Abstract
Rapid detection of antibiotic-resistant bacteria in blood cultures is critical for the timely treatment of patients with sepsis. The aim of this study was to develop a simple method for the rapid detection of drug-resistant bacteria from blood cultures and to evaluate its performance. We developed an optical microscopy-based microcolony detection method (MCD) for the rapid detection of antibiotic-resistant bacterial colonies in media. This method was tested using staphylococci resistant to methicillin and gram-negative bacilli resistant to third-generation cephalosporins and carbapenem. The results of the investigations of clinical samples using this method were compared with the drug susceptibility testing results for each of the 457 isolates, which included 134 staphylococci and 323 g-negative bacilli. The MCD was successful in detecting antibiotic-resistant bacterial growth from culture-positive blood samples in approximately 3 h. The sensitivity/specificity for methicillin-resistant staphylococci was 100%/97.2%. In the case of gram-negative bacilli, the sensitivity/specificity values for bacteria resistant to ceftriaxone, ceftazidime, and carbapenem were 100%/98.7%, 100%/89.3%, and 100%/90.9%, respectively. Therefore, MCD is a clinically useful screening method for the efficient and rapid detection of antibiotic-resistant bacteria and can be easily implemented in laboratories.
Keywords: Biotechnology, Microbiology, Genetics, Bacteria, Microbial genomics, Microorganism, Microbial biotechnology, Bloodstream infection, Antimicrobial therapy, Microcolony detection method, MRSA, Extended spectrum β-lactamase production in Enterobacterales, Carbapenem-resistant Enterobacterales
Biotechnology; Microbiology; Genetics; Bacteria; Microbial genomics; Microorganism; Microbial biotechnology; Bloodstream infection; Antimicrobial therapy; Microcolony detection method; MRSA; Extended spectrum β-lactamase production in Enterobacterales; Carbapenem-resistant Enterobacterales.
1. Introduction
Bloodstream infections (BSI) are an important cause of mortality and healthcare costs worldwide [1, 2]. Early treatment of sepsis with an appropriate antimicrobial drug has a positive influence on the prognosis [3]. Therefore, rapid detection of resistant bacteria in blood culture is extremely important in the treatment of infectious diseases. In this study, we have devised and evaluated a simple screening method for the rapid detection of antibiotic resistance in bacteria from blood culture.
2. Materials and methods
2.1. Proof-of-concept investigation
We first evaluated the performance of this method with respect to the rapid detection of methicillin resistance in Staphylococcus spp., specifically in strains of methicillin-resistant Staphylococcus aureus (MRSA), from culture-positive blood samples. We also utilized this method to detect resistance of gram-negative bacteria of the Enterobacterelaes towards third-generation cephalosporins (3GC), via the production of extended-spectrum β-lactamases (ESBL), and carbapenem, via the production of carbapenemases, as observed in carbapenemase-producing Enterobacterelaes (CPE). Specifically, the microcolony detection method (MCD) was evaluated using mecA-positive MRSA (30 strains), 30 ESBL-producing Enterobacterales (1 TEM-20, 1 TEM-52, 2 SHV-12, 12 CTX-M-1, 12 CTX-M-9, and 2 CTX-M-2), 30 plasmid-mediated AmpC β-lactamases (pAmpC)-producing Enterobacterales (17 CIT, 10 DHA, and 3 MOX), and 30 CPE (19 IMP, 2 VIM, 1 KPC, 1 NDM, 1 OXA-48, and 6 GES-4) strains. The strains used were either obtained from preserved clinical isolates or from the collection of the Study of Bacterial Resistance Kinki Region of Japan (SBRK). Confirmation of the mechanism of resistance was performed by PCR as previously reported [4, 5, 6, 7, 8, 9]. The amplified products were sequenced using an automated DNA sequencer (ABI 3100, Applied Biosystems, Foster City, CA). Clinical isolates, specifically, 30 g-positive and 30 g-negative bacterial strains that did not display antibiotic resistance, acted as negative controls.
2.2. Clinical investigation
Clinical investigations were conducted over a period of 36 months (April 2017 to March 2020), and 456 specimens that resulted in a positive blood culture were targeted. The isolates included 134 strains of cluster-forming gram-positive cocci and 323 strains of gram-negative bacilli. The microorganisms were identified using VITEK 2 (bioMérieux, Marcy-l'Étoile, France). The gram-positive bacteria were all strains of staphylococci, including 88 strains of S. aureus, 29 of Staphylococcus epidermidis, 5 of Staphylococcus lugdunensis, 6 of Staphylococcus hominis, 3 of Staphylococcus haemolyticus, 1 of Staphylococcus simulans, 1 of Staphylococcus warneri, and 1 of Staphylococcus schleiferi. The gram-negative bacteria included 289 Enterobacterales strains- 149 strains of Escherichia coli, 79 of Klebsiella pneumoniae, 18 of the Enterobacter cloacae complex, 14 of Klebsiella oxytoca, 10 of Proteus mirabilis, 6 of Klebsiella aerogenes (Enterobacter aerogenes), 5 of Citrobacter koseri, 4 of Citrobacter freundii, 2 of Serratia marcescens, 1 of Raoultella planticola, and 1 Salmonella spp. In addition, 3 strains of Aeromonas spp. and 31 strains of glucose-nofermenting Gram-negative rods (NF-GNR) including 17 strains of Pseudomonas aeruginosa, 8 of the Acinetobacter baumannii complex, 5 of Stenotrophomonas maltophilia, and 1 Shewanella algae were also analyzed. Anaerobes were excluded from the study. The study was approved by the Naga municipal hospital ethics committee.
2.3. Microcolony detection method (MCD)
We developed a novel test method for the rapid detection of bacterial microcolonies on selective media supplemented with antibiotics. Blood cultures were maintained using BD BACTEC FX (Becton Dickinson, Tokyo, Japan). When a positive signal of growth was observed, subcultures were made onto sheep blood agar (Kyokuto Pharmaceutical Co., Tokyo, Japan), modified Drigalski agar (BTB agar; Eiken Chemical, Tokyo, Japan), and an antibiotic-containing selective medium for the detection of possible antibiotic-resistant bacteria from the positive blood cultures. The selective medium used for gram-positive cocci was CHROMagar MRSA (CHROMagar, Paris, France), and that used for gram-negative bacilli included chromID ESBL medium (bioMérieux) and CHROMagar mSuperCARBA medium (CHROMagar). The subculture was performed by extracting the positive blood culture using a syringe and adding 1 μL of it to each culture medium. The subcultured media were incubated at 35 °C under aerobic conditions. After 2–4 h, antibiotic-containing selective media (with the lid on) were placed on the stage of an optical microscope, and formation of microcolonies at the base of the culture plate was monitored at 100x magnification using a 10x objective lens. We have termed this method MCD. Before growth in the selective bacterial culture medium can be confirmed, it is necessary to confirm bacterial growth in a nonselective medium. Accordingly, MCD was first used to determine the formation of microcolonies in BTB agar. The subsequent detection of similar microcolonies in selective bacterial culture media was considered positive for antibiotic-resistant bacteria and negative for the detection of no microcolonies (Figures 1 and 2). In the analysis of gram-negative bacilli, the appearance of microcolonies, which were either elliptic or had filamentation, were considered as signs of positive growth (Figure 3). In the chromID ESBL medium, the result was regarded as negative if there was no formation of microcolonies and a filament image could be confirmed (Figure 4c). If microcolonies did not form in the BTB agar within the stipulated time, it was judged as indeterminate.
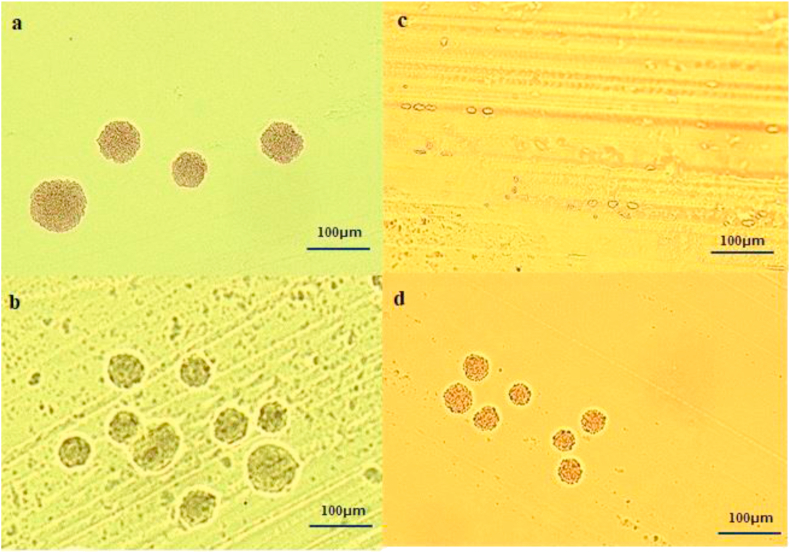
Figure 1.
Image of micro colonies of staphylococci formed on each medium by microcolony detection method (MCD) (×100) a:Methicillin-susceptible staphylococci on the BTB agar, b:Methicillin-resistant staphylococci on the BTB agar, c:Methicillin-susceptible staphylococci on the CHROMagar MRSA medium, there is no formation of small colonies, only red blood cells can be confirmed, d: Methicillin-resistant staphylococci on the CHROMagar MRSA medium.
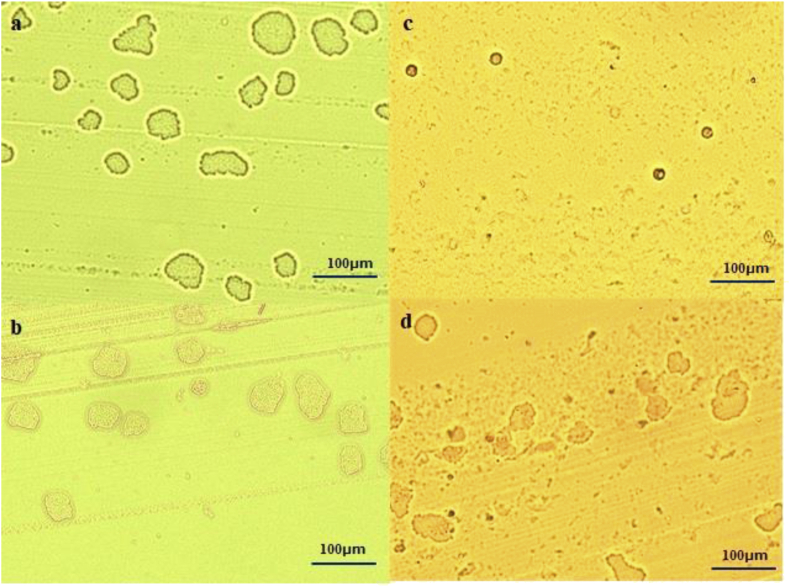
Figure 2.
Image of micro colonies of Enterobacterales formed on each medium by MCD (× 100) a: Carbapenem-susceptible Enterobacterales on the BTB agar, b: Carbapenem-resistant Enterobacterales (IMP-1 producing) on the BTB agar, c: Carbapenem-susceptible Enterobacterales on the CHROMagar mSuperCARBA medium, there is no formation of small colonies, only red blood cells can be confirmed, d: Carbapenem-resistant Enterobacterales (IMP-1 producing) on the CHROMagar mSuperCARBA medium.
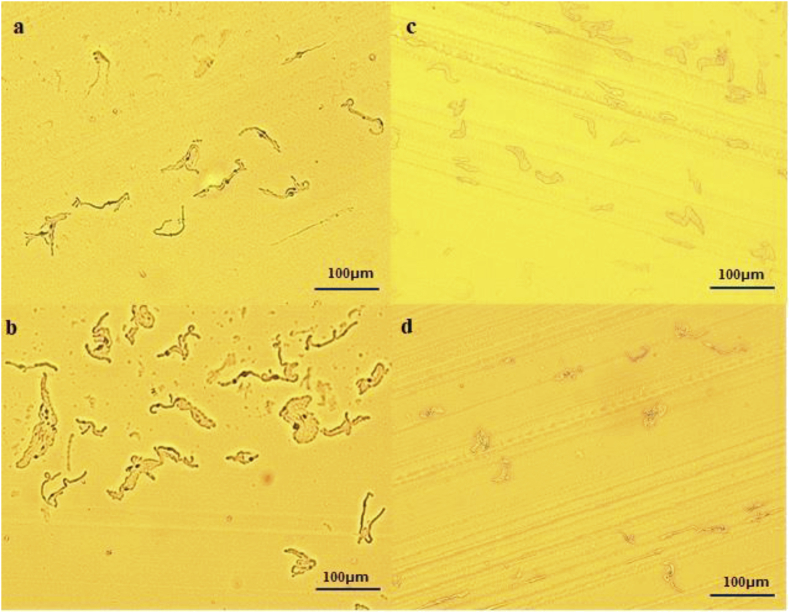
Figure 3.
Image of micro colonies with filamentization of gram-negative bacteria formed on each medium by MCD (× 100) a: Carbapenem-resistant Enterobacterales (IMP-6 producing) on the chromID ESBL medium, b: Carbapenem-resistant Enterobacterales (IMP-6 producing) on the CHROMagar mSuperCARBA medium. c: Pseudomonas aeruginosa on the chromID ESBL medium, d: Pseudomonas aeruginosa on the CHROMagar mSuperCARBA medium.
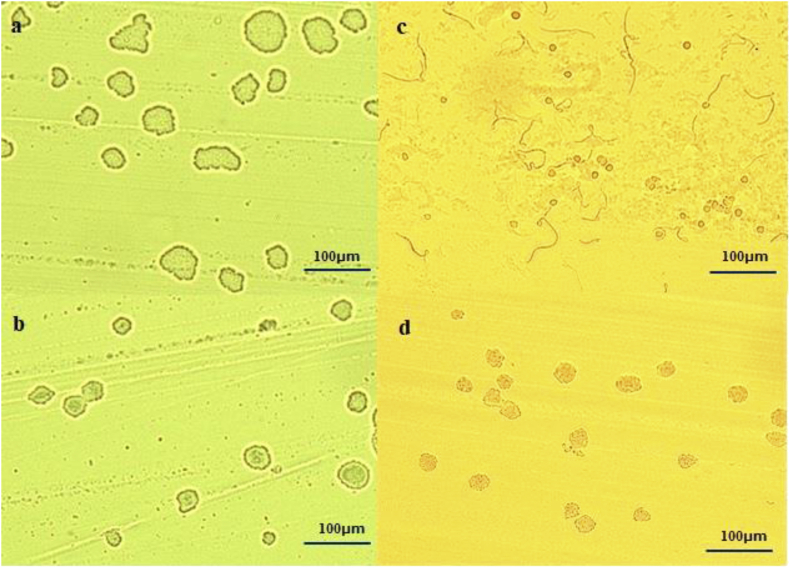
Figure 4.
Image of micro colonies of Enterobacterales formed on each medium by MCD (×100) a: Third generation cephalosporin (3GC)-susceptible Enterobacterales (non-ESBL producing) on the BTB agar, b: 3GC-resistant Enterobacterales (ESBL producing) on the BTB agar, c: 3GC-susceptible Enterobacterales (non-ESBL producing) forms filament type on the chromID ESBL medium, d: 3GC-resistant Enterobacterales (ESBL producing) on the chromID ESBL medium.
2.4. Evaluation of MCD
The MCD results obtained for strains used in the proof-of-concept investigation were compared to the search results of the resistance gene, whereas the MCD results from the clinical investigation were compared to those obtained by antimicrobial susceptibility testing carried out using VITEK 2. Antimicrobial susceptibility tests were carried out using CLSI M100 - S27 [10] with the use of oxacillin and cefoxitin for testing methicillin-resistance in staphylococci; ceftriaxone (CTRX) and ceftazidime (CAZ) for testing 3GC-resistance in gram-negative bacilli; and imipenem (IPM) and meropenem (MEPM) for testing carbapenem-resistance in gram-negative bacilli.
2.5. Quality control of MCD
Staphylococcus aureus ATCC 43300 was used as the positive control for methicillin-resistance, and Staphylococcus aureus ATCC 29213 was used as the negative control. Among the gram-negative bacilli, Klebsiella pneumoniae ATCC 700603 was used as a positive control for 3GC-resistance and Klebsiella pneumoniae ATCC BAA-1705 for carbapenem-resistance; E. coli ATCC 25922 was used as the negative control.
3. Results
The MCD analysis of all strains used in the proof-of-concept investigation revealed formation of microcolonies on BTB agar within 2–4 h (average 3 h) (Table 1). Hundred percent sensitivity and specificity was observed in the detection of the mecA-positive strains of Staphylococcus and the ESBL and carbapenemase gene-carrying strains of Enterobacterales by MCD. The pAmpC gene-carrying strains were detected with 56.7% sensitivity (Table 2). The antimicrobial susceptibility testing carried out as a part of the clinical investigation (Table 3) revealed that 17 isolates of S. aureus and 32 of coagulase-negative staphylococci were resistant to methicillin. However, 2 isolates of S. lugdunensis failed to form a clear colony on BTB agar within 4 h and were therefore judged as indeterminate. Among the gram-negative bacilli, 3GC-resistance was observed in 23 ESBL producers (17 E. coli, 5 K. pneumoniae, and 1 P. mirabilis), 4 AmpC β-lactamase producers (3 E. cloacae complex and 1 C. freundii), 5 NF-GNR, and 1 Aeromonas spp. Carbapenem-resistant strains included 1 non-carbapenemase-producing Enterobacter cloacae and 3 NF-GNR (Table 4). The results obtained for P. aeruginosa and A. baumannii complex showed discordance. When the results were divided by strain, the results obtained with Enterobacterales showed 100% sensitivity and specificity. Only one of the S. algae was indeterminate. Detection of antibiotic resistance using MCD was possible in most cases in an average of 3 h.
Table 1.
Judgment time of proof-of-concept investigation by comparison between MCD and each resistant bacteria.
| organism | Incubation time (h) |
average time (h) | ||||
|---|---|---|---|---|---|---|
| 2 | 3 | 4 | ||||
|
Staphylococcus aureus |
mecA gene |
positive (n = 30) | 0 | 29 | 1 | 3.03 |
| negative (n = 30) |
0 |
28 |
2 |
3.07 |
||
| total (n = 60) | 0 | 57 | 3 | 3.05 | ||
|
Enterobacterales |
ESBL gene |
positive (n = 30) | 6 | 24 | 0 | 2.80 |
| negative (n = 30) |
2 |
27 |
1 |
2.97 |
||
| total (n = 60) | 8 |
51 |
1 |
2.88 |
||
| pAmpC gene |
positive (n = 30) | 6 | 24 | 0 | 2.80 | |
| negative (n = 30) |
2 |
27 |
1 |
2.97 |
||
| total (n = 60) | 8 |
51 |
1 |
2.88 |
||
| Carbapenemase gene |
positive (n = 30) |
6 |
24 |
0 |
2.80 |
|
| negative (n = 30) |
2 |
27 |
1 |
2.97 |
||
| total (n = 60) | 8 | 51 | 1 | 2.88 | ||
MCD: Microcolony Detection method; ESBL: extended spectrum β-lactamase; pAmpC: plasmid mediated AmpC β-lactamase; The MCD used CHROMagar MRSA medium for comparison with mecA gene of staphylococci, chromID ESBL medium for comparison with ESBL and pAmpC gene of Enterobacterales, and CHROMagar mSuperCARBA medium for comparison with carbapenemase producing Enterobacterales.
Table 2.
Result of proof-of-concept investigation by comparison between MCD and each resistant bacteria.
|
Staphylococcus aureus |
Enterobacterales |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| mecA gene |
ESBL gene |
pAmpC gene |
Carbapenemase gene |
||||||
| Positive (n = 30) | Negative (n = 30) | Positive (n = 30) | Negative (n = 30) | Positive (n = 30) | Negative (n = 30) | Positive (n = 30) | Negative (n = 30) | ||
| MCD | Positive | 30 | 0 | 30 | 0 | 17 | 0 | 30 | 0 |
| Negative |
0 |
30 |
0 |
30 |
13 |
30 |
0 |
30 |
|
| sensitivity (%) | 100 | 100 | 56.7 | 100 | |||||
| specificity (%) | 100 | 100 | 100 | 100 | |||||
MCD: Microcolony Detection method; ESBL: extended spectrum β-lactamase; pAmpC: plasmid mediated AmpC β-lactamase; The MCD used CHROMagar MRSA medium for comparison with mecA gene of staphylococci, chromID ESBL medium for comparison with ESBL and pAmpC gene of Enterobacterales, and CHROMagar mSuperCARBA medium for comparison with carbapenemase producing Enterobacterales.
Table 3.
Results of comparative study between MCD and antimicrobial susceptibility of staphylococci using 134 clinical isolates.
| Staphylococci (n = 134) |
|||||
|---|---|---|---|---|---|
| MPIPC |
CFX |
||||
| Resistant (n = 62) | Susceptible (n = 72) | Resistant (n = 62) | Susceptible (n = 72) | ||
| MCD | Positive | 62 | 0 | 62 | 0 |
| Indeterminate | 0 | 2 | 0 | 2 | |
| Negative |
0 |
70 |
0 |
70 |
|
| sensitivity (%) | 100 | 100 | |||
| specificity (%) | 97.2 | 97.2 | |||
MCD: Microcolony Detection method; MPIPC: oxacillin; CFX: cefoxitin; The MCD used CHROMagar MRSA medium for comparison with methicillin sensitivity of staphylococci.
Table 4.
Results of comparative study between MCD and antimicrobial susceptibility of gram negative bacteria using 323 clinical isolates.
| Gram-negative bacteria (n = 323) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| CTRX |
CAZ |
IPM |
MEPM |
||||||
| Resistant (n = 86) | Susceptible (n = 237) | Resistant (n = 61) | Susceptible (n = 262) | Resistant (n = 6) | Susceptible (n = 317) | Resistant (n = 6) | Susceptible (n = 317) | ||
| MCD | Positive | 86 | 2 | 61 | 27 | 6 | 28 | 6 | 28 |
| Indeterminate | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | |
| Negative |
0 |
234 |
0 |
234 |
0 |
288 |
0 |
288 |
|
| sensitivity (%) | 100 | 100 | 100 | 100 | |||||
| specificity (%) | 98.7 | 89.3 | 90.9 | 90.9 | |||||
MCD: Microcolony Detection method; CTRX: ceftriaxone; CAZ: ceftazidime; IPM: imipenem; MEPM: meropenem; The MCD used chromID ESBL medium for comparison with susceptibility of third generation cephalosporin of gram-negative bacteria, and CHROMagar mSuperCARBA medium for comparison with susceptibility of carbapenem of gram-negative bacteria.
4. Discussion
Rapid detection of antimicrobial resistance in culture-positive blood specimens is critical to ensure timely and appropriate antibiotic therapy that can reduce mortality rates in patients with BSI [11]. In recent years, several methods have been developed to detect resistance genes in blood culture using PCR [12, 13, 14, 15]. Although these methods are being widely used, they have intrinsic limitations such as complex procedures, high costs and the necessity of a dedicated machine. MCD was developed with the aim of rapidly detecting antibiotic-resistant bacteria from blood cultures using a simple procedure. This method of observing the culture medium directly using a microscope to detect colonies is not a new one; it has previously been used for culturing mycoplasma using pleuropneumonia-like organism (PPLO) agar medium [16]. We believe that by using this method in combination with selective media containing antibiotics, it is possible to simplify and speed up the screening and detection of antibiotic-resistant bacteria in patients with sepsis.
Working on a simple principle, the MCD detects the formation of microcolonies that cannot be observed by the naked eye. Therefore, the accuracy of this method is a function of the quality of the agar medium. No special microscope or lens is required; therefore, MCD can be performed using an optical microscope routinely used in laboratories. We believe that it will be beneficial to investigate the use of other media in conjunction with MCD. The use of BTB agar is primarily to prevent false negatives; it is important to confirm the formation of microcolonies in a non-selective medium before results in a selective medium can be interpreted. It is easier to observe small colonies on BTB agar than on blood agar due to greater transmission of light in the former. In instances where BTB is not used in MCD analysis, we recommend the use of a non-selective agar medium such as Mueller-Hinton agar, which displays good light transmission. The detection of microcolonies is not difficult, and most researchers can discern bacterial growth in a short time. Although most bacterial species can be detected in about 3–4 h, MCD screening may be inappropriate for bacteria that show growth retardation or poor growth on non-selective agar medium.
MCD using chromID ESBL medium revealed filamentation of all Enterobacterales that were sensitive to 3GC; this was probably due to the antibacterial activity of the cefpodoxime added to this medium [17]. In line with this possibility, the bacteria that displayed filamentation were considered susceptible to 3GC. NF-GNR such as P. aeruginosa, however, displayed both microcolony formation and filamentation. Therefore, the analysis of gram-negative bacilli outside the Enterobacterales order should be performed very carefully. The chromID ESBL medium can detect ESBL-producing bacteria with high sensitivity, but false positives such as those reported for P. aeruginosa and chromosomal AmpC β-lactamase producing Enterobacterales are possible [17]. Nevertheless, the formation of colonies in this medium is indicative of bacterial resistance to 3GC (including in the cases of false positives), enabling the quick determination of the drug susceptibility of the strains. Since the detection sensitivity of pAmpC was low when the chromID ESBL medium was used, detection of AmpC-producing bacteria may require subculture in other media.
Carbapenem-susceptible strains were not found to exhibit filamentation on mSuperCARBA medium (the antimicrobial drugs in this medium were not disclosed by the company); however, it has been confirmed that CPE can form microcolonies while being filamentized in this medium. It is necessary to consider these instances as resistance to the drug. Cases of sepsis caused by CPE are extremely rare in Japan; therefore, only one carbapenem-resistant Enterobacterales (CRE) was used in this clinical study. It has been reported that the SuperCARBA and mSuperCARBA media have high sensitivity for the detection of CRE [18, 19]; the MCD method using these media can therefore be presumed to be useful for the rapid detection of CRE. Further investigation is, however, required for the detection of CPE.
NF-GNR detected in this study showed growth in chromID ESBL medium and mSuperCARBA media. The cause seems to be intrinsic resistance to the antibacterial drugs contained in these media. Growth detected by the MCD method can, therefore, be presumed to be NF-GNR in addition to CRE. It should be noted that although P. aeruginosa and A. baumannii complex displayed growth in the MCD method, these isolates were found to be susceptible to CAZ, IPM, and MEPM. Thus, the results of this method may not be consistent with those from susceptibility testing for NF–GNR.
Although the antimicrobial agent in CHROMagar MRSA is undisclosed, this medium has been reported to be very sensitive for the detection of methicillin-resistance [20]. Therefore, MCD using this medium was found to be useful for the rapid detection of methicillin-resistance.
A possible cause of false positives is the use of expired antibacterial media with reduced drug potency. In addition, false positives can occur even when observing only the first area where the blood culture positive fluid is applied in a concentrated manner. Therefore, colony observation should be judged in the area where independent colonies can be confirmed. False negatives may be due to bacterial growth failure. However, we speculate that false negatives can be prevented by observing the colonies of antibacterial-free media such as BTB agar and MH agar symmetrically. In this study, MCD showed high sensitivity and specificity. However, in cases where specially resistant bacteria or rare bacteria are contained, this may not always be the case, so further verification is required. Detection of antibiotic resistance conferring genes in culture-positive blood specimens using PCR is very useful; however, it has the disadvantage of being able to detect only known resistance genes. In other words, it is necessary to be aware that susceptibility to these antimicrobial drugs cannot be determined since this method does not consider any antimicrobial resistance mechanism other than that conferred by the gene of interest. The MCD method uses antibiotic-containing selective media to analyze the susceptibility or resistance of bacterial cultures regardless of the mechanism of resistance or the gene. We consider this to be an advantage over the existing method that detects resistance-conferring genes. Performing a combination of rapid detection of antimicrobial resistance genes in blood culture together with MCD could be clinically useful.
5. Conclusions
We have developed the MCD method for rapid screening and detection of antibiotic-resistant bacteria in positive blood cultures. It successfully detected the presence of drug-resistant bacteria in about 3 h with high sensitivity and specificity. Owing to its simplicity and non-reliance on specialized equipment, this method can be successfully implemented in many clinical microbiology laboratories.
Declarations
Author contribution statement
Tomokazu Kuchibiro: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Asami Hirano: Performed the experiments; Analyzed and interpreted the data.
Shirou Ogasawara: Performed the experiments.
Tatsuya Nakamura: Analyzed and interpreted the data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
This study was supported by the Study of Bacterial Resistance Kinki Region of Japan.
We would like to thank Editage (www.editage.com) for English language editing.
References
- 1.Álvaro-Meca A., Jiménez-Sousa M.A., Micheloud D., Sánchez-Lopez A., Heredia-Rodríguez M., Tamayo E., Resino S. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 2001;29:303–310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Goto M., Al-Hasan M.N. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin. Microbiol. Infect. 2013;19:501–509. doi: 10.1111/1469-0691.12195. [DOI] [PubMed] [Google Scholar]
- 3.Weinstein M.P., Towns M.L., Quartey S.M., Mirrett S., Reimer L.G., Parmigiani G., Reller L.B. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin. Infect. Dis. 1977;24:584–602. doi: 10.1093/clind/24.4.584. [DOI] [PubMed] [Google Scholar]
- 4.Ryffel C., Tesch W., Birch-Machin I., Reynolds P.E., Barberis-Maino L., Kayser F.H., Berger-Bachi B. Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Gene. 1900;94:137–138. doi: 10.1016/0378-1119(90)90481-6. [DOI] [PubMed] [Google Scholar]
- 5.Arlet G., Philippon A. Construction by polymerase chain reaction and use of intragenic DNA probes for three main types of transferableβ-lactamases (TEM, SHV, CARB) FEMS Microbiol. Lett. 1991;66:19–25. doi: 10.1016/0378-1097(91)90414-6. [DOI] [PubMed] [Google Scholar]
- 6.Queenan A.M., Bush K. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfeifer Y., Wilharm G., Zander E., Wichelhaus T.A., Götting S., Hunfeld K.P., Seifert H., Witte W., Higgins P.G. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 2011;66:1998–2001. doi: 10.1093/jac/dkr256. [DOI] [PubMed] [Google Scholar]
- 8.Dallenne C., Da Costa A., Decré D., Favier C., Arlet G. Development of a set of multi-plex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010;65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 9.Notake S., Matsuda M., Tamai k., Yanagisawa H., Hiramatsu K., Kikuchi K. Detection of IMP metallo-β-lactamase in carbapenem-nonsusceptible Enterobacteriaceae and non-glucose-fermenting Gram-negative rods by immunochromatography assay. J. Clin. Microbiol. 2013;51:1762–1768. doi: 10.1128/JCM.00234-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute (CLSI) Clinical and Laboratory Standards Institute; Wayne, P.A., CLSI: 2017. Performance Standards for Antimicrobial Susceptibility Testing 27th Ed CLSI Supplement M100-S27. [Google Scholar]
- 11.Leibovici L., Shraga I., Drucker M., Konigsberger H., Samra Z., Pitlik S.D. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J. Intern. Med. 1998;244:379–386. doi: 10.1046/j.1365-2796.1998.00379.x. [DOI] [PubMed] [Google Scholar]
- 12.Louie L., Goodfellow J., Mathieu P., Glatt A., Louie M., Simor A.E. Rapid detection of methicillin-resistant staphylococci from blood culture bottles by using a multiplex PCR assay. J. Clin. Microbiol. 2002;40:2786–2790. doi: 10.1128/JCM.40.8.2786-2790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wojewoda C.M., Sercia L., Navas M., Tuohy M., Wilson D., Hall G.S., Procop G.W., Richter S.S. Evaluation of the Verigene Gram-positive blood culture nucleic acid test for rapid detection of bacteria and resistance determinants. J. Clin. Microbiol. 2013;51:2072–2076. doi: 10.1128/JCM.00831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhatti M.M., Boonlayangoor S., Beavis K.G., Tesic V. Evaluation of FilmArray and Verigene systems for rapid identification of positive blood cultures. J. Clin. Microbiol. 2014;52:3433–3436. doi: 10.1128/JCM.01417-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodémont M., De Mendonça R., Nonhoff C., Roisin S., Denis O. Performance of the Verigene Gram-negative blood culture assay for rapid detection of bacteria and resistance determinants. J. Clin. Microbiol. 2014;52:3085–3087. doi: 10.1128/JCM.01099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chanock R.M., Hayflick L., Balile M.F. Growth on artificial medium of an agent associated with atypical pneumonia and its identification as a PPLO. Proc. Nat. Acad. Sci. U.S.A. 1962;48:41–49. doi: 10.1073/pnas.48.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Réglier-Poupet H., Naas T., Carrer A., Cady A., Adam J.M., Fortineau N., Poyart C., Nordmann P. Performance of chromID ESBL, a chromogenic medium for detection of Enterobacteriaceae producing extended-spectrum beta-lactamases. J. Med. Microbiol. 2008;57:310–315. doi: 10.1099/jmm.0.47625-0. [DOI] [PubMed] [Google Scholar]
- 18.Viau R., Frank K.M., Jacobs M.R., Wilson B., Kaye K., Donskey C.J., Perez F., Endimiani A., Bonomo R.A. Intestinal carriage of carbapenemase-producing organisms: current status of surveillance methods. Clin. Microbiol. Rev. 2016;29:1–27. doi: 10.1128/CMR.00108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Quintanilla M., Poirel L., Nordmann P. CHROMagar mSuperCARBA and RAPIDEC® Carba NP test for detection of carbapenemase-producing Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2018;90:77–80. doi: 10.1016/j.diagmicrobio.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Diederen B., van Duijn I., van Belkum A., Willemse P., van Keulen P., Kluytmans J. Performance of CHROMagar MRSA medium for detection of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2005;43:1925–1927. doi: 10.1128/JCM.43.4.1925-1927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]