Abstract
Msh homeobox (Msx) is a subclass of homeobox transcriptional regulators that control cell lineage development, including the early stage of vertebrate limb development, although the underlying mechanisms are not clear. Here, we demonstrate that Msx1 promotes the proliferation of myoblasts and mesenchymal stem cells (MSCs) by enhancing mitogen-activated protein kinase (MAPK) signaling. Msx1 directly binds to and upregulates the expression of fibroblast growth factor 9 (Fgf9) and Fgf18. Accordingly, knockdown or antibody neutralization of Fgf9/18 inhibits Msx1-activated extracellular signal-regulated kinase 1/2 (Erk1/2) phosphorylation. Mechanistically, we determined that the phosphorylation of Msx1 at Ser136 is critical for enhancing Fgf9 and Fgf18 expression and cell proliferation, and cyclin-dependent kinase 1 (CDK1) is apparently responsible for Ser136 phosphorylation. Furthermore, mesenchymal deletion of Msx1/2 results in decreased Fgf9 and Fgf18 expression and Erk1/2 phosphorylation, which leads to serious defects in limb development in mice. Collectively, our findings established an important function of the Msx1-Fgf-MAPK signaling axis in promoting cell proliferation, thus providing a new mechanistic insight into limb development.
INTRODUCTION
Vertebrate limb development relies on the activity of signaling that control patterning and growth of the limb bud along three orthogonal axes. Among them, fibroblast growth factor (Fgf) signaling is one of the dominant elements that control the elongation of limb bud along proximo–distal (P–D) axis, which promotes limb bud growth and progressive distalization (1). But how Fgf signaling is regulated remains to be further studied.
Extracellular signal-regulated kinase 1/2 (Erk1/2, also known as p44/42 mitogen-activated protein kinase, MAPK) can be activated by a variety of growth factors and mitogens (2–7). Growth factor-induced activation of the MAPK signaling pathway participates in most processes of vertebrate embryonic development, and in most cases, it functions in proliferation and differentiation regulation (8–11). For example, during myogenesis, MAPK signaling is crucial for the growth factor-induced cellular proliferation of myoblasts, and inactivation of MAPK is required for initiation of myogenesis (8,12,13). The way in which gene regulation of growth factors couples with MAPK activation during limb development is not yet well understood.
Homeoproteins are one of the major classes of transcriptional factors that regulate the development of tissues and organs in vertebrates (14). Msx (including Msx1, Msx2 and Msx3) comprises one of the subfamilies of homeoproteins that control cellular differentiation during development. In vertebrate, Msx is expressed in diverse spatial and temporal domains and participates in the formation of limbs, neurotubes, craniofacial glands, mammary glands and other structures.(15–25). Although Msx is important for diverse tissues during early development, it is mainly expressed in proliferating cells and is downregulated upon differentiation (17,23). For example, in the developing limb, Msx1 is expressed in a zone of undifferentiated proliferating mesenchymal cells destined to form structural elements of the limb but not in the differentiating cells forming these structures (15–18). These and other observations have led to the postulation that Msx1 may be responsible for driving the cellular proliferation (15,22,26–29), although the underlying mechanisms are not known.
In this study, we first observed that Msx1 is indeed able to promote the proliferation of mouse C2C12 myoblasts and C3H10T1/2 mesenchymal stem cells (MSCs). Significantly, the MAPK signaling pathway is markedly activated upon overexpression of Msx1. We then found that Msx1 directly binds to and upregulates Fgf9 and Fgf18 expression, which subsequently triggers MAPK signaling activation. Importantly, we identified a phosphorylation site of Msx1, Ser136, and observed that the mutation of Msx1 Ser136 to Ala (S136A) compromises its function, whereas the mutation of Ser136 to Asp (S136D) enhances its function in upregulating Fgf9 and Fgf18 expression and activating MAPK signaling, which is consistent with the role of the phosphorylation of Msx1 at Ser136 in promoting cell proliferation. Furthermore, we showed that cyclin-dependent kinase 1 (CDK1) is the kinase that phosphorylates Msx1 at Ser136. Significantly, in vivo, Fgf9, Fgf18 and p-Erk1/2 levels were downregulated in the developing limb buds when Msx1 and Msx2 were conditionally knocked out in bone, which resulted in developmental defects in limbs. In summary, our findings provide evidence of a novel mechanism of Msx1 involved in regulating gene expression and promoting cell proliferation and limb development.
MATERIALS AND METHODS
Plasmids and site-specific mutagenesis
The expression plasmid pcDNA3 (Invitrogen, Carlsbad, CA, USA) was used for transient transfection, and pLZRS-IRES-GFP was used for retroviral gene transfer. Sequences corresponding to mouse Flag-tagged Msx1 were cloned into pcDNA3 or pLZRS-IRES-GFP. Site-directed mutagenesis at Ser136, Ser152 and Ser160 was performed by overlap extension PCR with minor modifications (30–32). The point mutation primer information is shown in Supplementary Table S1. All plasmids used were sequenced for verification.
Cell culture and myogenic differentiation
Murine myoblast C2C12 cells were obtained from American Type Culture Collection (ATCC) and were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco) (growth medium). C3H10T1/2 (ATCC) cells as well as bone marrow-derived MSCs that extracted from femurs and tibiae of mice at 4–6 weeks after birth were cultured in α-MEM (Gibco) supplemented with 10% FBS.
For myoblasts differentiation assays, undifferentiated C2C12 cells were grown in growth medium, and differentiation procedure was induced by shifting medium with DMEM containing 2% horse serum (HS) (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) (differentiation medium) at 80% cell confluence for 1–7 days (33,34).
For retroviral gene transfer, replication-defective retroviruses were generated in Phoenix (35,36) ecotropic retroviral packaging cells (ATCC) by transfection of the relevant pLZRS-IRES-GFP plasmid derivatives using Lipofectamine 2000 reagent. The supernatants were collected 72 h later and filtered through 0.45 μm polyvinylidene fluoride (PVDF) membranes (Millipore, CA, USA). Cells were infected with the retroviruses plus 4 mg/ml Polybrene (Sigma-Aldrich).
5-Ethynyl-2′-deoxyuridine staining
The fraction of proliferating cells was determined using Click-iT TM EdU Alexa Fluor Imaging Kits (Thermo Fisher Scientific). Briefly, 20 000 C2C12 cells were seeded into each well of a 12-well plate and incubated for 48 h. Cells were incubated with 10 μM 5-ethynyl-2′-deoxyuridine (EdU) for 3 h prior to fixation with 4% paraformaldehyde (PFA) (Sangon Biotech, Shanghai, China) and permeabilized with 0.25% Triton X-100. The EdU-positive nuclei were labeled according to the manufacturer's protocol, which was followed by labeling of all nuclei with 500 ng/ml 4′6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific). Thereafter, the cells were observed, and images were obtained using a fluorescence microscope (ZEISS, Jena, Germany).
Cell cycle analysis
Differently treated cells were washed with phosphate-buffered saline (PBS). Resuspend cells with PBS containing propidium iodide (10 mg/mL), Triton-X 100 (3‰) (Sigma-Aldrich) and Rnase A (50 ug/ml) and incubate in the dark for 15 min. The fractions of viable cells in G1, S and G2 phases of cell cycle were measured with a FACStar flow cytometer (BD Biosciences, San Jose, CA, USA). The Flow cytometry was performed as previously described (37).
Cell proliferation assay
Cell proliferation assays were performed using Cell Counting Kit-8 (CCK-8) reagent (Dojindo Laboratories, Kumamoto, Japan). Briefly, differently treated cells were transferred into 96-well plates with 4 × 103 cells in 100 ul growth medium per well and examined at 0, 24, 48, 72 and 96 h respectively. At each time point, 10 ul CCK-8 reagent was added into each well and incubate in 37°C for 1 h. The absorbance at 450 nm was measured using a multimode microplate reader (BioTek, Vermont, USA).
Western blotting
For western blotting, cells were sonicated in lysis buffer (50 mM Tris–HCl, pH 7.5, containing 150 mM NaCl, 0.1 mM EDTA, 1% Triton X-100, 1 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM PMSF) and centrifuged at 20 000 × g for 10 min. The supernatants were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE), followed by western blotting analysis with indicated antibodies. Enhanced Chemiluminescence Plus Western Blotting Detection Kits (Bio-Rad) and a luminescent image analyzer (Tanon, Shanghai, China) were used to visualize protein bands according to the manufacturer's instructions. The list of antibodies used for western blotting is available in Supplementary Table S2.
Immunofluorescence
Cell immunofluorescence assays were performed as described previously (38). Briefly, cells were washed with PBS, fixed in 4% PFA, permeabilized with 0.2% Triton X-100 for 10 min, blocked with 2% bull serum albumin (BSA) in PBS for 1 h, and then stained with the indicated antibodies. The list of antibodies used for immunofluorescence staining is available in Supplementary Table S2.
Tissue immunofluorescence assays were performed as previously described (39). Freshly dissected bones were fixed in 4% PFA for 48 h and incubated in 15% DEPC–EDTA (pH 7.8) for decalcification. Then, the specimens were embedded in paraffin or OCT compound (SAKURA, CA, USA) and sectioned at a 10-μm thickness. Samples were blocked in PBS with 10% HS for 1 h and then incubated with mouse anti-Sox9 or rabbit anti-PCNA antibodies, followed by incubation with the corresponding conjugated secondary antibodies. Sections were counterstained with DAPI to visualize the nuclei.
RNA-seq
Total RNA was isolated with TRIzol from Msx1-overexpressing C2C12 cells (n = 3) or control cells (n = 3). cDNA sequencing libraries were prepared with an NEBNext® Ultra™ RNA Library Prep Kit for Illumina. The RNA-Seq FASTQ raw data were trimmed to remove low-quality reads and adapters using Trimmomatic (40). The trimmed reads were aligned to the mouse reference genome UCSC GRCm38/mm10 with HISAT2. Gene and transcript quantification was performed using StringTie. The results of the mapping of the RNA-seq reads, transcript assembly and abundance estimation were reported as fragments per kilobase of exon per million fragments mapped (FPKM). To identify genes that were differentially expressed, the fold changes of each gene were calculated by dividing the average FPKM for the case by the average FPKM for the control. To avoid infinite values, a value of 0.01 was added to the FPKM value of each gene before log2 transformation. Hierarchical clustering analysis (HCA) and principal component analysis (PCA) were performed using the relevant functions in R packages. Enrichment analysis of KEGG signaling pathways was conducted by using DAVID.
LC–ESI-MS/MS analysis
We performed the identification of Msx1 phosphorylation by overexpressing Msx1 in C2C12 cells. The protein samples were digested by the FASP method (41), followed by LC-ESI-MS/MS analysis using a nanoflow EASY-nLC 1000 system (Thermo Fisher) coupled to an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher). All analyses were performed with a two-column system. Samples were first loaded onto an Acclaim PepMap100 C18 Nano Trap Column (5 μm, 100 Å, 100 μm i.d. × 2 cm, (Thermo Fisher)) and then analyzed on an Acclaim PepMap RSLC C18 column (2 μm, 100 Å, 75 μm i.d. × 25 cm (Thermo Fisher)). The mobile phases consisted of Solution A (0.1% formic acid) and Solution B (0.1% formic acid in ACN). The peptides were eluted using the following gradients: 5–35% B for 0–58 min, 35–90% B for 10 min and 90% B for 5 min at a flow rate of 200 nl/min. The MS analysis was performed using data-dependent analysis; the 15 most abundant ions in each MS scan were automatically selected and fragmented in HCD mode. For data analysis, the raw data were analyzed by Proteome Discoverer (version 1.4, Thermo Fisher) using an in-house Mascot server (version 2.3, Matrix Science) (42). The mouse protein database (20170427) was downloaded from UniProt. Data were searched using the following parameters: trypsin/P as the enzyme; up to two missed cleavage sites were allowed; mass tolerances of 10 ppm for MS and 0.05 Da for MS/MS fragment ions; Carboxyamidomethylation of cysteine as a fixed modification; oxidation of methionine and phosphorylation of serine, threonine and tyrosine as variable modifications. The incorporated Target Decoy PSM Validator in Proteome Discoverer was used to validate the search results and identify the hits with a FDR ≤0.01.
Msx1 knockout cell line
Msx1 knockout cell lines were established using a previously reported CRISPR/Cas9 system (43). The gRNAs were designed at http://crispr.mit.edu/ website, and the relevant information is provided in Supplementary Table S3.
qRT-PCR analysis
Total RNA was prepared using a HiPure Total RNA Mini Kit (Magen, Guangzhou, China). RNA was reverse-transcribed using the All-in-One First-Strand cDNA Synthesis Super Mix for qPCR (One-Step gDNA Remover) Kit (TransGene Biotech, Beijing, China). qRT-PCR was performed with 2× SYBR UltraSYBR Mix (Cwbio, Beijing, China) using a Light Cycler 480 system (Roche, Basel, Switzerland). The amplification procedure was as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 10 s and 60°C for 20 s. The cycle threshold (Ct) of each sample was used for the calculation. The expression levels of mRNA were quantified using Relative Quantification Software with GAPDH as an internal control. Primer information is provided in Supplementary Table S4.
Enzyme-linked immunosorbent assay
The enzyme-linked immunosorbent assays (ELISA) were performed using the ELISA Kit for Mouse Fgf9 or Fgf18 (DL Develop, Wuxi, China). Briefly, C2C12 cells overexpressing Msx1 or the control were cultured overnight with the serum-free DMEM. The supernatants were obtained and Fgf9 and Fgf18 concentrations were quantified with the ELISA Kits respectively according to the instructions of the manufacture.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using cross-linked chromatin from C2C12 cells expressing exogenous Flag-Msx1 or its mutants with an EZ-CHIP Kit (Millipore). As described previously (33,34), 5 × 107 cells were lysed, and chromatin was sonicated in lysis buffer (1 mM EDTA, 0.5 mM EGTA, 10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.1% Na-deoxycholate, and 0.5% N-lauryl sarcosine) and then immunoprecipitated overnight with anti-Flag M2 antibody (Sigma-Aldrich). The immunoprecipitated protein-DNA complexes were recovered using protein G beads and washed with RIPA buffer (0.1% SDS, 1% Na-deoxycholate, 1% Triton X-100, 150 mM NaCl, 10 mM Tris–HCl [pH 7.5], 1 mM EDTA, and protease inhibitor cocktail), and the protein-DNA complexes were eluted and dissociated by heating at 65°C for 12 h. Following purification, the recovered DNA was prepared for ChIP-qPCR assays. The primer information is shown in Supplementary Table S5.
RNA interference
Small interfering RNAs (siRNAs) used for the knockdown of Fgf9 and Fgf18 and nonspecific siRNA negative controls were designed and synthesized by Genepharma (Suzhou, China). The oligonucleotide sequences used in this study are shown in Supplementary Table S6.
Transfection of siRNAs was performed in six-well plates. The cells were seeded into a six-well cell culture plate and cultured in growth medium on the day before transfection. The transfections of siRNAs were carried out using Lipofectamine RNAiMAX Reagent (Invitrogen) according to the manufacturer's instructions. Cells were harvested for qRT-PCR or western blotting 48 h later.
Animals
All procedures involving mice were approved by the Fudan University Animal Care and Use Committee. Mice were housed in the animal facility with free access to standard rodent chow and water. Germline Msx1 knockout mice were obtained from Richard Mass at Brigham and Women's Hospital (44). Prx1-Cre mice (45) were kindly provided by Professor Weiguo Zou at CAS. Msxflox/flox mice (46) were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). PCR genotyping was performed using protocols described by the supplier. Both male and female mice that were 0–6 months old were used. Embryos were collected after timed mating, and noon on the day of plug discovery was considered to be embryonic day 0.5 (E0.5).
Microcomputed tomography
Mouse scanning was performed with the microcomputed tomography (μCT) system SkyScan1176 (Bruker, Kartuizersweg, Belgium), and 50 slides (15 μm each) were used for quantifying bone parameters.
Analysis of bone phenotypes
Skeletal preparations were double-stained with alcian blue (Sigma) and Alizarin Red S (ARS) (47,48). Briefly, the carcasses were skinned and eviscerated, fixed with 95% ethanol for 3 days and stained with alcian blue for 3 days. Then, the skeletons were fixed with 95% ethanol three times for 1.5 h each, followed by clearing with 2% KOH for 3–4 h. After staining with ARS for 3–4 h, the skeletons were cleared in 1% KOH/20% glycerol and stored in glycerol. For histological analysis, bone tissues were fixed in 4% PFA and then embedded in paraffin. Tissue sections (10 μm) were used for alcian blue and alkaline phosphatase (ALP) staining.
Drug formulations
The MEK inhibitor PD0325901 was purchased from Selleck Chemicals. The CDK1 inhibitor RO-3306 was purchased from APExBio Technology. The drugs were prepared in dimethyl sulfoxide (DMSO) (10 mM stock) and diluted to their final concentrations in cell culture medium prior to in vitro assays.
Statistical analysis
At least three independent replicates were performed for each assay. The average values from the parallel experiments are given as the mean ± SD. The comparison of differences among the groups was carried out by Student's t-test. Significance was defined as P < 0.01 (***P < 0.0001, **P < 0.001, *P < 0.01).
RESULTS
Msx1 promotes the proliferation of myoblasts and MSCs
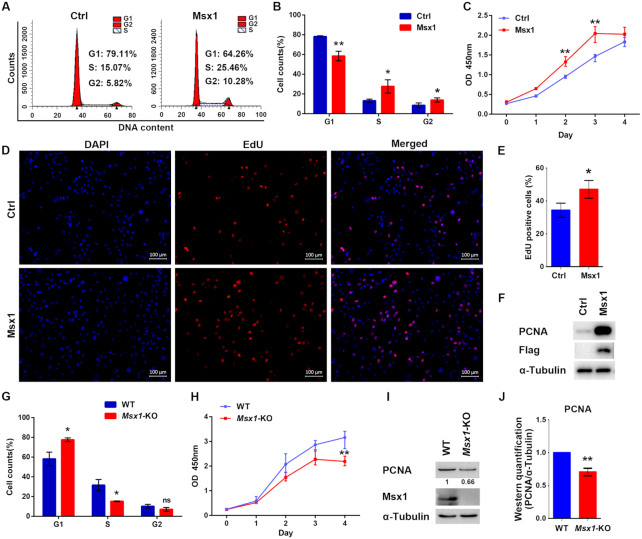
To understand the mechanisms underlying the role of Msx1 in limb development, we first analyzed the effects of Msx1 on myoblast cell growth. Flow cytometry analysis revealed that overexpressing Msx1 in C2C12 cells increased the proportion of cells in S and G2 phase and decreased the percentage of cells in G1 phase (Figure 1A and B), suggesting that Msx1 may promote C2C12 cell cycle progression. We further conducted cell proliferation assays and found that cells overexpressing Msx1 grew more rapidly compared with those with the empty vector control (Figure 1C). In addition, the EdU-positive cell numbers were increased in the Msx1 overexpression group compared with those in the control group (Figure 1D and E), and an increase in the proliferation marker PCNA was detected by western blotting in Msx1-overexpressing cells compared to that in control cells (Figure 1F). These data indicate that overexpression of Msx1 promoted C2C12 cell proliferation.
Figure 1.
Msx1 promotes C2C12 cell proliferation. (A) Flow cytometry assays to assess the impact of Msx1 on C2C12 cell cycle progression. (B) Statistical analysis of the cell cycle of C2C12 cells overexpressing Msx1 or the control. Values are the means ± SD. **P < 0.001, *P < 0.01. (C) CCK-8 assays to assess the proliferation rate of C2C12 cells overexpressing Msx1 or the control. Values are the means ± SD. **P < 0.001. (D) EdU assays to evaluate the effect of Msx1 on C2C12 cell proliferation. Scale bar = 100 μm. (E) Quantification of the ratio of proliferating cells to total cells in EdU assays. Values are the means ± SD. *P < 0.01. (F) Western blotting to detect the expression of PCNA in C2C12 cells overexpressing Msx1 and the control. (G) Statistical analysis of the cell cycle of Msx1-KO or wild-type C2C12 cells. Values are the means ± SD. *P < 0.01. (H) CCK-8 assays to assess the proliferation rate of Msx1-KO or wild-type C2C12 cells. Values are the means ± SD. **P < 0.001. (I) Western blotting to detect the expression of PCNA in Msx1-KO or wild-type C2C12 cells. Numbers under western blot bands represent relative quantifications over α-Tubulin quantified by ImageJ software. Statistical analysis of western blot gray scale values is shown in (J) (n = 3, wild-type was set as 1). Values are the means ± SD. **P < 0.001.
To further verify the effect of endogenous Msx1 on cell proliferation, a C2C12 cell line with Msx1 knock-out (KO) was established using a transient transfection-based CRISPR/Cas9 method (43). In contrast to the results of overexpressing Msx1, both cell cycle progression and cell proliferation were slowed down when Msx1 was deleted in C2C12 cells in comparison with the control group (Figure 1G and H). Accordingly, the level of PCNA was also lower in Msx1-KO than control C2C12 cells (Figure 1I, with quantification in Figure 1J).
Considering its important role in limb development, the function of Msx1 was also analyzed in the C3H10T1/2 mesenchymal stem cell line, which is another relevant multipotent cell line often used in musculoskeletal research. Similarly, PCNA expression was increased when Msx1 was overexpressed in C3H10T1/2 cells (Supplementary Figure S1A and B), and cells proliferated more rapidly as a result (Supplementary Figure S1C). Overall, the effects of Msx1 in C2C12 and C3H10T1/2 cells indicate that Msx1 promotes the proliferation of musculoskeletal progenitor cells.
Msx1 promotes cellular proliferation by activating the MAPK signaling pathway
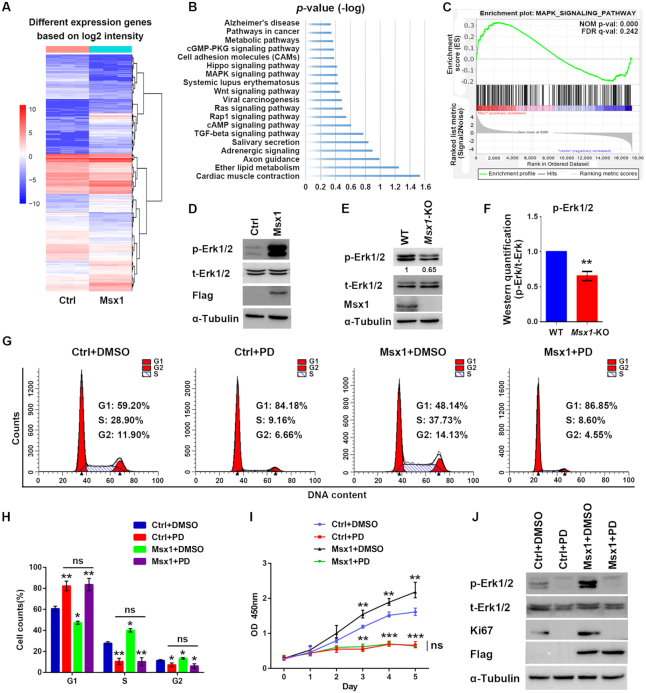
To investigate how Msx1 promoted myoblast proliferation, we carried out RNA profiling analysis to compare C2C12 cell lines overexpressing Msx1 with the control cell line. The down- or upregulated genes with >2-fold differential expression are shown in the heatmap (Figure 2A). In the KEGG enrichment analysis, we found that the differentially-expressed genes (DEGs) were involved in the Rap1, Ras and MAPK signaling pathways, which are all closely related to cell proliferation (8,12,49) (Figure 2B). We further performed gene set enrichment analysis (GSEA) of the DEGs, the MAPK signaling was again enriched and Msx1 expression is positively correlated with MAPK signaling activation (Figure 2C). Since MAPK signaling is common downstream of the Rap1 and Ras signaling pathways (50–52), we performed western blotting analysis and found that Msx1 greatly enhanced Erk1/2 phosphorylation in C2C12 cells, while the total Erk1/2 (t-Erk1/2) abundance was unchanged (Figure 2D). In contrast, the phosphorylated Erk1/2 (p-Erk1/2) level was decreased in the Msx1-KO C2C12 cell line (Figure 2E), with quantification of three experiments in Figure 2F. Therefore, Msx1 can activate the MAPK signaling pathway in myoblast cells.
Figure 2.
The MAPK signaling pathway is critical for the promotion of C2C12 cell proliferation by Msx1. (A) Heatmap of RNA-seq data from C2C12 cells overexpressing Msx1 or the control based on the log2 intensity (n = 3). (B) KEGG signaling pathway enrichment of DEGs in C2C12 cells overexpressing Msx1 or the control, ranked by the P value. (C) GSEA result of RNA-seq data of C2C12 cells overexpressing Msx1 and the control. The MAPK signaling pathway is enriched with FDR of 0.242. (D) Western blotting assays to detect the level of p-Erk1/2 in Msx1-overexpressing and the control C2C12 cells. (E) Western blotting assays to detect the level of p-Erk1/2 in Msx1-KO and the control C2C12 cells. Numbers under western blot bands represent relative quantifications over t-Erk1/2 quantified by ImageJ software. Statistical analysis of western blot grey scale values is shown in (F) (n = 3, wild-type was set as 1). Values are the means ± SD. **P < 0.001. (G) Flow cytometry assays to evaluate the impact of PD0325901 on C2C12 cell cycle progression promoted by Msx1. C2C12 cells overexpressing Msx1 or the control were treated with 1 μM PD0325901 or DMSO for 24 h, respectively. (H) Statistical analysis of the cell cycle of differently treated C2C12 cells in (G). Values are the means ± SD. **P < 0.001, *P < 0.01. (I) CCK-8 assays to assess the impact of PD0325901 on C2C12 cell proliferation promoted by Msx1. Values are the means ± SD. ***P < 0.0001, **P < 0.001. (J) Western blotting assays to detect the impact of PD0325901 on the expression of a proliferation marker promoted by Msx1.
To further verify whether Msx1 promotes C2C12 cell proliferation by activating the MAPK signaling pathway, we utilized the MAPK-specific inhibitor PD0325901 (53) to treat Msx1-overexpressing and control cells. Msx1 was unable to promote cell cycle progression when C2C12 cells were treated with PD0325901 (Figure 2G and H), and the increase in the proportion of cells in S and G2 driven by Msx1 also decreased to a level comparable to that found in the control cells treated with PD0325901 (Figure 2G and H). Meanwhile, the enhanced proliferation of C2C12 cells by Msx1 was reduced by PD0325901 to a rate similar to that of the control cells treated with the inhibitor (Figure 2I). Western blotting showed that when the phosphorylation of Erk1/2 was blocked by PD0325901, the proliferation marker Ki67 was no longer increased in Msx1-overexpressing cells (Figure 2J). These results demonstrated that MAPK signaling is likely the pathway through which Msx1 promotes C2C12 cell proliferation.
The importance of the MAPK signaling pathway was further verified in C3H10T1/2 cells. Once again, the p-Erk1/2 level was dramatically increased when Msx1 was overexpressed (Supplementary Figure S2A). In addition, inhibition of the MAPK signaling pathway by PD0325901 compromised the function of Msx1 in promoting C3H10T1/2 cell proliferation, reducing both the proliferation rate and PCNA level (Supplementary Figure S2B and C). Taken together, these results suggest that Msx1 promotes the cellular proliferation of both C2C12 and C3H10T1/2 cells by activating the MAPK signaling pathway.
Msx1 activates the MAPK signaling pathway by directly binding and upregulating Fgf9 and Fgf18 expression
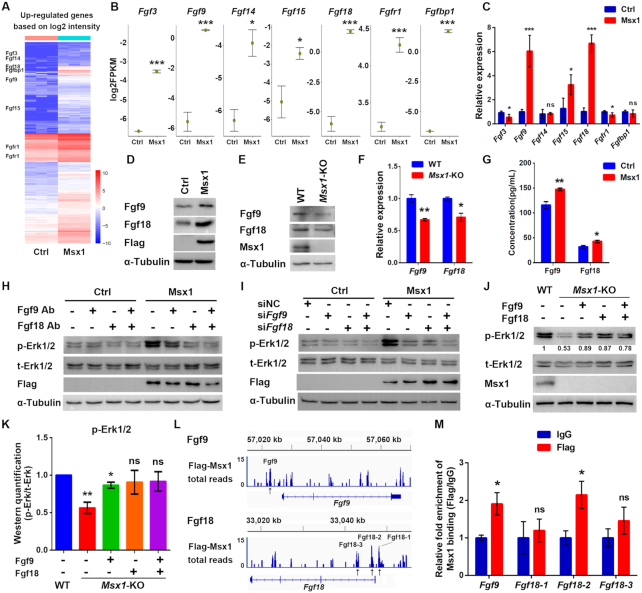
To understand the molecular mechanism through which Msx1 activates cellular proliferation, we analyzed the RNA-seq data of Msx1-expressing cells and found that a set of Fgf family genes, including Fgf3, Fgf9, Fgf14, Fgf15, Fgf18, Fgfr1 and Fgfbp1, were upregulated by Msx1 overexpression (Figure 3A and B). The mRNA levels of these genes were validated by qRT-PCR, and Fgf9 and Fgf18 were found to be most prominently upregulated when Msx1 was overexpressed (Figure 3C). The upregulation of Fgf9 and Fgf18 by Msx1 at the protein level was also confirmed by western blotting (Figure 3D). In contrast, the expression levels of Fgf9 and Fgf18 were decreased in the Msx1-KO C2C12 cell line (Figure 3E and F).
Figure 3.
Msx1 directly binds to and upregulates Fgf9 and Fgf18 to activate the MAPK signaling pathway. (A) Heatmap of upregulated genes based on the RNA-seq data (n = 3). Selected upregulated genes are indicated. (B) Diagrams of the upregulated genes that activate the MAPK signaling pathway based on the RNA-seq data. Values are the means ± SD. ***P < 0.0001, *P < 0.01. (C) qRT-PCR assays to validate the upregulated genes that activate the MAPK signaling pathway based on the RNA-seq data. Values are the means ± SD. ***P < 0.0001, *P < 0.01. (D) Western blotting to confirm the upregulation of Fgf9 and Fgf18 by Msx1. (E) Western blotting to confirm the downregulation of Fgf9 and Fgf18 by Msx1 knockout in C2C12 cells. (F) qRT-PCR assays to determine the mRNA levels of Fgf9 and Fgf18 in Msx1-KO C2C12 cells and the control cells. Values are the means ± SD. **P < 0.001, *P < 0.01. (G) ELISA assays to evaluate the impact of Msx1 on the secretion of Fgf9 and Fgf18 in C2C12 cells. C2C12 cells overexpressing Msx1 or the control were cultured overnight with serum-free DMEM and the supernatants were obtained and subjected to ELISA with the indicated kits. Values are the means ± SD. **P < 0.001, *P < 0.01. (H) Western blotting to detect the level of p-Erk1/2 when Fgf9 and/or Fgf18 protein was blocked. Antibodies against Fgf9 and/or Fgf18 were added to the medium of C2C12 cells at a dilution of 1:1000. After 24 h of incubation, cells were harvested and lysed for western blotting. (I) Western blotting to detect the effect of Fgf9 and/or Fgf18 knockdown on the level of p-Erk1/2. (J) Western blotting to examine the recovery of the p-Erk1/2 level induced by Fgf9 and Fgf18 in Msx1-KO C2C12 cells. Msx1-KO C2C12 cells treated with 50 ng/ml Fgf9 or/and Fgf18 recombinant protein for 24 h or control cells were harvested and lysed for western blotting. Numbers under western blot bands represent relative quantifications over t-Erk1/2 quantified by ImageJ software. Statistical analysis of western blot grey scale values is shown in (K) (n = 3, wild-type was set as 1). Values are the means ± SD. **P < 0.001, *P < 0.01. (L) Msx1 ChIP signal enrichment at Fgf9 and Fgf18 loci. ChIP assays were performed using C2C12 cells expressing Flag-tagged Msx1 or the control vector. The enriched regions from the Flag-Msx1 expressing cells were compared to an empirical background set of sequencing reads from a Flag immunoprecipitation performed on cells infected with an empty vector to exclude regions of the genome that may be non-specifically enriched by Flag immunoprecipitation. Black arrows indicate the possible Msx1 binding sites. (M) ChIP-qPCR analyses to validate the Msx1 binding at the Fgf9 and Fgf18 loci based on the ChIP-seq data. ChIP assays were performed with C2C12 cells overexpressing Flag-tagged Msx1 or control cells. ChIP-qPCR was then used to determine the relative enrichment of the Fgf9 and Fgf18 fragments based on the ChIP-seq performed with the same cell line C2C12 overexpressing Flag-tagged Msx1. The ChIP-qPCR data were analyzed to calculate the enrichment of Flag or IgG relative to input respectively, and IgG values were defined as 1 to calculate the fold enrichment of Flag over IgG. Values are the means ± SD. *P < 0.01.
At this point, we are not certain whether Msx1 activates MAPK signaling prior to Fgf upregulation or whether Msx1 activates the expression and secretion of Fgf first, which then leads to MAPK activation. So, we firstly examined the secretion of Fgf9 and Fgf18 by ELISA. The results showed that the abundance of Fgf9 and Fgf18 in the cell culture medium was both increased when overexpressing Msx1 in C2C12 cells (Figure 3G), suggesting that MAPK signaling could be activated by the secretion of Fgf9 and Fgf18. We then performed an experiment in which the activity of Fgf9 and/or Fgf18 was blocked by adding an antibody against Fgf9 or Fgf18 into the culture medium of C2C12 cells to determine the impact of the neutralization of Fgf activities. As a result, we observed that the increased p-Erk1/2 level induced by Msx1 was greatly reduced by the addition of Fgf9 and Fgf18 antibodies, to a level that was comparable to that of the control cells when equal amounts of IgG were added (Figure 3H). This result suggests that the Msx1-enhanced expression and secretion of Fgf9/18 likely precede and lead to the MAPK activation. For further validation, we used the synthesized siRNAs to knock down Fgf9 and Fgf18, and the level of p-Erk1/2 was examined. As expected, in Msx1-overexpressing cells, the level p-Erk1/2 decreased significantly when either Fgf9 or Fgf18 was knocked down; moreover, p-Erk1/2 almost decreased to the same level as that found in cells without Msx1 overexpression when both Fgf9 and Fgf18 were knocked down (Figure 3I), suggesting that Msx1 could not promote the phosphorylation of Erk1/2 without Fgf9 and Fgf18. On the other hand, the downregulation of p-Erk1/2 by Msx1 deletion in C2C12 cells was recovered by treating the cells with recombinant Fgf9 or/and Fgf18 protein (Figure 3J), and the quantification of three experiments was shown in Figure 3K. Thus, these results suggest that Msx1 activates the MAPK signaling pathway through upregulating Fgf9 and Fgf18 gene expression.
To further understand how Msx1 activates Fgf9/18, we re-examined our previous ChIP-seq data (GSE26711) (54), and found that Msx1 binds to the promoters of Fgf9 and Fgf18 (Figure 3L). Specifically, we found one Msx1 binding peak at Fgf9 and three at the Fgf18 gene promoter (Figure 3L). We accordingly designed primers for the ChIP-qPCR assays and found that the relative enrichment of Msx1 at the binding site of Fgf9 and the second binding site of Fgf18 was significantly higher than that in the control (Figure 3M). Therefore, Msx1 seems to bind to and upregulate Fgf9 and Fgf18 to activate MAPK signaling.
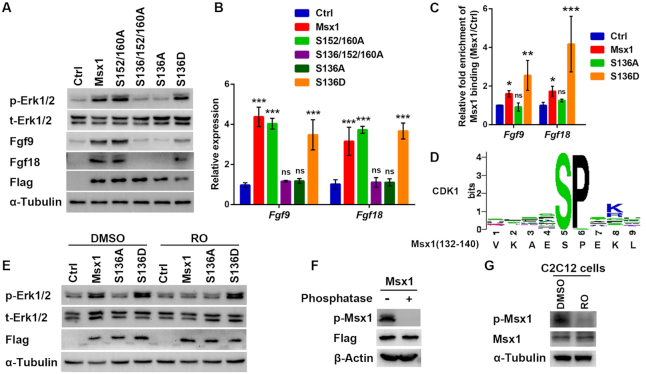
Phosphorylation of Ser136 is critical for Msx1 promotion of myoblast proliferation
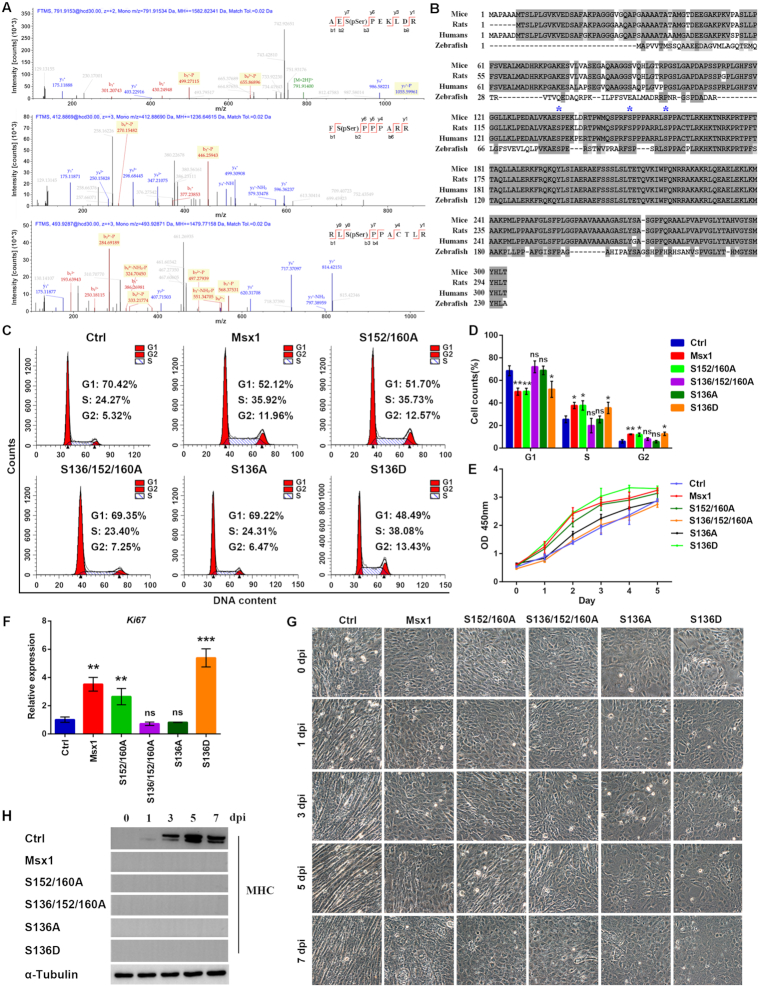
Increasing evidence suggests that posttranslational modifications, especially phosphorylation, impact transcriptional activities (55). To further understand the molecular mechanism through which Msx1 regulates cell growth, we performed mass spectrometry (MS) analysis and identified three novel phosphorylated serine residues, Ser136, Ser152 and Ser160, in Msx1 (Figure 4A). Multiple sequence alignment of amino acids showed that all three serine residues described above are conserved in mice, humans, rats and zebrafish (Figure 4B). We constructed various retroviral plasmids expressing Msx1 harboring a single mutation or a combination of mutations at the three phosphorylation sites and checked whether these phosphorylation sites may affect the capability of Msx1 to promote myoblast proliferation. Interestingly, the C2C12 cells expressing Msx1(S152/160A) show no change in cell cycle phases compared with the C2C12 cells expressing wild-type Msx1 (Figure 4C and D); however, the cells overexpressing Msx1(S136/152/160A) exhibit altered cell cycle progression; specifically, the number of cells in G1 phase increased to a level similar to that of the control cells (Figure 4C and D). This suggested that Ser136 phosphorylation may play a key role in the promotion of myoblast proliferation by Msx1. We then mutated Ser136 in Msx1 to either Ala or Asp to mimic the dephosphorylated and phosphorylated forms of Ser136. Remarkably, while the Ser136 to Ala (S136A) mutant compromised the function of Msx1 in promoting cell cycle progression, the Ser136 to Asp (S136D) mutant retained the cell cycle promotion function of Msx1 (Figure 4C and D).
Figure 4.
Msx1 promotes C2C12 cell proliferation via the phosphorylation of Ser136. (A) Tandem mass spectrometry of the Msx1 peptides modified by phosphorylation on the Ser136 (upper), Ser152 (middle) and Ser160 (lower) residues. (B) Alignment of Msx1 proteins across different species in chordates. The asterisks indicate Ser136, Ser152 and Ser160 respectively. (C) Flow cytometry assays to analyze the cell cycle of C2C12 cells overexpressing different Msx1 mutants. (D) Statistical analysis of the cell cycle of C2C12 cells overexpressing different Msx1 mutants. Values are the means ± SD. **P < 0.001, *P < 0.01. (E) CCK-8 assays to examine the proliferation of C2C12 cells overexpressing different Msx1 mutants. Values are the means ± SD. (F) qRT-PCR assays to detect Ki67 mRNA levels in C2C12 cells overexpressing different Msx1 mutants. Values are the means ± SD. ***P < 0.0001, **P < 0.001. (G) Micrographs of C2C12 cells at 0, 1, 3, 5 and 7 dpi with overexpression of different Msx1 mutants to assess the impact of Msx1 phosphorylation sites on the inhibition of differentiation. (H) Western blotting to detect the marker of terminal muscle differentiation MHC. C2C12 cells overexpressing different Msx1 mutants were harvested and lysed separately at 0, 1, 3, 5 and 7 dpi. Western blotting assays were performed to detect MHC expression.
When analyzing cell proliferation, the C2C12 cells overexpressing the Msx1 S152/160A double mutant produced no changes compared with the cells expressing wild-type Msx1, but the cells with Msx1 in which the three phosphorylation sites were mutated to Ala (S136/152/160A) showed much slower growth (Figure 4E). Significantly, Msx1 with the single mutation S136A did not promote C2C12 cell proliferation compared to wild-type Msx1, whereas Msx1 with the single mutation S136D did promote C2C12 cell proliferation at a similar level as wild-type Msx1 (Figure 4E). Consistently, the proliferation marker Ki67 was also upregulated in C2C12 cells expressing wild-type Msx1, Msx1 (S152/160A) or Msx1 (S136D) but not in cells expressing Msx1 (S136/152/160A) or Msx1 (S136A) (Figure 4F). These results demonstrated that the phosphorylation of Msx1 Ser136, but not Ser152 or Ser160, is critically required for the promotion of cell proliferation.
We sought to determine whether these newly identified phosphorylation sites of Msx1 may have any effects on myoblast differentiation. Therefore, C2C12 cells expressing Msx1 with various combinations of the three mutated phosphorylation sites were subjected to myogenic differentiation. As the microscopy data showed in Figure 4G, C2C12 cells in the control group started to fuse to each other 3 days postinduction (dpi) and to form complete myotubes at 7 dpi. By contrast, C2C12 cells expressing either wild-type Msx1 or any mutant (S152/160A, S136/152/160A, S136A or S136D) failed to differentiate during the induction period. Consistently, the level of the C2C12 differentiation marker, myosin heavy chain (MHC) increased upon induction in the control cells, but failed to do so in cells expressing Msx1 or any of the mutants during differentiation (Figure 4H). While the three phosphorylation sites of Msx1 show distinctive effects on cell proliferation, they all show inhibitory effect on C2C12 differentiation. Thus, the ability of Msx1 to regulate proliferation or differentiation seems to be uncoupled, and the two functions appear to be independent of each other.
Ser136 phosphorylation of Msx1 is required for MAPK signaling activation and Fgf9/18 expression
We next examined how p-Erk1/2 and Fgf expression levels were regulated by different Msx1 mutants. Similar to wild-type Msx1, Msx1(S152/160A) or Msx1(S136D) was able to increase the p-Erk1/2 level in C2C12 cells; however, Msx1(S136/152/160A) and Msx1(S136A) failed to do so (Figure 5A). Consistent with the p-Erk1/2 levels, Fgf9 and Fgf18 expression was also upregulated in cells expressing wild-type Msx1, Msx1(S152/160A) or Msx1(S136D), but was not upregulated in cells expressing Msx1(S136/152/160A) or Msx1(S136A) (Figure 5A and B). Thus, the phosphorylation of Msx1 at Ser136 seems to be critical for the upregulation of Fgf9 and Fgf18 and the further activation of MAPK signaling.
Figure 5.
Phosphorylation of Msx1 at Ser136 is essential for its binding to and upregulation of Fgf9 and Fgf18 and further activation the MAPK signaling pathway, and CDK1 appears to be the kinase that performs this phosphorylation. (A) Western blotting to assess the effect of Msx1 phosphorylation sites on increases in the levels of p-Erk1/2, Fgf9 and Fgf18 by wild-type Msx1. (B) qRT-PCR assays to assess the effect of Msx1 phosphorylation sites on increases in the levels of Fgf9 and Fgf18 by wild-type Msx1. Values are the means ± SD. ***P < 0.0001. (C) ChIP-qPCR analysis to determine the impact of Ser136 mutants of Msx1 on Fgf9/18 binding. ChIP assays were performed with C2C12 cells overexpressing wild-type Msx1, Msx1 (S136A), Msx1 (S136D) or the control. ChIP-qPCR was then used to determine the relative enrichment of the Msx1 binding fragments of the Fgf9 and Fgf18 genes. The ChIP-qPCR data were analyzed to calculate the enrichment of the control or different Msx1 mutants relative to input respectively, and the control values were defined as 1 to calculate the fold enrichment of Msx1 or its mutants over the control. Values are the means ± SD. ***P < 0.0001, **P < 0.001, *P < 0.01. (D) Diagram of the conserved residues of the CDK1 catalytic domain and the corresponding phosphorylated peptide sequence of Msx1. (E) Western blotting to examine the effect of Msx1 or its mutants and Ro3306 on the level of p-Erk1/2 in C2C12 cells. C2C12 cells overexpressing wild-type Msx1, Msx1(S136A), Msx1(S136D) or the control were treated with 10 μM Ro3306 or DMSO. Twenty hours later, the cells were harvested and lysed for western blotting, and the level of p-Erk1/2 was checked. (F) Western blotting to verify the specificity of Ser136 phosphorylated Msx1 (p-Msx1) antibody. Lysates from C2C12 cells overexpressing Flag-tagged Msx1 were incubated with calf intestinal alkaline phosphatase (CIAP) at 37°C for half an hour and subjected to immunoblot analysis with the indicated antibodies. (G) Western blotting to examine the effect of Ro3306 on the level of endogenous p-Msx1 in C2C12 cells. C2C12 cells were treated with 10 μM Ro3306 or DMSO. Twenty hours later, the cells were harvested and lysed for western blotting with the indicated antibodies.
To understand how the phosphorylation of Msx1 Ser136 activate Fgf-MAPK signaling, we further performed ChIP assays in C2C12 cells overexpressing wild-type Msx1, Msx1(S136A), and Msx1(S136D). By examining the Msx1 binding of Fgf9 and Fgf18, we detected that the enrichment of Msx1 at the Fgf9 and Fgf18 gene promoters was dramatically decreased in cells expressing Msx1(S136A) but remained comparable to that of wild-type Msx1 in cells expressing Msx1(S136D) (Figure 5C). Therefore, the binding of Msx1 to the Fgf9 and Fgf18 genes was dramatically weakened when Ser136 of Msx1 was not in its phosphorylated state. These observations suggest that Msx1 may promote C2C12 cell proliferation by phosphorylating Ser136 to enhance its binding to Fgf9 and Fgf18, upregulating the expression of Fgf9 and Fgf18 and consequently activating MAPK signaling.
Similar observations were made in C3H10T1/2 cells. Likewise, overexpression of Msx1 upregulated the expression of Fgf9 and Fgf18 in C3H10T1/2 cells (Supplementary Figure S3A and B), while phosphorylation of Msx1 at Ser136 seems to be a key mechanism that upregulates Fgf-MAPK signaling (Supplementary Figure S3C). As a result, the proliferation of C3H10T1/2 cells was increased by Msx1(S136D) (Supplementary Figure S3C and D).
CDK1 is the kinase that phosphorylates Msx1 at Ser136
We further investigated which kinase phosphorylates Msx1 at Ser136. The online software KinasePhos predicted that the only kinase that may phosphorylate Ser136 of Msx1 is CDK1 (Supplementary Table S7). The substrate peptide sequence, which CDK1 is inclined to catalyze, corresponds to the sequence of residues 132–140 in Msx1, as shown in Figure 5D. To verify this prediction, we utilized the specific inhibitor of CDK1, Ro3306 (56), to analyze its effect on C2C12 cells. By western blotting, we found that the phosphorylation of Erk1/2 in C2C12 cells was reduced by Ro3306 treatment compared with that in control cells treated with DMSO (Figure 5E). Thus, Msx1 could not promote Erk1/2 phosphorylation when the activity of CDK1 was blocked. Remarkably, the cell line expressing the Msx1(S136D) mutant, a mimic form for constitutive phosphorylation, showed increased p-Erk1/2 levels, which is independent of Ro3306 treatment (Figure 5E); however, the cell line harboring the Msx1(S136A) mutant exhibited no increase in Erk1/2 phosphorylation regardless of the presence or absence of Ro3306 (Figure 5E). This is because the Msx1(S136A) is resistant to the activity of CDK1, thus unable to promote the phosphorylation of Erk1/2. For further validation, we have successfully made the antibody specifically recognizing the Ser136 phosphorylation form of Msx1. To prove this antibody works, we first demonstrate that this antibody does recognize the phosphorylation form of Msx1. We observed that the phosphorylation signal is missing when Msx1 was dephosphorylated by the calf intestinal alkaline phosphatase (CIAP) treatment (Figure 5F). Using this antibody, we further found that in C2C12 cells, the phosphorylation at Msx1 Ser136 (p-Msx1) was reduced by Ro3306 treatment compared with that in control cells treated with DMSO (Figure 5G). These results suggest that CDK1 is apparently responsible for phosphorylation of Msx1 Ser136.
The molecular Msx1-Fgf9/18-MAPK axis is required for limb development
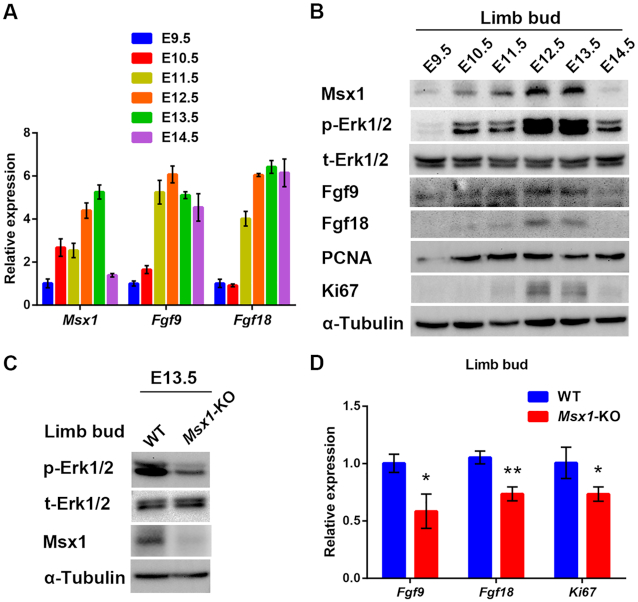
Based on these new molecular insights into the promotion of cell proliferation by Msx1, we further explored whether these mechanisms may apply to limb development. The expression profiles of Msx1, Fgf9, Fgf18, Ki67 and PCNA and the level of p-Erk1/2 in forelimb buds of mouse embryos from E9.5–14.5 were determined by qRT-PCR analysis or western blotting. As shown in Figure 6A and B, the variation patterns in the expression of all these molecules were strikingly similar during early limb development. In brief, they all gradually increased from E9.5 to E12.5, peaked at E12.5 and E13.5 and began to decrease at E14.5 (Figure 6A and B).
Figure 6.
In vivo validation of Msx1-Fgf9/18-MAPK signaling in limb bud development. (A) qRT-PCR analysis to examine the expression profiles of Msx1, Fgf9, and Fgf18 in embryo limb buds during E9.5–14.5. Forelimb buds truncated from embryonic day 9.5–14.5 embryos were lysed. Total RNA was obtained and reverse-transcribed for qRT-PCR and RNA levels of Msx1, Fgf9 and Fgf18 were determined respectively. (B) Western blotting to detect the expression profiles of Msx1, Fgf9, Fgf18, PCNA, Ki67 and p-Erk1/2 in embryo limb buds during E9.5–14.5. (C) Western blotting to detect the impact of Msx1 knockout on the level of p-Erk1/2 in embryo limb buds. (D) qRT-PCR to detect the impact of Msx1 knockout on the mRNA levels of Fgf9, Fgf18 and Ki67 in mouse embryo limb buds. Values are the means ± SD. **P < 0.001, *P < 0.01.
To determine the developmental regulation of the Fgf9/18-MAPK signaling axis by Msx1 during in vivo development, we next generated germline Msx1-KO mice by heterozygote inbreeding. The forelimb buds of embryos at E13.5, when Msx1 is mostly expressed, were collected for further analysis. In the limb buds of the Msx1-KO mice, the levels of Fgf9, Fgf18, Ki67 and p-Erk1/2 were all decreased to different degrees in comparison with those in the buds of the wild-type mice (Figure 6C and D). Thus, the in vivo results found in the developmental embryos agreed with our observations in vitro, indicating that Msx1 indeed participates in promoting cell proliferation in limb development by upregulating Fgf9/18 and activating MAPK signaling.
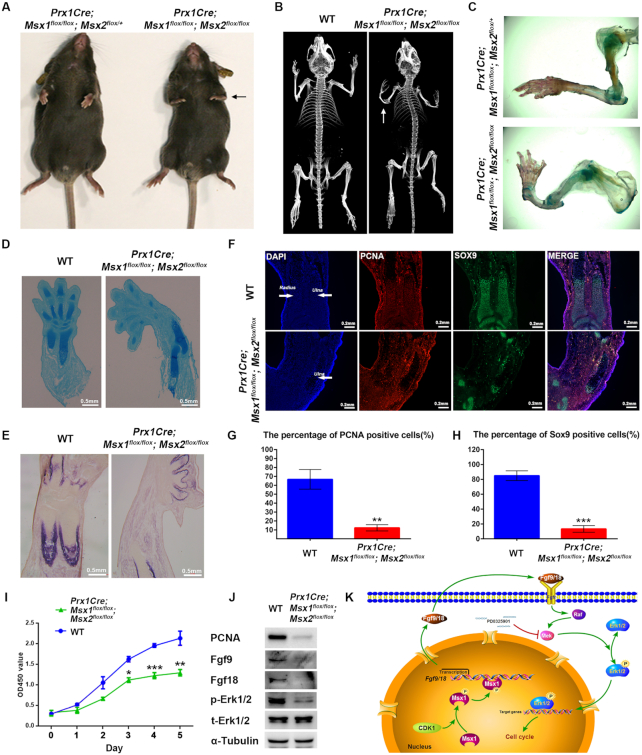
To further verify the role of Msx1 in the commitment of MSCs to an osteoblast fate in limb development, Msx1 MSC-specific knockout mice were generated by crossing Prx1-Cre mice with Msx1flox/flox mice. As reported, due to the functional redundancy of Msx1 and Msx2, Msx1 or Msx2 homozygous mutants (Msx1–/– or Msx2–/–) do not display gross limb abnormalities (21,44,57). We further generated Msx1/2 MSC-specific double-knockout mice using Prx1-Cre mice and Msx1/2flox/flox mice (Supplementary Figure S4A). As a result, unlike germline Msx1/2 KO, which was perinatally lethal (21), Msx1/2 MSC-specific knockout produced mice (Prx1-Cre; Msx1flox/flox; Msx2flox/flox) that were viable but relatively smaller in size at both 2 days and 6 weeks after birth compared with the controls (wild-type and Msx1 or Msx2 MSC-specific knockout mice; Supplementary Figure S4B–D). The Msx1/2 MSC-specific knockout mice had an approximately 30% lower body weight at 3 weeks after birth compared with the controls (Supplementary Figure S4D). In addition, the Msx1/2 MSC-specific knockout mice displayed more severe defects in forelimb development. In appearance, the forelimbs of Msx1/2 MSC-specific knockout mice were shorter and smaller than those of the controls and could not function normally (Figure 7A). Using μCT, we found the complete absence of the radius and a decreased ulna size in Msx1/2 MSC-specific knockout mice compared with those in wild-type mice (Figure 7B and C). Defects were not limited to the radius and ulna, as finger truncation, polydactyly, and oligodactyly were also observed (Figure 7C).
Figure 7.
Deficiency of Msx in the lateral plate mesoderm reduced bone formation. (A) Representative views of 4-week-old control and Msx1/2 MSC-specific knockout littermates. The black arrow indicates the forelimb. (B) μCT images of bones from 4-week-old wild-type and Msx1/2 MSC-specific knockout mice. (C) Alcian blue and ARS staining in the forelimbs from 4-week-old control and Msx1/2 MSC-specific knockout mice. (D, E) Alcian blue (D) and ALP (E) staining of limb sections from wild-type and Msx1/2 MSC-specific knockout mice at postnatal day 2 (P2), scale bar = 0.5 mm. (F) IF assays of limb sections from wild-type and Msx1/2 MSC-specific knockout mice at P2. Expression of Sox9 (green) and PCNA (red) was detected. DAPI was used to counterstain the nuclei. Scale bar = 0.2 mm. (G, H) Diagram summarizing the immunofluorescence data. The proportions of PCNA (G)- and Sox9 (H)-positive cells (n = 400) were calculated. Values are the means ± SD. ***P < 0.0001, **P < 0.001. (I) CCK-8 assays to evaluate the proliferation of primary bone marrow MSCs from wild-type and Msx1/2 MSC-specific knockout mice. Values are the means ± SD. ***P < 0.0001, **P < 0.001, *P < 0.01. (J) Western blotting to detect PCNA, p-Erk1/2, Fgf9 and Fgf18 levels in primary bone marrow MSCs from wild-type and Msx1/2 MSC-specific knockout mice. (K) Model of the promotion of cellular proliferation by Msx1. In the nucleus, Ser136 of Msx1 is phosphorylated by CDK1. Phosphorylated Msx1 binds to Fgf9 and Fgf18 to activate their transcription. Upregulated Fgf9 and Fgf18 are shuttled to the extracellular matrix, where they bind to Fgfrs to activate the MAPK signaling pathway and promote the phosphorylation of Erk1/2. p-Erk1/2 then promotes cell proliferation in a variety of ways.
To determine whether the abnormality of the limbs in Msx1/2 MSC-specific knockout mice resulted from a primary defect in osteoblast or chondrocyte development, we analyzed limbs isolated from Msx1/2 MSC-specific knockout mice and compared them in terms of the corresponding elements of the wild-type. The staining results showed that chondrogenesis (Figure 7D) and osteogenesis (Figure 7E) were both reduced in the limbs of Msx1/2 MSC-specific knockout mice compared with those in wild-type mice. The stem cells in the marrow cavities are key for osteochondrogenesis. To explore the cause of the reduction of osteochondrogenesis, we evaluated the levels of the proliferation marker PCNA and the chondrogenic marker Sox9 in marrow cavities using immunofluorescence staining. The results showed that both the Sox9 and PCNA levels were much lower in the studied tissues of Msx1/2 MSC-specific knockout mice compared with those in wild-type mice (Figure 7F), and the numbers of both PCNA- and Sox9-positive cells in the marrow cavities were decreased by >80% in double knockout mice compared with those in controls (Figure 7G and H). These observations suggested that MSC-specific knockout of Msx1/2 impaired the proliferation of bone marrow stem cells and further led to a reduction in osteochondrogenesis.
For further validation, primary bone marrow MSCs from Msx1/2 MSC-specific knockout mice and wild-type mice were obtained and cultured in vitro, and cellular proliferation was observed by CCK-8 assays. As shown in Figure 7I, compared with that of wild-type bone marrow MSCs, the proliferation of bone marrow MSCs with Msx1/2 double-knockout was significantly slowed down. Meanwhile, the levels of PCNA, p-Erk1/2 and Fgf9/18 were dramatically decreased in Msx1/2 double-knockout bone marrow MSCs (Figure 7J). Thus, in developing limbs, Msx1 and Msx2 function redundantly to promote bone marrow MSC proliferation by upregulating Fgf9/18 and further activating MAPK signaling.
In conclusion, our studies demonstrate a coupling mechanism of Msx with Fgf/MAPK signaling to promote cell proliferation and embryonic limb development, thus providing novel molecular insights.
DISCUSSION
As shown in the working model in Figure 7K, CDK1 first phosphorylates Msx1 at Ser136, empowering Msx1 with the capability to bind and upregulate the Fgf9/18 genes. The increased Fgf9 and Fgf18 proteins are exocytosed to the extracellular matrix, where they activate the MAPK signaling pathway by binding to Fgfrs in an autocrine or paracrine fashion, leading to the increased phosphorylation of Erk1/2. Phosphorylated Erk1/2 then promotes myoblast proliferation in various ways.
The well-established function of Msx1 in development is the inhibition of the differentiation of cells, such as those of the myogenic lineage (33,54). Our current findings focus on another function of Msx1 in promoting cell proliferation. In mice, limb development first begins with the induction of the forelimb buds at E9.5, followed by the formation of the hindlimb buds at E10 on both flanks of the embryo (marking the future forelimbs and hindlimbs, respectively) (58,59). During mouse embryonic limb development, the period from E9.5–13.5 is the fast-growing phase for the limbs, which exactly corresponds to the time period of Msx1 expression. As we have shown in this study, Msx may play a key role in driving cell proliferation during this period, allowing for the rapid growth of limb buds.
We show here that Msx1 enhances cell proliferation by activating the MAPK signaling pathway by targeting Fgf9 and Fgf18 gene expression. Evidence has suggested a critical role for Erk1/2 in myoblast proliferation (60,61). Erk1/2 activity can be stimulated by a variety of growth factors in myoblasts, including Fgf, hepatocyte growth factor (Hgf) and insulin-like growth factor (Igf) (5,7–10,60). Here, we reveal a particular way by which Erk1/2 is stimulated by Fgfs that are activated by the phosphorylated Msx1 in myoblasts. Regarding how p-Erk1/2 promotes myoblast proliferation, it has been reported that p-Erk1/2 prevents cell cycle exit during G1 (49) and promotes entry into S phase (8,62). In addition, only when p-Erk1/2 is shuttled into the nucleus can it promote cell proliferation (63).
For Msx1 to regulate Fgf9 and Fgf18 gene expression, the phosphorylation of Msx1 at Ser136 was carefully examined. We demonstrated that a non-phosphorylated form of Msx1 mutant S136A is unable to stimulated Fgf/MAPK activation, whereas a phosphorylation mimic form of Msx1 mutant S136D constitutively activates Fgf-MAPK signaling pathway and promotes cell proliferation. Interestingly, a previous study showed that phosphorylation of Msx2 at Thr135 and Thr141 is the key mechanism allowing its regulation of target genes (64). Together, these studies demonstrated that phosphorylation of homeoproteins could be key to their functions in controlling transcription and development.
In addition to skeletal muscle and bone, Msx1 is also expressed in many other tissues and organs, such as the heart, craniofacial derivations, neurotube, mammary gland and a few types of tumors (65–72). It was recently found that Msx1 plays an important role in human odontogenesis. Specifically, a frame-shift mutation in Msx1 in human dental pulp stem cells weakened the activity of the MAPK signaling pathway and decreased the proliferation of cells, leading to the developmental deficiency of teeth (73). This mutation disrupted the Msx1 protein at the amino acid residue Met43; therefore, Ser136 was also disrupted. In addition, we have obtained preliminary results that indicate that Msx1 promotes the proliferation of ZR-75-30 breast cancer cells and PC-3 prostate cancer cells. In these cancer cells, MAPK signaling is also activated by Msx1 overexpression, and a cell line with a Msx1(S136A) knock-in mutation showed reduced Fgf-MAPK signaling and cellular proliferation. Overall, the mechanisms by which Msx1 promotes cell proliferation may function in different tissues and organs, which deserves to be further investigated.
DATA AVAILABILITY
The RNA-seq datasets used in this study have been deposited with the NCBI GEO under accession number GSE150349.
Supplementary Material
Notes
Present address: Zhangjing Ma, Department of Chemical Pathology, Prince of Wales Hospital, The Chinese University of Hong Kong, Hong Kong 999077, China.
Contributor Information
Yenan Yang, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Xiaoli Zhu, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Anhui 230001, China.
Xiang Jia, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Wanwan Hou, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Guoqiang Zhou, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Zhangjing Ma, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Bin Yu, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Yan Pi, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Xumin Zhang, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Jingqiang Wang, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Gang Wang, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31671543, 91740110 to G.W., 31972885 to J.W.] (in part); Ministry of Science and Technology of China [2017YFA0102700 to G.W.]. Funding for open access charge: National Natural Science Foundation of China [31671543, 91740110 to G.W., 31972885 to J.W.]; Ministry of Science and Technology of China [2017YFA0102700 to G.W.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Fernandez-Teran M., Ros M.A.. The Apical Ectodermal Ridge: morphological aspects and signaling pathways. Int. J. Dev. Biol. 2008; 52:857–871. [DOI] [PubMed] [Google Scholar]
- 2. Ray L.B., Sturgill T.W.. Rapid stimulation by insulin of a serine/threonine kinase in 3T3-L1 adipocytes that phosphorylates microtubule-associated protein 2 in vitro. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:1502–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boulton T.G., Yancopoulos G.D., Gregory J.S., Slaughter C., Moomaw C., Hsu J., Cobb M.H.. An insulin-stimulated protein kinase similar to yeast kinases involved in cell cycle control. Science. 1990; 249:64–67. [DOI] [PubMed] [Google Scholar]
- 4. Adi S., Bin-Abbas B., Wu N.Y., Rosenthal S.M.. Early stimulation and late inhibition of extracellular signal-regulated kinase 1/2 phosphorylation by IGF-I: A potential mechanism mediating the switch in IGF-I action on skeletal muscle cell differentiation. Endocrinology. 2002; 143:511–516. [DOI] [PubMed] [Google Scholar]
- 5. Coolican S.A., Samuel D.S., Ewton D.Z., McWade F.J., Florini J.R.. The mitogenic and myogenic actions of insulin-like growth factors utilize distinct signaling pathways. J. Biol. Chem. 1997; 272:6653–6662. [DOI] [PubMed] [Google Scholar]
- 6. Jo C., Kim H., Jo I., Choi I., Jung S.C., Kim J., Kim S.S., Jo S.A.. Leukemia inhibitory factor blocks early differentiation of skeletal muscle cells by activating ERK. Biochim. Biophys. Acta. 2005; 1743:187–197. [DOI] [PubMed] [Google Scholar]
- 7. Milasincic D.J., Calera M.R., Farmer S.R., Pilch P.F.. Stimulation of C2C12 myoblast growth by basic fibroblast growth factor and insulin-like growth factor 1 can occur via mitogen-activated protein kinase-dependent and -independent pathways. Mol. Cell. Biol. 1996; 16:5964–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jones N.C., Fedorov Y.V., Rosenthal R.S., Olwin B.B.. ERK1/2 is required for myoblast proliferation but is dispensable for muscle gene expression and cell fusion. J. Cell. Physiol. 2001; 186:104–115. [DOI] [PubMed] [Google Scholar]
- 9. Tortorella L.L., Milasincic D.J., Pilch P.F.. Critical proliferation-independent window for basic fibroblast growth factor repression of myogenesis via the p42/p44 MAPK signaling pathway. J. Biol. Chem. 2001; 276:13709–13717. [DOI] [PubMed] [Google Scholar]
- 10. Nagata Y., Honda Y., Matsuda R.. FGF2 induces ERK phosphorylation through Grb2 and PKC during quiescent myogenic cell activation. Cell Struct. Funct. 2010; 35:63–71. [DOI] [PubMed] [Google Scholar]
- 11. Spizz G., Roman D., Strauss A., Olson E.N.. Serum and fibroblast growth factor inhibit myogenic differentiation through a mechanism dependent on protein synthesis and independent of cell proliferation. J. Biol. Chem. 1986; 261:9483–9488. [PubMed] [Google Scholar]
- 12. Knight J.D., Kothary R.. The myogenic kinome: protein kinases critical to mammalian skeletal myogenesis. Skelet Muscle. 2011; 1:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bennett A.M., Tonks N.K.. Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science. 1997; 278:1288–1291. [DOI] [PubMed] [Google Scholar]
- 14. Holland P.W., Takahashi T.. The evolution of homeobox genes: implications for the study of brain development. Brain Res. Bull. 2005; 66:484–490. [DOI] [PubMed] [Google Scholar]
- 15. Bendall A.J., Ding J., Hu G., Shen M.M., Abate-Shen C.. Msx1 antagonizes the myogenic activity of Pax3 in migrating limb muscle precursors. Development. 1999; 126:4965–4976. [DOI] [PubMed] [Google Scholar]
- 16. Catron K.M., Wang H., Hu G., Shen M.M., Abate-Shen C.. Comparison of MSX-1 and MSX-2 suggests a molecular basis for functional redundancy. Mech. Dev. 1996; 55:185–199. [DOI] [PubMed] [Google Scholar]
- 17. Davidson D. The function and evolution of Msx genes: pointers and paradoxes. Trends Genet. 1995; 11:405–411. [DOI] [PubMed] [Google Scholar]
- 18. Davidson D.R., Crawley A., Hill R.E., Tickle C.. Position-dependent expression of two related homeobox genes in developing vertebrate limbs. Nature. 1991; 352:429–431. [DOI] [PubMed] [Google Scholar]
- 19. Houzelstein D., Auda-Boucher G., Cheraud Y., Rouaud T., Blanc I., Tajbakhsh S., Buckingham M.E., Fontaine-Perus J., Robert B.. The homeobox gene Msx1 is expressed in a subset of somites, and in muscle progenitor cells migrating into the forelimb. Development. 1999; 126:2689–2701. [DOI] [PubMed] [Google Scholar]
- 20. Hu G., Lee H., Price S.M., Shen M.M., Abate-Shen C.. Msx homeobox genes inhibit differentiation through upregulation of cyclin D1. Development. 2001; 128:2373–2384. [DOI] [PubMed] [Google Scholar]
- 21. Lallemand Y., Nicola M.A., Ramos C., Bach A., Cloment C.S., Robert B.. Analysis of Msx1; Msx2 double mutants reveals multiple roles for Msx genes in limb development. Development. 2005; 132:3003–3014. [DOI] [PubMed] [Google Scholar]
- 22. Woloshin P., Song K., Degnin C., Killary A.M., Goldhamer D.J., Sassoon D., Thayer M.J.. MSX1 inhibits myoD expression in fibroblast x 10T1/2 cell hybrids. Cell. 1995; 82:611–620. [DOI] [PubMed] [Google Scholar]
- 23. Bendall A.J., Abate-Shen C.. Roles for Msx and Dlx homeoproteins in vertebrate development. Gene. 2000; 247:17–31. [DOI] [PubMed] [Google Scholar]
- 24. Cornelison D.D., Olwin B.B., Rudnicki M.A., Wold B.J.. MyoD(-/-) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev. Biol. 2000; 224:122–137. [DOI] [PubMed] [Google Scholar]
- 25. Odelberg S.J., Kollhoff A., Keating M.T.. Dedifferentiation of mammalian myotubes induced by msx1. Cell. 2000; 103:1099–1109. [DOI] [PubMed] [Google Scholar]
- 26. Dodig M., Tadic T., Kronenberg M.S., Dacic S., Liu Y.H., Maxson R., Rowe D.W., Lichtler A.C.. Ectopic Msx2 overexpression inhibits and Msx2 antisense stimulates calvarial osteoblast differentiation. Dev. Biol. 1999; 209:298–307. [DOI] [PubMed] [Google Scholar]
- 27. Mina M., Gluhak J., Rodgers B.. Downregulation of Msx-2 expression results in chondrogenesis in the medial region of the avian mandible. Connect. Tissue Res. 1996; 35:79–84. [DOI] [PubMed] [Google Scholar]
- 28. Pavlova A., Boutin E., Cunha G., Sassoon D.. Msx1 (Hox-7.1) in the adult-mouse uterus - cellular interactions underlying regulation of expression. Development. 1994; 120:335–345. [DOI] [PubMed] [Google Scholar]
- 29. Song K., Wang Y., Sassoon D.. Expression of Hox-7.1 in myoblasts inhibits terminal differentiation and induces cell transformation. Nature. 1992; 360:477–481. [DOI] [PubMed] [Google Scholar]
- 30. Qiu J., Huang T., Xu J., Bi C., Chen C., Zhou M.. beta-Tubulins in Gibberella zeae: their characterization and contribution to carbendazim resistance. Pest Manag Sci. 2012; 68:1191–1198. [DOI] [PubMed] [Google Scholar]
- 31. Urban A., Neukirchen S., Jaeger K.E.. A rapid and efficient method for site-directed mutagenesis using one-step overlap extension PCR. Nucleic Acids Res. 1997; 25:2227–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R.. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989; 77:51–59. [DOI] [PubMed] [Google Scholar]
- 33. Lee H., Habas R., Abate-Shen C.. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004; 304:1675–1678. [DOI] [PubMed] [Google Scholar]
- 34. Lee H., Quinn J.C., Prasanth K.V., Swiss V.A., Economides K.D., Camacho M.M., Spector D.L., Abate-Shen C.. PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev. 2006; 20:784–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Swift S., Lorens J., Achacoso P., Nolan G.P.. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr. Protoc. Immunol. 2001; 31:10.17.14–10.17.29. [DOI] [PubMed] [Google Scholar]
- 36. Pear W.S., Nolan G.P., Scott M.L., Baltimore D.. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Riccardi C., Nicoletti I.. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat. Protoc. 2006; 1:1458–1461. [DOI] [PubMed] [Google Scholar]
- 38. Zhu X., Li M., Jia X., Hou W., Yang J., Zhao H., Wang G., Wang J.. The homeoprotein Msx1 cooperates with Pkn1 to prevent terminal differentiation in myogenic precursor cells. Biochimie. 2019; 162:55–65. [DOI] [PubMed] [Google Scholar]
- 39. Li G., Jin D., Zhong T.P.. Tubgcp3 is required for retinal progenitor cell proliferation during zebrafish development. Front Mol Neurosci. 2019; 12:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wisniewski J.R., Zougman A., Nagaraj N., Mann M.. Universal sample preparation method for proteome analysis. Nat. Methods. 2009; 6:359–362. [DOI] [PubMed] [Google Scholar]
- 42. Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S.. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999; 20:3551–3567. [DOI] [PubMed] [Google Scholar]
- 43. Xie Y., Wang D., Lan F., Wei G., Ni T., Chai R., Liu D., Hu S., Li M., Li D. et al.. An episomal vector-based CRISPR/Cas9 system for highly efficient gene knockout in human pluripotent stem cells. Sci. Rep. 2017; 7:2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Satokata I., Maas R.. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat. Genet. 1994; 6:348–356. [DOI] [PubMed] [Google Scholar]
- 45. Logan M., Martin J.F., Nagy A., Lobe C., Olson E.N., Tabin C.J.. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002; 33:77–80. [DOI] [PubMed] [Google Scholar]
- 46. Fu H., Ishii M., Gu Y., Maxson R.. Conditional Alleles of Msx1 and Msx2. Genesis. 2007; 45:477–481. [DOI] [PubMed] [Google Scholar]
- 47. Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R.T., Gao Y.H., Inada M. et al.. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997; 89:755–764. [DOI] [PubMed] [Google Scholar]
- 48. Otto F., Thornell A.P., Crompton T., Denzel A., Gilmour K.C., Rosewell I.R., Stamp G.W.H., Beddington R.S.P., Mundlos S., Olsen B.R. et al.. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997; 89:765–771. [DOI] [PubMed] [Google Scholar]
- 49. Heller H., Gredinger E., Bengal E.. Rac1 inhibits myogenic differentiation by preventing the complete withdrawal of myoblasts from the cell cycle. J. Biol. Chem. 2001; 276:37307–37316. [DOI] [PubMed] [Google Scholar]
- 50. Subramanian J., Dye L., Morozov A.. Rap1 signaling prevents L-type calcium channel-dependent neurotransmitter release. J. Neurosci. 2013; 33:7245–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lin A.W., Barradas M., Stone J.C., van Aelst L., Serrano M., Lowe S.W.. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Gene Dev. 1998; 12:3008–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oh Y.T., Yue P., Zhou W., Balko J.M., Black E.P., Owonikoko T.K., Khuri F.R., Sun S.Y.. Oncogenic Ras and B-Raf proteins positively regulate death receptor 5 expression through co-activation of ERK and JNK signaling. J. Biol. Chem. 2012; 287:257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barrett S.D., Bridges A.J., Dudley D.T., Saltiel A.R., Fergus J.H., Flamme C.M., Delaney A.M., Kaufman M., LePage S., Leopold W.R. et al.. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg. Med. Chem. Lett. 2008; 18:6501–6504. [DOI] [PubMed] [Google Scholar]
- 54. Wang J., Kumar R.M., Biggs V.J., Lee H., Chen Y., Kagey M.H., Young R.A., Abate-Shen C.. The Msx1 homeoprotein recruits polycomb to the nuclear periphery during development. Dev. Cell. 2011; 21:575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Holmberg C.I., Tran S.E., Eriksson J.E., Sistonen L.. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem. Sci. 2002; 27:619–627. [DOI] [PubMed] [Google Scholar]
- 56. Vassilev L.T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D.C., Chen L.. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl Acad. Sci. U.S.A. 2006; 103:10660–10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Satokata I., Ma L., Ohshima H., Bei M., Woo I., Nishizawa K., Maeda T., Takano Y., Uchiyama M., Heaney S. et al.. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat. Genet. 2000; 24:391–395. [DOI] [PubMed] [Google Scholar]
- 58. Lu P., Yu Y., Perdue Y., Werb Z.. The apical ectodermal ridge is a timer for generating distal limb progenitors. Development. 2008; 135:1395–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Danopoulos S., Parsa S., Al Alam D., Tabatabai R., Baptista S., Tiozzo C., Carraro G., Wheeler M., Barreto G., Braun T. et al.. Transient Inhibition of FGFR2b-ligands signaling leads to irreversible loss of cellular beta-catenin organization and signaling in AER during mouse limb development. PLoS One. 2013; 8:e76248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Volonte D., Liu Y., Galbiati F.. The modulation of caveolin-1 expression controls satellite cell activation during muscle repair. FASEB J. 2005; 19:237–239. [DOI] [PubMed] [Google Scholar]
- 61. Kook S.H., Son Y.O., Choi K.C., Lee H.J., Chung W.T., Hwang I.H., Lee J.C.. Cyclic mechanical stress suppresses myogenic differentiation of adult bovine satellite cells through activation of extracellular signal-regulated kinase. Mol. Cell. Biochem. 2008; 309:133–141. [DOI] [PubMed] [Google Scholar]
- 62. Lathrop B., Thomas K., Glaser L.. Control of myogenic differentiation by fibroblast growth factor is mediated by position in the G1 phase of the cell cycle. J. Cell Biol. 1985; 101:2194–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Michailovici I., Harrington H.A., Azogui H.H., Yahalom-Ronen Y., Plotnikov A., Ching S., Stumpf M.P., Klein O.D., Seger R., Tzahor E.. Nuclear to cytoplasmic shuttling of ERK promotes differentiation of muscle stem/progenitor cells. Development. 2014; 141:2611–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jeong H.M., Jin Y.H., Choi Y.H., Yum J., Choi J.K., Yeo C.Y., Lee K.Y.. PKC signaling inhibits osteogenic differentiation through the regulation of Msx2 function. BBA-Mol. Cell Res. 2012; 1823:1225–1232. [DOI] [PubMed] [Google Scholar]
- 65. Garin E., Lemieux M., Coulombe Y., Robinson G.W., Jeannotte L.. Stromal Hoxa5 function controls the growth and differentiation of mammary alveolar epithelium. Dev. Dyn. 2006; 235:1858–1871. [DOI] [PubMed] [Google Scholar]
- 66. Raman V., Martensen S.A., Reisman D., Evron E., Odenwald W.F., Jaffee E., Marks J., Sukumar S.. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000; 405:974–978. [DOI] [PubMed] [Google Scholar]
- 67. Stasinopoulos I.A., Mironchik Y., Raman A., Wildes F., Winnard P. Jr, Raman V.. HOXA5-twist interaction alters p53 homeostasis in breast cancer cells. J. Biol. Chem. 2005; 280:2294–2299. [DOI] [PubMed] [Google Scholar]
- 68. Chen H., Chung S., Sukumar S.. HOXA5-induced apoptosis in breast cancer cells is mediated by caspases 2 and 8. Mol. Cell. Biol. 2004; 24:924–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dai J., Mou Z., Shen S., Dong Y., Yang T., Shen S.G.. Bioinformatic analysis of Msx1 and Msx2 involved in craniofacial development. J. Craniofac. Surg. 2014; 25:129–134. [DOI] [PubMed] [Google Scholar]
- 70. Zhang W., Qu H.C., Zhang Y.. Association of MSX1 and TGF-beta1 genetic polymorphisms with hypodontia: meta-analysis. Genet. Mol. Res. 2014; 13:10007–10016. [DOI] [PubMed] [Google Scholar]
- 71. Nassif A., Senussi I., Meary F., Loiodice S., Hotton D., Robert B., Bensidhoum M., Berdal A., Babajko S.. Msx1 role in craniofacial bone morphogenesis. Bone. 2014; 66:96–104. [DOI] [PubMed] [Google Scholar]
- 72. Reddy N.A., Adusumilli G., Devanna R., Pichai S., Rohra M.G., Arjunan S.. MSX1 gene variant - its presence in tooth absence - a case control genetic study. J Int Oral Health. 2013; 5:20–26. [PMC free article] [PubMed] [Google Scholar]
- 73. Xin T., Zhang T., Li Q., Yu T., Zhu Y., Yang R., Zhou Y.. A novel mutation of MSX1 in oligodontia inhibits odontogenesis of dental pulp stem cells via the ERK pathway. Stem Cell Res. Ther. 2018; 9:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq datasets used in this study have been deposited with the NCBI GEO under accession number GSE150349.