Abstract
Huntington’s disease (HD) is a fatal, inherited neurodegenerative disease that causes neuronal death, particularly in medium spiny neurons. HD leads to serious and progressive motor, cognitive and psychiatric symptoms. Its genetic basis is an expansion of the CAG triplet repeat in the HTT gene, leading to extra glutamines in the huntingtin protein. HD is one of nine genetic diseases in this polyglutamine (polyQ) category, that also includes a number of inherited spinocerebellar ataxias (SCAs). Traditionally it has been assumed that HD age of onset and disease progression were solely the outcome of age-dependent exposure of neurons to toxic effects of the inherited mutant huntingtin protein. However, recent genome-wide association studies (GWAS) have revealed significant effects of genetic variants outside of HTT. Surprisingly, these variants turn out to be mostly in genes encoding DNA repair factors, suggesting that at least some disease modulation occurs at the level of the HTT DNA itself. These DNA repair proteins are known from model systems to promote ongoing somatic CAG repeat expansions in tissues affected by HD. Thus, for triplet repeats, some DNA repair proteins seem to abandon their normal genoprotective roles and, instead, drive expansions and accelerate disease. One attractive hypothesis—still to be proven rigorously—is that somatic HTT expansions augment the disease burden of the inherited allele. If so, therapeutic approaches that lower levels of huntingtin protein may need blending with additional therapies that reduce levels of somatic CAG repeat expansions to achieve maximal effect.
Keywords: DNA synthesis and repair, genome integrity, mutation, neurodegeneration
The main purpose of this review is to provide an overview of recent developments in the role of DNA repair in Huntington’s disease (HD) and other triplet repeat diseases. The review is primarily intended for neurobiologists, physicians and others working in the field, or those interested in the general topic, but who are unfamiliar with DNA repair. For additional analysis, the reader is directed to other recent review articles [1–8].
HD
HD (OMIM #143100) is named after George Huntington, an American physician who first reported its familial nature in 1872 [9]. HD typically affects adults [8], although there are cases of juvenile onset at ages as young as 2 years [10]. Initially, the major recognized symptom of HD was progressive chorea, usually presenting as involuntary and irregular movement of hips, shoulders and face [1]. Additional motor symptoms include failure of voluntary motor control (especially gait and swallowing), dystonia, bradykinesia and rigidity. However, it is now clear that the disease manifestations of HD are more complex. There are also substantial cognitive and psychiatric symptoms which often precede motor dysfunction, sometimes by many years [11–13]. Cognitive issues affect attention, memory and speech, which progress gradually to dementia [14]. Cognitive decline is a critical concern for HD patients and families, profoundly affecting quality of life [15]. Psychiatric symptoms are most often depression, anxiety, apathy and irritability, although some patients also exhibit aggression [11]. Together, the cognitive, psychiatric and motor symptoms lead to debilitating disease that typically lasts approximately 15 years and results in premature death. The gradual appearance of symptoms creates a challenge in unambiguously discerning age of onset, even for motor dysfunction, which is usually the most obvious change and is used to assign age of onset. As described later, the challenge in accurately identifying age of onset partially obscured the effects of genetic modifiers of disease.
Multiple brain regions are affected in HD [13,16], mainly due to neuronal death by apoptosis [17,18]. The striatum is the region affected most strongly, exhibiting gross atrophy. Medium spiny neurons are especially sensitive in HD [13], although the precise reason for this sensitivity remains obscure [19]. Disease progression is scored with the Vonsattel grading system of 0–4 based on striatal morphology of post-mortem brain [20]. The cerebral cortex is also visibly affected by neuronal loss and shrinkage [21]. Later disease stages also affect numerous other brain regions; because of these widespread effects, the brains of advanced stage HD patient weigh 25–30% less than normal [22]. Thus, advanced HD can be regarded as whole brain disease [13,16].
HD genetics
Despite the complex nature of its symptoms, HD results from mutation of a single gene, HTT [23]. The mutation is autosomal dominant, meaning that one mutant copy of HTT leads to disease regardless of the presence of a second, normal allele. The worldwide HD prevalence is approximately 3 per 100000, but this value increases to ∼10 per 100000 in European and North American populations [24]. A well-known hotspot for HD is Lake Maracaibo, Venezuela with prevalence of 700 per 100000 [25], due to founder effects. Despite the devastation caused by HD, this Venezuelan population turned out to be a significant resource in mapping the mutated gene.
The genetic locus causing HD was mapped in 1983 to the tip of chromosome 4 [26]. The gene itself, now called HTT, was identified and sequenced in 1993 [23]. Its protein product is called huntingtin. Examination of HTT DNA sequence revealed a long CAG repeat in exon 1 whose expansion is now known to cause the disease. HD became the fourth triplet repeat expansion disease identified, after X-linked spinal and bulbar muscular atrophy (SBMA) [27], Fragile X syndrome [28–30] and myotonic dystrophy type 1 [31]. Normal individuals have 9–26 copies of CAG in HTT, with alleles of 17–20 repeats being most common [32]. Thus, the repeat itself is a natural part of HTT and only becomes problematic upon expansion. Individuals with 27–34 CAG repeats are not generally affected by HD but they are prone to passing longer repeat tracts to their offspring [33,34], a process known as genetic anticipation. A CAG range of 36–39 is pathogenic but exhibits reduced penetrance [35,36], meaning that not every individual in this range is affected. Alleles of 40 repeats or more are pathogenic with full penetrance [37]. Most adult-onset HD patients have alleles from 40 to ∼55 repeats. Alleles longer than ∼60 repeats are usually associated with the juvenile onset form of the disease [38]. For HTT, the CAG repeat falls in the coding region, where it encodes a polyglutamine (polyQ) tract [23]. HD is member of polyQ family of nine inherited diseases, which includes SBMA and several of the inherited spinocerebellar ataxias (SCAs) [6]. PolyQ diseases are protein-mediated, expressing a toxic protein that leads to disease [6].
One of the most remarkable features of HD is that its complex symptomology is due to addition of a few extra CAG repeats to one copy of one gene. An individual with 30 repeats is normal but another person with 40 repeats suffers the full scope of debilitating symptoms [37]. Surprisingly, this implies that adding a few more glutamine residues to those already present in huntingtin is the sole event necessary to cause this multisymptom disease. As described later, recent findings indicate that additional genetic changes in HTT are thought to add substantially to disease burden [39,40].
What aberrant properties does the mutant huntingtin protein convey? Among the many cellular and molecular changes that occur in the presence of mutant huntingtin [41], the protein causes transcriptional dysfunction, leading to altered expression signatures in many genes [41–43]. This disruption of normal expression patterns is one feasible explanation for the wide-ranging effects associated with the mutation. Both wildtype and mutant huntingtin are ubiquitously expressed in all tissues, including all brain regions albeit at different levels [44,45]. One function of the wildtype protein is in cellular trafficking [41]. It also contains domains called HEAT repeats that facilitate interactions with other proteins [41,46]. Proteomic analysis showed that mutant huntingtin is known to interact with hundreds of proteins [41,47]. One possibility is that the expanded polyQ tract exacerbates inappropriate protein–protein interactions, including the sequestration of transcription factors, which could explain part of the widespread transcriptional dysfunction. Mutant huntingtin is well known to cause persistent nuclear inclusions, which are aggregates visible by microscopy and containing many different proteins [48]. However, it is not clear that these inclusions are causal for HD or merely a byproduct.
One of the key demonstrations that the expanded HTT gene causing HD was the development of mouse models that closely mimic the human disease [49–52]. Initial experiments described a transgenic mouse with a fragment of the human HTT gene harboring exon 1 and the expanded CAG repeat. These animals developed progressive neurological phenotypes similar to the human condition [53]. The CAG repeat in these animals were also shown to be genetically unstable in both somatic tissues and during transmission to offspring [54]. Since these pioneering studies, a number of mouse models have been developed, including knock in animals, where the endogenous mouse Htt gene has been modified to harbor expanded CAG tracts [50]. Nearly all these mouse lines exhibit some or most disease aspects of HD, and most of them also show expansions of the CAG tract.
Somatic expansions in HTT as a potential modulator of disease onset and progression
The traditional interpretation of HD is that exposure of neurons and other brain cells to the toxic mutant huntingtin over the course of years leads to age-related neuronal death (Figure 1A). This viewpoint is consistent with several key observations about HD. First, the CAG repeat expansion in HTT is the only mutation necessary to cause disease [23]. Second, the length of the inherited expansion is the major factor (∼60–70%) that determines age of disease onset [55–58]. Finally, mutant huntingtin exhibits toxic effects in many experimental systems [50,53]. However, the traditional viewpoint is less satisfactory in explaining why huntingtin with 30 glutamines is safe but protein with 40 glutamines is deadly. Also, there is significant evidence that ∼30–40% of the age of onset is determined by other features besides inherited CAG length [56–58]. What other elements should be considered?
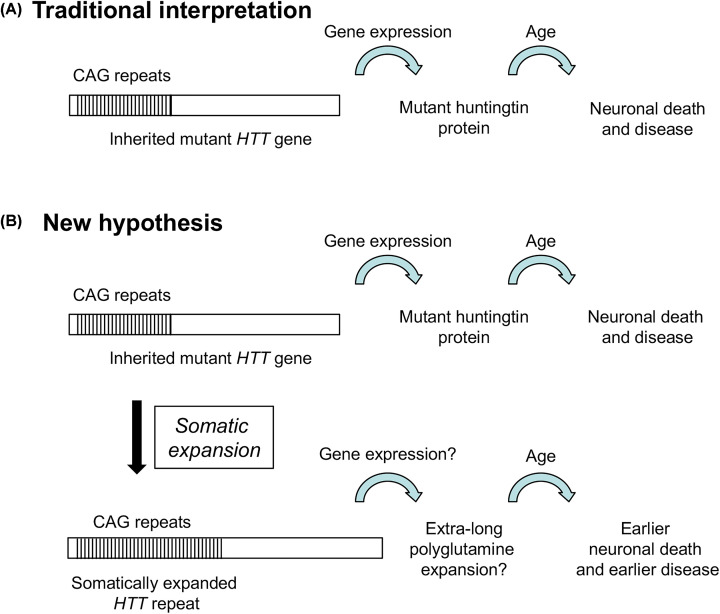
Figure 1. Models for neuronal death and HD.
(A) Traditional viewpoint of HD: inherited allele expresses mutant huntingtin protein. With age, neurons die due to toxic effect of mutant huntingtin. (B) New model. In addition to (A), ongoing somatic expansions add to toxic burden in neurons and other brain cells.
One additional feature of HD is the existence of somatic expansions. In addition to inherited CAG repeat expansions, HD individuals also undergo somatic expansions that are now believed to impact disease onset and progression. In some somatic tissues, particularly in brain, the inherited HTT mutation continues to expand during the lifetime of the individual. Dramatic striatal expansions, up to 1000 repeats, were observed in HD brains [59,60]. Moreover, expansions were observed in striatal neurons of both humans and mice [61,62], where they occur early in the disease process and continue to accumulate [61].
What causes somatic instability and is it important? The inherited CAG length was shown to be a major modifier of somatic instability, although additional genetic and environmental modifiers were also deduced but not initially specified [55–58]. Somatic instability in HTT was shown to be a significant predictor of disease age of onset, with longer somatic expansions linked to earlier onset [63]. Together, these studies verified two key predictions of the hypothesis that somatic expansions modify HD onset and progression. First, somatic expansions occur in the tissues affected by HD, thus fulfilling the spatial requirement. Second, somatic expansions precede disease symptoms [60], supporting the temporal requirement. A third prediction is that genetic modifiers of somatic instability would also modify disease. These early groundbreaking studies helped lay the foundation for the recent identification of genetic modifiers of HD, described in the next section.
If somatic instability helps determine HD age of onset, then a new hypothesis for the disease can be formulated (Figure 1B) [2,5,8]. This new hypothesis retains the traditional thinking about the age-dependent toxic effects of inherited polyQ length in huntingtin but adds a second branch where somatic CAG repeat expansions hasten disease onset and progression. The somatically-expanded version of HTT is predicted to encode a mutant huntingtin protein with extra-long polyQ tract. By this model, the extra-long huntingtin adds to the disease burden and exacerbates onset and progression. This hypothesis can also be extended to the subset of SCAs where somatic expansions occur [2,7]. Somatic expansions are the linchpin of this model and recent genome-wide association studies (GWAS) pointed directly at DNA repair factors as novel modifiers of age of onset and progression of HD and in some of the SCAs.
GWAS identify DNA repair factors as modulators of HD
GWAS search among a population for genetic variants that are associated with a particular trait, such as inherited disease. In the case of HD, GWAS sought factors that, by themselves, do not confer risk of HD but which modify the course of the disorder [3,39,40]. This approach is based on the idea that the expanded CAG repeat in HD patients provides a genetically sensitized background to find modifying factors [39]. The two major advantages of GWAS are first, that this approach makes no assumptions about disease mechanism. Therefore, GWAS is not influenced by any pre-existing expectation for factors that drive the outcome, in this case, HD age of onset. Second, GWAS takes advantage of existing information among HD cohorts of thousands of patients. In effect, nature has already performed the experiment and GWAS looks for the result. For HD, effective GWAS required the creation of large consortia of HD patients with accurate clinical and genetic information. TRACK-HD and Enroll-HD are examples of such consortia. Subsequent GWAS reports have also included information from several SCA consortia (SCA1, SCA2, SCA3, SCA6, SCA7 and SCA17) to extend the findings beyond HD [2,64]. Most HD GWAS have focused on age of motor onset (AOO) because it is quantifiable [39,40]. Additional GWAS reports stem from assessing disease progression [65] and CAG repeat instability in blood samples from patients [66].
The unexpected outcome from these GWAS is that DNA repair genes comprise many, although not all, modifiers for HD and other polyQ diseases [2,4,39,40,64–66]. Genetic loci were clearly identified that contain DNA repair genes MSH3, MLH1, PMS1, PMS2, MLH3 and FAN1, which are described in more detail in the following section. The effect of polymorphisms in these loci was surprisingly large, accounting for alterations of up to 6 years in age of onset [39]. In addition, SNPs in some DNA repair genes are also associated with changes in progression of HD [67]. While the loci from GWAS often contain numerous genes and therefore cannot unambiguously identify a specific gene, a process called pathway analysis looks for commonalities among candidate genes in independent loci. The pathway analysis is clear that DNA repair, and particularly one particular activity called DNA mismatch repair (MMR), is very tightly associated with AOO, providing strong secondary proof of the correct gene assignments [39]. A third correlation is that polymorphisms that favor MMR expression accelerate HD age of onset, whereas alternative SNPs that reduce expression slow AOO [65,67]. Thus, GWAS supports the idea that high levels of MMR proteins and repair activity accelerate disease onset in HD. A fourth point is that HD onset tracks with CAG repeat length, not the number of glutamine codons [40,68]. This finding helped refocus attention from huntingtin back to the HTT DNA itself. A final supporting line of evidence is that DNA repair factors in HD mice are well known to influence somatic CAG repeat expansions [69] and to modulate disease [70]. Knockouts of Msh2 or Msh3 (encoding MutSβ) [70–74] or of Mlh1 or Mlh3 (encoding MutLα and MutLγ) [75] eliminate nearly all somatic and inherited Htt expansions. In total, this evidence provides a very strong case that at least some modulation of HD occurs at the level of maintaining the HTT DNA itself [4].
DNA repair loci implicated in HD and other triplet repeat expansion disorders
The strong connection between DNA repair and HD age of onset and disease progression has led to significant new thinking about maintenance of the CAG repeat within HTT as a major modifier of disease. This DNA-centric view has major implications for both scientific mechanism, which is considered in this section, and therapeutic approaches to HD and related polyQ diseases that are considered later.
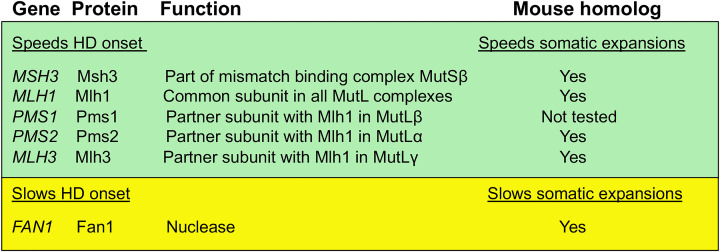
The relevant DNA repair genes and the functions of their proteins are summarized in Figure 2. The first panel includes factors that speed onset of HD. MSH3 encodes a protein that identifies DNA damage, specifically mismatched DNA that normally arises from errors in DNA replication. The Msh3 protein partners with a related but distinct protein called Msh2 to form the functional complex, MutSβ [76]. Both Msh3 and Msh2 are required for MutSβ activity. Additional MMR proteins are encoded by MLH1, PMS1, PMS2 and MLH3 (Figure 2), referred to collectively as the MutL homologs, after the bacterial prototype. The eukaryotic MutL proteins function as heterodimers, with Mlh1 protein being the common partner. Inclusion of either Pms2, Pms1 or Mlh3 yields MutLα, MutLβ and MutLγ, respectively [76]. Mouse studies identified a key role in expansions for both MutLα [77] and MutLγ [75]. While each of these MutL complexes has unique roles in MMR, a recent finding in cultured mouse Fragile X stem cells surprisingly suggested that all three MutL complexes are required for CCG repeat expansions [78]. It is not yet known if all three MutL complexes are also required for expansions of CAG repeats. The potential mechanistic role of MMR in expansions is considered in the next section.
Figure 2. DNA repair genes and their proteins identified by GWAS.
GWAS analysis also identified a prominent signal for another gene, FAN1, whose protein product Fan 1 acts independently of MMR (Figure 2). GWAS analysis suggest that the key SNP in FAN1 is associated with high expression of the Fan1 protein and is also associated with later age of onset. Thus, in contrast with MMR which accelerates disease, the presence of Fan1 acts in a disease-slowing process [39,40,79]. Fan1 is a nuclease, an enzyme that cleaves DNA in a distinct pathway called interstrand cross-link repair. Mouse and stem cell knockout studies of FAN1 are consistent with a protective role for Fan1 in blocking triplet repeat expansions [80–82]. One possibility is that Fan1 might remove DNA intermediates before they become fully expanded, thereby stabilizing the repeat sequence. A recent mouse study looking at double knockouts of Fan1 and Mlh1 found that functional Mlh1 protein was required to see the CAG repeat destabilization that occurs due to Fan1 knockout [83], suggesting that normally Mlh1 is required to allow Fan1 stabilization of the triplet repeat. This effect could be due to protein–protein interactions that were reported between Fan1 and the MutL homologs Mlh1, Pms1, Pms2 and Mlh3 [84].
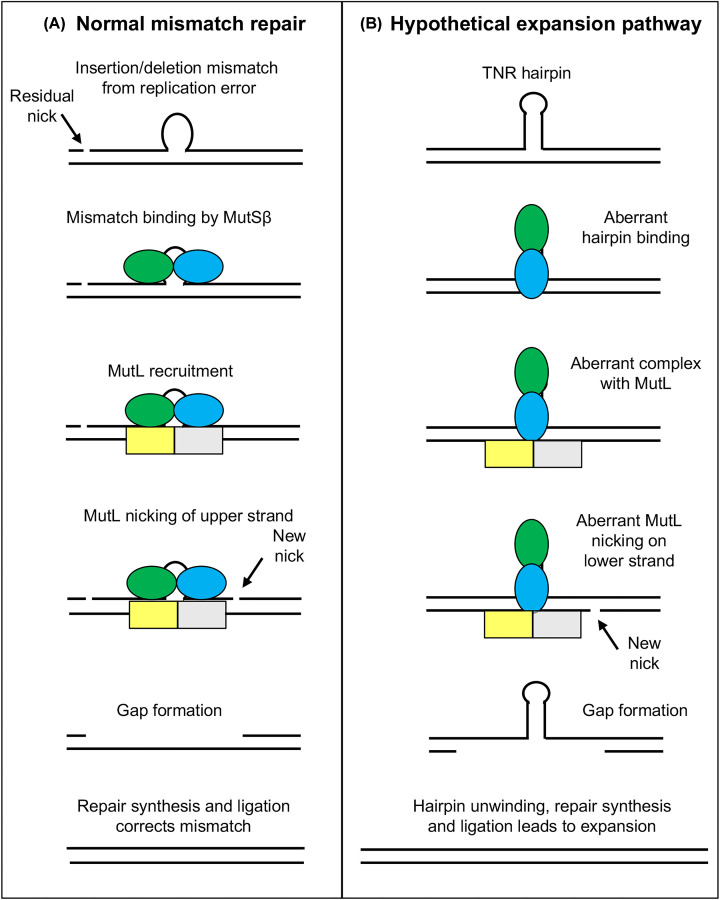
What does MMR normally do and what goes wrong to cause triplet repeat expansions? The major role of MMR is as a genetic spellchecker to correct errors made during DNA replication (Figure 3A) [76,85–87]. Although replication is very precise, the size of mammalian genome leads to inevitable errors. For human cells, a few hundred mismatches or so are left behind after each round of replication [88]. Some of these mismatches are in repetitive sequences and involve synthesis of too many or too few copies of the repeat, referred to as insertion/deletion mispairs (loop symbol in Figure 3A). The MMR protein MutSβ recognizes these insertion/deletion mismatches and triggers their repair [89]. Thus, MutSβ provides the first key function in MMR, finding the ‘needle’ (mismatch) per ‘haystack’ of ∼1 million correctly synthesized base pairs. The second required step in MMR is identifying which strand has the incorrect information and therefore must be targeted for repair. This is primarily the function of the MutL homolog proteins [90]. Since both strands of the mismatch comprise normal Watson and Crick bases—A, C, T and G—there is no chemical signal to direct MMR to the strand with the incorrect sequence. Instead, MMR uses residual strand breaks (nicks) left over from DNA replication to identify the newly synthesized strands (Figure 3A) [91,92]. Using protein–protein interactions between the MutS and MutL homologs and other factors, the nicks are used to direct repair to the strand bearing the incorrect information [93]. In some repair events, the MutL proteins introduce a second nick into the newly synthesized strand (Figure 3A) [94]. Subsequent processing by additional nucleases creates a single-strand gap that removes the mismatch and a few hundred neighboring base pairs. The gap is then filled by DNA synthesis and the strand is sealed by DNA ligase to complete the repair process [94].
Figure 3. Model for how non-canonical DNA MMR could drive expansions.
(A) A simplified scheme for how normal MMR correctly repairs ‘top’ strand to remove replication error. Ovals, MutSβ subunits Msh2 (green) and Msh3 (blue). Rectangles, MutL homolog subunits Mlh1 (yellow) and either Pms2 or Mlh3 (grey). (B) Hypothetical scheme for how non-canonical MMR aberrantly repairs ‘bottom’ strand to cause expansion.
There are several key differences in MMR activity on triplet repeats (Figure 3B). First, mismatched TNRs are known to form DNA secondary structures, such as hairpins (stem-loops) [95]. Hairpins can occur in post-mitotic neurons, since hairpin formation does not require DNA replication. Instead, experiments in model systems show that hairpins can occur during gene transcription. The requirement for strand separation during transcription allows transiently single-stranded TNR DNA to fold on itself prior to reannealing with the complementary strand [96–99]. Biochemical experiments with model DNA substrates showed that MutSβ binds significantly differently to TNR hairpins compared with normal insertion/deletion mispairs [73,100]. Although not to be taken literally, this difference in binding is shown schematically as a ‘vertical’ alignment of MutSβ proteins (Figure 3B). Second, somatic expansions occur in postmitotic neurons [61,62] where there is no DNA replication. This means that there is no nick to serve as a strand signal for MMR. Instead, this model suggests that MutL homolog proteins interact aberrantly with the unusual TNR hairpin–MutSβ complex. This aberrant interaction leads to MutL-mediated incision of the DNA on the incorrect strand, across from the hairpin [101,102] (Figure 3B). Subsequent processing of the nick by additional nucleases opens up a gap, which is then filled in by DNA repair synthesis and sealed by a DNA ligase. The overall outcome, in this hypothetical situation, is to cause an expansion (Figure 3B). The model is also consistent with the fact that DNA repair is functional in non-dividing cells [4]. Finally, this model has the virtue of simplicity—it only requires that MMR incises the wrong strand, but otherwise utilizes most of its normal mechanism. An interesting challenge for DNA repair experts will be to develop assays to test this model.
Post-GWAS: a new model for understanding HD age of onset and progression
The important revelations about somatic CAG repeat expansions in HTT suggest that the traditional model of disease may need refinement to add somatic expansions as part of the pathogenic process (Figure 1B). While neurons with the inherited mutant HTT gene continue to express mutant huntingtin with its toxic effects, any cells with a somatic expansion are predicted to express an extra-long version of mutant huntingtin protein with even more glutamines than encoded by the inherited allele. If huntingtin with extra glutamine residues is more toxic than the inherited version, then somatic expansions would add to neuron toxicity and therefore cause earlier neuronal death and disease pathogenesis. This is one model to explain the GWAS findings (Figure 1B). A competing model is that neurons with inherited expansions undergo exposure to DNA damaging agents, such as oxidative damage, or to toxicity induced by somatic expansions. By this model, the key role of DNA repair protects neurons after damage. Genetic variants in DNA repair are predicted to result in sensitization to this damage, leading to earlier cell death [5]. The key difference is that model 1 predicts that somatic expansions contribute significantly to disease, whereas model 2 does not. The best available data to distinguish the two models used an HD mouse model in which a DNA repair gene called OGG1 was inactivated [103]. Although OGG1 has not shown up in GWAS reports on triplet repeat diseases, the gene is known to promote somatic CAG repeat expansions in some mouse models of HD [103,104] and therefore provides a useful experimental tool. Loss of OGG1 in this system selectively block somatic expansions, and these animals showed a delay in onset of HD-like symptoms compared with control littermates. While these results support model 1, they can still be interpreted in light of model 2.
An even better test to distinguish model 1 from model 2 would be to create an HD mouse model with an inherited expansion that encodes mutant huntingtin but where the Htt gene cannot undergo somatic expansions. If model 1 is correct, disease onset should be slowed due to loss of the somatic expansions, whereas model 2 predicts no change in disease onset. How could such an experiment be designed? One way is to take advantage of the genetic redundancy in glutamine codons. Both CAG and CAA encode glutamine, but normally the human HTT sequence is nearly all CAG codons, with a CAA codon or two near the 3′ end [66,68,105]. Artificially changing this sequence to include more CAA codons (‘interruptions’) scattered through the repeat would still encode huntingtin with the same number of glutamines. However, interrupted triplet repeats are known to be much more genetically stable, with fewer expansions [40,66,68]. The prediction of this experiment is that the interrupted version of mouse Htt would retain the ability to encode the inherited form of huntingtin, but the gene itself would undergo few somatic expansions and thereby greatly reduce abundance of any putative ‘extra-long’ polyQ huntingtin. If model 1 is correct, disease onset should occur later in mice with the interrupted Htt gene compared with animals with the uninterrupted version. Model 2 predicts no significant difference in disease onset. The idea was tested in one mouse model where the polyQ tract of huntingtin is encoded by mixed CAA-CAG repeats and is genetically stable. The result suggests that, in this mouse model, somatic instability does not play a necessary role in the selective neuropathogenesis [106]. Unfortunately, no comparison was available with a perfect CAG repeat control animal, suggesting that this result should be viewed with caution.
HD homozygotes provide another interesting evaluation of model 1. Although rare, some individuals harbor two mutant HTT alleles, which can be of different CAG repeat length. Age of disease onset correlates with the longer of the two alleles [68,107]. If model 1 is correct, why does the shorter expanded allele seem not to have much effect on age of onset? One possibility is that the longer allele is primarily targeted for somatic expansions, perhaps due to the greater number of repeats. A useful experiment would be to look for allele-specific somatic expansions and see if they primarily stem from the longer allele. A second possibility is that both alleles undergo somatic expansions at similar frequencies but that disease onset is somehow particularly sensitive to changes in the longer repeat tract. Testing these possibilities and any other theories will require additional experimentation.
Connections to therapy
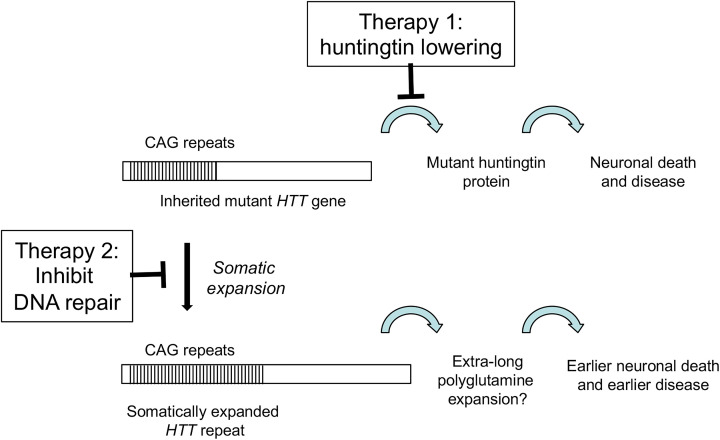
How might this new information about DNA repair affect therapeutic efforts for HD and other triplet repeat expansion diseases? A major effort is currently underway to treat HD by lowering the levels of huntingtin protein [108,109] (Figure 4). The idea is that less huntingtin—especially less of the mutant version of the protein—will reduce HD symptoms and relieve suffering. As an example, antisense oligonucleotides (ASOs) have been developed that inhibit translation of huntingtin by targeting its messenger RNA [108,109]. One such ASO was reported in a Phase I/IIa clinical study to be safely tolerated and to reduce huntingtin levels in spinal fluid by up to 40% [110]. This ASO is now proceeding to a Phase III trial. These huntingtin-lowering approaches are a welcome addition and hopefully they will prove safe and efficacious for HD patients. In principal, the protein-lowering approach could be used to reduce other expanded polyQ proteins that cause additional diseases [111]. An obvious drawback is that individual ASOs must be designed and tested for each disease. A second unanswered question is whether ASO technology would be effective against the putative ‘extra-long’ version of HTT. While there is no reason a priori to believe otherwise, this point will require experimental proof.
Figure 4. Potential therapies against HD.
Therapy 1 involves huntingtin lowering strategies. Therapy 2 seeks to inhibit specific DNA repair factors to reduce somatic CAG repeat expansions.
A second approach to therapy was opened by the discovery that genetic variants in DNA repair proteins, particularly MMR proteins, help drive disease [39,40]. The possibility that somatic expansions are important for disease onset and progression means that MMR proteins might be additional druggable targets that would impact HD and any other triplet repeat expansion diseases with somatic instability (Figure 4) [112]. As an example, supportive evidence from an HD mouse model indicates that knockout of the MSH2 subunit of MutSβ eliminated striatal expansions and also delayed nuclear accumulation of mutant huntingtin [70,113]. Since the mice contain ∼111 uninterrupted CAG repeats, this result suggests effectiveness against the putative extra-long version of mutant huntingtin alluded to above. Another reason to be attracted to this idea is that loss of MSH3, which encodes the unique subunit of MutSβ, has a low impact on cancer predisposition [106]. Thus, inhibiting MutSβ to treat triplet repeat expansion diseases may minimize any complications in tumorigenesis. This line of thinking has led to vigorous efforts to identify and test MutSβ inhibitors for efficacy in HD. Several entities have initiated very active efforts to screen for small molecules that inhibit MutSβ (for example, see https://chdifoundation.org/dna-repair-handling/). In principle, these interventions could target MutSβ in several ways [108]. One way is to inhibit its enzymatic activity, particularly the ATPase function that is crucial for expansions [114], perhaps using novel small molecule inhibitors that would necessarily need to be selective for MutSβ. A second approach would be to disrupt the interactions of MutSβ with the MutL homologs. This will require clear identification of which MutL homolog is most important for driving expansions, development of a clear understanding of the relevant protein–protein interactions and a suitable screen for disrupting agents with good specificity. A third idea is to reduce the abundance of Msh3, one of the subunits of MutSβ, through ASO or similar technology. Perhaps ASO against Msh3 could be added to the huntingtin ASOs already being tested. Studies in mice and human cells show that lower Msh3 abundance leads to fewer CAG repeat expansions [114–117]. Msh3 levels in humans also correlate positively with disease progression [65,67]. A final approach, which already has some positive preclinical support, is to inhibit enzymes that activate MutSβ. One such enzyme is the histone deacetylase, HDAC3, which was recently demonstrated to directly deacetylate MutSβ and stimulate expansions [118]. Potent inhibitors already exist that selectively block HDAC3 activity [119–123]. Several of these HDAC3 inhibitors have been shown in mouse studies to alleviate motor and cognitive symptoms of HD, and also to inhibit striatal Htt expansions [120,124,125]. Any of these approaches can, in principal, be blended with huntingtin-lowering therapy to potentially provide a more potent therapy against HD and related triplet repeat expansion diseases.
Conclusions and future perspectives
It was a revelation when GWAS identified DNA repair proteins as modifiers of HD age of onset and disease progression [2,4,39,40,65,66]. Perhaps the biggest surprise was the extent of this modification—polymorphisms in DNA repair genes can mean the difference of up to 6 years of healthy living for HD patients [39]. Moreover, the modifier genes discovered by GWAS were highly consistent with mouse studies and cellular experiments that had identified many of the same DNA repair proteins as causal for somatic CAG repeat expansions. Together, these findings open two important avenues for ongoing studies. The first is mechanistic: do somatic expansions add to disease burden in HD and, if so, how? The second is therapeutic: regardless of the mechanism of how DNA repair modifies HD, can DNA repair be used as a therapeutic target? Both these avenues offer exciting new opportunities to better understand HD and some of the related triplet repeat diseases, and they open a potential new therapeutic landscape for what had been untreatable conditions.
Funding
The author is a paid consultant of LoQus23 Therapeutics and serves on their scientific advisory board; and Science Foundation Ireland [grant number 16/BBSRC/3395].
Acknowledgements
The author thanks Eilís Dowd and Elaine Lahue for thoughtful comments on this manuscript.
Abbreviations
- AOO
age of motor onset
- ASO
antisense oligonucleotide
- GWAS
genome-wide association studies
- HD
Huntington’s disease
- HEAT
huntingtin/elongation factor 3/protein phosphatase 2A/Tor1
- MMR
mismatch repair
- polyQ
polyglutamine
- SBMA
X-linked spinal and bulbar muscular atrophy
- SCA
spinocerebellar ataxia
- TNR
trinucleotide repeat
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Bates G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R. et al. (2015) Huntington disease. Nat. Rev. Dis. Primers 1,15005 10.1038/nrdp.2015.5 [DOI] [PubMed] [Google Scholar]
- 2.Bettencourt C., Hensman-Moss D., Flower M., Wiethoff S., Brice A., Goizet C. et al. (2016) DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases. Ann. Neurol. 79, 983–990 10.1002/ana.24656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmans P.A., Massey T.H. and Jones L. (2017) Genetic modifiers of Mendelian disease: Huntington’s disease and the trinucleotide repeat disorders. Hum. Mol. Genet. 26, 83–90 10.1093/hmg/ddx261 [DOI] [PubMed] [Google Scholar]
- 4.Jones L., Houlden H. and Tabrizi S.J. (2017) DNA repair in the trinucleotide repeat disorders. Lancet Neurol. 16, 88–96 10.1016/S1474-4422(16)30350-7 [DOI] [PubMed] [Google Scholar]
- 5.Massey T.H. and Jones L. (2018) The central role of DNA damage and repair in CAG repeat diseases. Dis. Model Mech. 11, pii: dmm031930 10.1242/dmm.031930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoyas C.A. and La Spada A.R. (2018) The CAG-polyglutamine repeat diseases: a clinical, molecular, genetic, and pathophysiologic nosology. Handb. Clin. Neurol. 147, 143–170 10.1016/B978-0-444-63233-3.00011-7 [DOI] [PubMed] [Google Scholar]
- 7.Yau W.Y., O’Connor E., Sullivan R., Akijian L. and Wood N.W. (2018) DNA repair in trinucleotide repeat ataxias. FEBS J. 285, 3669–3682 10.1111/febs.14644 [DOI] [PubMed] [Google Scholar]
- 8.Tabrizi S.J., Flower M.D., Ross C.A. and Wild E.J. (2020) Huntington disease: new insights into molecular pathogenesis and therapeutic opportunities. Nat. Rev. Neurol. 16, 529–546 10.1038/s41582-020-0389-4 [DOI] [PubMed] [Google Scholar]
- 9.Huntington G. (1872) On chorea. Med. Sur. Rep. 26, 317–321 [Google Scholar]
- 10.Nance M.A. and Myers R.H. (2001) Juvenile onset Huntington’s disease—clinical and research perspectives. Ment. Retard. Dev. Disabil. Res. Rev. 7, 153–157 10.1002/mrdd.1022 [DOI] [PubMed] [Google Scholar]
- 11.Julien C.L., Thompson J.C., Wild S., Yardumian P., Snowden J.S., Turner G. et al. (2007) Psychiatric disorders in preclinical Huntington’s disease. J. Neurol. Neurosurg. Psychiatry 78, 939–943 10.1136/jnnp.2006.103309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paulsen J.S., Langbehn D.R., Stout J.C., Aylward E., Ross C.A., Nance M. et al. (2008) Detection of Huntington’s disease decades before diagnosis: the Predict-HD study. J. Neurol. Neurosurg. Psychiatry 79, 874–880 10.1136/jnnp.2007.128728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross C.A. and Tabrizi S.J. (2011) Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet North Am. Ed. 10, 83–98 [DOI] [PubMed] [Google Scholar]
- 14.Paulsen J.S. (2011) Cognitive impairment in Huntington disease: diagnosis and treatment. Curr. Neurol. Neurosci. Rep. 11, 474–483 10.1007/s11910-011-0215-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Begeti F., Schwab L.C., Mason S.L. and Barker R.A. (2016) Hippocampal dysfunction defines disease onset in Huntington’s disease. J. Neurol. Neurosurg. Pschiatry 87, 975–981 10.1136/jnnp-2015-312413 [DOI] [PubMed] [Google Scholar]
- 16.Henley S.M.D., Wild E.J., Hobbs N.Z., Frost C., MacManus D.G., Barker R.A. et al. (2009) Whole-brain atrophy as a measure of progression in premanifest and early Huntington’s disease. Mov. Disord. 24, 932–936 10.1002/mds.22485 [DOI] [PubMed] [Google Scholar]
- 17.Thomas L.B., Gates D.J., Richfield E.K., O’Brien T.F., Schweitzer J.B. and Steindler D.A. (1995) DNA end labeling (TUNEL) in Huntington's disease and other neuropathological conditions. Exp. Neurol. 133, 265–272 10.1006/exnr.1995.1029 [DOI] [PubMed] [Google Scholar]
- 18.Hickey M.A. and Chesselet M.F. (2003) Apoptosis in Huntington's disease. Prog. Neuropsychopharmacol. Biol. Psychiatry 27, 255–265 10.1016/S0278-5846(03)00021-6 [DOI] [PubMed] [Google Scholar]
- 19.Zuccato C., Valenza M. and Cattaneo E. (2010) Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol. Rev. 90, 905–981 10.1152/physrev.00041.2009 [DOI] [PubMed] [Google Scholar]
- 20.Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D. and Richardson E.P. Jr (1985) Neuropathological classification of Huntington's disease. J. Neuropathol. Exp. Neurol. 44, 559–577 10.1097/00005072-198511000-00003 [DOI] [PubMed] [Google Scholar]
- 21.Sotrel A., Paskevich P.A., Kiely D.K., Bird E.D., Williams R.S. and Myers R.H. (1991) Morphometric analysis of the prefrontal cortex in Huntington’s disease. Neurology 41, 1117–1123 10.1212/WNL.41.7.1117 [DOI] [PubMed] [Google Scholar]
- 22.Sharp A.H. and Ross C.A. (1996) Neurobiology of Huntington’s disease. Neurobiol. Dis. 3, 3–15 10.1006/nbdi.1996.0002 [DOI] [PubMed] [Google Scholar]
- 23.Group THsDCR (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72, 971–983 10.1016/0092-8674(93)90585-E [DOI] [PubMed] [Google Scholar]
- 24.Pringsheim T., Wiltshire K., Day L., Dykeman J., Steeves T. and Jette N. (2012) The incidence and prevalence of Huntington’s disease: a systematic review and meta-analysis. Mov. Disord. 27, 1083–1091 10.1002/mds.25075 [DOI] [PubMed] [Google Scholar]
- 25.Okun M.S. and Thommi N. (2004) Americo Negrette (1924 to 2003): diagnosing Huntington disease in Venezuela. Historical Neurol. 63, 340–343 [DOI] [PubMed] [Google Scholar]
- 26.Gusella J.F., Wexler N.S., Conneally P.M., Naylor S.L., Anderson M.A., Tanzi R.E. et al. (1983) A polymorphic DNA marker genetically linked to Huntington’s disease. Nature 306, 234–238 10.1038/306234a0 [DOI] [PubMed] [Google Scholar]
- 27.La Spada A.R., Wilson E.M., Lubahn D.B., Harding E.E. and Fischbeck K.H. (1991) Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 352, 77–79 10.1038/352077a0 [DOI] [PubMed] [Google Scholar]
- 28.Fu Y.-H., Kuhl D.P.A., Pizzuti A., Pieretti M., Sutcliffe J.S., Richards S.R. et al. (1991) Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 67, 1047–1058 10.1016/0092-8674(91)90283-5 [DOI] [PubMed] [Google Scholar]
- 29.Kremer E.J., Pritchard M., Lynch M., Yu S., Holman K., Baker E. et al. (1991) Mapping of DNA instability at the fragile X to a trinucleotide repat sequences p(CCG)n. Science 252, 1711–1714 10.1126/science.1675488 [DOI] [PubMed] [Google Scholar]
- 30.Verkerk A.J.M.H., Pieretti M., Sutcliffe J.S., Fu Y.-H., Kulh D.P.A., Pizzuti A. et al. (1991) Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in Fragile X syndrome. Cell 65, 905–914 10.1016/0092-8674(91)90397-H [DOI] [PubMed] [Google Scholar]
- 31.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H. et al. (1992) Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 68, 799–808 10.1016/0092-8674(92)90154-5 [DOI] [PubMed] [Google Scholar]
- 32.Kremer B., Goldberg P., Andrew S.E., Theilmann J., Telenius H., Zeisler J. et al. (1994) A worldwide study of the Huntington’s disease mutation: the sensitivity and specificity of measuring CAG repeats. N. Engl. J. Med. 330, 1401–1406 10.1056/NEJM199405193302001 [DOI] [PubMed] [Google Scholar]
- 33.Kelly T.E., Allinson P., McGlennen R.C., Baker J. and Bao Y. (1999) Expansion of a 27 CAG repeat allele into a symptomatic Huntington disease‐producing allele. Am. J. Hum. Genet. 87, 91–92, [DOI] [PubMed] [Google Scholar]
- 34.Semaka A., Kay C., Doty C., Collins J.A., Bijlsma E.K., Richards F. et al. (2013) CAG size-specific risk estimates for intermediate allele repeat instability in Huntington disease. J. Med. Genet. 50, 696–703 10.1136/jmedgenet-2013-101796 [DOI] [PubMed] [Google Scholar]
- 35.Rubinsztein D.C., Leggo J., Coles R., Almqvist E., Biancalana V., Cassiman J.-J. et al. (1996) Phenotypic characterization of individuals with 30-40 CAG repeats in the Huntington Disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36-39 repeats. Am. J. Hum. Genet. 59, 16–22 [PMC free article] [PubMed] [Google Scholar]
- 36.McNeil S.M., Novelletto A., Srinidhi J., Barnes G., Kornbluth I., Altherr M.R. et al. (1997) Reduced penetrance of the Huntington's disease mutation. Hum. Mol. Genet. 6, 775–779 10.1093/hmg/6.5.775 [DOI] [PubMed] [Google Scholar]
- 37.Nance M.A., Seltzer W., Ashizawa T., Bennet R., McIntosh N., Myers R.H. et al. (1998) ACMG/ASHG Statement. Laboratory guidelines for Huntington disease genetic testing. Am. J. Hum. Genet. 62, 1243–1247 10.1086/301846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Telenius H., Kremer H.P.H., Thellmann J., Andrew S.E., Almqvist E., Anvret M. et al. (1993) Molecular analysis of juvenile Huntington disease: the major influence on (CAG)n repeat length is the sex of the affected parent. Hum. Mol. Genet. 2, 1535–1540 10.1093/hmg/2.10.1535 [DOI] [PubMed] [Google Scholar]
- 39.Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium (2015) Identification of genetic factors that modify clinical onset of Huntington’s disease. Cell 162, 516–526 10.1016/j.cell.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium (2019) CAG repeat not polyglutamine length determines timing of Huntington’s disease onset. Cell 178, 887–900 10.1016/j.cell.2019.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saudou F. and Humbert S. (2016) The biology of Huntingin. Neuron 89, 910–926 10.1016/j.neuron.2016.02.003 [DOI] [PubMed] [Google Scholar]
- 42.Hodges A., Strand A.D., Aragaki A.K., Kuhn A., Sengstag T., Hughes G. et al. (2006) Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet. 15, 965–977 10.1093/hmg/ddl013 [DOI] [PubMed] [Google Scholar]
- 43.Hensman Moss D.J., Flower M.D., Lo K.K., Miller J.R.C., van Ommen G.-J.B., ’t Hoen P.A.C. et al. (2017) Huntington’s disease blood and brain show a common gene expression pattern and share an immune signature with Alzheimer’s disease. Sci. Rep. 7, 44489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharp A.H., Loev S.J., Schilling G., Li S.H., Li X.J., Bao J. et al. (1995) Widespread expression of Huntington’s disease gene (IT15) protein product. Neuron 14, 1065–1074 10.1016/0896-6273(95)90345-3 [DOI] [PubMed] [Google Scholar]
- 45.Schilling G., Sharp A.H., Loev S.J., Wagster M.V., Li S.H., Stine O.C. et al. (1995) Expression of the Huntington’s disease (IT15) protein product in HD patients. Hum. Mol. Genet. 4, 1365–1371 10.1093/hmg/4.8.1365 [DOI] [PubMed] [Google Scholar]
- 46.Yoshimura S.H. and Hirano T. (2016) HEAT repeats - versatile arrays of amphiphilic helices working in crowded environments? J. Cell Sci. 129, 3963–3970 [DOI] [PubMed] [Google Scholar]
- 47.Wanker E.E., Ast A., Schindler F., Trepte P. and Schnoegl S. (2019) The pathobiology of perturbed mutant huntingtin protein-protein interactions in Huntington’s disease. J. Neurochem. 151, 507–519 10.1111/jnc.14853 [DOI] [PubMed] [Google Scholar]
- 48.DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P. and Aronin N. (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 10.1126/science.277.5334.1990 [DOI] [PubMed] [Google Scholar]
- 49.Crook Z.R. and Housman D. (2011) Huntington’s disease: can mice lead the way to treatment? Neuron 69, 423–435 10.1016/j.neuron.2010.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pouladi M.A., Morton A.J. and Hayden M.R. (2013) Choosing an animal model for the study of Huntington’s disease. Nat. Rev. Neurosci. 14, 708–721 10.1038/nrn3570 [DOI] [PubMed] [Google Scholar]
- 51.Brooks S.P. and Dunnett S.B. (2015) Mouse models of Huntington’s disease. Curr. Top. Behav. Neurosci. 22, 101–133 10.1007/7854_2013_256 [DOI] [PubMed] [Google Scholar]
- 52.Farshim P.P. and Baters G.P. (2018) Mouse models of Huntington’s disease. Methods Mol. Biol. 1780, 97–120 10.1007/978-1-4939-7825-0_6 [DOI] [PubMed] [Google Scholar]
- 53.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C. et al. (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506 10.1016/S0092-8674(00)81369-0 [DOI] [PubMed] [Google Scholar]
- 54.Mangiarini L., Sathasivam K., Mahal A., Mott R., Seller M. and Bates G.P. (1997) Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nat. Genet. 15, 197–200 10.1038/ng0297-197 [DOI] [PubMed] [Google Scholar]
- 55.Djousse L., Knowlton B., Hayden M., Almqvist E.W., Brinkman R., Ross C.A. et al. (2003) Interaction of normal and expanded CAG repeat sizes influences age at onset of Huntington disease. Am. J. Hum. Genet. 119A, 279–282 [DOI] [PubMed] [Google Scholar]
- 56.The U.S.-Venezuela Collaborative Research Project, Wexler NS (2004) Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc. Natl. Acad. Sci. U.S.A. 101, 3498–3503 10.1073/pnas.0308679101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veitch N.J., Ennis M., McAbney J.P., Project U-VCR, Shelbourne P.F. and Monckton D.G. (2007) Inherited CAG.CTG allele length is a major modifier of somatic mutation length variability in Huntington disease. DNA Rep. (Amst.) 6, 789–796 10.1016/j.dnarep.2007.01.002 [DOI] [PubMed] [Google Scholar]
- 58.Gusella J.F. and MacDonald M.E. (2009) Huntington’s disease: the case for genetic modifiers. Genome Med. 1, 80 10.1186/gm80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kennedy L. and Shelbourne P.F. (2000) Dramatic mutation instability in HD mouse striaturm: does polyglutamine load contribute to cell-specific vulnerability in Huntington’s disease? Hum. Mol. Genet. 9, 2539–2544 10.1093/hmg/9.17.2539 [DOI] [PubMed] [Google Scholar]
- 60.Kennedy L., Evans E., Chen C.M., Craven L., Detloff P.J., Ennis M. et al. (2003) Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet. 12, 3359–3367 10.1093/hmg/ddg352 [DOI] [PubMed] [Google Scholar]
- 61.Shelbourne P.F., Keller-McGandy C., Bi W.L., Yoon S.R., Dubeau L., Veitch N.J. et al. (2007) Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum. Mol. Genet. 16, 1133–1142 10.1093/hmg/ddm054 [DOI] [PubMed] [Google Scholar]
- 62.Gonitel R., Moffitt H., Sathasivam K., Woodman B., Detloff P.J., Faull R.L.M. et al. (2008) DNA instability in postmitotic neurons. Proc. Natl. Acad. Sci. U.S.A. 105, 3467–3472 10.1073/pnas.0800048105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swami M., Hendricks A., Gillis T., Massood T., Mysore J., Myers R.H. et al. (2009) Somatic expansion of the Huntington’s disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum. Mol. Genet. 18, 3039–3047 10.1093/hmg/ddp242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mergener R., Furtado G.V., de Mattos E.P., Leotti V.B., Jardin L.B. and Saraiva-Pereira M.L. (2020) Variation in DNA repair system gene as an additional modifier of age at onset in spinocerebellar ataxia type 3/Machado-Joseph disease. Neuromolecular Med. 22, 133–138 [DOI] [PubMed] [Google Scholar]
- 65.Hensman Moss D.J., Pardinas A.F., Langbehn D., Lo K., Leavitt B.R., Roos R. et al. (2017) Identification of genetic variants associated with Huntington’s disease progression: a genome-wide association study. Lancet Neurol. 16, 701–711 10.1016/S1474-4422(17)30161-8 [DOI] [PubMed] [Google Scholar]
- 66.Ciosi M., Maxwell A., Cumming S.A., Hensman-Moss D.J., Alshammari A.M., Flower M.D. et al. (2019) A genetic association study of glutamine-encoding DNA sequence structures, somatic CAG expansion, and DNA repair gene variants, with Huntington disease clinical outcomes. EBioMedicine 48, 568–580 10.1016/j.ebiom.2019.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flower M., Lomeikaite V., Ciosi M., Cumming S., Morales F., Lo K. et al. (2019) MSH3 modifies somatic instability and disease severity in Huntington’s and myotonic dystrophy type 1. Brain 142, 1876–1886 10.1093/brain/awz115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wright G.E.B., Collins J.A., Kay C., McDonald C., Dolzhenko E., Xia Q. et al. (2019) Length of uninterrupted CAG, independent of polyglutamine size, results in increased somatic instability, hastening onset of Huntington disease. Am. J. Hum. Genet. 104, 1116–1126 10.1016/j.ajhg.2019.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dandelot E. and Tome S. (2017) Genetic modifiers of CAG.CTG repeat instability in Huntington’s disease mouse models. In Huntington’s Disease-Molecular Pathogenesis and Current Models (Tunah N.E., ed.), IntechOpen, https://www.intechopen.com/books/howtoreference/huntington-s-disease-molecular-pathogenesis-and-current-models/genetic-modifiers-of-cag-ctg-repeat-instability-in-huntington-s-disease-mouse-models [Google Scholar]
- 70.Wheeler V.C., Lebel L.-A., Vrbanac V., Teed A., te Riele H. and MacDonald M.E. (2003) Mismatch repair gene Msh2 modifies the timing of early disease in HdhQ111 striatum. Hum. Mol. Genet. 12, 273–281 10.1093/hmg/ddg056 [DOI] [PubMed] [Google Scholar]
- 71.Manley K., Shirley T.L., Flaherty L. and Messer A. (1999) Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 23, 471–473 10.1038/70598 [DOI] [PubMed] [Google Scholar]
- 72.Kovtun I.V. and McMurray C.T. (2001) Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 27, 407–411 10.1038/86906 [DOI] [PubMed] [Google Scholar]
- 73.Owen B.A.L., Yang Z., Lai M., Gajek M., Badger J.D. II, Hayes J.J. et al. (2005) (CAG)n-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol. 12, 663–670 10.1038/nsmb965 [DOI] [PubMed] [Google Scholar]
- 74.Dragileva E., Hendricks A., Teed A., Gillis T., Lopez E.T., Friedberg E.C. et al. (2009) Intergenerational and striatal CAG repeat instability in Huntington’s disease knock-in mice involve different DNA repair genes. Neurobiol. Dis. 33, 37–47 10.1016/j.nbd.2008.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pinto R.M., Dragileva E., Kirby A., Lloret A., Lopez E.T., St Claire J. et al. (2013) Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington's disease mice: genome-wide and candidate approaches. PLoS Genet. 9, e1003930 10.1371/journal.pgen.1003930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Modrich P. (2016) Mechanisms in E. coli and human mismatch repair (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 55, 8490–8501 10.1002/anie.201601412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gomes-Pereira M., Fortune M.T., Ingram L., McAbney J.P. and Monckton D.G. (2004) Pms2 is a genetic enhancer of trinucleotide CAG•CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum. Mol. Genet. 13, 1815–1825 10.1093/hmg/ddh186 [DOI] [PubMed] [Google Scholar]
- 78.Miller C.A., Kim G.-Y., Zhao X.-N. and Usdin K. (2020) All three mammalian MutL complexes are required for repeat expansion in a mouse cell model of the Fragile X-related disorders. PLoS Genet. 16, e1008902 10.1371/journal.pgen.1008902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Long J.D., Lee J.-M., Aylward E.H., Gillis T., Mysore J.S., Elneel K.A. et al. (2018) Genetic modification of Huntington disease acts early in the prediagnosis phase. Am. J. Hum. Genet. 103, 349–357 10.1016/j.ajhg.2018.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao X.-N. and Usdin K. (2018) FAN1 protects against repeat expansions in a Fragile X mouse model. DNA Rep. (Amst.) 69, 1–5 10.1016/j.dnarep.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goold R., Flower M., Hensman-Moss D., Medway C., Wood-Kaczmar A., Andre R. et al. (2019) FAN1 modifies Huntington’s disease progression by stabilizing the expanded HTT CAG repeat. Hum. Mol. Genet. 28, 650–661 10.1093/hmg/ddy375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim K.-H., Hong E.P., Shin J.W., Chao M.J., Loupe J., Gillis T. et al. (2020) Genetic and functional analyses point to FAN1 as the source of multiple Huntington disease modifier effects. Am. J. Hum. Genet. 107, 96–110 10.1016/j.ajhg.2020.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Loupe J.M., Mouro Pinto R., Kim K.-H., Gillis T., Mysore J.S., Andrew M.A. et al. (2020) Promotion of somatic CAG repeat expansion by Fan1 knock-out in Huntington’s disease knock-in mice is blocked by Mlh1 knock-out. Hum. Mol. Genet. 10.1093/hmg/ddaa196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smogorzewska A., Desetty R., Saito T.T., Schlabach M., Lach F.P., Sowa M.E. et al. (2010) A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell 39, 36–47 10.1016/j.molcel.2010.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Modrich P. and Lahue R.S. (1996) Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem. 65, 101–133 10.1146/annurev.bi.65.070196.000533 [DOI] [PubMed] [Google Scholar]
- 86.Li G.-M. (2008) Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 10.1038/cr.2007.115 [DOI] [PubMed] [Google Scholar]
- 87.Hsieh P. and Zhang Y. (2017) The devil is in the details for DNA mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 114, 3552–3554 10.1073/pnas.1702747114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Parsons R., Li G.M., Longley M.J., Fang W.H., Papadopoulos N., Jen J. et al. (1993) Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 75, 1227–1236 10.1016/0092-8674(93)90331-J [DOI] [PubMed] [Google Scholar]
- 89.Genschel J., Littman S.J., Drummond J.T. and Modrich P. (1998) Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J. Biol. Chem. 273, 19895–19901 10.1074/jbc.273.31.19895 [DOI] [PubMed] [Google Scholar]
- 90.Kadyrova L.Y. and Kadyrov F.A. (2016) Endonuclease activities of MutLα and its homologs in DNA mismatch repair. DNA Rep. (Amst.) 38, 42–49 10.1016/j.dnarep.2015.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holmes J., Clark S. and Modrich P. (1990) Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc. Natl Acad. Sci. U.S.A. 87, 5837–5841 10.1073/pnas.87.15.5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thomas D.C., Roberts J.D. and Kunkel T.A. (1991) Heteroduplex repair in extracts of human HeLa cells. J. Biol. Chem. 266, 3744–3751, https://www.jbc.org/content/266/6/3744.abstract [PubMed] [Google Scholar]
- 93.Pluciennik A., Dzantiev L., Iyer R.R., Constantin N., Kadyrov F.A. and Modrich P. (2010) PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 107, 16066–16071 10.1073/pnas.1010662107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kadyrov F.A., Dzantiev L., Constantin N. and Modrich P. (2006) Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126, 297–308 10.1016/j.cell.2006.05.039 [DOI] [PubMed] [Google Scholar]
- 95.Gacy A.M., Goellner G., Juranic N., Macura S. and McMurray C.T. (1995) Trinucleotide repeats that expand in human disease form hairpin structure in vitro. Cell 81, 533–540 10.1016/0092-8674(95)90074-8 [DOI] [PubMed] [Google Scholar]
- 96.Pearson C.E., Wang Y.-H., Griffith J.D. and Sinden R.R. (1998) Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n-(CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 26, 816–823 10.1093/nar/26.3.816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iyer R.R., Pluciennik A., Napierala M. and Wells R.D. (2015) DNA triplet repeat expansion and mismatch repair. Annu. Rev. Biochem. 84, 199–226 10.1146/annurev-biochem-060614-034010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schmidt M.H.M. and Pearson C.E. (2016) Disease-associated repeat instability and mismatch repair. DNA Rep. (Amst.) 38, 117–126 10.1016/j.dnarep.2015.11.008 [DOI] [PubMed] [Google Scholar]
- 99.Khristich A.N. and Mirkin S.M. (2020) On the wrong DNA track: molecular mechanisms of repeat-mediated genome instability. J. Biol. Chem. 295, 4134–4170 10.1074/jbc.REV119.007678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lang W.H., Coats J.E., Majka J., Hura G.L., Lin Y., Rasnik I. et al. (2011) Conformational trapping of mismatch recognition complex MSH2/MSH3 on repair-resistant DNA loops. Proc. Natl. Acad. Sci. U.S.A. 108, E837–E844 10.1073/pnas.1105461108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pluciennik A., Burdett V., Baitinger C., Iyer R.R. and Modrich P. (2013) Extrahelical (CAG)/(CTG) triplet repeat elements support proliferating cell nuclear antigen loading and MutLα endonuclease activation. Proc. Natl. Acad. Sci. U.S.A. 110, 12277–12282 10.1073/pnas.1311325110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kadyrova L.Y., Gujar V., Burdett V., Modrich P.L. and Kadyrov F.A. (2020) Human MutLgamma, the MLH1-MLH3 heterodimer, is an endonuclease that promotes DNA expansion. Proc. Natl. Acad. Sci. U.S.A. 117, 3535–3542 10.1073/pnas.1914718117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Budworth H., Harris F.R., Williams P., Lee D.Y., Holt A., Pahnke J. et al. (2015) Suppression of somatic expansions delays the onset of pathophysiology in a mouse model of Huntington’s disease. PLoS Genet. 11, e1005267 10.1371/journal.pgen.1005267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kovtun I.V., Liu Y., Bjoras M., Klungland A., Wilson S.H. and McMurray C.T. (2007) OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 447, 447–452 10.1038/nature05778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wright G.E.B., Findlay Black H., Collins J.A., Gail-Duncan T., Caron N.S., Pearson C.E. et al. (2020) Interrupting sequence variants and age of onset in Huntington’s disease: clinical implications and emerging therapies. Lancet Neurol. 19, 930–939 10.1016/S1474-4422(20)30343-4 [DOI] [PubMed] [Google Scholar]
- 106.Valle L., de Voer R.M., Goldber Y., Sjursen W., Forsti A., Ruiz-Ponte C. et al. (2019) Update on genetic predisposition to colorectal cancer and polyposis. Mol. Aspects Med. 69, 10–26 10.1016/j.mam.2019.03.001 [DOI] [PubMed] [Google Scholar]
- 107.Squitieri F., Gellera C., Cannella M., Mariotti C., Cislaghi G., Rubinsztein D.C. et al. (2003) Homozygosity for CAG mutation in Huntington disease is associated with a more severe clinical course. Brain 126, 946–955 10.1093/brain/awg077 [DOI] [PubMed] [Google Scholar]
- 108.Tabrizi S.J., Ghosh R. and Leavitt B.R. (2019) Huntingtin lowering strategies for disease modification in Huntington’s disease. Neuron 101, 801–819 10.1016/j.neuron.2019.01.039 [DOI] [PubMed] [Google Scholar]
- 109.Marxreiter F., Stemick J. and Kohl Z. (2020) Huntingtin lowering strategies. Int. J. Mol. Sci. 21, 2146 10.3390/ijms21062146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tabrizi S.J., Leavitt B.R., Landwehrmeyer G.B., Wild E.J., Saft C., Barker R.A. et al. (2019) Targeting Huntingtin expression in patients with Huntington’s disease. N. Engl. J. Med. 380, 2307–2316 10.1056/NEJMoa1900907 [DOI] [PubMed] [Google Scholar]
- 111.Silva A.C., Lobo D.D., Martins I.M., Lopes S.M., Henriques C., Duarte S.P. et al. (2020) Antisense oligonucleotide therapeutics in neurodegenerative diseases: the case of polyglutamine disorders. Brain 143, 407–429 10.1093/brain/awz328 [DOI] [PubMed] [Google Scholar]
- 112.Lopez Castel A., Cleary J.D. and Pearson C.E. (2010) Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 11, 165–170 10.1038/nrm2854 [DOI] [PubMed] [Google Scholar]
- 113.Kovalenko M., Dragileva E., St Claire J., Gillis T., Guide J.R., New J. et al. (2012) Msh2 acts in medium-spiny striatal neurons as an enhancer of CAG instability and mutant huntingtin phenotypes in Huntington’s disease knock-in mice. PLoS ONE 7, e44273 10.1371/journal.pone.0044273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Keogh N., Chan K.Y., Li G.-M. and Lahue R.S. (2017) MutSβ abundance and Msh3 ATP hydrolysis activity are important drivers of CTG•CAG repeat expansions. Nucleic Acids Res. 45, 10068–10078 10.1093/nar/gkx650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Foiry L., Dong L., Savouret C., Hubert L., te Riele H., Junien C. et al. (2006) Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum. Genet. 119, 520–526 10.1007/s00439-006-0164-7 [DOI] [PubMed] [Google Scholar]
- 116.Tome S., Manley K., Simard J.P., Clark G.W., Slean M.M., Swami M. et al. (2013) MSH3 polymorphisms and protein levels affect CAG repeat instability in Huntington’s disease mice. PLoS Genet. 9, e1003280 10.1371/journal.pgen.1003280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nakatani R., Nakamori M., Fujimura H., Mochizuki H. and Takahashi M.P. (2015) Large expansion of CTG•CAG repeats is exacerbated by MutSβ in human cells. Sci. Rep. 5,11020 10.1038/srep11020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Williams G.M., Paschalis V., Ortega J., Muskett F.W., Hodgkinson J.T., Li G.-M. et al. (2020) HDAC3 deacetylates the DNA mismatch repair factor MutSβ to stimulate triplet repeat expansions. Proc. Natl. Acad. Sci. U.S.A. 117, 23596–23605 10.1073/pnas.2013223117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gottesfeld J.M. and Pandolfo M. (2009) Development of histone deacetylase inhibitors as therapeutics for neurological disease. Future Neurol. 4, 775–784 10.2217/fnl.09.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Thomas E.A. (2014) Involvement of HDAC1 and HDAC3 in the pathology of polyglutamine disorders: therapeutic implications for selective HDAC1/HDAC3 inhibitors. Pharmaceuticals 7, 634–661 10.3390/ph7060634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thomas E.A. and D’Mello S.R. (2018) Complex neuroprotective and neurotoxic effects of histone deacetylases. J. Neurochem. 145, 96–110 10.1111/jnc.14309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rodrigues D.A., Pinheiro P.D.M., Sagrillo F.S., Bolognesi M.L. and Fraga C.A.M. (2020) Histone deacetylases as targets for the treatment of neurodegenerative disorders: challenges and future opportunities. Med. Res. Rev. 40, 2177–2211 10.1002/med.21701 [DOI] [PubMed] [Google Scholar]
- 123.Ho T.C.S., Chan A.H.Y. and Ganesan A. (2020) Thirty years of HDAC inhibitors: 2020 insight and hindsight. J. Med. Chem. 10.1021/acs.jmedchem.0c00830 [DOI] [PubMed] [Google Scholar]
- 124.Jia H., Morris C.D., Williams R.M., Loring J.F. and Thomas E.A. (2015) HDAC inhibition imparts beneficial transgenerational effects in Huntington’s disease mice via altered DNA and histone methylation. Proc. Natl. Acad. Sci. U.S.A. 112, E56–E64 10.1073/pnas.1415195112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Suelves N., Kirkham-McCarthy L., Lahue R.S. and Gines S. (2017) An HDAC3-selective inhibitor delivers concurrent benefits in Huntington’s disease mice by preventing cognitive decline and suppressing somatic CAG repeat expansions. Sci. Rep. 7, 6082 10.1038/s41598-017-05125-2 [DOI] [PMC free article] [PubMed] [Google Scholar]