Abstract
Rheum palmatum L. (RPL) is a known traditional herbal medicine with the functions of “heat-clearing and damp-drying” in traditional Chinese medicine. Its anti-cancer effect against lung cancer has been confirmed previously, but the related mechanisms and active substances for its action has been little studied. This study adopted the network pharmacology, built the network map of drug ingredients and disease targets (DDN), and discussed the effective components of RPL and its possible mechanisms. All constituents of RPL were collected through database search and literature mining, and the potential active constituents were screened. The inverse pharmacophore matching model was used to predict the targets of active ingredients, and the method was supplemented by database retrieval and literature mining. Compounds-target data were inputted into Cytoscape software to build the DDN of RPL, and functional annotation analysis and pathway enrichment analysis were carried out. Finally, 20 active compounds were screened, which acted on 817 targets. A total of 22,418 lung cancer-related targets were collected, and 761 overlapped with drug targets. By bioinformatics annotation of these overlapping genes, a total of 235 gene ontology (GO) functional annotation analyses and 46 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were obtained. It was found that the enrichment of GO and KEGG was associated with apoptosis, suggesting RPL plays an anti-lung cancer role via inducing cell apoptosis. Subsequent cell experiment results showed RPL and its active constituents inhibited the proliferation of A549 cells and reduced clone formation rate of A549 cells via induction of apoptosis. In this study, the pharmacodynamic basis and mechanism of RPL against lung cancer were studied from the perspective of systematic pharmacology, which would be beneficial for further elucidating the anticancer effect of RPL on lung cancer.
Keywords: network pharmacology, traditional Chinese medicine, lung cancer, apoptosis, Rheum palmatum L. (Dahuang)
Introduction
Lung cancer, as a malignant tumor of respiratory system, is also one of the most common tumors worldwide and the leading cause of cancer death in both men and women (Peng et al., 2015; Bray et al., 2018). Although surgery, chemotherapy, radiotherapy and targeted drugs have achieved great progress in the prevention and treatment of lung cancer, the death rate of lung cancer patients is still very high. It is reported that the 5-year survival rate of patients with early stage lung cancer is 45–65%, while the 5-year survival rate of patients with advanced lung cancer is only 14% (Syed et al., 2016). Increasing evidences have suggested that herbal medicines are good resources for finding novel drugs, and previous investigations have also demonstrated that lots of extracts or monomers isolated from herbal medicines have the potentials for prevention or treatment of lung cancers (Ma et al., 2012; Peng et al., 2015). Rheum palmatum L. (RPL), belonging to the Polygonaceae family, is a known herbal medicine in traditional Chinese medicine (TCM), and modern pharmacological studies have found that RPL has various pharmacological activities including anti-inflammatory, anti-tumor, bacteriostatic, hemostatic, lipid-lowering, and hypotensive effects (Lee et. al., 2012; Jeong et al., 2017; Park et al., 2018). In addition, a growing number of studies have found that RPL extract and its monomers can induce apoptosis in various human cancer cells (Chiu et al., 2009; Ma et al., 2012). Emodin, which is an important natural constituent in plants of Polygonaceae family, such as Rheum palmatum, Polygonum cuspidatum, and Rumex japonicas can inhibit the proliferation of human lung adenocarcinoma A549 cells (Zhang and Guo, 2011; Peng et al., 2013; Li Z. et al., 2019). Nho et al. found that RPL extract can inhibit breast cancer cell migration, movement, and invasion in a concentration-dependent manner in vitro (Nho et al., 2015). In 2008, Su et al. found that emodin, a natural anthraquinone derivative from RPL, can induce apoptosis of human lung adenocarcinoma cells (Su et al., 2005).
In recent years, TCM has been widely accepted as a supplement or alternative medicine for the treatment of diseases due to its good therapeutic effect and low toxic and side effects. However, TCM has the characteristics of multiple components, multiple targets, and multiple pathways, which leads to its unclear therapeutic mechanism and medicinal substance basis, and limits its promotion and development. Network pharmacology can systematically analyze the interaction network of drug components, protein targets, diseases, genes, and other elements, which coincides with the overall concept of TCM treatment. Therefore, the application of network pharmacology to the study of TCM is scientific and necessary. At present, more and more scholars have applied network pharmacology to the research on the material basis and mechanism of action of TCM on various diseases (Jiang et al., 2019; Xiong et al., 2019). In this study, network pharmacology was used to study the active ingredients and mechanism of action of RPL against lung cancer and in combination with in vitro experiments to provide scientific evidence for the anti-lung cancer effect of RPL.
Materials and Methods
Active Components Screening
All the chemical components of RPL were retrieved from TCM systematic pharmacology database (TCMSP; http://lsp.nwu.edu.cn/tcmsp.php), integrated pharmacology-based research platform of TCM V2.0 (TCMIP; www.tcmip.cn/TCMIP/index.php/Home/Index/All) and TCM integrated pharmacology database (TCMID, https://omictools.com/tcmid-tool), and supplemented by literature mining. Oral bioavailability (OB) is an important parameter to measure the pharmacokinetic process (absorption, distribution, metabolism and excretion) of drugs in vivo as well as druggability. Drug-Likeness (DL) refers to the physical and chemical properties related to good clinical efficacy, such as solubility, stability, biological properties, etc. Therefore, it is also known as the similarity between the molecule to be tested and the drug molecule, which has a considerable indicator role in the development of new drugs. Compounds with high properties of OB and DL are not drugs but have the potential to become drugs, so these two parameters are often used to screen for active compounds. The compounds of RPL were screened for activity using the TCMSP platform and the DL prediction tool provided by the molsoft website (http://www.molsoft.com/docking.html), and compounds meeting DL ≥ 0.18 and OB ≥30% were selected as candidate active ingredients. Searching for the standard names and specific structures of the active candidate compounds in SciFinder (https://scifinder.cas.org/), PubChem (https://pubchem.ncbi.nlm.nih.gov/), and ChemSpider (http://www.chemspider.com/), and use ChemDraw 14.0 to draw the structures of these compounds, which are stored in MOL format.
Establishment of Target Library
Information on target proteins of candidate compounds was collected from the TCMSP database and TCMID, and the target proteins of these compounds were obtained by searching Pubmed database. In addition, in order to retrieve the completeness, we also use Pharmmapper online database (http://www.lilab-ecust.cn/pharmmapper/) and Swiss Target Prediction database (http://www.swisstargetprediction.ch/) looking for compound rhubarb targets. In simple terms, the candidate compounds are uploaded to an online database, the species are restricted to human sources, and potential targets are identified through high-throughput screening. These targets were combined and the repeated target sets of small molecular compounds were deleted. Online Mendelian Inheritance in Man (OMIM; https://omim.org/), DisGeNET (http://www.disgenet.org/), TTD (http://db.idrblab.net/ttd/), and GeneCards (https://www.genecards.org/) were used to search the therapeutic targets related to lung cancer, and the repeated targets were removed to construct the target set of the disease. Finally, the two targets were collected by R software and the intersection was taken as the protein target library of this study. Standard names for all protein targets were identified through UniProt (http://www.uniprot.org/).
Network Construction
Cytoscape is a software that graphically displays the network, analyzes and edits the data, and visualizes it. At the same time, it contains a large number of functional plug-ins for in-depth analysis of the network, adding rich annotation information. Based on the previous steps, we prepared the data pairs of active compounds and disease target genes with R software, imported the data pairs into cytoscape software, and constructed the network map of drug ingredients and disease targets (DDN). In the network diagram, the nodes represent drugs, active ingredients, target genes, and diseases, and the edges represent the interrelationships among the nodes. In addition, the network analyzer plug-in is used to calculate Degree, Closeness, Node-betweenness, K-coreness, Edge-betweenness, and other important parameters to assess the topological properties of each node in the networks in order to make clear the more important composition of RPL and their targets, further scientific and rational interpretation of its molecular mechanisms for the treatment of lung cancer.
Protein-Protein Interaction Network
The regulation of various biological processes in the body is not regulated by a single protein or gene, but through a complex regulatory network. There is a certain degree of signal transduction between different signaling pathways and targets, so the effective components of drugs are more likely to exert their effects by acting on multiple proteins or multiple signaling pathways. String (https://string-db.org/) is an open source online database that can be used to analyze known and predicted interactions between proteins. The target proteins were uploaded to the string database, and the study species were limited as human sources. Download the protein interaction relationship, save it as TSV format, import it into the Cytoscape software, and construct the protein-protein interaction network diagram (PPI) in which each node represents a protein, and the connection between nodes represents the interaction between two proteins. Meanwhile, cytoHubba tool is used to analyze the core regulatory genes of PPI network.
Bioinformatic Annotation
Proteins from the combined target library were uploaded to Funrich or R software for functional annotation analysis and pathway enrichment analysis, including gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The GO enrichment analysis includes cellular component (CC), biological process (BP), and molecular function (MF), showing 10 remarkably rich terms in each category, with bar charts drawn by Funrich software. The results of KEGG pathway enrichment were used to explain the potential molecular mechanism of RPL for lung cancer and the bar and bubble diagrams of the significant KEGG pathway were drawn by the R language tool. P < 0.05 was considered statistically significant in this study.
Experimental Validation
Chemicals and Reagents
Fetal bovine serum (FBS), phosphate buffered saline (PBS), penicillin-streptomycin, trypsin-EDTA and dulbecco modified eagle medium (DMEM) were purchased from GIBCO (Grand Island, NY, USA). Dimethyl sulfoxide (DMSO) was provided by Sigma-Aldrich Co. (St. Louis, MO, USA). The 4 ,6-diamidino-2- phenylindole (DAPI) and Annexin V-FITC/PI apoptosis kits were obtained from the US Everbright® Inc (Suzhou, China). Cell Counting Kit-8 (CCK8) detection kit was obtained from the Beijing 4A Biotech Co., Ltd (Beijing, China). Annexin V-FITC/PI apoptosis kits were purchased from BOSTER biological technology company (Wuhan, China). The paraformaldehyde and crystal violet staining solution were obtained from Chengdu Kelong Chemical reagent factory (Chengdu, China). All the used reference substances were purchased from the PUSH Bio-Technology Co. (https://www.push-herbchem.com/, Chengdu, China) with the purity over 98%.
Preparation of the Lyophilized Powder of RPL Extracts
The herbal medicine was purchased from the Neautus Chinese Herbal Pieces Ltd. Co. (Chengdu, China), and identified by Prof. Chunjie Wu (School of Pharmacy, Chengdu University of TCM). A voucher specimen for future reference has been deposited in Department of Chinese Medicinal Processing, School of Pharmacy, Chengdu University of TCM, Chengdu, China (no. 2019091701#).
The crude herbal medicine (126 g) was powered and refluxed with eight times 70% ethanol (1:8, w/v) for 30 min, then the filtrates were concentrated, and the ethanol was recovered using a rotary evaporator, subsequently the extracts were freeze-dried by a LGJ-12B freeze dryer (Shanghai GIPP Co. Ltd, Shanghai, China). Furthermore, the HPLC assay of the RPL extracts was carried out, and the results showed the main constituents in RPL are anthraquinones such as Sennoside A, B, Rhein and Aloe-emodin and its glucoside, emodin and its glucoside, Chrysophanol and its glucoside, etc ( Figure S1 ).
Cell Culture
Human lung cancer cell line A549 and human umbilical vein endothelial cells (HUVEC) were purchased from the Beina Biological Co. (Beijing, China), and cultured at 37°C in a humidified atmosphere of 5% CO and 95% air in sterile DMEM medium with 10% FBS and supplemented with 100 U/mL penicillin, 100 U/mL streptomycin.
Cell Activity Detection
CCK-8 assays were carried out to detect the effect of RPL on A549 cell and HUVEC cell activity. Briefly, the cells were seeded into 96-well plate (1×104/well) and cultured for 12h at 37°C. After that, cells were treated with different does of RPL for 24h in a 5% CO2 incubator at 37°C, 10 µL CCK-8 was added to each well and incubated for 1h. The optical density (OD) values were measured at 450 nm using a microtablet reader (Bio-RAD, USA). Each experiment was repeated three times individually.
Plate Clone Formation Assay
Cells were seeded in 6 well plates with 1000 cells per well for 24 h of drug intervention (RPL 0.2, 0.4, and 0.6 mg/mL). Continue incubation for 14 days, then wash twice with PBS, and stained with Crystal Violet Staining (Sangon Biotech, China). The colony formation was observed under a microscope, and five fields were randomly selected for counting. Each experiment was repeated three times.
DAPI Staining
DAPI staining was used to determine whether the activity inhibition of RPL on A549 cells was related to apoptosis induction in accordance with previous reported methods. The cells were incubated with different concentrations of RPL (0.2, 0.4, and 0.6 mg/mL) for 24 h and fixed with 4% paraformaldehyde at room temperature for 15 min. The cells were then stained with DAPI solution at room temperature for 10 min and washed in PBS, and the image data were observed and recorded under an Olympus IX71 inverted fluorescence microscopy (Olympus, Tokyo, Japan). Each experiment was repeated three times.
Apoptosis Assay by Flow Cytometer
Cell apoptosis was detected by flow cytometry (CytoFLEX FCM, Beckman Coulter Inc., Atlanta, Georgia, USA). The A549 cells were seeded on six-well plates for 12 h and treated with different concentrations of RPL (0.2, 0.4, and 0.6 mg/mL) for 24 h. After that, the cells were collected, washed, and centrifuged. According to the manufacturer’s instructions of Annexin V/PI apoptosis kit, the cells were resuscitated with 500 μL of binding buffer, mixed with 5μL of Annexin V-FITC and PI, and incubated at room temperature in dark for 15 min. The cell apoptosis was detected by flow cytometry analysis. Each experiment was repeated three times.
Statistical Analysis
Results were expressed in terms of mean and standard deviation (SD), and all statistical comparisons were evaluated using one-way analysis of variance (ANOVA) and significant differences between the mean values were measured using the Duncan’s multiple-range tests.
P < 0.01 means the difference is statistically significant.
Results
Screening of Active Components
A total of 1380 compounds in RPL were found through database and literature searching. According to the screening results of OB and DL, the OB and DL values of 16 compounds were considered to be “Qualified”, indicating that these 16 compounds had good proprietary properties and were potential active ingredients of RPL in the treatment of lung cancer. Furthermore, some important monmers with low properties of OB and OL in RPL which were reported to possess anti-lung cancer tumors were also included in this study, such as the emodin, resveratrol, chrysophanol, and physcion. At the end of, we obtained 20 potential active ingredients in the RPL, including one terpenoids (Mutatochrome), two steroids (Daucosterol, β-sitosterol), four flavonoids (Eupatin, Catechin, Procyanidin B-5,3’-O-gallate, gallic acid-3-O-(6’-O-galloyl)-glucoside), 13 anthraquinones (Toralactone, Torachrysone-8-O-β-D-(6’-oxayl)-glucoside, Sennoside E, Sennoside D, Rhein, Physciondiglucoside, Palmidin A, Emodin-1-O-β-D-glucopyranoside, Aloe-emodin, emodin, resveratrol, chrysophanol, and physcion), and the detail information of these compounds is listed in Table 1 .
Table 1.
The main molecular descriptors for the potential active components.
| ID | Compound name | Molecule ID | PubChem CID | MW | AlogP | OB | DL |
|---|---|---|---|---|---|---|---|
| C1 | Toralactone | MOL002281 | 5321980 | 272.27 | 2.25 | 46.46 | 0.24 |
| C2 | Torachrysone-8-O-β-D-(6’-oxayl)-glucoside | MOL002280 | 5321979 | 480.46 | 0.64 | 43.02 | 0.74 |
| C3 | Sennoside E | MOL002276 | NA | 524.5 | 3.91 | 50.69 | 0.61 |
| C4 | Sennoside D | MOL002293 | NA | 524.5 | 3.91 | 61.06 | 0.61 |
| C5 | Rhein | MOL002268 | 10168 | 284.23 | 1.88 | 47.07 | 0.28 |
| C6 | Procyanidin B-5,3’-O-gallate | MOL002260 | NA | 730.67 | 4.6 | 31.99 | 0.32 |
| C7 | Physciondiglucoside | MOL002259 | 442762 | 608.6 | -0.91 | 41.65 | 0.63 |
| C8 | palmidin A | MOL002303 | 5320384 | 510.52 | 4.52 | 32.45 | 0.65 |
| C9 | Mutatochrome | MOL002251 | 5281246 | 552.96 | 10.9 | 48.64 | 0.61 |
| C10 | gallic acid-3-O-(6’-O-galloyl)-glucoside | MOL000554 | 11972353 | 484.4 | -0.03 | 30.25 | 0.67 |
| C11 | Eupatin | MOL002235 | 5317287 | 360.34 | 1.99 | 50.8 | 0.41 |
| C12 | Emodin-1-O-β-D-glucopyranoside | MOL002288 | 11968447 | 432.41 | 0.59 | 44.81 | 0.8 |
| C13 | Daucosterol | MOL002297 | NA | 386.73 | 7.67 | 35.89 | 0.7 |
| C14 | β-sitosterol | MOL000359 | 12303645 | 414.79 | 8.08 | 36.91 | 0.75 |
| C15 | Aloe-emodin | MOL000471 | 10207 | 270.25 | 1.67 | 83.38 | 0.24 |
| C16 | (-)-catechin | MOL000096 | 73160 | 290.29 | 1.92 | 49.68 | 0.24 |
| C17 | Emodin | MOL000472 | 3220 | 270.25 | 2.49 | 24.40 | 0.24 |
| C18 | Resveratrol | MOL012744 | 445154 | 228.26 | 3.01 | 19.07 | 0.11 |
| C19 | Chrysophanol | MOL001729 | 10208 | 254.25 | 2.76 | 18.64 | 0.21 |
| C20 | Physcion | MOL000476 | 10639 | 284.28 | 2,74 | 22.29 | 0.27 |
Screening of Potential Targets
A total of 22,418 targets related to lung cancer were obtained from OMIM, DisGeNET, TTD, and GeneCards database, and a total of 817 potential targets of 20 active compounds of RPL were collected. As shown in the Venn diagram in Figure 1 , a total of 761 potential anti-lung cancer targets were obtained through a combined collection of common targets. Part of the target information is shown in Table 2 . Interestingly, most of the targets of the 20 active compounds in RPL are contained in these 761 targets.
Figure 1.

Venn diagram of the potential anti-lung cancer targets.
Table 2.
Degree of core regulatory genes analyzed by Cytoscape.
| Name | Symbol | Degree | Average Shortest PathLength | Betweenness Centrality | Closeness Centrality | Neighborhood Connectivity | |
|---|---|---|---|---|---|---|---|
| Insulin | INS | 291 | 1.64760638 | 0.06648305 | 0.60694108 | 60.62199313 | |
| RAC-alpha serine/threonine-protein kinase | AKT1 | 275 | 1.66888298 | 0.04719193 | 0.59920319 | 65.13454545 | |
| Cellular tumor antigen p53 | TP53 | 269 | 1.67952128 | 0.05452844 | 0.59540776 | 64.11152416 | |
| Serum albumin | ALB | 258 | 1.70611702 | 0.05336222 | 0.58612627 | 62.71705426 | |
| Interleukin-6 | IL6 | 229 | 1.76329787 | 0.02780752 | 0.56711916 | 66.6419214 | |
| Epidermal growth factor receptor | EGFR | 223 | 1.75664894 | 0.02834682 | 0.56926571 | 70.00896861 | |
| Vascular endothelial growth factor A | VEGFA | 216 | 1.7712766 | 0.01954464 | 0.56456456 | 71.61574074 | |
| Myc proto-oncogene protein | MYC | 207 | 1.76994681 | 0.02037922 | 0.56498873 | 73.23671498 | |
| Proto-oncogene tyrosine-protein kinase Src | SRC | 203 | 1.80585106 | 0.02519928 | 0.55375552 | 71.49261084 | |
| Tumor necrosis factor | TNF | 201 | 1.80452128 | 0.02042326 | 0.5541636 | 69.76119403 | |
| Caspase-3 | CASP3 | 197 | 1.80053191 | 0.01560464 | 0.55539143 | 75.9035533 | |
| Heat shock protein HSP 90-alpha | HSP90AA1 | 192 | 1.79920213 | 0.02821964 | 0.55580192 | 70.609375 | |
| Signal transducer and activator of transcription 3 | STAT3 | 175 | 1.84973404 | 0.01091074 | 0.54061826 | 78.70857143 | |
| Estrogen receptor | ESR1 | 174 | 1.83909574 | 0.02209485 | 0.54374548 | 76.94252874 | |
| Mitogen-activated protein kinase 8 | MAPK8 | 172 | 1.84308511 | 0.00999066 | 0.54256854 | 79.03488372 | |
| Catenin beta-1 | CTNNB1 | 162 | 1.86835106 | 0.01984121 | 0.53523132 | 74.83950617 | |
| Serine/threonine-protein kinase mTOR | MTOR | 159 | 1.86303191 | 0.00875725 | 0.53675946 | 82.40880503 | |
| G1/S-specific cyclin-D1 | CCND1 | 155 | 1.87898936 | 0.00860106 | 0.53220099 | 83.10322581 | |
| Receptor tyrosine-protein kinase erbB-2 | ERBB2 | 154 | 1.86702128 | 0.00959932 | 0.53561254 | 79.12987013 | |
| Amyloid beta A4 precursor protein-binding family A member 3 | APP | 144 | 1.91888298 | 0.02165668 | 0.52113652 | 66.77777778 | |
PPI and Hub Genes of RPL Against Lung Cancer
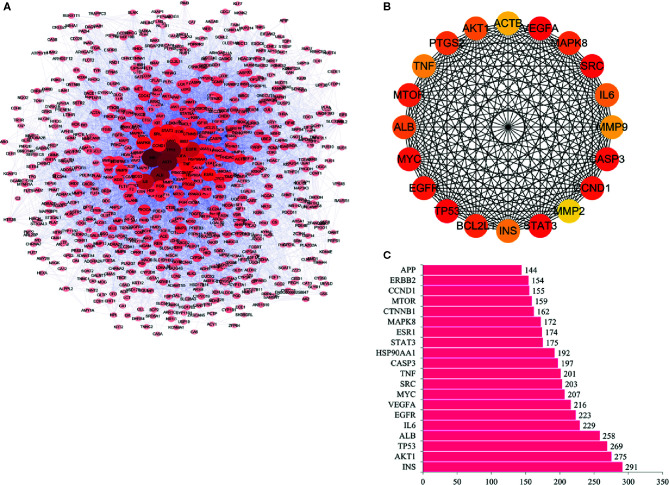
According to the predictions results of the String, and with the help of cytoscape software, the interaction between proteins was visualized, including 761 nodes and 6840 edge ( Figure 2A ). The top 20 hub genes were screened out according to the degree of nodes, including INS, AKT1, TP53, ALB, IL6, EGFR, VEGFA, MYC, SRC, TNF, CASP3, HSP90AA1, STAT3, ESR1, MAPK8, CTNNB1, MTOR, CCND1, ERBB2, and APP. The interaction between these genes is shown in Figure 2B . Among these genes, INS, AKT1,TP53 and ALB have the highest node degrees, which are 291, 275, 269, and 258, respectively ( Figure 2C and Table 2 ). It is suggested that INS, AKT1, TP53, and ALB may be three key targets for anti-cancer activity of RPL for lung cancer.
Figure 2.
PPI and hub genes of RPL against lung cancer. (A) Interaction between proteins. (B) Interaction between these genes. (C) Genes with highest node degrees.
Network Analysis of Targets
A total of data pairs of active compounds and disease target genes were prepared by R software, and the DDN constructed is shown in Figure 3 . This network contains 783 nodes (761 genes, 20 chemicals, 1 drug, and 1 disease) and 2047 edges. In this network, the yellow and red hexagons represent drug and disease, the magenta circle is the protein target, and the green red diamond is the compound. The degree of a node is the number of edges connected to the node, and the higher the degree, the more nodes in the network are directly related to the node, indicating that the node is more important in the network. According to our results, C18, C17, C6, C2, C10, C9, C20, C19, and C16 are linked to more than 100 genes, which are considered to be the main active components for RPL against lung cancer ( Table 3 ).
Figure 3.
Network analysis of targets. A total of data pairs of active compounds and disease target genes were prepared by R software, and the DDN was constructed.
Table 3.
Degree of 16 active components analyzed by Cytoscape.
| Name | Degree | AverageShortestPathLength | BetweennessCentrality | ClosenessCentrality | NeighborhoodConnectivity |
|---|---|---|---|---|---|
| C18 | 178 | 2.5390525 | 0.02319971 | 0.39384771 | 4.02352941 |
| C17 | 117 | 2.68245839 | 0.00987261 | 0.37279236 | 4.5 |
| C6 | 117 | 2.67989757 | 0.00976497 | 0.37314859 | 5.47826087 |
| C2 | 114 | 2.60947503 | 0.00911454 | 0.38321884 | 4.9380531 |
| C10 | 111 | 2.69270166 | 0.00824008 | 0.37137423 | 4.6 |
| C9 | 110 | 2.69270166 | 0.01003758 | 0.37137423 | 4.10909091 |
| C20 | 109 | 2.6978233 | 0.00836353 | 0.3706692 | 4.2962963 |
| C19 | 107 | 2.70038412 | 0.00822794 | 0.37031769 | 4.44859813 |
| C16 | 100 | 2.7234315 | 0.00714986 | 0.36718383 | 5.28571429 |
| C14 | 84 | 2.75928297 | 0.00520247 | 0.36241299 | 4.85714286 |
| C5 | 82 | 2.76696543 | 0.00482567 | 0.36140676 | 5.20987654 |
| C15 | 79 | 2.77464789 | 0.00443887 | 0.36040609 | 5.1025641 |
| C13 | 79 | 2.77208707 | 0.00428062 | 0.36073903 | 5.20253165 |
| C3 | 71 | 2.79257362 | 0.00360904 | 0.35809262 | 5.92957746 |
| C7 | 68 | 2.66709347 | 0.00362509 | 0.37493999 | 6.86764706 |
| C8 | 66 | 2.80537772 | 0.00272869 | 0.35645824 | 5.90909091 |
| C1 | 65 | 2.80793854 | 0.00300304 | 0.35613315 | 5.58461538 |
| C4 | 63 | 2.81306018 | 0.00258953 | 0.35548475 | 6.3968254 |
| C11 | 59 | 2.82330346 | 0.00263375 | 0.35419501 | 5.06779661 |
| C12 | 9 | 2.95134443 | 0.00002801 | 0.33882863 | 10.66666667 |
GO Functional Enrichment Analysis
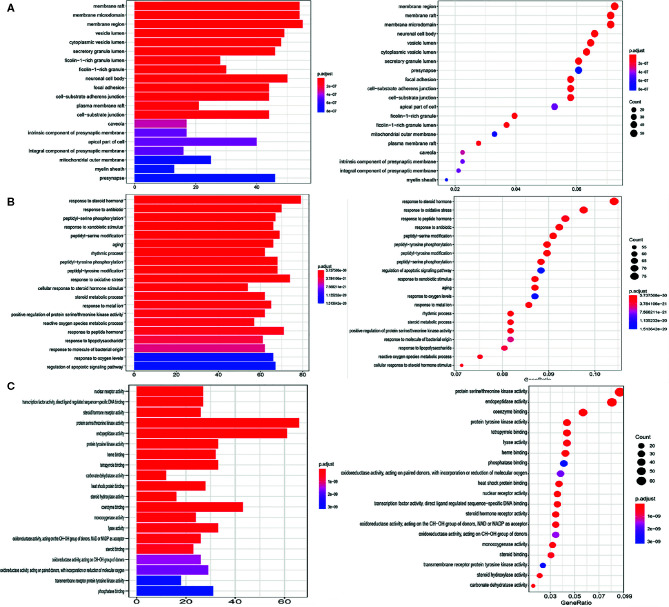
Three types of GO functional annotation analyses were performed for these potential target genes, and the cellular component (CC), biological process (BP) and molecular function (MF) were included. The analysis results as shown in Figure 4 , the images show only the top 20 related functions. The enriched GO functions for target genes included the membrane raft, membrane microdomain, membrane region, vesicle lumen and cytoplasmic vesicle lumen in the CC category ( Figure 4A ); response to steroid hormone, response to antibiotic, response to oxidative stress, reactive oxygen species metabolic process and regulation of apoptotic signaling pathway in the BP category ( Figure 4B ); kinase activity, endopeptidase activity, protein tyrosine kinase activity and heme binding in the MF category ( Figure 4C ).
Figure 4.
GO functional enrichment analysis. (A) Cellular component category. (B) Biological process category. (C) Molecular function category.
KEGG Pathway Enrichment Analysis
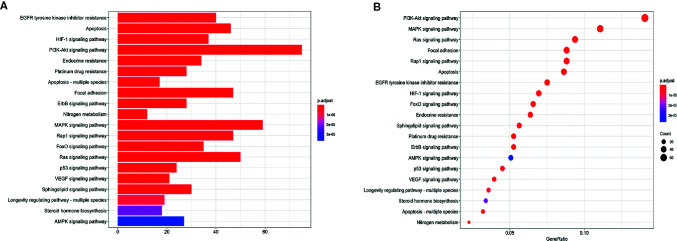
The enrichment analysis of the KEGG pathway involved in these target genes was carried out by R language software. The results showed that a total of 533 genes were involved in the enrichment, and 46 pathways significantly correlated with target genes were finally obtained (P ≤ 0.05). The top 20 pathways are shown in Figure 5 and Table 4 , mainly including EGFR tyrosine kinase inhibitor resistance, Apoptosis, PI3K-Akt signaling pathway, Apoptosis-multiple species, MAPK signaling pathway, and p53 signaling pathway. Through extensive literature search, these signaling pathways are directly or indirectly related to the occurrence and development of lung cancer, suggesting that these signaling pathways may be closely related to the mechanism of anticancer effect of RPL in lung cancer.
Figure 5.
(A) Histogram of KEGG enrichment results; (B) Bubble diagram of KEGG enrichment results.
Table 4.
KEGG pathway enrichment analysis.
| ID | Description | P value | P.adjust | Q value | Count |
|---|---|---|---|---|---|
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 5.14E-22 | 8.28E-20 | 4.87E-20 | 40 |
| hsa04210 | Apoptosis | 1.89E-16 | 1.52E-14 | 8.95E-15 | 46 |
| hsa04066 | HIF-1 signaling pathway | 1.68E-13 | 8.77E-12 | 5.16E-12 | 37 |
| hsa04151 | PI3K-Akt signaling pathway | 2.18E-13 | 8.77E-12 | 5.16E-12 | 75 |
| hsa01522 | Endocrine resistance | 8.16E-13 | 2.63E-11 | 1.55E-11 | 34 |
| hsa01524 | Platinum drug resistance | 5.45E-12 | 1.46E-10 | 8.60E-11 | 28 |
| hsa04215 | Apoptosis - multiple species | 1.78E-10 | 4.09E-09 | 2.40E-09 | 17 |
| hsa04510 | Focal adhesion | 2.11E-10 | 4.24E-09 | 2.50E-09 | 47 |
| hsa04012 | ErbB signaling pathway | 3.62E-10 | 6.48E-09 | 3.82E-09 | 28 |
| hsa00910 | Nitrogen metabolism | 9.18E-10 | 1.48E-08 | 8.69E-09 | 12 |
| hsa04010 | MAPK signaling pathway | 1.16E-09 | 1.70E-08 | 1.00E-08 | 59 |
| hsa04015 | Rap1 signaling pathway | 1.45E-09 | 1.81E-08 | 1.07E-08 | 47 |
| hsa04068 | FoxO signaling pathway | 1.46E-09 | 1.81E-08 | 1.07E-08 | 35 |
| hsa04014 | Ras signaling pathway | 1.69E-09 | 1.94E-08 | 1.14E-08 | 50 |
| hsa04115 | p53 signaling pathway | 5.16E-09 | 5.54E-08 | 3.26E-08 | 24 |
| hsa04370 | VEGF signaling pathway | 1.26E-08 | 1.27E-07 | 7.46E-08 | 21 |
| hsa04071 | Sphingolipid signaling pathway | 9.42E-08 | 8.92E-07 | 5.25E-07 | 30 |
| hsa04213 | Longevity regulating pathway - multiple species | 9.21E-07 | 8.24E-06 | 4.85E-06 | 19 |
| hsa00140 | Steroid hormone biosynthesis | 2.52E-06 | 2.14E-05 | 1.26E-05 | 18 |
| hsa04152 | AMPK signaling pathway | 4.54E-06 | 3.66E-05 | 2.15E-05 | 27 |
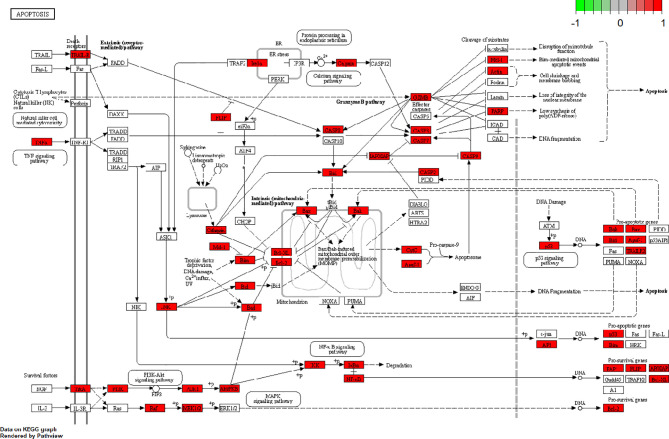
More and more evidence shows that the abnormal regulation of apoptosis gene plays an important role in the occurrence and development of cancer. Therefore, it is generally accepted that inducing apoptosis of tumor cells is a feasible and most direct treatment for cancer. Interestingly, consistent with the results of the previous GO functional annotation, the KEGG pathway also enriched the apoptotic pathways, suggesting that RPL may play an anti-lung cancer role by inducing apoptosis in lung cancer cells ( Figure 6 ). Based on the results of these predicted molecular mechanisms and network analyses, we designed experiments to test our hypothesis at the cellular level.
Figure 6.
The potential apoptotic pathway for anticancer effects of RPL against lung cancer.
RPL Inhibits the Proliferation of A549 Cell
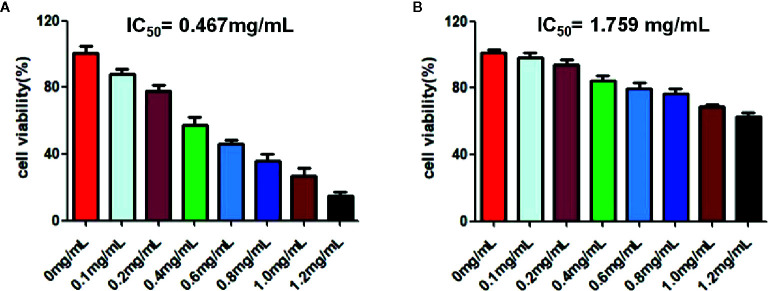
CCK-8 was used to investigate the effect of RPL on the activity of A549 cells. As can be seen from Figure 7A , RPL at series concentrations (0–1.2 mg/mL) showed the ability to inhibit the proliferation of A549 cells with an obvious concentration-dependent manner. More importantly, RPL did not significantly inhibit HUVEC cells, suggesting that it did not seem to have a significant effect on the viability of normal cells ( Figure 7B ).
Figure 7.
Cytotoxic effects of RPL on A549 cells (A) and HUVEC cells (B). Cells were treated with RPL (0, 0.75, 1.5, 3.0, 6.0, 12, and 24 µg/mL) for 24 h. Finally, CCK-8 assays were carried out to detect the cytotoxic effects of candidate drugs. The IC50 values were calculated for evauation of the cytotoxic effects of candidate drugs.
Cloning Formation Assay
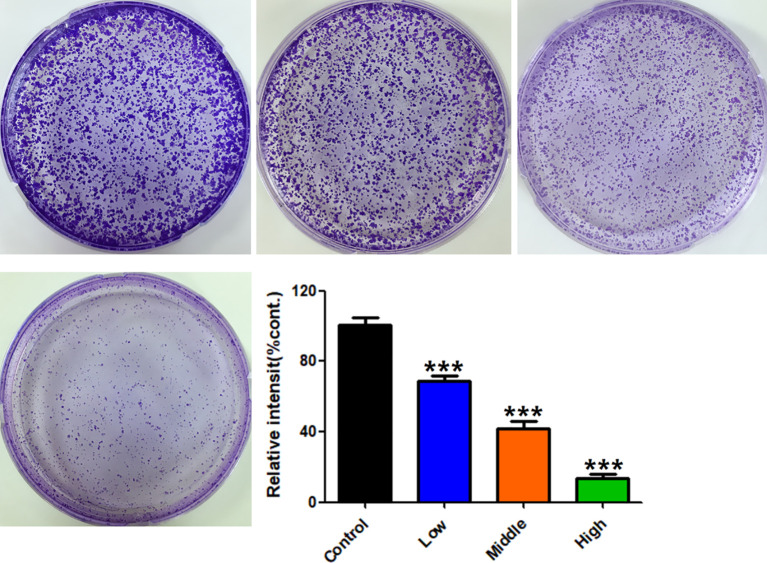
Consistent with CCK-8 results, RPL significantly inhibited the cloning formation of A549 cells compared with the control cells ( Figure 8 ), and the inhibition was concentration-dependent (0.2, 0.4, and 0.6 mg/mL). Taken together, these results suggest that RPL could inhibit the proliferation of A549 cells.
Figure 8.
Effects of RPL on cloning formation of A549 cells. Cells were treated with RPL (0.2, 0.4, and 0.6 mg/mL) for 14 days and then stained with crystal violet. The colony formation was observed under a microscope, and five fields were randomly selected for counting. Data were expressed as mean ± SD (n = 3), and asterisk indicated significant difference, ***p < 0.001 vs. control.
RPL Induces Aapoptosis in A549 Cells
Previous results suggested that RPL had a significant anti-proliferation effect on A549 cells. To determine whether this anti-proliferation effect was related to apoptosis, DAPI staining and flow cytometry were used to detect the apoptosis-inducing effect of RPL on A549 cells. As shown in Figure 9A , the experimental results showed that weak fluorescence could be observed in the control A549 cells (intact nucleus) after DAPI staining. On the contrary, A549 cells treated with RPL (0.2, 0.4, and 0.6 mg/mL) showed significantly increased fluorescence brightness, suggesting that the nucleus underwent morphological changes of apoptosis, such as nuclear condensation and nuclear shrinkage. Subsequent flow cytometric analysis further confirmed the pro-apoptotic effect of RPL. As can be seen from the Figure 9B , with the increase of RPL concentration ranging from 0.2 to 0.6 mg/mL, the apoptosis rate of A549 cells increased gradually (P < 0.01, P < 0.01, P < 0.01, respectively, vs. untreated A549 cells). These results suggest that RPL can induce apoptosis of human lung cancer cells.
Figure 9.
Apoptotic induction acitivities of RPL on A549 cells. Cells were treated with RPL (0.2, 0.4, and 0.6 mg/mL) for 24 h, and the apoptotic effects of RPL were detected by (A) DAPI staining assay with a fluorescence microscope and (B) flow cytometric analysis with Annexin V-FITC and PI staining. ***p < 0.001 vs. control.
Monmers in RPL Possess Cytotoxic Effects on A549 Cells via Induction of Apoptosis
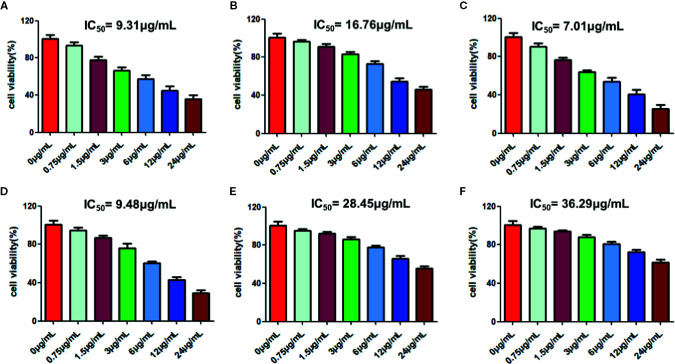
Furthermore, we studied the cototoxic effects of some avialiable monmers in RPL on the lung cancer cell line of A549, and our present results showed that these monmers in RPL including emodin, resveratrol, aloe-emodin, rhein, chrysophanol, and physcion show significant inhibitory effects on the proliferation of A549 cells with the IC50 values below 40μg/mL (9.31, 16.76, 7.01, 9.48, 28.45, and 36.29 μg/mL for emodin, resveratrol, aloe-emodin, rhein, chrysophanol, and physcion, respectively) ( Figure 10 ).
Figure 10.
Cytotoxic effects of emodin (A), resveratrol (B), aloe-emodin (C), rhein (D), chrysophanol (E), and physcion (F) on A549 cells. Cells were treated with the candidate drugs (0, 0.75, 1.5, 3.0, 6.0, 12, and 24 µg/mL) for 24 h. Finally, CCK-8 assays were carried out to detect the cytotoxic effects of candidate drugs. Each experiment was repeated three times individually. The IC50 values were calculated for evauation of the cytotoxic effects of candidate drugs.
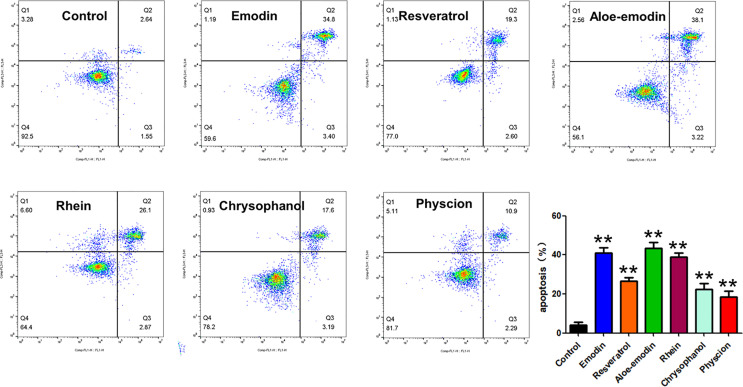
Then, we also investigated whether the anti-proliferative effects of these monmers are related to indcution of apoptosis or not. Interesting, our results suggested that all the testing monmers from RPL showed obvious pro-apoptic potentials on A549 cells ( Figure 11 ).
Figure 11.
Apoptotic induction activities of emodin, resveratrol, aloe-emodin, rhein, chrysophanol, and physcion on A549 cells. Cells were treated with candidate drugs (6 µg/mL) for 24 h, and the apoptotic cells were detected by staining with Annexin V-FITC/PI followed by flow cytometry analysis. Data were expressed as mean ± SD (n = 3), and asterisk indicated significant difference, **p < 0.01 vs. control.
Discussion
It’s no doubt that TCMs can be used to prevent or treat various difficult miscellaneous diseases (Newman and Cragg, 2016; Long et al., 2020), and also promising resources for discovering more candidate drugs for treating tumors. Due to the TCMs are characterized as “multi-components”, “multi-targets,” and “multi-approaches,” it would take a lot of manpower, material resources, and time to study the effects and mechanisms of herbal medicines by routine pharmacological experiments, which is also the big gap for wide acceptance and use of herbal medicine in clinic.
Network pharmacology enables rapid screening of drugs and targets, as well as analysis of pathways of action, greatly reducing the time and cost of drug development. In addition, network pharmacology can systematically analyze the interaction between drugs and diseases, which is basically consistent with the overall concept of TCMs. Meanwhile, it also provides new ideas and technical means to study the material basis and mechanism of action of TCMs. In this study, network pharmacology was used to study the potential active compounds and possible targets of RPL in the treatment of lung cancer, combined with in vitro experiments for verification, to scientifically and systematically clarify the material basis and mechanism of RPL in the treatment of lung cancer.
RPL is a famous TCM which has been used for thousands of years with the functions of “attacking stagnation, clearing damp and heat, purging fire, cooling blood, removing stasis and detoxifying” and is currently used for treating various diseases, such as high fever, constipation, abdominal pain, and tumor, etc (Aichner and Ganzera, 2015; Liu et al., 2016). In our present results, 20 potential compounds from RPL were screened, and interestingly these 20 compounds have appeared in the drug ingredients and disease targets (DDN), indicating that anti-tumor effects of RPL might be closely related to these 20 compounds mentioned above, including one terpenoids, two steroids, six flavonoids, and seven anthraquinones. According to the degree of each compound in DDN, the compounds numbered C2 (Torachrysone-8-O-β-D-(6’-oxayl)-glucoside), C6 (Procyanidin B-5,3’-O-gallate), C9 (Mutatochrome), C10 (gallic acid-3-O-(6’-O-galloyl)-glucoside), C15 (Aloe-emodin), C17 (Emodin), C18 (Resveratrol), C19 (Chrysophanol), and C20 (Physcion) are considered as the main active components of RPL for anti-lung cancer. Some of these mentioned compounds, such as aloe-emodin, emodin, and resveratrol have been reported to can be used for treating lung cancer (Peng et al., 2013; Dong et al., 2016; Dong et al., 2020). In addition, Mao et al. reported that Procyanidins can play an anti-lung cancer activity by inhibiting inflammation and inducing tumor cell apoptosis, and the mechanism of action is related to the inhibition of cyclooxygenase-2 (COX-2)/prostaglandin E2 (PGE2) eicosan-like pathway and the initiation of caspase-3 apoptosis pathway, suggesting that Procyanidins can be used as a potential preventive and therapeutic drug against lung cancer, which has been also supported by a recent clinical study (Mao et al., 2016; Mao et al., 2019).
Furthermore, our results also suggest that INS, AKT1, EGFR and TP53 are the key targets of anticancer activities of active compounds in RPL. INS, which encoded insulin in the body, plays a crucial role in regulating carbohydrate and lipid metabolism. Growing reports have suggested that insulin may increase the risk of certain cancers, such as prostate, rectal, breast and pancreatic cancers (Hsing et al., 2001; Goodwin et al., 2002; Stolzenberg-Solomon et al., 2005; Limburg et al., 2006; Albanes et al., 2009). A case cohort study in 2017 found that higher insulin levels and insulin resistance may increase the risk of lung cancer (Argirion et al., 2017). In addition, Hayakawa et al. found that patients with non-small cell lung cancer (NSCLC) would further develop resistance to the available treatments by activating insulin-like growth factor-1 receptor (IGF1R) (Hayakawa et al., 2020). In addition, it’s reported that INS plays an important role in the proliferation and metastasis of NSCLC cells (Li S. et al., 2019). AKT is an important kinase in the PI3k-AKT pathway, which is catalyzed by PI3K to migrate to the cell membrane and activate (Zhang et al., 2019). Activated Akt regulates the proliferation and growth of cells and is also involved in the apoptosis and glucose metabolism. In addition, insulin also activates the Akt signaling pathway to conservatively execute the insulin metabolic response in tumor cells, driving the metabolism of tumor cells. Thus, AKT has become an attractive intervention target for the development of antitumor drugs. Alpha subunit protein kinase B (AKT1) is an important member of the AKT family, and current studies have confirmed that AKT1, as an important regulatory factor, is in a state of disorder in various tumors, such as prostate cancer, osteosarcoma, ovarian cancer, and endometrial cancer, leading to uncontrolled proliferation of tumor cells, apoptosis defects, and enhanced metastasis and invasion ability of tumor (Alwhaibi et al., 2019; Huo et al., 2019; Wang J. et al., 2019). Rao et al. found that AKT1 is a key metastasis regulator in NSCLC cells, and in vitro inhibition of AKT1 can promote invasion and metastasis of NSCLC cells with K-RAS or EGFR mutations (Rao et al., 2017). It has been reported that long non-coding RNAs can inhibit the proliferation and invasion of NSCLC cells by targeting the AKT1 signaling pathway (Zhang et al., 2018). Another study also found that MicroRNAs can inhibit cell proliferation, migration and invasion of lung cancer, regulate cell cycle distribution and epithelial-mesenchymal transition (EMT), the mechanism of which is related to down-regulation of AKT1 expression (Wu et al., 2019). Epidermal growth factor receptor (EGFR) is a glycoprotein that plays an important role in physiological processes such as proliferation, apoptosis and differentiation of cell by activating the calcium signaling pathway, MAPK signaling pathway and PI3K/AKT signaling pathway (Carlisle et al., 2007). As an important anticancer gene, the inactivation of p53 plays an important role in tumor formation, and increasing studies have found that the p53 plays a crucial role in retarding tumor cell cycle, promoting tumor cell apoptosis and inhibiting tumor angiogenesis (Bi et al., 2018; Li P. et al., 2019; Wang et al., 2020).
For analysis of the GO function annotation of the target genes, it revealed that the related target cell components for anticancer activity of this herbal medicine include nucleus cytoplasm, exosomes, cytosol, lysosome, and extracellular region; its biological processes are involved in metabolism, energy pathways, apoptosis, neurotransmitter metabolism, and protein metabolism and others; and the related molecular functions are associated with catalytic activity, protein serine/threonine kinase activity, ligand-dependent nuclear receptor activity, cysteine-type peptidase activity, and carboxy-lyase activity. What’s more, KEGG pathway analysis revealed these genes were significantly enriched in a variety of cancer pathways, nitrogen metabolism, neurotrophin signaling pathway, EGFR tyrosine kinase inhibitor resistance, platinum drug resistance, and apoptosis pathways.
Collectively, the GO and KEGG pathway analysis suggested that induction of apoptosis appeared to be a potential way for the anticancer activities of RPL against lung cancer. Abnormal proliferation and insufficient apoptosis in tumor cells are the main causes of lung cancer. Therefore, inhibiting the proliferation and inducing apoptosis of lung cancer has become a feasible strategy for the treatment of lung cancer. A large number of studies have also found that mechanisms of many natural anticancer drugs against lung cancer are related to the regulation of different signaling pathways to inhibit the proliferation of lung cancer cells and induce the apoptosis of lung cancer cells (Wang S. et al., 2019; Han et al., 2020; Hu et al., 2020). Therefore, we subsequently designed corresponding in vitro experiments in order to prove the prediction results of network pharmacology. CCK-8 results showed that RPL could inhibit the proliferation of A549 cells with a concentration-dependent manner. Furthermore, we carried out the CCK-8 assays on a normal human cell line of HUVECs to evaluate the toxicological effect RPL. Importantly, RPL does not appear to inhibit the growth of normal HUVEC cells. Furthermore, RPL treatment can also reduce the clone formation rate of A549 cells, further proving that this TCM can inhibit the proliferation ability of A549 cells. The results of DAPI staining and flow cytometry results showed that the anti-proliferation effect of RPL on lung cancer cells was related to the induction of apoptosis. Furthermore, we also determined the apoptotic effects of some characteristic constituents in RPL against lung cancer cells of A549, including emodin, resveratrol, aloe-emodin, rhein, chrysophanol, and physcion. From the results of CCK-8 and flow cytometry assays, it’s showed that all of the 6 monomers exhibited the IC50 values below 50μg/mL, and could induce significant apoptosis in A549 cells. All these experimental results confirmed the prediction results of network pharmacology.
Based on the experimental evidences of previous works and our studies, it’s suggested that RPL can play an anticancer role by inducing apoptosis of lung cancers. However, some works are also needed to be devoted in the further mechanisms of this plant for anti-lung cancers via induction of apoptosis. In addition, we also reported some active constituents with anti-lung cancer properties in this plant, however the in vivo experiments for confirming their activities are also necessary. Besides, from the results of our study, it’s also suggested that the application of network pharmacology to the study of TCM is a scientific and feasible method.
Conclusion
This study is the first to apply network pharmacology method to study the material basis and action mechanism of RPL against lung cancer. The results showed that 16 active ingredients in RPL had potential anti-lung cancer activity, involving 563 target genes related to lung cancer. Our results showed that C2 (Torachrysone-8-O-β-D-(6’-oxayl)-glucoside), C6 (Procyanidin B-5,3’-O-gallate), C9 (Mutatochrome), C10 (gallic acid-3-O-(6’-O-galloyl)-glucoside), C15 (Aloe-emodin), C17 (Emodin), C18 (Resveratrol), C19 (Chrysophanol), and C20 (Physcion) are the main active components of RPL in the treatment of lung cancer. INS, AKT1, EGFR, and TP53 are the hub genes of RPL in the treatment of lung cancer. The mechanism of RPL against lung cancer is related to 134 signaling pathways, and the key mechanism of RPL against lung cancer may be related to inducing lung cancer cell apoptosis. These network pharmacological predictions were verified in subsequent cell experiments. This study studied the pharmacodynamic basis and mechanism of RPL on lung cancer from the perspective of systematic pharmacology, providing a scientific basis for further elucidating the effect of rhubarb on lung cancer.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding authors.
Author Contributions
All authors contributed to the article and approved the submitted version. CW and WJ conceived and designed this paper. QZ, JL, RL, RZ, and DR completed the experiments. MZ and SW analyzed the data. QZ and JL wrote the original draft of the paper. CW reviewed and edited the paper.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Jiayi Sun (Innovative Institute of Chinese Medicine and Pharmacy, Chengdu University of Traditional Chinese Medicine, Chengdu, China) for his technological assistances in fluorescence microscope and Flow cytometry analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.528308/full#supplementary-material
Abbreviations
RPL, Rheum palmatum L.; TCM, traditional Chinese medicine; OB, Oral bioavailability; DL, Drug-Likeness; DDN, drug ingredients and disease targets; PPI, Protein-Protein Interaction Network; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; CC, cellular component; BP, biological process; MF, molecular function; FBS, Fetal bovine serum; PBS, phosphate buffered saline; DMEM, dulbecco modified eagle medium; DMSO, Dimethyl sulfoxide; DAPI, 4′,6-diamidino2- phenylindole; CCK8, Cell Counting Kit-8; HUVEC, human umbilical vein endothelial cells; OD, optical density; SD, standard deviation; ANOVA, one-way analysis of variance; COX-2, cyclooxygenase-2; PGE2, prostaglandin E2; NSCLC, non-small cell lung cancer; IGF1R, insulin-like growth factor-1 receptor; EMT, epithelial-mesenchymal transition; EGFR, Epidermal growth factor receptor.
References
- Aichner D., Ganzera M. (2015). Analysis of anthraquinones in rhubarb (Rheum palmatum and Rheum officinale) by supercritical fluid chromatography. Talanta 144, 1239–1244. 10.1016/j.talanta.2015.08.011 [DOI] [PubMed] [Google Scholar]
- Albanes D., Weinstein S. J., Wright M. E., Männistö S., Limburg P. J., Snyder K., et al. (2009). Serum insulin, glucose, indices of insulin resistance, and risk of prostate cancer. J. Natl. Cancer Inst. 101, 1272–1279. 10.1093/jnci/djp260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwhaibi A., Verma A., Artham S., Adil M. S., Somanath P. R. (2019). Nodal pathway activation due to Akt1 suppression is a molecular switch for prostate cancer cell epithelial-to-mesenchymal transition and metastasis. Biochem. Pharmacol. 168, 1–13. 10.1016/j.bcp.2019.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argirion I., Weinstein S. J., Männistö S., Mondul A. M. (2017). Serum insulin, glucose, indices of insulin resistance, and risk of lung cancer. Cancer Epidemiol. Biomarkers Prev. 10, 1519–1524. 10.1158/1055-9965.EPI-17-0293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Liu D., Li Y., Mao X., Wang A., Wang J. (2018). Oridonin induces growth inhibition and apoptosis in human gastric carcinoma cells by enhancement of p53 expression and function. Braz. J. Med. Biol. Res. 51, e7599. 10.1590/1414-431x20187599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Carlisle D. L., Liu X., Hopkins T. M., Swick M. C., Dhir R., Siegfried J. M. (2007). Nicotine activates cell-signaling pathways through muscle-type and neuronal nicotinic acetylcholine receptors in non-small cell lung cancer cells. Pulm. Pharmacol. Ther. 20, 629–641. 10.1016/j.pupt.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Chiu T. H., Lai W. W., Hsia T. C., Yang J. S., Lai T. Y., Wu P. P., et al. (2009). Aloe-emodin induces cell death through S-phase arrest and caspase-dependent pathways in human tongue squamous cancer SCC-4 cells. Anticancer Res. 29, 4503–4511. [PubMed] [Google Scholar]
- Dong X., Fu J., Yin X., Cao S., Li X., Lin L., et al. (2016). Emodin: a review of its pharmacology, toxicity and pharmacokinetics. Phytother. Res. 30, 1207–1218. 10.1002/ptr.5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Zeng Y., Liu Y., You L., Yin X., Fu J., et al. (2020). Aloe-emodin: a review of its pharmacology, toxicity, and pharmacokinetics. Phytother. Res. 34, 270–281. 10.1002/ptr.6532 [DOI] [PubMed] [Google Scholar]
- Goodwin P. J., Ennis M., Pritchard K. I., Trudeau M. E., Koo J., Madarnas Y., et al. (2002). Fasting insulin and outcome in early-stage breast cancer: results of a prospective cohort study. J. Clin. Oncol. 20, 42–51. 10.1200/JCO.2002.20.1.42 [DOI] [PubMed] [Google Scholar]
- Han B., Wu J., Huang L. (2020). Induction of apoptosis in lung cancer cells by viburnum grandiflorum via mitochondrial pathway. Med. Sci. Monit. 26, e920265. 10.12659/MSM.920265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa D., Takahashi F., Mitsuishi Y., Tajima K., Hidayat M., Winardi W., et al. (2020). Activation of insulin-like growth factor-1 receptor confers acquired resistance to osimertinib in non-small cell lung cancer with EGFR T790M mutation. Thorac. Cancer 1, 140–149. 10.1111/1759-7714.13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing A. W., Chua S., Gao Y. T., Gentzschein E., Chang L., Deng J. (2001). Prostate cancer risk and serum levels of insulin and leptin: a population-based study. J. Natl. Cancer Inst. 93, 783–789. 10.1093/jnci/93.10.783 [DOI] [PubMed] [Google Scholar]
- Hu Y., Liu P., Kang L., Li J., Li R., Liu T. (2020). Mechanism of Marsdenia tenacissima extract promoting apoptosis of lung cancer by regulating Ca2+/CaM/CaMK signaling. J. Ethnopharmacol. 8, 112535. 10.1016/j.jep.2019.112535 [DOI] [PubMed] [Google Scholar]
- Huo X., Sun H., Liu Q., Ma X., Peng P., Yu M. (2019). Clinical and expression significance of AKT1 by co-expression network analysis in endometrial cancer. Front. Oncol. 9, 1147. 10.3389/fonc.2019.01147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong B., Choi S. Y., Jang H. S., Yoo G., Kim S. H., Kim J. H. (2017). Optimization of extraction conditions for active compounds of herbal medicinal formula, DF, by response surface methodology. Nat. Prod. Sci. 23, 9–15. 10.20307/nps.2017.23.1.9 [DOI] [Google Scholar]
- Jiang Y., Zhong M., Long F., Yang R., Zhang Y., Liu T. (2019). Network pharmacology-based prediction of active ingredients and mechanisms of Lamiophlomis rotata (Benth.) Kudo against rheumatoid arthritis. Front. Pharmacol. 10, 1435. 10.3389/fphar.2019.01435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. W., Hwang B. S., Kim M. H., Park C. S., Lee W. S., Oh H. M., et al. (2012). Inhibition of LFA-1/ICAM-1-mediated cell adhesion by stilbene derivatives from Rheum undulatum . Arch. Pharm. Res. 35, 1763–1770. 10.1007/s12272-012-1008-8 [DOI] [PubMed] [Google Scholar]
- Li P., Lv X., Zhang Z., Xie S. (2019). S100A6/miR193a regulates the proliferation, invasion, migration and angiogenesis of lung cancercells through the P53 acetylation. Am. J. Transl. Res. 11, 4634–4649. [PMC free article] [PubMed] [Google Scholar]
- Li S., Niu X., Li H., Liang Y., Sun Z., Yan Y. (2019). Circ_0000003 promotes the proliferation and metastasis of non-small cell lung cancer cells via miR-338-3p/insulin receptor substrate 2. Cell Cycle 24, 3525–3539. 10.1080/15384101.2019.1690883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Lin Y., Zhang S., Zhou L., Yan G., Wang Y., et al. (2019). Emodin regulates neutrophil phenotypes to prevent hypercoagulation and lung carcinogenesis. J. Transl. Med. 17, 90. 10.1186/s12967-019-1838-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limburg P. J., Stolzenberg-Solomon R. Z., Vierkant R. A., Roberts K., Sellers T. A., Taylor P. R., et al. (2006). Insulin, glucose, insulin resistance, and incident colorectal cancer in male smokers. Clin. Gastroenterol. Hepatol. 4, 1514–1521. 10.1016/j.cgh.2006.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li L., Xiao Y. Q., Yao J. Q., Li P. Y., Yu D. R., et al. (2016). Global metabolite profiling and diagnostic ion filtering strategy by LC-QTOF MS for rapid identification of raw and processed pieces of Rheum palmatum L. Food Chem. 192, 531–540. 10.1016/j.foodchem.2015.07.013 [DOI] [PubMed] [Google Scholar]
- Long Y., Yang Q., Xiang Y., Zhang Y., Wan J., Liu S., et al. (2020). Nose to brain drug delivery - a promising strategy for active components from herbal medicine for treating cerebral ischemia reperfusion. Pharmacol. Res. 159, 104795. 10.1016/j.phrs.2020.104795 [DOI] [PubMed] [Google Scholar]
- Ma Y. S., Weng S. W., Lin M. W., Lu C. C., Chiang J. H., Yang J. S., et al. (2012). Antitumor effects of emodin on LS1034 human colon cancer cells in vitro and in vivo: roles of apoptotic cell death and LS1034 tumor xenografts model. Food Chem. Toxicol. 50, 1271–1278. 10.1016/j.fct.2012.01.033 [DOI] [PubMed] [Google Scholar]
- Mao J. T., Smoake J., Park H. K., Lu Q. Y., Xue B. (2016). Grape seed procyanidin extract mediates antineoplastic effects against lung cancer via modulations of prostacyclin and 15-HETE eicosanoid pathways. Cancer Prev. Res. (Phila). 12, 925–932. 10.1158/1940-6207.CAPR-16-0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. T., Lu Q. Y., Xue B., Neis P., Zamora F. D., Lundmark L., et al. (2019). A pilot study of a grape seed procyanidin extract for lung cancer chemoprevention. Cancer Prev. Res. (Phila). 8, 557–566. 10.1158/1940-6207.CAPR-19-0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M. (2016). Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661. 10.1021/acs.jnatprod.5b01055 [DOI] [PubMed] [Google Scholar]
- Nho K. J., Jin M. C., Lee A. Y., Kim H. K. (2015). Anti-metastatic effects of Rheum Palmatum L. extract in human MDA-MB-231 breast cancer cells. Environ. Toxicol. Pharmacol. 40, 30–38. 10.1016/j.etap.2015.05.006 [DOI] [PubMed] [Google Scholar]
- Park S., Kim Y. N., Kwak H. J., Jeong E. J., Kim S. H. (2018). Estrogenic activity of constituents from the rhizomes of Rheum undulatum Linné, Bioorg. Med. Chem. Lett. 28, 552–557. 10.1016/j.bmcl.2018.01.063 [DOI] [PubMed] [Google Scholar]
- Peng W., Qin R. X., Li X. L., Zhou H. (2013). Polygonum cuspidatum Sieb. et Zucc.: a review of its botany, phytochemistry, pharmacology, and potential applications. J. Ethnopharmacol. 2013 (148), 729–745. 10.1016/j.jep.2013.05.007 [DOI] [PubMed] [Google Scholar]
- Peng W., Wu J. G., Jiang Y. B., Liu Y. J., Sun T., Wu N., et al. (2015). Antitumor activity of 4-O-(2″-O-acetyl-6″-O-p-coumaroyl-β-D-glucopyranosyl)-p-coumaric acid against lung cancers via mitochondrial-mediated apoptosis. Chem. Biol. Interact. 233, 8–13. 10.1016/j.cbi.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Rao G., Pierobon M., Kim I. K., Hsu W. H., Deng J., Moon Y. W., et al. (2017). Inhibition of AKT1 signaling promotes invasion and metastasis of non-small cell lung cancer cells with K-RAS or EGFR mutations. Sci. Rep. 1, 7066. 10.1038/s41598-017-06128-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzenberg-Solomon R. Z., Graubard B., II, Chari S., Limburg P., Taylor P. R., Virtamo J., et al. (2005). Insulin, glucose, insulin resistance, and pancreatic cancer in male smokers. Jama 294, 2872–2878. 10.1001/jama.294.22.2872 [DOI] [PubMed] [Google Scholar]
- Su Y. T., Chang H. L., Shyue S. K., Hsu S. L. (2005). Emodin induces apoptosis in human lung adenocarcinoma cells through a reactive oxygen species-dependent mitochondrial signaling pathway. Biochem. Pharmacol. 70, 229–241. 10.1016/j.bcp.2005.04.026 [DOI] [PubMed] [Google Scholar]
- Syed S. R., Mohan M. J., Sandeep S., Christos S. (2016). Sub-lobar Resection Is Equivalent To Lobectomy for T1a Non-Small Cell Lung Cancer In The Elderly: A Surveillance, Epidemiology and End Results (SEER) Database Analysis. J. Surg. Res. 200, 683–689. 10.1016/j.jss.2015.08.045 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhu W., Chen X., Wei G., Jiang G., Zhang G. (2020). Selenium-binding protein 1 transcriptionally activates p21 expression via p53-independent mechanism and its frequent reduction associates with poor prognosis in bladder cancer. J. Transl. Med. 18, 17. 10.1186/s12967-020-02211-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ni J., Song D., Ding M., Huang J., Li W., et al. (2019). MAT1 facilitates the lung metastasis of osteosarcoma through upregulation of AKT1 expression. Life Sci. 234, 116771. 10.1016/j.lfs.2019.116771 [DOI] [PubMed] [Google Scholar]
- Wang S., Xu X., Hu Y., Lei T., Liu T. (2019). Sotetsuflavone induces autophagy in non-small cell lung cancer through blocking PI3K/Akt/mTOR signaling pathway in vivo and in vitro. Front. Pharmacol. 10, 1460. 10.3389/fphar.2019.01460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Liu H. Y., Liu W. J., Shi Y. L., Bao D. (2019). miR-377-5p inhibits lung cancer cell proliferation, invasion, and cell cycle progression by targeting AKT1 signaling. J. Cell Biochem. 10.1002/jcb.28091 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Yang Y., Xiong W., Yao Y., Wu H., Zhang M. (2019). Network pharmacology-based research on the active component and mechanism of the antihepatoma effect of rubia cordifolia L. J. Cell Biochem. 8, 12461–12472. 10.1002/jcb.28513 [DOI] [PubMed] [Google Scholar]
- Zhang L. L., Guo J. K. (2011). Effects of Rheum palmatum L. and its active components on A549 cells of human lung adenocarcinoma. Shanxi Tra Chine Med. 05, 51–53. 10.3969/j.issn.1000-7156.2011.05.031 [DOI] [Google Scholar]
- Zhang C., Su C., Song Q., Dong F., Yu S., Huo J. (2018). LncRNA PICART1 suppressed non-small cell lung cancer cells proliferation and invasion by targeting AKT1 signaling pathway. Am. J. Transl. Res. 12, 4193–4201. [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Liu J., Zhang M. M., Wei S. J., Li R. L., Gao Y. X., et al. (2019). Apoptosis induction of fibroblast-like synoviocytes is an important molecular mechanism for herbal medicine and its active components to treat rheumatoid arthritis. Biomolecules 9, 795. 10.3390/biom9120795 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study are available on request to the corresponding authors.