzik L Functional characterization of the rat chemokine KC and its importance in neutrophil recruitment in a rat model of pulmonary inflammation. Journal of Immunology. 1995;154(1):335–344. [PubMed] [Google Scholar]
- 11.Guo L, Feng K, Wang YC, Mei JJ, Ning RT, Zheng HW, et al Critical role of CXCL4 in the lung pathogenesis of influenza (H1N1) respiratory infection. Mucosal Immunology. 2017;10(6):1529–1541. doi: 10.1038/mi.2017.1. [DOI] [PubMed] [Google Scholar]
- 12.Huang CL, Wang YM, Li XW, Ren LL, Zhao JP, Hu Y, et al Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang RD, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR, et al Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182(1):50–58.e8. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li WH, Moore MJ, Vasilieva N, Sui JH, Wong SK, Berne MA, et al Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu SY, Zhao Y, Yu WH, Yang Y, Gao JH, Wang JB, et al Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduction and Targeted Therapy. 2020;5(1):157. doi: 10.1038/s41392-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukacs NW, Hogabaom C, Campbell E, Kunkel SL Chemokines: function, regulation and alteration of inflammatory responses. Chemical Immunology. 1999;72:102–120. doi: 10.1159/000058729. [DOI] [PubMed] [Google Scholar]
- 17.McCray PB Jr, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, et al Lethal infection of K18-hACE2 mice infected with wevere acute respiratory sundrome coronavirus . Journal of Virology. 2007;81(2):813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mei JJ, Liu YH, Dai N, Favara M, Greene T, Jeyaseelan S, et al CXCL5 regulates chemokine scavenging and pulmonary host defense to bacterial infection. Immunity. 2010;33(1):106–117. doi: 10.1016/j.immuni.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore JB, June CH Cytokine release syndrome in severe COVID-19. Science. 2020;368(6490):473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 20.Puneet P, Moochhala S, Bhatia M Chemokines in acute respiratory distress syndrome. American Journal of Physiology - Lung Cellular and Molecular Physiology. 2005;288(1):L3–L15. doi: 10.1152/ajplung.00405.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin C, Zhou LQ, Hu ZW, Zhang SQ, Yang S, Tao Y, et al Dysregulation of immune response in patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clinical Infectious Diseases. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Read LJ, Muench H A simple method of estimating fifty percent endpoints. American Journal of Epidemiology. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 23.Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Oude Munnink BB, et al Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368(6949):1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shan C, Yao YF, Yang XL, Zhou YW, Gao G, Peng Y, et al Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in Rhesus macaques . Cell Research. 2020;30(8):670–677. doi: 10.1038/s41422-020-0364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi JZ, Wen ZY, Zhong GX, Yang HL, Wang C, Huang BY, et al Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368(6494):1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song TZ, Zheng HY, Han JB, Jin L, Yang X, Liu FL, et al Delayed severe cytokine storm and immune cell infiltration in SARS-CoV-2-infected aged Chinese rhesus macaques. Zoological Research. 2020;41(5):503–516. doi: 10.24272/j.issn.2095-8137.2020.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J, Zhuang Z, Zheng J, Li K, Wong RLY, Liu DL, et al Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182(3):734–743. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tate MD, Deng YM, Jones JE, Anderson GP, Brooks AG, Reading PC Neutrophils ameliorate lung injury and the development of severe disease during influenza infection. The Journal of Immunology. 2009;183(11):7441–7450. doi: 10.4049/jimmunol.0902497. [DOI] [PubMed] [Google Scholar]
- 29.Tseng CTK, Huang C, Newman P, Wang N, Narayanan K, Watts DM, et al Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. Journal of Virology. 2007;81(3):1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu L, Yu DD, Ma YH, Yao YL, Luo RH, Feng XL, et al COVID-19-like symptoms observed in Chinese tree shrews infected with SARS-CoV-2. Zoological Research. 2020a;41(5):517–526. doi: 10.24272/j.issn.2095-8137.2020.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z, Shi L, Wang YJ, Zhang JY, Huang L, Zhang C, et al Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet Respiratory Medicine. 2020b;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XH, Deng W, Tong Z, Liu YX, Zhang LF, Zhu H, et al Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comparative Medicine. 2007;57(5):450–459. [PubMed] [Google Scholar]
- 33.Zhang YN, Li XD, Zhang ZR, Zhang HQ, Li N, Liu J, et al A mouse model for SARS-CoV-2 infection by exogenous delivery of hACE2 using alphavirus replicon particles. Cell Research. 2020 doi: 10.1038/s41422-020-00405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.