Abstract
Objective
This study exploits the intersection between molecular-targeted therapies and immune-checkpoint inhibition to define new means to treat pancreatic cancer.
Design
Patient-derived cell lines and xenograft models were used to define the response to CDK4/6 and MEK inhibition in the tumour compartment. Impacts relative to immunotherapy were performed using subcutaneous and orthotopic syngeneic models. Single-cell RNA sequencing and multispectral imaging were employed to delineate effects on the immunological milieu in the tumour microenvironment.
Results
We found that combination treatment with MEK and CDK4/6 inhibitors was effective across a broad range of PDX models in delaying tumour progression. These effects were associated with stable cell-cycle arrest, as well as the induction of multiple genes associated with interferon response and antigen presentation in an RB-dependent fashion. Using single-cell sequencing and complementary approaches, we found that the combination of CDK4/6 and MEK inhibition had a significant impact on increasing T-cell infiltration and altering myeloid populations, while potently cooperating with immune checkpoint inhibitors.
Conclusions
Together, these data indicate that there are canonical and non-canonical features of CDK4/6 and MEK inhibition that impact on the tumour and immune microenvironment. This combination-targeted treatment can promote robust tumour control in combination with immune checkpoint inhibitor therapy.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) has a universally poor prognosis that has proven resistant to multiple-targeted interventions.1,2 KRAS mutations dominate the genetic landscape of pancreatic cancer and exist in concert with a number of high-potency genetic events (eg, CDKN2A loss, TP53 mutation and SMAD4 loss) that contribute to the aggressive nature of PDAC.3–5 Clinical trials of agents targeting RAS pathway in pancreatic cancer via MEK or other effector pathway inhibitors have largely failed to demonstrate improved outcome.6,7 Additionally, even genetic strategies to ablate KRAS in preclinical models of pancreatic cancer only lead to transient remission.8 Thus, targeting multiple pathways to control oncogenic signalling is likely important for limiting adaptation that emerge after inhibiting critical genetic nodes in PDAC.
Many oncogenes, including KRAS, impinge on the cell cycle through the activation of CDK4 and CDK6 kinases.9–11 In PDAC, deletion or mutation of the CDKN2A gene leads to the loss of p16ink4a that is an endogenous CDK4/6 inhibitor. In the context of KRAS-driven tumourigenesis, loss of p16ink4a is critical for the bypassing oncogene-induced senescence12 and thus effective suppression of CDK4/6 activity could be particularly relevant in the context of pancreatic cancer.13 Interestingly, in preclinical studies, PDAC models are surprisingly resistant to CDK4/6 inhibition.14,15 However, combination with MTOR or MEK inhibitors have been reported to enhance response to CDK4/6 inhibition in PDAC and other RAS-driven tumour types.15–17 In spite of the potency of such combinations in enforcing cell-cycle withdrawal, tumour cells continue to survive and, as a result, acquired resistance represents a significant challenge with such cytostatic combinations.
Immunotherapy has emerged as an important treatment modality in a number of tumour types including those harbouring RAS mutation (eg, lung cancer and melanoma).18,19 However, single-agent immunotherapy treatments have generally not been successful in pancreatic cancer.20–22 This lack of response may in part be attributed to the presence of dense desmoplastic stroma, the relatively low level of quality neoepitopes and the activation of multiple immune suppressive features of the tumour microenvironment.21–25 Understanding the complex interplay between tumour cell and stromal compartment and agents that target these components have been utilised in combination with immune modulating treatments and have shown promising results.26,27 Rationally combining targeted and immune-modulatory agents for the treatment of PDAC could represent an important advance in yielding long-term disease control for pancreatic cancer patients.
METHODS
Cell culture
Pancreatic ductal adenocarcinoma cells were grown in Keratinocyte SFM with 0.2 ng/mL EFG, 30 μg/mL bovine pituitary extract (Life Technologies, 10744019), and 2% foetal bovine serum on collagen coated (Millipore 08–115), tissue culture-treated plates as we published.24,28 The 4662 model was provided by Dr Robert Vonderheide, and cultured in DMEM+10% FBS as reported.29,30
Immunoblot analysis
Primary antibodies for immunoblot analysis were purchased from Cell Signaling Technology include p-RB (S807/S811) (8516S), RB (9313S) (pAkt (S473) (4070S), Akt (4691), pS6 (S235/236) (2211), S6 (2217), cyclin E1 (4129S) and p27 (2552S). Actin (SC-47778), pERK (Y204) (SC-7383), ERK (SC-514302), cyclin D1 (SC-20044) and cyclin A (SC-271682) antibodies were purchased from Santa Cruz Biotechnology. The whole-cell extracts were prepared by lysing the cells with RIPA lysis buffer (Santa Cruz Biotechnology, SC-24948A) in the presence of 1X Halt protease inhibitor (Thermo Fisher) and 1 mM PMSF (Sigma). The extracted proteins (20 μg) were resolved by SDS-PAGE and transferred to PVDF membranes, which were then incubated with primary antibodies at 4°C overnight, followed by incubation with HRP-tagged antimouse or antirabbit secondary antibodies at room temperature up to 1 hour. An enhanced chemiluminescence kit (Thermo Fisher, 34076) was used to detect the immunoreactive bands.
CDK2 kinase assay
To analyse the CDK2 kinase activity, primary PDAC cells were lysed using the kinase lysis buffer (50 mM HEPES-KOH pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% Tween-20) in the presence of 1X Halt protease inhibitor (Thermo Fisher) and 1 mM PMSF (Sigma). Active CDK2 complex was immunoprecipitated by incubating 300 μg of the lysate with 5 μg of anti-CDK2 (Santa Cruz Biotechnology, SC-6248) overnight at 4°C. Normal mouse IgG1 (Cell Signaling Technology, 5415) was used as a control. Protein G agarose-beads were added to each IP samples and incubated up to 4 hour at 4°C. Protein immuno-complexes were washed three times with the kinase lysis buffer and two times with kinase reaction buffer (40 mM Tris-HCl pH 8, 20 mM MgCl2, 0.1 mg/mL BSA, 50 μM DTT). Kinase reactions were carried out in 100 μL of kinase buffer in the presence of 100 μM ATP and 0.5 μg of RB protein as substrate by gently shaking at room temperature up to 30 min. The resulting phosphorylated RB protein was detected by immunoblotting using anti-p-Rb (S807/S811) antibody (Cell Signaling Technology, 8516S).
Cell proliferation assay
Cell proliferation following different treatments was determined using chemiluminescent BrdU ELISA kit (Roche, 11669915001) as described by the manufacturer. Luminescence was read on a Biotek Synergy 2 plate reader.
FuCCI (fluorescent, ubiquitination-based cell-cycle indicator) analyses
ES-FUCCI, a gift from Pierre Neveu (Addgene plasmid #62451), was transfected into 226 cell line using Lipofectamine 3000 Reagent (Invitrogen, LSL3000001) by manufacturer’s protocol. Transfected cells were selected using Hygromycin and sorted with FACSAria to generate a homogeneous population. The FUCCI activity was determined by flow-cytometry detecting the GFP and RFP signals simultaneously.
Drug screen
IncuCyte S3 Live-Cell Analysis System (Essen Biosciences) was utilised for live cell analysis. Primary PDAC cells were labelled with H2B-GFP and seeded in 384-well collagen-coated plates. Cells were treated with DMSO or palbociclib (100 nM) for 24 hours prior to library screen of over 300 cancer drugs at 100 nM. Two phase and fluorescent images per well were captured hourly for 72 hours at 10× magnification. Essen Bioscience software was used to quantify number of cells per well and normalised to DMSO-treated cell proliferation. Data were exported to Prism 7 (GraphPad) for statistical analyses.
Gene expression analysis
RNA was isolated from cells or snap frozen tumour tissue using RNeasy Plus Kit (Qiagen). Resultant RNA was employed for RNA sequencing using previously described procedures.31 To selectively evaluate the expression within the PDX tumour tissue and exclude reads from the mouse genome, the alignment of reads was performed over a combined genome, so that human selective reads were captured. Triplicate reads were grouped by cell lines or respective PDX model, then normalised using the package edgeR. Log fold-changes and Student two-tailed t-test p-values (assuming equal variance) were calculated for each treatment from the normalised reads. Volcano plots used cut-offs for log fold change as specified in the figure legends. Genes that were upregulated and downregulated in the palbo-ciclib+tramentinib condition were used for gene ontology analysis using ENRICHR (amp.pharm.mssm.edu/Enrichr/). Gene set enrichment analysis was performed as previously published. For the analysis of TCGA analyses, expressed genes in palboci-clib+tramentinib related to immune function and cell cycle were applied to the pancreatic cohort and clustered based on k-means. Kaplan-Meier analysis for overall survival comparing the clusters was performed using the survival package in R.
Immunohistochemical, multispectral imaging
Immunohistochemistry on mouse tissues was performed using standard procedures with antibodies and dilutions indicated CD8 (ab209775–1:1000) opal 540 nm 1:300, CD163 (ab182422–1:250) opal 620 nm 1:300 and Ki67 (RM-9106S1–1:150) opal 690 nm 1:100. Staining was performed on a Leica auto-stainer. Multispectral panels including the above antibodies were developed and images captured on a Vectra Polaris Instrument (Perkin Elmer). Phenotype counts were determined using InFORM software (Perkin Elmer) according to the manufacture’s protocol.
Mice and patient-derived xenografts
NSG (Jackson Laboratories) mice were maintained in the University of Arizona animal care facility. Mice were both male and female and used between 10 and 20 weeks of age for engraftment. All animal care, treatment and sacrifice were approved by the University of Arizona Institutional Animal Care and Use Committee (IACUC) in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. Mice were implanted subcutaneously with early passage PDX tumour fragments that have never been placed into culture as we have published.28,31 When tumours reached a volume of ~200 mm3, they were randomised to treatment cohorts such that the starting volume across the cohorts was similar. Mice were treated for 3 weeks by gastric gavage with single agent or a combination. Palbociclib (PD-0332991, 100 mg/kg) diluted in 50 mM lactate buffer at pH 4.0, trametinib (0.5 mg/kg) diluted in 0.5% hydroxypropyl cellulose and 0.2% tween 80. Mice were treated daily for 5 days, followed by a 2-day break during first week, and then every other day during second and third weeks for a total treatment time of 3 weeks as previously published.32 Tumour size was assessed every 2 days using digital callipers. Tumour measures were carried out independently by multiple laboratory members. Progression on therapy was indicated by a 50% increase in tumour volume while on treatment. Progression-free survival was analysed in Prism (Graph-Pad).
Syngeneic tumour model
C57BL/6J mice were subcutaneously injected with 4662 cells (3×106/mouse). All animal care, treatment and sacrifice were approved by the University of Arizona and Roswell Park Cancer Center IACUC in accordance with the NIH Guide for the Care and Use of Laboratory Animals. When tumour volumes reached 150 mm3, mice were randomised to the following treatment groups that include (a) palbociclib (100 mg/kg) diluted in 50 mM lactate buffer, pH 4.0 was administered orally for 5 days during the first week followed by a 2-day break and then administered orally (75 mg/kg) during the second and third weeks; (b) trametinib (0.5 mg/kg) was diluted in in 0.5% hydroxypropyl cellulose and 0.2% tween 80, was administered orally for 5 days per week and the treatment continued up to 3 weeks; (c) in the combination arm palbociclib (100 mg/kg) and trametinib (0.5 mg/kg) were diluted in 50 mM lactate buffer, pH 4.0 and administered orally for 5 days during the first week followed by a 2-day break and then palbociclib (75 mg/kg) in combination with trametinib (0.5 mg/kg) was administered every other day orally during the second and third weeks. The anti-PD-L1 Clone 10F.9G2 (BioX-Cell, BE0101) was prepared in the dilution buffer (pH 6.5) (BioXcell, IP0065) and administered by intraperitoneal injection for every other day (200 μg/mouse, 3 times/week) up to 3 weeks. In vivo MAB rat IgG2b isotype (Bioxcell, BE0090) was used as a negative control. Tumours were measured every other day with calliper and volume was calculated with the following equation: V=0.5 ((greatest diameter) x (shortest diameter)2). Body weight was measured every other day. All the treatments lasted for 21 days or until tumour volumes reached 2000 mm3. In mice where the tumour completely regressed, treatment was ceased and the mice were monitored up to 30 days for tumour outgrowth. At the end of 30th day, mice were rechallenged with 3×105 4662 cells and the tumour growth were monitored up to 45 days. Orthotopic models were performed as previously published,33 mice were randomised at ~100 mm3 as determined by MRI.34
Replicates and mouse numbers
All mouse studies involved at least five tumour-bearing mice, unless otherwise specified (eg, exceptional responders). The RNA sequencing analysis is from at least three independent cell culture or at least three independent PDX models. All biochemical studies have been replicated and include internal controls for the validation of results. Immunohistochemical staining was confirmed on multiple tissue sections and independent regions of interest employed for quantification.
Single-cell sequencing and analysis
C57BL/6J mice harbouring 4662 tumours were sacrificed on treatment and dissociated by mincing and digestions with liberase (Sigma # 541020001) (2×15’). Resultant cells were stained with antimouse CD45 BUV395-conjugated antibody (BD Biosciences, 564616) and viability dye eFluor780 (Thermo Fisher,65–0865-14) and subjected to sorting. Single-cell libraries were generated using the 10X Genomics platform. Cell suspensions were first assessed with Trypan Blue using a Countess FL automated cell counter (ThermoFisher), to determine concentration, viability and the absence of clumps and debris that could interfere with single-cell capture. Cells were loaded into the Chromium Controller (10X Genomics) where they were partitioned into nanoliter-scale Gel Beads-in-emulsion with a single barcode per cell. Reverse transcription was performed and the resulting cDNA amplified. The full-length amplified cDNA was used to generate gene expression libraries by enzymatic fragmentation, end-repair, a-tailing, adapter ligation and PCR to add Illumina compatible sequencing adapters. The resulting libraries were evaluated on D1000 screentape using a TapeStation 4200 (Agilent Technologies), and quantitated using Kapa Biosystems qPCR quantitation kit for Illumina. They were then pooled, denatured and diluted to 300pM with 1% PhiX control library added. The resulting pool was then loaded into the appropriate NovaSeq Reagent cartridge and sequenced on a NovaSeq6000 following the manufacturer’s recommended protocol (Illumina). The raw sequencing data were processed using Cell Ranger software with mouse mm10 reference genome.35 The filtered gene-barcode matrices which contain barcodes with the Unique Molecular Identifier counts that passed the cell detection algorithm were used for Seurat single-cell data analysis R package.36 Cells with low RNA features (<200) or high RNA feature (>5000) or high mitochondrial RNA contents (10%) were filtered out from the analysis. These filters resulted in the following cell counts for each treatment: vehicle control (n=8509 cells), palbociclib+trametinib (n=1065 cells) and palbociclib+trametinib+anti-PD-L1 (n=8639 cells). Data from all samples were merged and normalised using the SCTransform. Dimension reductions (PCA and UMAP) using the highly variable genes were calculated for clustering analysis using the first 30 principal components resulting in 12 clusters. The SingleR package was utilised to identify the predominant cell types. The Seurat soft-ware package was used to generate gene expression analysis based on treatment conditions. RNA velocity analysis were carried out using velocyto R package.37,38
RESULTS
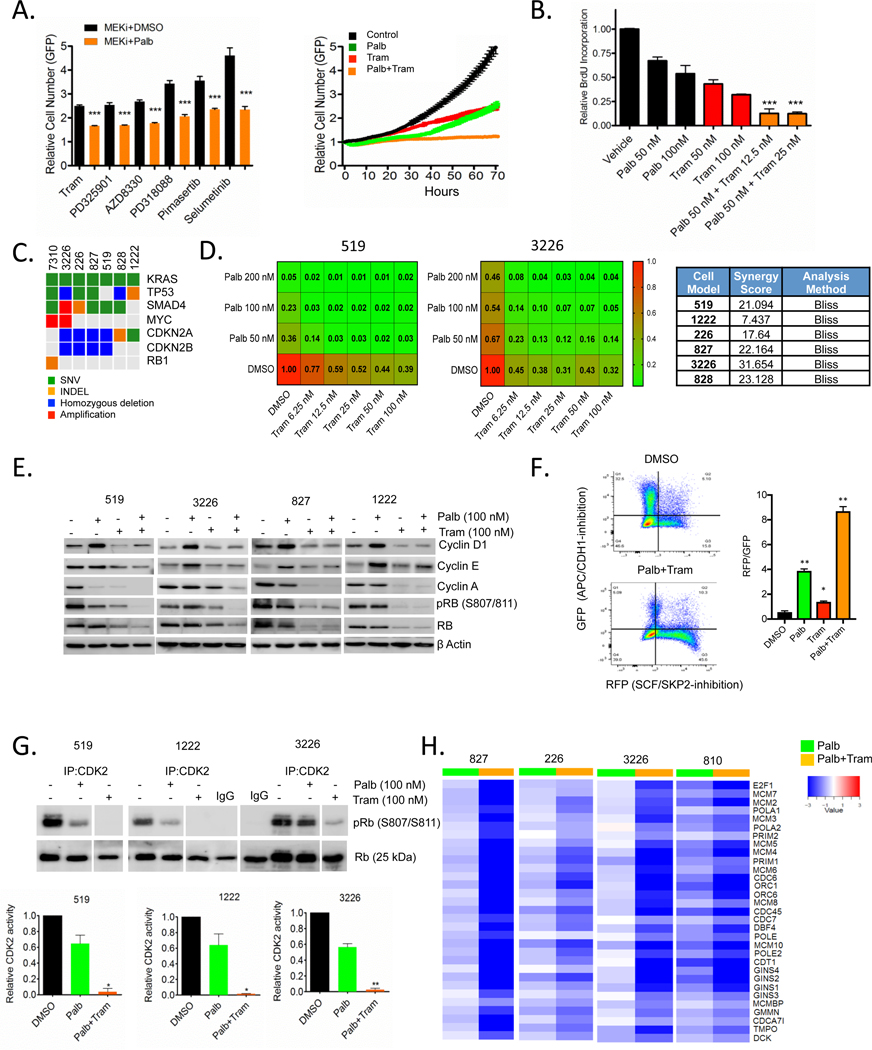
In spite of genetic features that would be expected to yield sensitivity to CDK4/6 inhibitors, pancreatic cancer cell lines are surprisingly resistant to this treatment.16,32 To systematically define cooperating agents, we carried out live cell imaging-based drug screens where the suppression of proliferation is directly monitored by evaluating cell number (online supplementary figure 1A and S1). In this screen, MEK inhibitors were highly enriched for cooperating with the CDK4/6 inhibitor palbociclib. Validation studies with the MEK inhibitor trametinib demonstrated that these effects are due to cooperation relative to cell-cycle inhibition as determined by BrdU incorporation (figure 1B). Furthermore, dose response analysis showed that the drug interaction is synergistic as determined by Bliss analysis in multiple patient-derived cell models24,31 that reflect genetic variability of pancreatic cancer (figure 1C and D, and S1). To interrogate the mechanisms related to response, initially we evaluated how CDK4/6 inhibition may impact on features of signalling downstream from KRAS (online supplementary figure S1). These data showed that CDK4/6 inhibition had no effect related to canonical signalling through ERK, AKT or MTOR pathways as determined by phosphorylation of ERK, AKT and S6, respectively (online supplementary figure S1). However, treatment with trametinib resulted in expected suppression of ERK activity and in several models also suppressed activity through the MTOR and AKT pathways (online supplementary figure S1), as has been recently reported in other RAS-driven tumours.17 To determine the features of therapeutic cooperation, we interrogated canonical determinants of cell-cycle control. Treatment with CDK4/6 inhibitors lead to the adaptive upregulation of cyclin D1 and cyclin E in the pancreatic cancer cell lines (figure 1E), consistent with prior findings.32,39 These adaptive features of CDK4/6 inhibition were ameliorated with the combined treatment with trametinib that yielded potent blockade of RB phosphorylation and suppression of cyclin A expression (figure 1E). Flow cytometry analysis using an FUCCI reporter for APC/CDH1 and SCF/SKP2 activity was used to determine the action of drug treatment on cell-cycle regulatory activities. These data showed that the combination of CDK4/6 and MEK inhibition yielded a potent suppression of SCF/SKP2, yet APC/CDH1 remained active during the G1 arrest (online supplementary figure 1F and S2). In parallel with these results, we found that MEK inhibition induced p27Kip1 (online supplementary figure S2), which is an important determinant of response to CDK4/6 inhibition.10 While CDK4/6 inhibition had a modest effect on CDK2 activity, MEK inhibition cooperated with CDK4/6 inhibition to decrease CDK2 kinase activity (figure 1G and online supplementary figure S2). The coordination between CDK4/6 inhibition and the suppression of CDK2 activity is critically important for the cytostatic action of CDK4/6 inhibitors.40,41 Consistent with the effects on CDK2 and RB activity, gene expression analysis illustrated significant suppression of E2F-target genes in multiple cell models with the combination treatment (figure 1H). Thus, these data support the impact of simultaneously targeting both a KRAS effector pathway (MEK1/2) and cell cycle (CDK4/6) to elicit pronounced cell-cycle exit.
Figure 1.
Cooperation between MEK and CDK4/6 inhibition in PDAC cell model. (A) The 3226 pancreatic cancer cell line, expressing H2B-GFP was treated with palbociclib (100 nM)±the indicated MEK inhibitors (250 nM) and relative proliferation was determined by live cell imaging. Representative data from the screen is shown in the growth curve. Statistical significance was determined by Student t-test (two-sided) ***p<0.001, mean and SD are shown. (B) Relative BrdU incorporation was determined following 48 hours exposure to palbociclib±trametinib at the indicated concentrations in the 3226 cell line. Statistical significance was determined by Student t-test (two-sided) ***p<0.001 relative to the single agents, mean and SD are shown. (C) The genetics of the employed cell models are summarised in the oncoprint (somatic single nucleotide variant: green, homozygous deletion: blue, small insertion/deletion: orange, amplification: red). (D) Isobologram analyses were performed as indicated in the heatmaps of relatively CDK4/6 inhibitor sensitive (519) and resistant (3226) models. Heatmaps represent relative BrdU incorporation after 48 hours treatment and synergy was determined by the BLISS method. (E) Immunoblot analysis of the indicated cell-cycle proteins in 519, 3226, 827 and 1222 cell lines that were treated with palbociclib (100 nM)±trametinib (100 nM) for 48 hours. Data representative of multiple experiments is shown. (F) 226 cells were labelled with FUCCI and treated with DMSO, palbociclib (200 nM), trametinib (50 nM) and combination for 48 hours prior to flow cytometry analysis. Cells were counter stained with DAPI for DNA content. Panel shows GFP (APC/CDH1-inhibition) on Y-axis and RFP (SCF/ SKP2-inhibition) on X-axis. Graph shows quantitation of RFP to GFP ratio of indicated treatments. *P-value<0.05, **p-value<0.01 Student t-test (two-sided), mean and SD are shown. (G) In vitro CDK2 kinase assays were performed in 519, 1222 and 3226 cell lines following the treatment with palbociclib (100 nM)±trametinib (100 nM). Specific kinase activity was determined based on the phosphorylation of an exogenous RB substrate at S807/811 and the band intensities were quantified. Representative blot images (interspersed lanes have been cropped as indicated) and mean and SD are shown (*p<0.5, **p<0.01, as determined by Student t-test two-sided). (G) Heatmap showing the log-fold change of the indicated cell-cycle regulatory genes from cells treated with palbociclib alone or the combination with trametinib. The RNA sequencing was performed in triplicate. PDAC, pancreatic ductal adenocarcinoma.
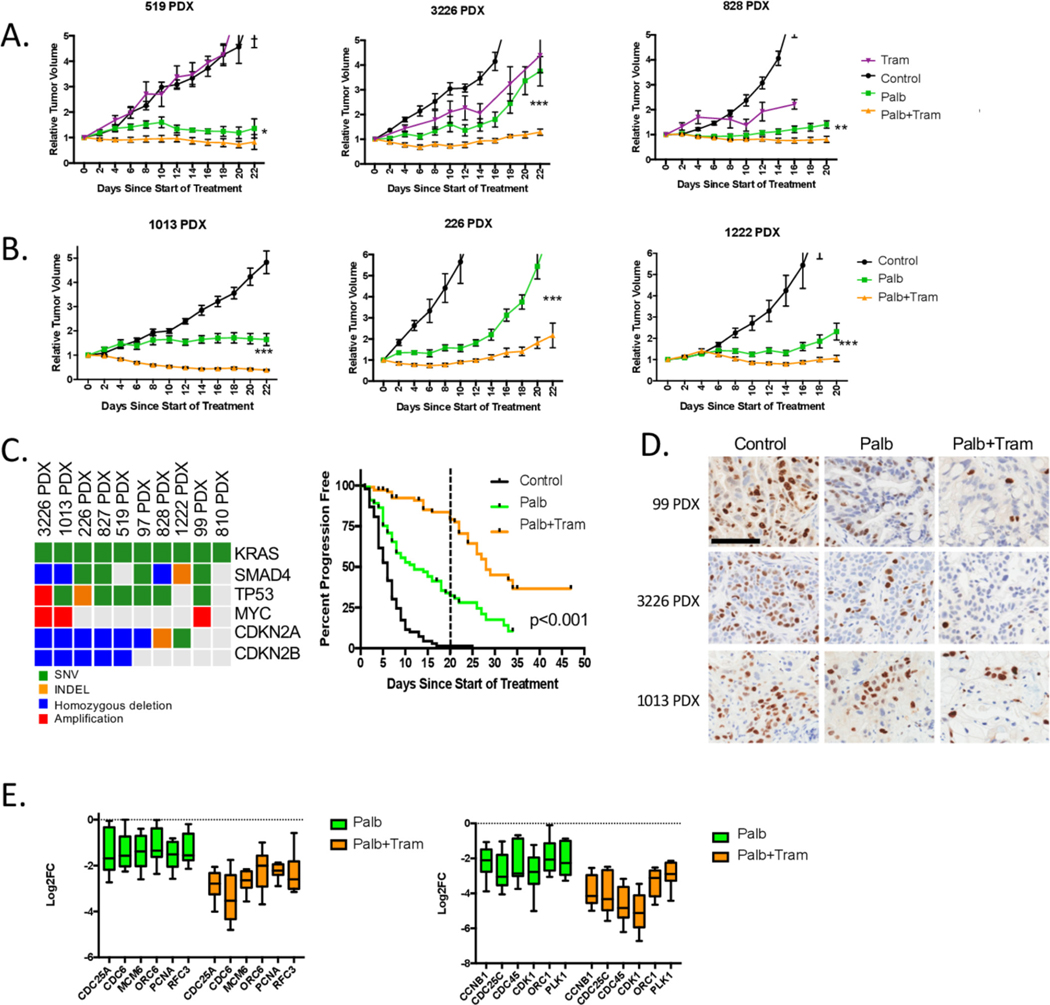
We utilised a panel of pancreatic cancer PDX models to delineate in vivo response to CDK4/6 and MEK inhibition. These models have been sequenced28,31 and exhibit genetic drivers consistent with the clinical diversity of PDAC (figure 2). In specific models, treatment with single-agent MEK or CDK4/6 inhibitor had a transient impact on tumour growth (figure 2A and B). However, the combination significantly reduced tumour growth. Across the panel of individual 10 PDX models (n=368 individual tumours), we found that the combination significantly increased progression-free survival relative to CDK4/6 inhibition alone (figure 2C). However, with the cessation of therapy at 21 days, tumours progressed indicating the reversible cytostatic nature of the treatment. These responses were associated with enhanced suppression of the Ki67 proliferation marker by immunohistochemistry, and the repression of E2F-target genes by RNA sequencing (figure 2D and E). Notably, in the PDX models (as in the cell models), the combination treatment limited phosphorylation of RB, and the expression of cyclin D1 and E (online supplementary figure S3). Together, this work shows that the combination with CDK4/6 and MEK inhibition provides a putative therapeutic opportunity for the treatment of PDAC that is associated with profound cell-cycle exit.
Figure 2.
Cooperation between MEK and CDK4/6 inhibition in PDX models of pancreatic cancer. (A) The indicated PDX models were treated with vehicle, palbociclib or trametinib and the combination. The tumour volume was measured by calliper and the relative tumour volume for the cohorts is plotted as a function of time (*p<0.05, **p<0.01, ***p<0.001, Student t-test two-sided). There are at least five mice per condition. (B) Additional PDX models were treated with vehicle, palbociclib or palbociclb+trametinib combination. The relative tumour volume for the cohorts is plotted as a function of time (*p<0.05, **p<0.01, ***p<0.001, Student t-test two-sided). There are at least five mice per condition. (C) 10 different PDX models with the genetic features summarised in the oncoprint were treated with vehicle (n=90), palbociclib (n=101) or palbociclib+trametinib (n=97). Progression-free survival (where progression represents a 50% change in tumour volume) is plotted. Treatment was for 21 days (marked by dashed line), and subsets of animals were followed with the cessation of therapy (***p<0.001 determined by log-rank test). (D) Immunohistochemical staining for Ki67 in three different PDX models treated with palbociclib or the combination with trametinib. Representative fields are shown (scale bar=100 μm) (E) Expression of the indicated cell-cycle regulatory genes is shown from the panel of treated 10 PDX models. For each PDX model, RNA sequencing was performed in triplicate and the Log2FC was determined. The average Log2FC is graphed for each of the indicated genes involved in DNA replication (top panel) or mitosis (bottom panel). The box and whisker plot shows minimum, first quartile, median, third quartile and maximum.
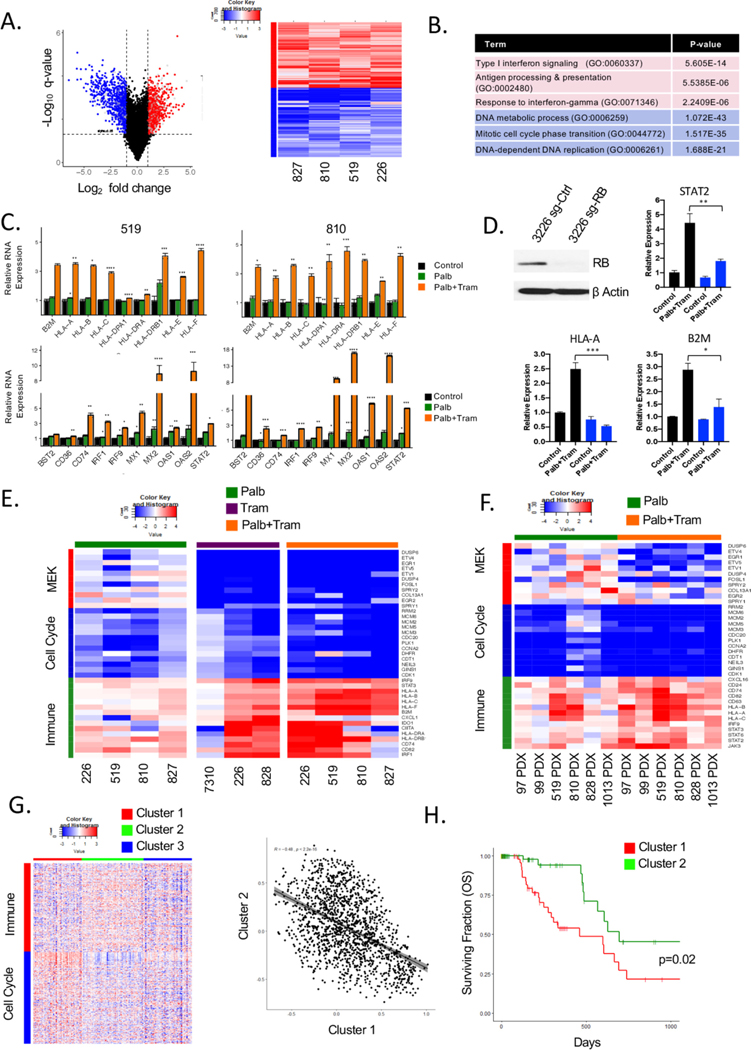
In order to understand means to expand on the cytostatic efficacy of the CDK4/6 and MEK inhibitor combination, we evaluated RNA sequencing data from treated PDAC models. In cell culture models, we found that the combination of CDK4/6 and MEK inhibition elicited both the consistent suppression of genes, and also induced an equivalent number of genes (figure 3A). While the suppressed genes were strongly associated with cell cycle, the upregulated genes were associated with antigen presentation and features of interferon signalling (figure 3A and B and online supplementary figure S4). This signature of immune response is similar to that observed either with single-agent MEK or CDK4/6 inhibition in other models.26,42,43 In particular, we observed that the combination induced the expression of multiple MHC genes (eg, HLA-A, HLA-C) and genes involved in interferon signalling (eg, STAT2 and IRF9) (online supplementary figure 3C and S4). The induction of immune-related proteins was dependent on the presence of RB, as the 7310 cell line which is RB-deficient31 failed to elicit this response (online supplementary figure S4). These findings were further supported through the use of CRISPR-mediated deletion of RB in the 3226 cell line. In this context, both the transcriptional repression and gene activation observed with palbociclib and trametinib treatment are mitigated (figure 3D). Further evaluating independent treatments illustrated that MEK inhibition suppresses canonical target genes (eg, DUSP6 and ETV1), as well as cell cycle (eg, CCNA2 and PLK1), while eliciting the induction of the genes associated with interferon response and antigen presentation (figure 3E). In this context, the suppression of MEK genes is RB independent, while cell cycle and induced genes require RB, as noted by the RB-deficient cell line 7310 (online supplementary figure 3E and S4). The observed changes in gene expression induce secretion of CCL5 and CXCL10 that are downstream from interferon signalling and associated with enhanced T-cell infiltration (online supplementary figure S5). Gene expression findings were also recapitulated in PDX models (online supplementary figure 3F and S5), where tumour selective transcripts were evaluated through the use of a hybrid mouse/human genome.44 The interferon-like response has been suggested to represent a senescence-associated secretory phenotype (SASP). However, in our data, there was no induction of IL6, IL8 and IL1β and little positive enrichment for the classical SASP signature (online supplementary figure S6). In addition to MEK inhibition, MTOR inhibitors can cooperate with CDK4/6 inhibitors in driving cell-cycle exit.32 The TORC1/2 inhibitor TAK228 results in potent cooperation with CDK4/6 inhibition in the suppression of cell-cycle-regulated genes in PDX models and cell lines (online supplementary figure S7). However, the combination of palbociclib and TAK228 did not recapitulate the induction of the antigen-presentation genes (online supplementary figure S7). These data validate that the combination of CDK4/6 and MEK inhibition is distinct in mediating both profound cell-cycle exit and immunological response features in pancreatic cancer models. It has been reported in several tumour types that there is an inverse relationship between cell cycle and interferon-related gene expression signatures.45 Using TCGA data, we found that those tumours with high-index for proliferation, but low for immune response are associated with poor prognosis (figure 3G and H and online supplementary figure S8). Thus, in principle, shifting the transcriptional programme toward that induced by MEK and CDK4/6 inhibition would be associated with improved survival coupling dual biological effects.
Figure 3.
Induction of immunological genes in an RB-dependent manner in PDAC models treated with MEK and CDK4/6 inhibitors. (A) Common genes significantly induced or repressed in palbociclib and trametinib-treated cell lines were determined using an average log 2 fold-change greater than 1 and a false-discovery rate less than 5% were considered to be differentially expressed. The blue symbols denote repressed cell-cycle genes and the red symbols denote induced genes associated with the immune system. (right panel) Heatmap comparing the log 2 fold change of genes related to immune function (red-colour bar) and cell cycle (blue-colour bar) in palbociclib+trametinib-treated cell PDAC cell line models. (B) Top gene ontology terms related to the immune system and cell cycle ranked based on p-value. (C) Relative mRNA expression of palbociclib (green) and palbociclib+trametinib (orange) treated cells for a subset of genes related to antigen presentation and Interferon signalling (*p≤0.05,**p≤0.01, ***p≤0.001,****p≤0.0001, determined by Student t-test two-sided, the average and SD are shown). (D) qRT-PCR for the immune-related genes were performed in 3226 cell line with CRISPR-mediated deletion of RB treated with palbociclib+trametinib (*p<=0.05,**p<=0.01,***p<=0.001, determined by Student t-test two-sided, the average and SD are shown). (E) Heatmap showing the log fold change of representative MEK target genes (red colour-bar), cell cycle (blue colour-bar) and immune function (green colour-bar) with the indicated treatments across cell line models. (F) Heatmap showing the log fold change of representative MEK target genes (red colour-bar), cell cycle (blue colour-bar) and immune function (green colour-bar) with the indicated treatments across PDX models. (G) Heatmap showing K-means clustering of TCGA pancreatic cancer cohort based on significant genes altered with palbociclib+trametinib treatment related to immune function (red colour-bar) or cell cycle (blue colour-bar). Pearson correlation between cluster 1 and cluster 2 with a negative correlation of −0.48 and significance p<2.2e−16. (H) Kaplan-Meier analysis for overall survival comparing cluster 1 and cluster 2 from the heatmap. PDAC, pancreatic ductal adenocarcinoma.
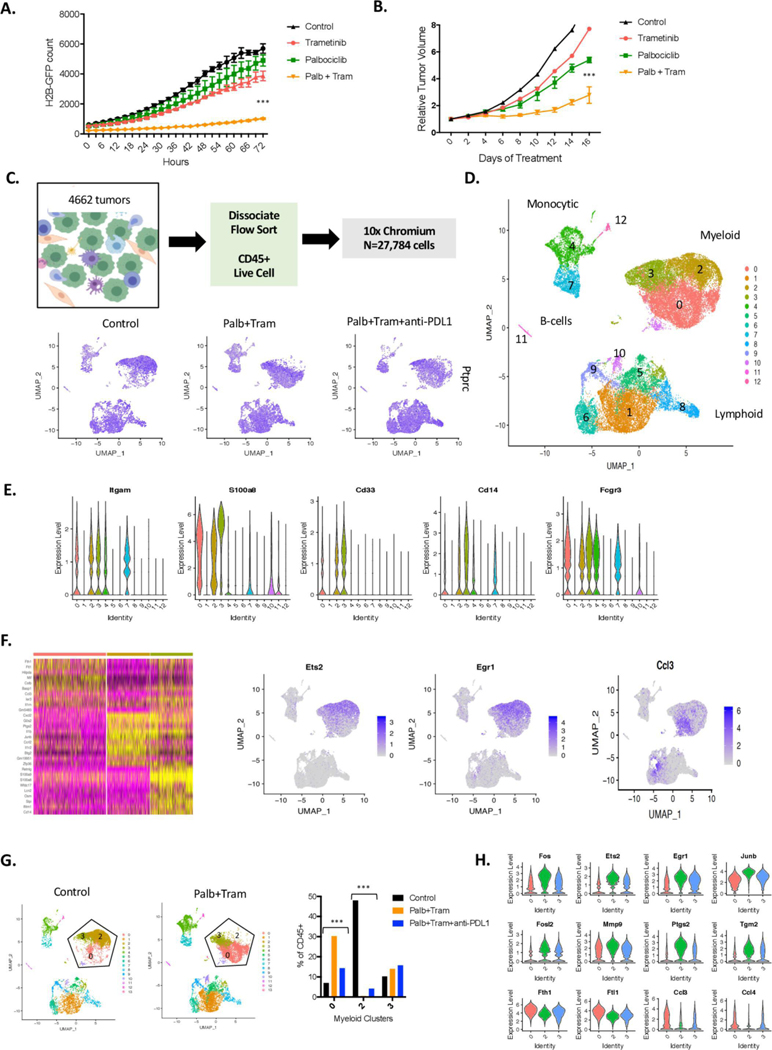
To functionally assess how the combination of CDK4/6 and MEK inhibition impacts on the immune system, we employed a syngeneic pancreatic cancer model derived from the KPC mouse model (the 4662 model).29 This model is highly aggressive and neither CDK4/6 inhibition nor MEK inhibition alone had a significant impact on either cell growth or tumour growth in C57BL/6J mice (figure 4A and B). However, consistent with the findings in the patient-derived models, the combination treatment with MEK and CDK4/6 inhibition delayed the progression of this model, although even within 14 days there was tumour growth on treatment (figure 4B). To define how CDK4/6 and MEK inhibition impacts on the immunological tumour microenvironment and immune-checkpoint inhibitor therapy, single-cell sequencing was performed. Mice bearing established 4662 tumours were treated with palbociclib and trametinib, or a triple combination with anti-PD-L1. The triple combination was well-tolerated based on mouse weight (online supplementary figure S9) and minimal systemic effects on the bone-marrow of the mice (not shown). Tumours from all treatment groups were dissociated and the CD45+ fraction was isolated by fluorescent-activated cell sorting (figure 4C). Single-cell RNA sequencing was performed using the 10x chromium method. Consistent with the enrichment approach, all cells were positive for the CD45 gene (PTPRC) irrespective of treatment (figure 4C). Dimensional clustering algorithms were applied to all cells that passed quality-control metrics (n=27 784) and defined 12 populations of cells that capture lymphoid, myeloid, monocytic and B-cell components of the tumour (figure 4D and online supplementary S10). The myeloid compartment was defined by classical phenotyping markers (CD33, S100A8 and ITGAM), with two dominant myeloid-macrophage populations (clusters 0 and 2) (figure 4E). Treatment with palbociclib and trametinib lead to a switch in the dominant myeloid-macrophage population present in the control tumours (figure 4F and G). Specifically, the myeloid-macrophages were suppressed for immediate early genes (eg, EGR1, JUNB, ETS2 and FOS) that are downstream from MEK/ ERK signalling.46 These findings were accompanied by downregulation of PTGS2, VEGFA and MMP9 that are associated with immune-suppressive M2-like macrophages (figure 4H and online supplementary S10).47 The observed changes were also apparent when all cells were interrogated based on treatment (online supplementary figure S11). Concordantly, with treatment there was induction of genes associated with iron-metabolism (FTL1 and FLH1) and macrophage functions (BNIP3L and CTSD) that are typically associated with immune-activating M1-like macrophages (figure 4H).47 Additionally, top-induced genes included CCL3, CCL4 and CCL6 that are involved in T-cell, dendritic cell, B-cell and NK-cell infiltration (figure 4H). This shift in macrophage populations was also observed in tumours treated with the triplet that includes anti-PD-L1 (figure 4G and H and online supplementary figure S10). Across treatments, there was only a modest alteration in the neutrophil population (cluster 3).
Figure 4.
Impact of CDK4/6 and MEK inhibition on the tumour microenvironment. (A) The KPC-derived 4662 cell line labelled with H2B-GFP was treated with palbociclib, trametinib or the combination. Cellular proliferation was determined using IncuCyte live cell imaging over the indicated time frame (***p<0.001, by Student t-test relative to the single-agent controls). (B) The 4662 model was introduced into C57BL/6J mice subcutaneously, mice were randomised at approximately 200 mm3 and tumour volume was measured by callipers with mice on treatment (***p<0.001 for the combination relative to vehicle control by Student t-test). (C) Schematic workflow for the single-cell sequencing. Feature plots denote the expression of PTPRC (CD45 gene) in all of the captured cells with treatment. (D) All of the cells from the treated mice were filtered for quality and clustered using Seurat software in the dimension plot. The general clustering of different immunological subtypes is indicated. (E) The violin plots denote gene markers associated with myeloid, neutrophil and macrophage populations. (F) Seurat heatmap identifying top differences between the myeloid clusters (0, 2 and and representative feature plots are shown). (G) Distribution of cells in the indicated myeloid clusters, changes in clusters 0 and 2 with treatments are highly significant by χ2statistic (***p<1E−100). (H) Genes differentially expressed in cells between clusters 2 and 0 are highlighted in the violin plot (all genes are highly significant between the clusters p<1E−5).
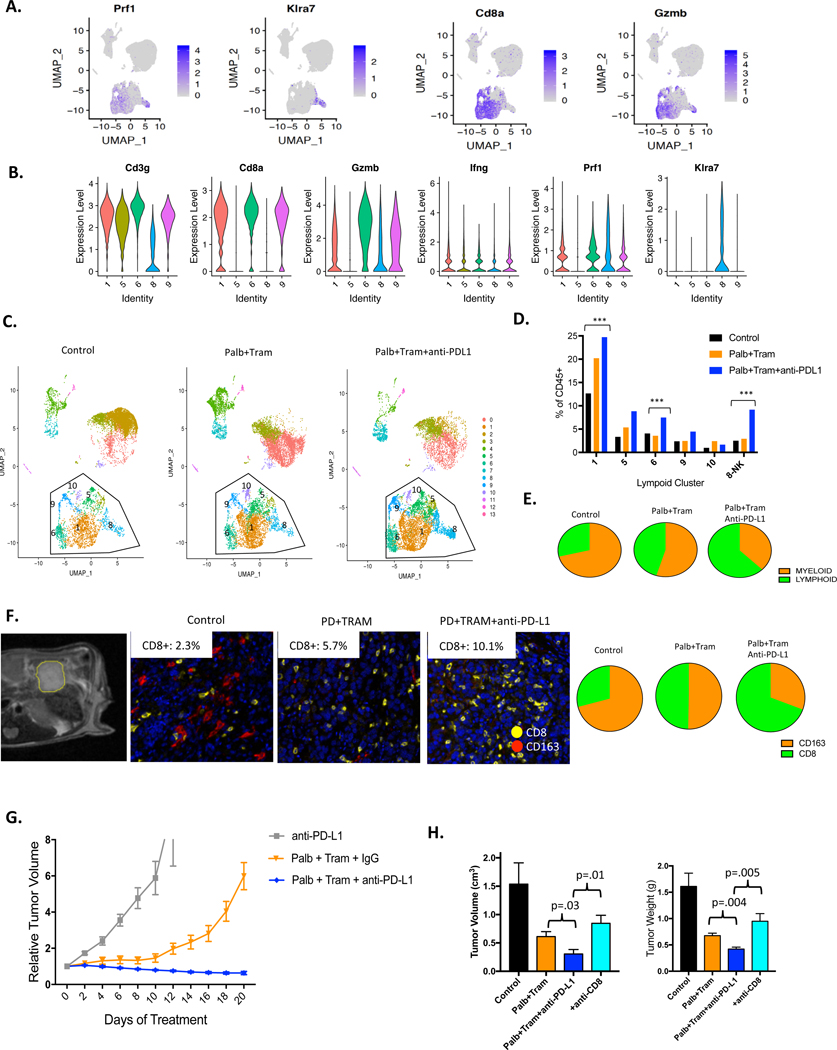
The lymphoid compartment of the tumour encompassed multiple subtypes of T-cells and NK cells (cluster 8) as indicated by conventional markers (figure 5A and B, online supplementary figure S12). The lymphoid populations were clearly enhanced with both CDK4/6 and MEK inhibition as well as the triplet combining these inhibitors with anti-PD-L1 (figure 5C). The most common CD8+ T-cell population (cluster 1) almost doubled with treatment of palbociclib and trametinib and was further enhanced with anti-PD-L1. This T-cell population is GZMB and PRF1 positive, indicative of cytotoxic function. Interestingly, other populations were more modestly induced or did not change with the doublet (figure 5D). This increase in T-cell infiltrate could also be clearly detected by differential expression analysis based on treatment (online supplementary figure S13). The inclusion of anti-PD-L1 treatment further enhanced the T-cell infiltrate with significant increases in veritably all populations, most notably the NK-cell population which is also positive for TCF7 that is a mediator of NK activity/ survival48 (online supplementary figure 5D and S12). It is known that a subset of active T-cells are proliferative, and this cluster (cluster 9) did not diminish with treatment, but in fact increased with combined treatment (figure 5D). This was confirmed by multispectral imaging and single-cell feature analysis (online supplementary figure 5D and S14).
Figure 5.
Cooperation between CDK4/6-MEK inhibition and immune-checkpoint inhibition. (A) Predominant T-cell clusters were defined using feature plots of classical markers. (B) Violin plots demonstrating the expression of the indicated markers across the lymphoid clusters. (C) Dimension cluster plots illustrating the accumulation of T-cell (clusters 1, 5, 6, 9, 10) and NK (cluster 8) populations with the indicated treatments. (D) Quantitation of cells present in each of the T-cell and NK cell clusters is shown. Changes indicated are statistically significant as determined by χ2 analysis (p<1E−10). (E) Myeloid to lymphoid relationship as determined by single-cell sequencing analysis is presented in the pie-charts. (F) Orthotopic tumours were monitored by MRI with representative axial T2-weighted magnetic resonance image showing engrafted 4662 tumour model (outlined in yellow) in the pancreas. Representative multispectral staining of orthotopic tumours treated with the indicated regimens (CD8-yellow, CD163-red). The percent of CD8+ cells in multiple independent regions of interest is displayed in the inset. CD163 to CD8 relationship as determined by multispectral imaging of orthotoopic tumours is presented in the pie-charts. (G) Mice harbouring subcutaneous tumours were treated as indicated. The mean and SD are shown with at least five mice per treatment group. (H) Orthotopic tumours were treated with the indicated regimens where anti-CD8 was employed to neutralise the T-cell response in the triplet combination. Mice were sacrificed with 21 days of treatment. The mean and SD are shown for tumour volume. Statistical analysis is by Student t-test, **p<0.01, *p<0.05.
In the control tumours, there was a significant skewing toward myeloid infiltration, while treatment with CDK4/6 and MEK inhibitors increased the lymphoid infiltrate which, was further enhanced with anti-PD-L1 (online supplementary figure 5E and S13). To confirm these findings, orthotopic 4662 tumours were employed, wherein tumour burden was assessed by MRI. Combined anti-CD8 and CD163 multispectral imaging was employed to quantify T-cells versus M2-like macrophages (figure 5F). These data largely mirrored the findings from the single-cell sequencing of subcutaneous tumours. The monocytic populations were more limited in the treatment-naïve tumours, but augmented with treatment, as were relatively rare BATF3+ dendritic cell populations (online supplementary figure S15). Velocity analysis suggests that the monocytic pools were transiting towards the dendritic cell populations as would be expected based on the differentiation paradigm (online supplementary figure S14). In addition, while limited, the number of B-cells within the tumour increased with the triple combination. Together, these data suggest an overall enhancement of immune response with CDK4/6 and MEK inhibition, which is further stimulated with immune checkpoint inhibition.
The 4662 syngeneic model is largely resistant to single-agent anti-PD-L1 treatment consistent with other studies;29 however, the combination of CDK4/6 and MEK inhibition with anti-PD-L1 was highly effective yielding regression of the tumour (online supplementary figure 5G and S16). The combination of both CDK4/6 and MEK inhibitor was required for the cooperation with anti-PD-L1, as neither treatment alone provided pronounced disease control (online supplementary figure S16). In exceptional cases, tumours became undetectable and were followed for long-term tumour and resistance to subsequent challenge was also observed (online supplementary figure S16). To confirm the antitumour activity, orthotopic models where also employed (figure 5H). Mice were randomised to treatment groups, and as in the subcutaneous models, there was more profound disease control with the triplet combining CDK4/6 and MEK inhibitors and anti-PD-L1 (figure 5H). To interrogate the significance of the CD8+T cells for this therapeutic response, mice were coordinately treated with anti-CD8, which depleted CD8+ cells in the tumour microenvironment (online supplementary figure S16), and reversed the impact of anti-PD-L1 (figure 5H). Together, these studies underscore the potency of combining targeted and immunotherapy for the treatment of PDAC and illuminate previously unrecognised contributions of targeted therapies on the tumour-immune microenvironment.
DISCUSSION
As pancreatic cancer represents a therapy recalcitrant disease, combinatorial means to cancer treatment will assuredly be required. While CDK4/6 inhibitors have been found to be effective in ER+breast cancer and FDA-approved for that indication, clinical advances in additional indications have yet to mature.40 Pancreatic cancer is driven by activated KRAS and CDKN2A loss; thus, it would be expected that CDK4/6-inhibition would have dominant effects. The data herein reinforce the finding that cell-cycle plasticity enables escape from CDK4/6 inhibitors. However, in unbiased drug screens, MEK inhibitors emerged as key cooperative agents to block that plasticity and couple CDK4/6 inhibition to the activation of RB, suppression of CDK2 and repression of E2F target genes. These interactions are synergistic in cell culture and highly potent in all PDX models tested derived from 10 different patients. While the combination is highly effective, with cessation of treatment cells re-enter the cell cycle (not shown) and tumours ultimately progress. Additionally, in very fast-growing tumours for example, 4662 model, progression occurs on therapy and reinforces the need for an adjuvant strategies to complement the cell-cycle inhibition.
In evaluating the effect of CDK4/6 and MEK inhibition on gene expression, we observed a large number of genes that are repressed or activated and could yield a specific vulnerability beyond the cell cycle. For example, there is potent suppression of EZH2 and additional chromatin modifiers as well as inhibition of DNA repair mechanisms that could confer sensitivity to diverse therapeutics. A noted feature of the response to CDK4/6 and MEK inhibition is the upregulation of interferon and antigen presentation that has been observed in other tumour types.27,49 This signature is related to cell-cycle arrest and is RB dependent; however, whether it is related to senescence is unclear in this setting. The gene expression analysis of cell lines occurred at 48–72 hours before the morphological changes associated with senescence, additionally the signature defined here shares little similarity with the classical SASP signature defined in replicative senescence.26,50 Additionally, while the combination of CDK4/6 and MTOR inhibitors induces potent cell-cycle arrest, the induction of immune-related pathways is not present. We favour the model that aberrant chromatin/transcripts elicited as a consequence of RB activation in the presence of oncogenic signals drive the interferon-like response, which is highly related to that mediated by knockdown of EZH2 or loss of DNMT1 which are both repressed with MEK and CDK4/6 inhibition.26,51 Irrespective of these mechanistic nuances, the induction of immunological gene expression and cell-cycle inhibition are dependent on RB. From genetic analysis in both resected and metastatic pancreatic cancer, RB loss is relatively infrequent (<3%).52,53 Therefore, the treatments herein could be relevant to the majority of PDAC cases and potentially germane to a number of RAS-driven tumours that retain the RB tumour suppressor (eg, lung adenocarcinoma and colorectal cancer).
The impact of targeted agents on the immune system is becoming progressively important with the success of immune-checkpoint therapies. Since both MEK and CDK4/6 have important roles in hematopoiesis understanding the intersection with the immunological milieu within the tumour is clearly significant.54 Using single-cell sequencing of CD45+ cells provided a highly detailed analysis of the immune repertoire in a KPC-based tumour model. These tumours are dominated by myeloid populations, which is consistent with data from mouse models and clinical pancreatic cancer cases.24,55 Treatment with CDK4/6 and MEK inhibition had a pronounced effect on the immune cells within the tumour with the most notable changes occurring within the myeloid-macrophage population. In this context, the combinatorial treatment limited immediate early genes that are associated with angiogenesis and protumourigenic elements of M2-macrophages. These changes were linked to the acquisition of a macrophage population that exhibited enhanced iron-metabolism and chemokine secretion promoting recruitment of lymphocytes and antigen-presenting cells. These proximal alterations on immediate early signalling were associated with increased recruitment of selected T-cell populations. Importantly, the combination treatment with CDK4/6 and MEK inhibitors did not suppress the proliferative T-cell population within the tumour microenvironment. This finding is consistent with the observation that CDK4/6 inhibition can stimulate NFAT activity to promote as opposed to inhibit T-cell activation and yield limited effects on proliferation;56 in this cellular context p38 or other kinases could be initiating RB-phosphorylation which remains poorly understood.57 However, anti-PD-L1 was required for potent engagement of NK populations into the tumour. These data are consistent with the cooperative effect of CDK4/6-MEK inhibition with anti-PD-L1. This cooperation was T-cell mediated as anti-CD8 treatment limits the effectiveness of therapy. Furthermore, in mice that were cured by the triplet therapy, there was subsequent antitumour immunity. These data suggest that the targeted therapies utilised herein can serve as a significant adjunct to immune-checkpoint inhibition in RAS-driven tumours.
Supplementary Material
Significance of this study.
What is already known on this subject?
Pancreatic cancer is a therapy recalcitrant disease largely resistant to the effects of CDK4/6 and MEK inhibitors.
Combination therapies can elicit more potent effects to limit therapeutic resistance that emerge commonly in pancreatic cancer.
Immune checkpoint inhibitors are largely ineffective in the context of pancreatic cancer.
What are the new findings?
MEK inhibition cooperates with CDK4/6 inhibition to limit cell-cycle plasticity and invoke stable cell-cycle arrest in patient-derived models of pancreatic cancer.
The combination of MEK and CDK4/6 inhibition induces interferon and antigen presentation genes in a cell autonomous fashion in the tumour compartment.
The treatment with MEK and CDK4/6 inhibitors has a profound impact on the myeloid and T-cell populations within the tumour compartment.
The combined targeting of signalling pathways elicits sensitivity to anti-PD-L1 therapy in immune competent models.
How might it impact on clinical practice in the foreseeable future?
These studies support the use of MEK and CDK4/6 inhibitors to change the tumour microenvironment to drive sensitivity to anti-PD-L1 therapy that can be tested in clinical trials.
Acknowledgements
We thank all members of the laboratory and our administrators for assistance with assembling the manuscript. Dr Robert Vonderheide (University of Pennsylvania) kindly provided the 4662 syngeneic model that was used in this study. Dr. Rod Bremner and Dr. Joel Pearson (Lunenfeld Tannenbaum Research Institute) provided the lentiviral vector for RB deletion. The multispectral imaging was performed in the Center for Personalized Medicine, drug screening was performed through the Small Molecule Screening Facility and the MRI was supported by the Translational Imaging Shared Resource at the Roswell Park Comprehensive Cancer Center which is supported in part by the NCI Cancer Center Support Grant 5P30 CA016056 by NCI P30CA16056.
Funding CA211878 from the National Institutes of Health.
Footnotes
Competing interests None declared.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting or dissemination plans of this research.
Patient consent for publication Not required.
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability statement Data are available upon reasonable request. The gene expression data in this manuscript will be made available through Gene Expression Omnibus under the title of the manuscript. Any other primary data will be made available upon reasonable request.
Additional material is published online only. To view please visit the journal online (http://dx.doi.org/10.1136/gutjnl-2020-321000).
RefeRences
- 1.Knudsen ES, O’Reilly EM, Brody Jr, et al. Genetic diversity of pancreatic ductal adenocarcinoma and opportunities for precision medicine. Gastroenterology 2016;150:48–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleeff J, Korc M, Apte M, et al. Pancreatic cancer. Nat Rev Dis Primers 2016;2:16022. [DOI] [PubMed] [Google Scholar]
- 3.Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey P, chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. [DOI] [PubMed] [Google Scholar]
- 5.Waddell N, Pajic M, Patch A-M, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Infante JR, somer BG, Park JO, et al. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer 2014;50:2072–81. [DOI] [PubMed] [Google Scholar]
- 7.Bodoky G, Timcheva C, Spigel DR, et al. A phase II open-label randomized study to assess the efficacy and safety of selumetinib (AZD6244 [ARRY-142886]) versus capecitabine in patients with advanced or metastatic pancreatic cancer who have failed first-line gemcitabine therapy. Invest New Drugs 2012;30:1216–23. [DOI] [PubMed] [Google Scholar]
- 8.Kapoor A, Yao W, Ying H, et al. Yap1 activation enables bypass of oncogenic KRAS addiction in pancreatic cancer. Cell 2014;158:185–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherr CJ. Cell cycle control and cancer. Harvey Lect 2000;96:73–92. [PubMed] [Google Scholar]
- 10.Knudsen ES, Witkiewicz AK. The strange case of Cdk4/6 inhibitors: mechanisms, resistance, and combination strategies. Trends Cancer 2017;3:39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filmus J, Robles AI, Shi W, et al. Induction of cyclin D1 overexpression by activated Ras. Oncogene 1994;9:3627–33. [PubMed] [Google Scholar]
- 12.Serrano M, Gómezlahoz E, DePinho RA, et al. Inhibition of Ras-induced proliferation and cellular transformation by p16inK4. Science 1995;267:249–52. [DOI] [PubMed] [Google Scholar]
- 13.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993;366:704–7. [DOI] [PubMed] [Google Scholar]
- 14.Heilmann AM, Perera RM, Ecker V, et al. Cdk4/6 and IGF1 receptor inhibitors synergize to suppress the growth of p16INK4a-deficient pancreatic cancers. Cancer Res 2014;74:3947–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franco J, Witkiewicz AK, Knudsen ES. Cdk4/6 inhibitors have potent activity in combination with pathway selective therapeutic agents in models of pancreatic cancer. Oncotarget 2014;5:6512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franco J, Balaji U, Freinkman E, et al. Metabolic reprogramming of pancreatic cancer mediated by cDK4/6 inhibition elicits unique vulnerabilities. Cell Rep 2016;14:979–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haines E, Chen T, Kommajosyula N, et al. Palbociclib resistance confers dependence on an FGFR-MAP kinase-mTOR-driven pathway in KRAS-mutant non-small cell lung cancer. Oncotarget 2018;9:31572–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fares CM, Van allen EM, Drake CG, et al. Mechanisms of resistance to immune checkpoint blockade: why does checkpoint inhibitor immunotherapy not work for all patients? Am Soc Clin Oncol Educ Book 2019;39:147–64. [DOI] [PubMed] [Google Scholar]
- 19.Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov 2018;8:1069–86. [DOI] [PubMed] [Google Scholar]
- 20.Das S, Berlin J, Cardin D. Harnessing the immune system in pancreatic cancer. Curr Treat Options Oncol 2018;19:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Bosch N, Vinaixa J, Navarro P. Immune evasion in pancreatic cancer: from mechanisms to therapy. Cancers 2018;10. doi: 10.3390/cancers10010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu AA, Jaffee E, Lee V. Current status of immunotherapies for treating pancreatic cancer. Curr Oncol Rep 2019;21:60. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y, Knolhoff BL, Meyer MA, et al. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res 2014;74:5057–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knudsen E, Vail P, Balaji U, et al. Stratification of pancreatic ductal adenocarcinoma: combinatorial genetic, stromal, and immunological markers. Clin Cancer Res 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balachandran VP, Łuksza M, Zhao JN, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017;551:512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goel S, Decristo MJ, Watt AC, et al. Cdk4/6 inhibition triggers anti-tumour immunity. Nature 2017;548:471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaer DA, Beckmann RP, Dempsey JA, et al. The CDK4/6 inhibitor Abemaciclib induces a T cell inflamed tumor microenvironment and enhances the efficacy of PD-L1 checkpoint blockade. Cell Rep 2018;22:2978–94. [DOI] [PubMed] [Google Scholar]
- 28.Witkiewicz AK, Balaji U, Eslinger C, et al. Integrated patient-derived models delineate individualized therapeutic vulnerabilities of pancreatic cancer. Cell Rep 2016;16:2017–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans RA, Diamond MS, Rech AJ, et al. Lack of immunoediting in murine pancreatic cancer reversed with neoantigen. JCI Insight 2016;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winograd R, Byrne KT, Evans RA, et al. Induction of T-cell immunity overcomes complete resistance to PD-1 and CTLA-4 blockade and improves survival in pancreatic carcinoma. Cancer Immunol Res 2015;3:399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knudsen ES, Balaji U, Mannakee B, et al. Pancreatic cancer cell lines as patient-derived avatars: genetic characterisation and functional utility. Gut 2018;67:508–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knudsen ES, Kumarasamy V, Ruiz A, et al. Cell cycle plasticity driven by mTOR signaling: integral resistance to CDK4/6 inhibition in patient-derived models of pancreatic cancer. Oncogene 2019;38:3355–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung S, Vail P, Witkiewicz AK, et al. Coordinately targeting cell-cycle checkpoint functions in integrated models of pancreatic cancer. Clin Cancer Res 2019;25:2290–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma R, Buitrago S, Pitoniak R, et al. Influence of the implantation site on the sensitivity of patient pancreatic tumor xenografts to Apo2L/TRAIL therapy. Pancreas 2014;43:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sl Wolock, Lopez R Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst 2019;8:281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aran D, Looney AP, Liu L, et al. Reference-Based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019;20:163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Street K, Risso D, Fletcher RB, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 2018;19:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.La Manno G, Soldatov R, Zeisel A, et al. Rna velocity of single cells. Nature 2018;560:494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vilgelm AE, Saleh N, Shattuck-Brandt R, et al. Mdm2 antagonists overcome intrinsic resistance to CDK4/6 inhibition by inducing p21. Sci Transl Med 2019;11. doi: 10.1126/scitranslmed.aav7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knudsen ES, Pruitt SC, Hershberger PA, et al. Cell cycle and beyond: exploiting new Rb1 controlled mechanisms for cancer therapy. Trends Cancer 2019;5:308–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrera-Abreu MT, Palafox M, Asghar U, et al. Early adaptation and acquired resistance to CDK4/6 inhibition in estrogen receptor-positive breast cancer. Cancer Res 2016;76:2301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu-Lieskovan S, Mok S, Homet Moreno B, et al. Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAF(V600E) melanoma. Sci Transl Med 2015;7:279ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruscetti M, Leibold J, Bott MJ, et al. Nk cell-mediated cytotoxicity contributes to tumor control by a cytostatic drug combination. Science 2018;362:1416–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruna A, Rueda OM, Greenwood W, et al. A Biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell 2016;167:260–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hutcheson J, Witkiewicz AK, Knudsen ES. The Rb tumor suppressor at the intersection of proliferation and immunity: relevance to disease immune evasion and immunotherapy. Cell Cycle 2015;14:3812–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy lO MacKeigan JP, Blenis J. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol Cell Biol 2004;24:144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guerriero JL. Macrophages: the road less traveled, changing anticancer therapy. Trends Mol Med 2018;24:472–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeevan-Raj B, Gehrig J, Charmoy M, et al. The transcription factor TCF1 contributes to normal NK cell development and function by limiting the expression of granzymes. Cell Rep 2017;20:613–26. [DOI] [PubMed] [Google Scholar]
- 49.Goel S, Wang Q, Watt AC, et al. Overcoming therapeutic resistance in HER2-positive breast cancers with CDK4/6 inhibitors. Cancer Cell 2016;29:255–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hernandez-Segura A, de Jong TV, Melov S, et al. Unmasking transcriptional heterogeneity in senescent cells. Curr Biol 2017;27:2652–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cañadas I, Thummalapalli R, Kim JW, et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med 2018;24:1143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Witkiewicz a K, McMillan E, Balaji U, et al. Whole exome sequencing of Pancreatic cancer:genetic Diversity, Prognostic Features, and Potential Therapeutic Targets. Nature Communication 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Connor AA, Denroche RE, Jang GH, et al. Integration of genomic and transcriptional features in pancreatic cancer reveals increased cell cycle progression in metastases. Cancer Cell 2019;35:267–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malumbres M, Sotillo R, Santamaría D, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases cDK4 and cDK6. Cell 2004;118:493–504. [DOI] [PubMed] [Google Scholar]
- 55.Ene-Obong A, Clear AJ, Watt J, et al. Activated pancreatic stellate cells sequester cD8+ T cells to reduce their infiltration of the juxtatumoral compartment of pancreatic ductal adenocarcinoma. Gastroenterology 2013;145:1121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng J, Wang ES, Jenkins RW, et al. Cdk4/6 inhibition augments antitumor immunity by enhancing T-cell activation. Cancer Discov 2018;8:216–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomás-Loba A, Manieri E, gonzález-Terán B, et al. p38γ is essential for cell cycle progression and liver tumorigenesis. Nature 2019;568:557–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.