Summary
The tuberculosis vaccine bacillus Calmette-Guérin (BCG) protects against some heterologous infections, probably via induction of non-specific innate immune memory in monocytes and natural killer (NK) cells, a process known as trained immunity. Recent studies have revealed that the induction of trained immunity is associated with a bias toward granulopoiesis in bone marrow hematopoietic progenitor cells, but it is unknown whether BCG vaccination also leads to functional reprogramming of mature neutrophils. Here, we show that BCG vaccination of healthy humans induces long-lasting changes in neutrophil phenotype, characterized by increased expression of activation markers and antimicrobial function. The enhanced function of human neutrophils persists for at least 3 months after vaccination and is associated with genome-wide epigenetic modifications in trimethylation at histone 3 lysine 4. Functional reprogramming of neutrophils by the induction of trained immunity might offer novel therapeutic strategies in clinical conditions that could benefit from modulation of neutrophil effector function.
Keywords: innate immune memory, trained immunity, neutrophil, polymorphonuclear leukocytes, BCG, nonspecific effects of vaccines, epigenetics
Graphical Abstract
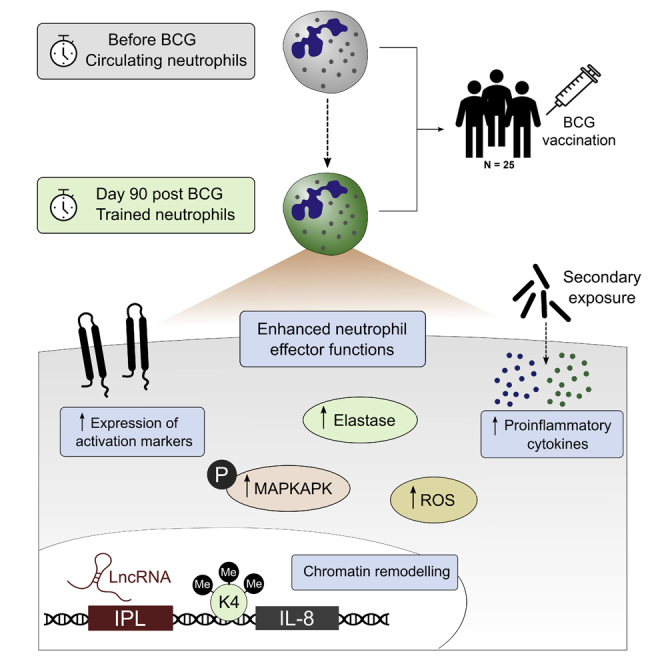
Highlights
-
•
BCG vaccination of humans induces long-term immunophenotypic changes in neutrophils
-
•
BCG increases antimicrobial activity of neutrophils against unrelated pathogens
-
•
BCG-induced functional changes associate with modifications in histone methylation
-
•
Trained immunity may be a therapeutic target in neutrophil-mediated diseases
Moorlag et al. show that BCG vaccination induces long-lasting functional changes in human neutrophils, characterized by increased expression of activation markers and enhanced antimicrobial function upon secondary stimulation. These findings highlight the potential of trained immunity as a therapeutic target to modulate neutrophil effector function.
Introduction
Bacillus Calmette-Guérin (BCG) vaccine not only protects against infection with Mycobacterium tuberculosis, but has also been shown to decrease overall morbidity and mortality to other non-related infections (Aaby et al., 2011; Biering-Sørensen et al., 2012; Roth et al., 2005). A growing body of evidence has indicated that these effects are mediated by the ability of innate immune cells to build a heterologous immunological memory after certain infections or vaccinations via a process termed “trained immunity,” resulting in heightened innate immune responses upon exposure to secondary infections (Netea et al., 2016). Various studies have shown that vaccination of healthy adults with BCG enhances proinflammatory cytokine responses of monocytes and natural killer (NK) cells upon ex vivo stimulation with various unrelated pathogens (Arts et al., 2018; Kleinnijenhuis et al., 2012, 2014; Walk et al., 2019). These functional changes persisted for at least 3 months—even up to 1 year—and were associated with increased expression of activation markers. Importantly, a recent study revealed that the increase in cytokine production capacity protects against an experimental viral infection in humans (Arts et al., 2018). Epigenetic reprogramming of monocytes, characterized by the deposition of chromatin marks and changes in DNA methylation status that facilitate expression of proinflammatory genes, as well as metabolic rewiring underlie the long-term altered immune responses (Arts et al., 2016b; Kleinnijenhuis et al., 2012).
Recent studies indicate that induction of trained immunity not only alters the functional program of mature monocytes/macrophages, but also reprograms the hematopoietic stem and progenitor cells (HSPCs) in the bone marrow. Remarkably, analysis of HSPCs after BCG vaccination in mice revealed a strong long-term bias toward granulopoiesis (Kaufmann et al., 2018). This effect was not specific for BCG, as similar alterations in the bone marrow have been shown for other inducers of trained immunity, such as β-glucan (Chavakis et al., 2019; Mitroulis et al., 2018) and systemic inflammation induced by a Western-type diet (Christ et al., 2018). In addition to these experimental studies in mice, emerging data suggest that a similar lineage bias in favor of enhanced neutrophil production is induced in humans. A recent randomized human BCG vaccination study revealed that the genes in HSPCs that were most differentially expressed 3 months after vaccination were strongly enriched for pathways involved in neutrophil activation and degranulation (Cirovic et al., 2020). In line with these findings, results of a randomized controlled trial performed in 1400 children demonstrated an increased number of circulating neutrophils in the group that received the BCG vaccine shortly after birth (Cirovic et al., 2020). Interestingly, these effects were dependent on the genotype of the host, specifically on polymorphisms in IL32 (Dos Santos et al., 2019).
Neutrophils play an essential antimicrobial function in the innate immune response, providing a crucial first line of defense against invading pathogens. They are the most abundant circulating leukocytes in humans, making up 50%–70% of all leukocytes in the circulation. About 1011 neutrophils are produced each day in the bone marrow (Furze and Rankin, 2008) from a bone marrow precursor that also gives rise to monocytes (Akashi et al., 2000). From the bone marrow, neutrophils are released into the circulation, from which they migrate to tissues. Although neutrophils are considered to be short-lived, some studies have challenged this paradigm, indicating that neutrophils can live up to 5.4 days (Pillay et al., 2010).
Neutrophils use various effector mechanisms to kill invading pathogens, including phagocytosis; the production of reactive oxygen species (ROS); the release of genomic DNA in the form of neutrophil extracellular traps (NETs); and the production of proinflammatory cytokines, chemokines, and cytotoxic granules (Nicolás-Avila et al., 2017). Because of their short lifespan, the pre-formed mediators that are released upon stimulation, and the relatively low RNA content, neutrophils were considered to be a homogeneous population, unable to adapt to changing environments. However, it has recently become clear that neutrophils are a highly heterogeneous group, showing large differences in phenotype and functional versatility (Ng et al., 2019). Furthermore, an increasing body of evidence indicates that they respond to environmental changes with significantly altered gene expression, both in health and disease. Various studies have recently shown that these transcriptional changes are partially controlled by epigenetic reprogramming (Chen et al., 2016; Ecker et al., 2017).
Although recent studies suggest a transcriptional bias toward granulopoiesis in the myeloid progenitors in the bone marrow after BCG vaccination, it remains unknown whether these alterations subsequently result in long-term functional changes in mature neutrophils. Here, we investigated whether BCG vaccination in humans alters the function of mature neutrophils in vivo in healthy individuals. We show that BCG vaccination induces trained immunity in neutrophils, characterized by increased expression of activation markers, downregulation of immunosuppressive markers, and increased responsiveness to heterologous stimulation. Furthermore, we demonstrate that BCG vaccination enhances key antimicrobial functions in neutrophils and confers an enhanced capacity to kill the heterologous fungal pathogen Candida albicans. Importantly, these findings highlight the potential for the induction of trained immunity as a therapeutic and/or prophylactic strategy to enhance neutrophil function and resistance against infections by vaccination.
Results
BCG Vaccination Induces Long-Term Changes in Neutrophil Phenotype Associated with Increased Activation
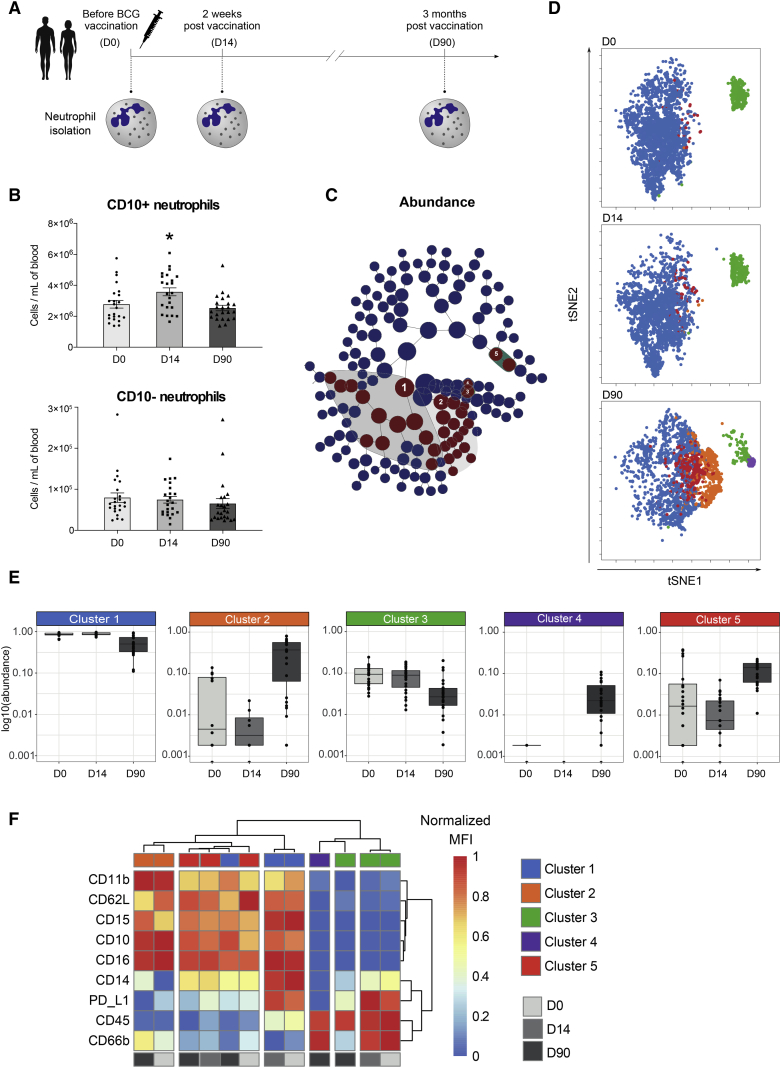
To study the effect of BCG vaccination on neutrophils in vivo, blood was collected from 25 healthy human individuals, and neutrophils were immunophenotyped by flow cytometry before, 2 weeks after, and 3 months after vaccination with BCG (InterVax, Canada) (Figures 1A and S1). While the proportion of neutrophils within the total leukocyte population did not change (Figure S2), the absolute number of neutrophils was increased 2 weeks after vaccination and returned to baseline after 3 months. This increase was accounted for mainly by mature neutrophils (Figure 1B), marked by the surface expression of CD10 (Marini et al., 2017). To further investigate the effect of BCG on granulocyte populations, particularly on neutrophil subpopulations, we assessed the expression of nine surface markers (CD16, CD10, CD11b, CD14, CD62L, PD-L1, CD66b, CD15, and CD45) by flow cytometry. We used CITRUS (cluster identification, characterization, and regression) (Bruggner et al., 2014) analysis to generate clusters that predicted differences among the three time points measured and viSNE (visualization of high-dimensional single-cell data based on the t-Distributed Stochastic Neighbor Embedding (t-SNE) algorithm) (Amir et al., 2013) to visualize cells on a 2D map. CITRUS generated around 150 stratifying clusters and identified a subset of 34 clusters that differed in abundance between the time points (FDR < 1%) (Figure 1C; Data S1). From those 34, the 5 parental clusters were chosen for further examination. When we visualized these clusters in the viSNE 2D map, we observed a transition of clusters 1 and 3 into clusters 2, 4, and 5 three months post-vaccination (Figure 1D). Three months upon BCG vaccination, the relative abundance of clusters 1 and 3 was significantly decreased (FDR < 1%), while clusters 2, 4, and 5 were increased (Figure 1E). Upon examination of the expression of the surface markers, we observed that clusters 1, 2, and 5 had a high expression of CD10, CD15, and CD16, which are specific markers for mature neutrophils. Clusters 3 and 4 lacked expression of CD15 and CD16 and had a high expression of CD45, resembling eosinophils (Figures 1D–1F; Data S1). Comparison of the functional marker expression of all identified clusters revealed that BCG significantly increased the expression of various well-known activation markers on neutrophils, including CD11b and CD66b, and decreased the expression of CD62L (Figure 1F; Data S1). In parallel, the expression of PD-L1, a key immune checkpoint protein associated with immunosuppression, was significantly decreased upon BCG vaccination. Together, these data show that circulating neutrophils increase the expression of immune activation markers upon BCG vaccination, which indicates a change in phenotype that resembles trained innate immune cells. Additionally, we observed significant changes in the eosinophil phenotype upon vaccination, characterized by significantly higher levels of the activation marker CD66b and lower levels of PD-L1 as compared to levels before vaccination (Figure 1F; Data S1). No significant changes in eosinophil count were observed upon vaccination (Figure S2).
Figure 1.
BCG Vaccination Alters the Phenotype of Circulating Neutrophils
(A) Schematic representation of the BCG vaccination trial. Neutrophils were isolated from 25 healthy volunteers before (D0, n = 25), 2 weeks after (D14, n = 25) and 3 months after (D90, n = 23) vaccination with BCG.
(B) Absolute counts of CD10+ mature and CD10− immature neutrophils before, 2 weeks after, and 3 months after BCG vaccination (mean ± SEM, n = 25, ∗p < 0.05, Wilcoxon signed-rank test).
(C) CITRUS hierarchy plot with stratifying features using FDR of <1%.
(D) Overlay of the five parental stratifying CITRUS clusters on the viSNE plot.
(E) Relative abundance of the clusters before, 2 weeks after, and 3 months after vaccination.
(F) Heatmaps showing the differential expression of the surface markers in the clusters of neutrophils (clusters 1, 2, and 5) and eosinophils (clusters 3 and 4) (normalized MFI clustered with Ward’s method).
See also Figures S1 and S2 and Data S1.
BCG Vaccination Induces an Enhanced Immunophenotype in Neutrophils Exposed to Bacterial and Fungal Pathogens
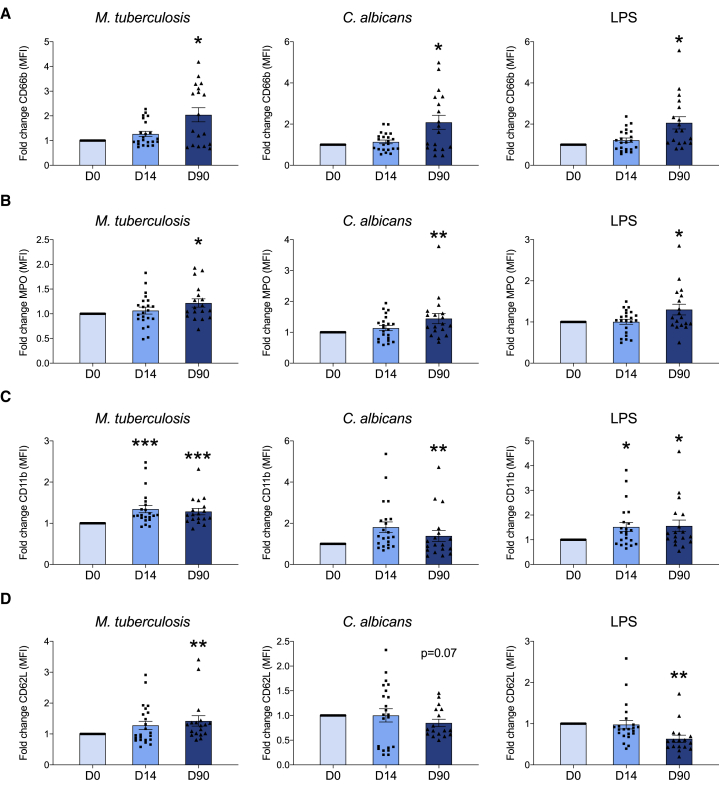
To test whether BCG vaccination also alters the expression of cell-surface activation markers in response to a secondary stimulus, we incubated neutrophils ex vivo for 17 h with M. tuberculosis, C. albicans, lipopolysaccharide (LPS), phorbol 12-myristate 13-acetate (PMA), or RPMI medium (control) before, as well as 2 weeks and 3 months after vaccination. We found significant differences in the expression of cell activation markers on ex-vivo-stimulated neutrophils sampled after vaccination as compared to baseline (ratio on top of medium control, median fluorescence intensity levels are shown in Figure S3). Three months after BCG vaccination, the expression of CD66b and myeloperoxidase (MPO) on neutrophils upon exposure to either M. tuberculosis, C. albicans, or LPS was significantly enhanced, as compared to neutrophils before vaccination, indicating increased degranulation (Figures 2A and 2B). In addition, an increase in the expression of the integrin alphaM (CD11b) was observed upon vaccination, suggesting enhanced activation 2 weeks and 3 months after BCG vaccination (Figure 2C). We also observed that the expression of CD62L was significantly decreased upon stimulation with LPS, indicative of active shedding of L-selectin (Figure 2D). Remarkably, there was an increase in the fold change of CD62L expression upon stimulation with M. tuberculosis post-BCG. The precise mechanism explaining the difference between LPS and M. tuberculosis stimulation remains to be deciphered in future studies, but one hypothesis is that it might be explained by the differential stimulation of pattern recognition receptors by the two stimuli (TLR4 for LPS versus NOD2/TLR2 for M. tuberculosis). Overall, these results suggest that ex-vivo-stimulated neutrophils display a distinct phenotype upon BCG vaccination, which is characterized by enhanced expression of activation and degranulation markers, suggesting increased functional capacity.
Figure 2.
Neutrophil Phenotype upon Exposure to Unrelated Pathogens Is Modified by BCG
Cell-surface expression molecules associated with neutrophil activation were analyzed by flow cytometry upon ex vivo stimulation with M. tuberculosis, C. albicans, or LPS before, 2 weeks after, and 3 months after vaccination. (A) CD66b, (B) MPO, (C) CD11b, and (D) CD62L. Fold change of MFI as compared to medium control, upon vaccination versus baseline (mean ± SEM, n = 22 [D14], n = 18 [D90], CD62L_Mtb_D14 [n = 23], CD62L_Candida/LPS_D14 [n = 17]; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Wilcoxon signed-rank test).
See also Figures S3 and S4.
BCG Vaccination Increases Nonspecific Antimicrobial Activity of Human Neutrophils
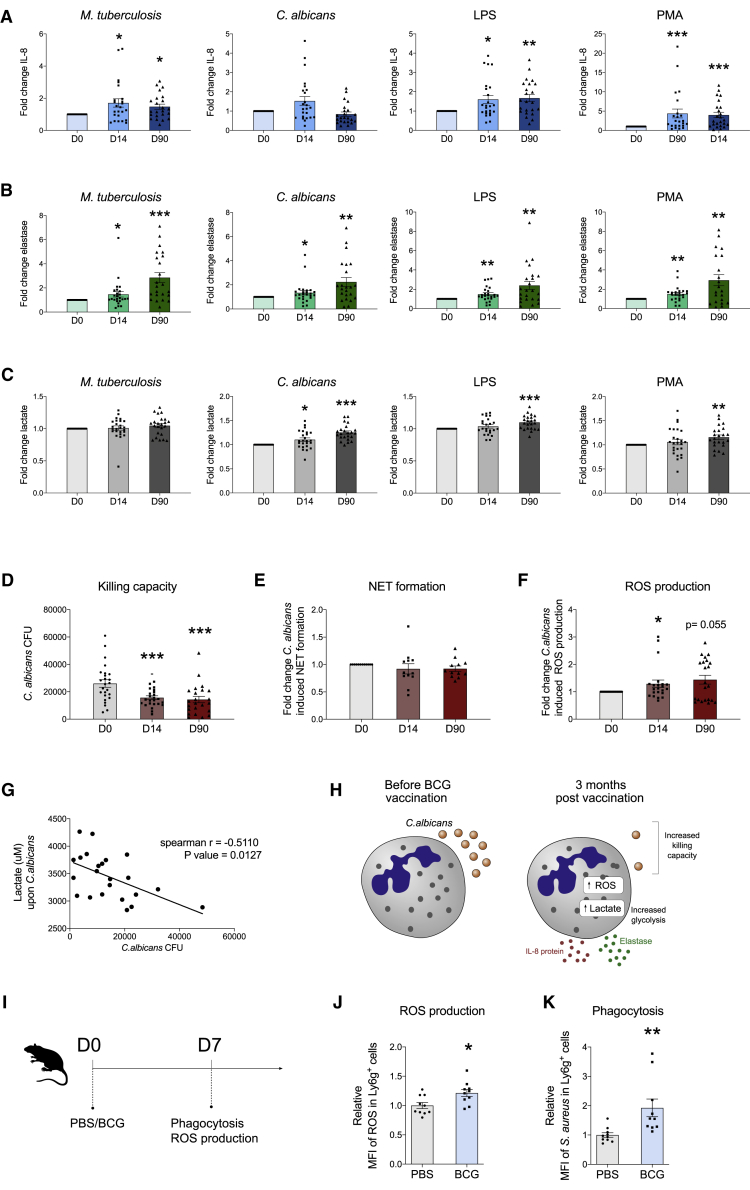
Previous studies have shown that BCG vaccination induces functional changes in human monocytes and NK cells in vivo, as shown by enhanced ex vivo cytokine production to both target (mycobacterial) and unrelated pathogens upon BCG vaccination (Arts et al., 2018; Kleinnijenhuis et al., 2012, 2014). Consistent with our previous findings in monocytes (Arts et al., 2018), BCG vaccination enhanced production of the chemokine interleukin (IL)-8 in neutrophils stimulated with either M. tuberculosis or unrelated stimuli as compared to concentrations before vaccination (Figures 3A and S4). Except for C. albicans stimulation, this effect remained for at least 3 months. Potential differences in IL-1β and tumor necrosis factor alpha (TNF-α) production could not be determined, as the concentration of these cytokines in the supernatants remained below the detection threshold (data not shown). Besides the production of cytokines, neutrophils store an arsenal of antimicrobial enzymes in granules, including the serine protease elastase. Interestingly, release of elastase in response to stimulation was significantly increased in neutrophils 2 weeks and 3 months post-vaccination, compared to the levels before vaccination (Figure 3B).
Figure 3.
BCG Vaccination Induces Trained Immunity In Vivo in Neutrophils
(A–C) Neutrophils were isolated before (n = 25) and after vaccination (n = 23) with BCG and restimulated ex vivo. Production of IL-8, elastase, and lactate was measured in the supernatants. Fold increases, as compared to medium control upon vaccination versus baseline, of IL-8 (A), elastase (B), and lactate (C) to sonicated M. tuberculosis, heat-killed C. albicans, LPS, and PMA are shown.
(D) Quantification of C. albicans colony-forming units (CFUs) after 17 h ex vivo incubation with neutrophils before and after BCG vaccination (mean ± SEM, ∗∗∗p < 0.001, Wilcoxon signed-rank test).
(E) Fold change (compared to levels before BCG vaccination) in C. albicans-induced NET formation (n = 12).
(F) Fold change in the production of ROS in response to C. albicans before and after vaccination.
(G) Correlation plot showing the relationship between C. albicans-induced lactate production and the remaining number of C. albicans CFU after 17 h ex vivo incubation with neutrophils.
(H) BCG vaccination increases the killing capacity of neutrophils, which may be partially mediated by enhanced glycolysis and the release of antimicrobial molecules.
(I) Wild-type (WT) mice were injected with BCG or PBS, and ROS production and phagocytosis were assessed in splenic neutrophils 7 days later.
(J) ROS production in CD11b+ Ly6g+ cells.
(K) Phagocytosis of S. aureus BioParticles in CD11b+ Ly6g+ cells. Data are presented as mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, n = 10 mice per group, unpaired t test.
See also Figure S4.
Earlier work demonstrated a crucial role for the induction of glycolysis in BCG-induced trained immunity in monocytes (Arts et al., 2016b). In order to investigate whether the BCG-induced changes in the release of IL-8 and elastase were associated with alterations in intracellular metabolism, we assessed lactate production 17 h after ex vivo stimulation. Indeed, neutrophils isolated 3 months after vaccination produced a slightly higher amount of lactate after stimulation with C. albicans, LPS, or PMA compared to levels before vaccination, suggestive of an increased glycolytic rate (Figure 3C).
BCG Vaccination Enhances the Killing Capacity of Neutrophils and Results in Increased ROS Production and Phagocytosis
One of the main functions of neutrophils is to kill invading microorganisms. As such, they are considered to be the most important effector cells in the killing of pathogenic fungi such as C. albicans (Fulurija et al., 1996). Therefore, we next assessed whether BCG vaccination would affect neutrophil killing capacity toward C. albicans. The number of C. albicans colonies that survived incubation with neutrophils was significantly decreased after vaccination, as compared to the number before vaccination (Figure 3D), indicating a significant impact of BCG vaccination on the killing capacity of neutrophils. To investigate the mechanisms underlying the enhanced killing capacity, we first investigated whether the ability to form NETs contributed to this effect. However, ex vivo NET formation was not affected by BCG vaccination (Figures 3E and S4).
Since previous work demonstrated increased production of ROS in monocytes trained in vitro with BCG (Bekkering et al., 2016), and the production of ROS plays an important role in microbial clearance by neutrophils (Mittal et al., 2014), we next hypothesized that BCG vaccination may enhance the ability of neutrophils to generate ROS. Indeed, the ex vivo production of ROS by neutrophils upon C. albicans stimulation increased after vaccination (Figure 3F). Interestingly, the production of lactate upon C. albicans stimulation correlated with the killing activity (Spearman ρ = −0.51, p = 0.0127) (Figure 3G). These findings might suggest that an increase in glycolytic capacity contributes to the observed enhanced killing of C. albicans in neutrophils upon BCG vaccination (Figure 3H).
To study the effects of BCG vaccination on neutrophil function in an additional context, we performed a murine study in which we assessed the effect of BCG-mediated training on neutrophils. Specifically, splenic neutrophils were isolated from mice that had received BCG or PBS as control 7 days earlier (Figure 3I). Neutrophils from trained BCG-trained mice displayed enhanced ROS production, as compared to neutrophils from control-treated mice (Figure 3J), which was consistent with our findings in human neutrophils upon C. albicans stimulation. Furthermore, we observed a profound increase in the capacity of neutrophils from BCG-trained mice to phagocytose S. aureus BioParticles, as compared to neutrophils from mice that received PBS (Figure 3K).
BCG Vaccination Enhances MAPK Phosphorylation in Neutrophils
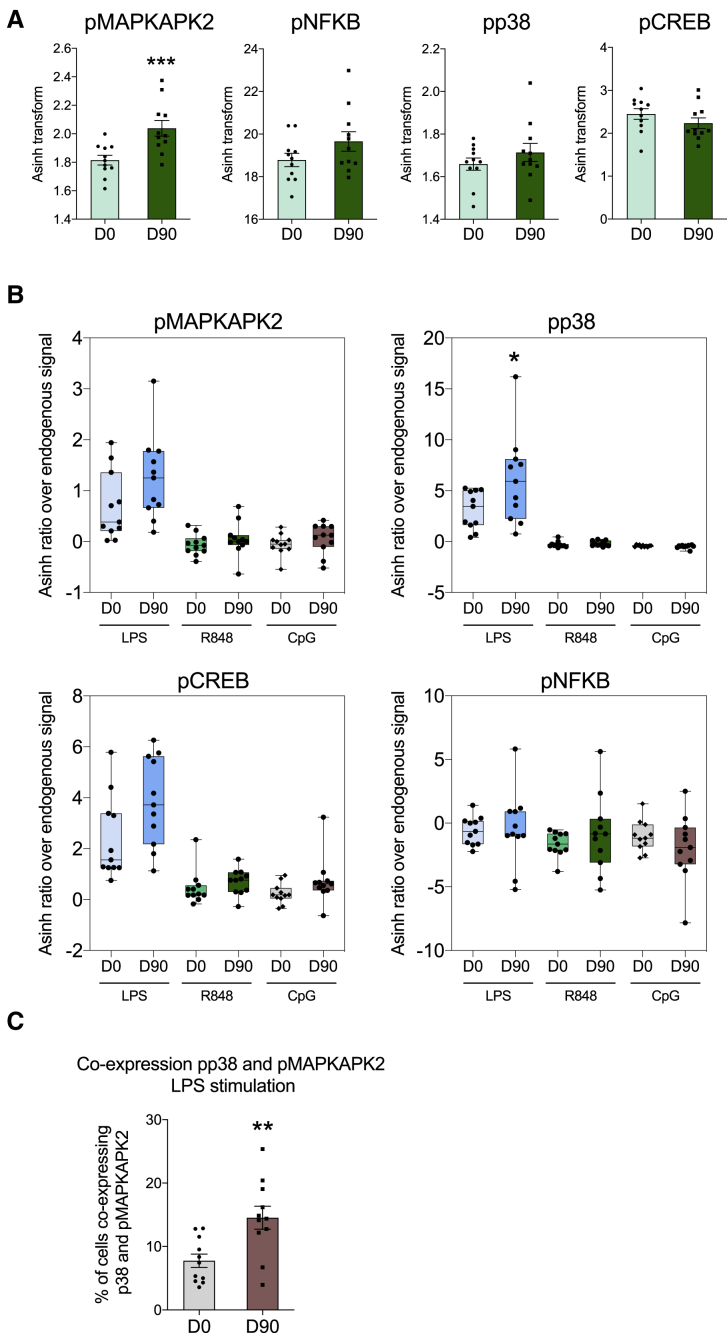
The nuclear factor κB (NF-κB) signaling pathway and mitogen-activated protein kinase (MAPK) pathways are major regulators of the transcription and post-transcriptional regulation of inflammation in innate immune cells (Arthur and Ley, 2013). Therefore, to identify mechanisms that may contribute to the enhanced antimicrobial function of neutrophils upon BCG vaccination, we investigated the activation of key signaling pathways in neutrophils before and 3 months after BCG vaccination. Whole blood from 11 vaccinated donors was left unstimulated (endogenous signal) or stimulated with one of three Toll-like receptor (TLR) ligands (LPS, R848, and CpG). Subsequently, phosphoprotein phosphorylation of neutrophils was quantified by mass cytometry (see Figures S5 and S6 for gating strategy). Three months after BCG vaccination, basal expression of phosphorylated MAPK-activated protein kinase 2 (pMAPKAPK2) was significantly increased as compared to expression levels before BCG vaccination (Figure 4A). Upon ex vivo stimulation with TLR agonists, neutrophils responded strongest to LPS stimulation. Interestingly, neutrophils displayed significantly enhanced p38 phosphorylation in response to LPS stimulation at 3 months post-BCG vaccination as compared to levels before vaccination (Figure 4B). A similar pattern was observed for MAPKAPK2 and cAMP-response element binding protein (CREB) but did not reach statistical significance. The responsiveness of neutrophils to stimulation with R848 or CpG was low, which could be explained by previous reports describing low expression of TLR7 in neutrophils and the requirement of GM-CSF for TLR9-mediated responses (Hayashi et al., 2003). In line with these findings, co-expression of pMAPKAPK2 and pp38 was significantly enhanced in response to LPS stimulation 3 months upon BCG vaccination (Figure 4C). Together, these findings point to an enhanced activation of the MAPK signaling pathway upon BCG vaccination in neutrophils, in particular upon activation of TLR4.
Figure 4.
MAPKAPK2, p38, CREB, and NF-κB Phosphorylation in Neutrophils before and 3 Months after BCG Vaccination
(A) Endogenous signal for the indicated phosphoproteins before BCG vaccination (D0) and 3 months post-BCG vaccination (D90).
(B) Phosphoprotein phosphorylation in response to the indicated stimuli.
(C) pMAPKAPK2 and pp38 co-expression upon LPS restimulation. Data are presented as mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, n = 11, Wilcoxon signed-rank test.
See Figures S5 and S6 for gating strategy.
BCG Induces Epigenetic Modifications in Histone Methylation in Neutrophils
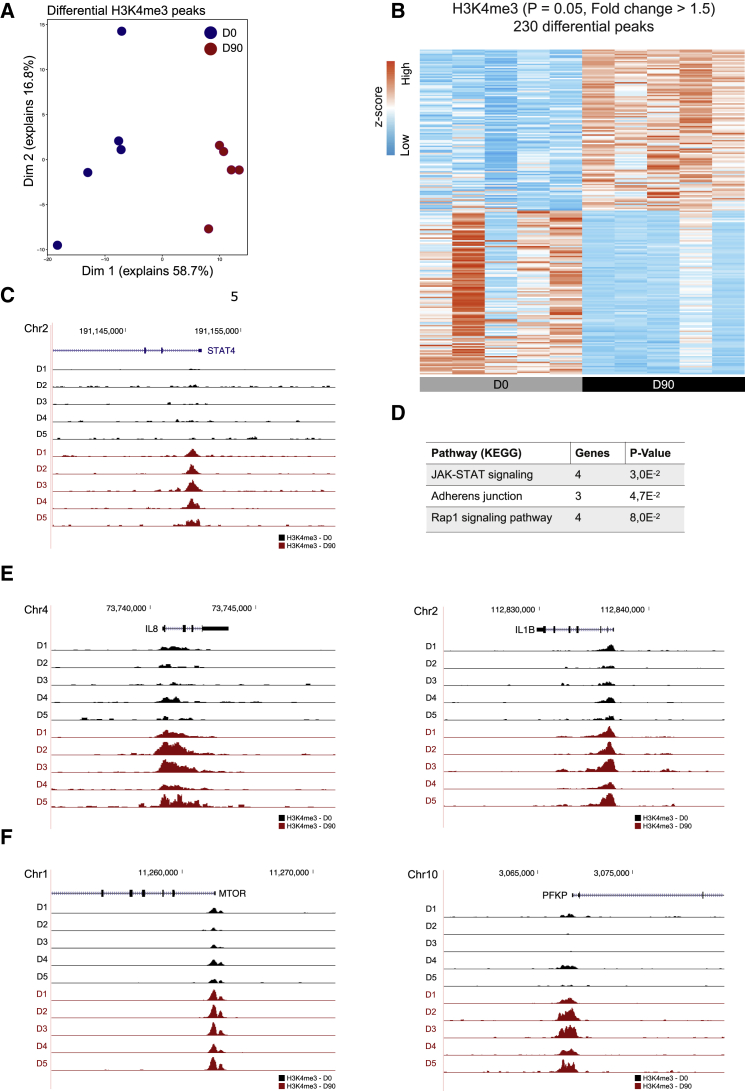
BCG vaccination induces genome-wide chromatin remodeling in human monocytes in vivo (Arts et al., 2018), and epigenetic mechanisms have been shown to control gene expression in human neutrophils (Chen et al., 2016; Ecker et al., 2017). Therefore, to further elucidate the mechanisms through which BCG induces the observed changes in neutrophil effector function, we performed RNA sequencing (RNA-seq) and a whole-genome assessment of the distribution of trimethylation of lysine 4 at histone 3 (H3K4me3) by chromatin immunoprecipitation (ChIP) sequencing in neutrophils before and 3 months after BCG vaccination (GEO: GSE153729). Whole-genome epigenetic analysis revealed significant differences in the levels of H3K4me3, a marker of increased gene transcription (Figures 5A and 5B). BCG induced increased H3K4me3 at promoter sites of genes involved in JAK-STAT signaling, including signal transducer and activator of transcription 4 (STAT4) (Figures 5C and 5D), which has recently been shown to play a critical role in proinflammatory neutrophil function and is required for antimicrobial defense in mice (Keeter et al., 2018, 2020). In addition, BCG vaccination induced significantly higher levels of H3K4me3 in promoter regions of various genes encoding proinflammatory cytokines (Figure 5E), consistent with our previous observations of a trained immunity profile in neutrophils upon BCG vaccination. Moreover, we found a significant increase in H3K4me3 at the promoter regions of genes that play a key role in glycolysis, including mammalian target of rapamycin (mTOR) and phosphofructokinase platelet (PFKP) (Figure 5F), supporting our previous observations showing a different metabolic profile in stimulated neutrophils upon BCG vaccination. These findings were validated by ChIP and quantitative real-time PCR (ChIP-qPCR) experiments (Figure S7). Furthermore, ChIP-qPCR data showed that higher levels of H3K4me3 corresponded with increased killing capacity (Figure S7). However, only seven individuals were assessed, and these correlations did not reach statistical significance. Collectively, these data indicate that BCG-induced long-term epigenetic reprogramming of neutrophils at the level of histone methylation may underlie their altered protective state.
Figure 5.
Vaccination with BCG Induces Epigenetic Reprogramming of Neutrophils
(A) PCA of ChIP-seq data of human neutrophils isolated before and 3 months after BCG vaccination (p < 0.05, fold change > 1.5, n = 5).
(B) Heatmap of genome-wide H3K4me3 changes before and 3 months after BCG vaccination.
(C) Tracks of H3K4me3 peaks near STAT4.
(D) Pathways associated with genes near the 230 differential peaks.
(E and F) Tracks of H3K4me3 peaks near CXCL8 and IL-1β (E) and mTOR and PFKB (F).
See also Figure S7.
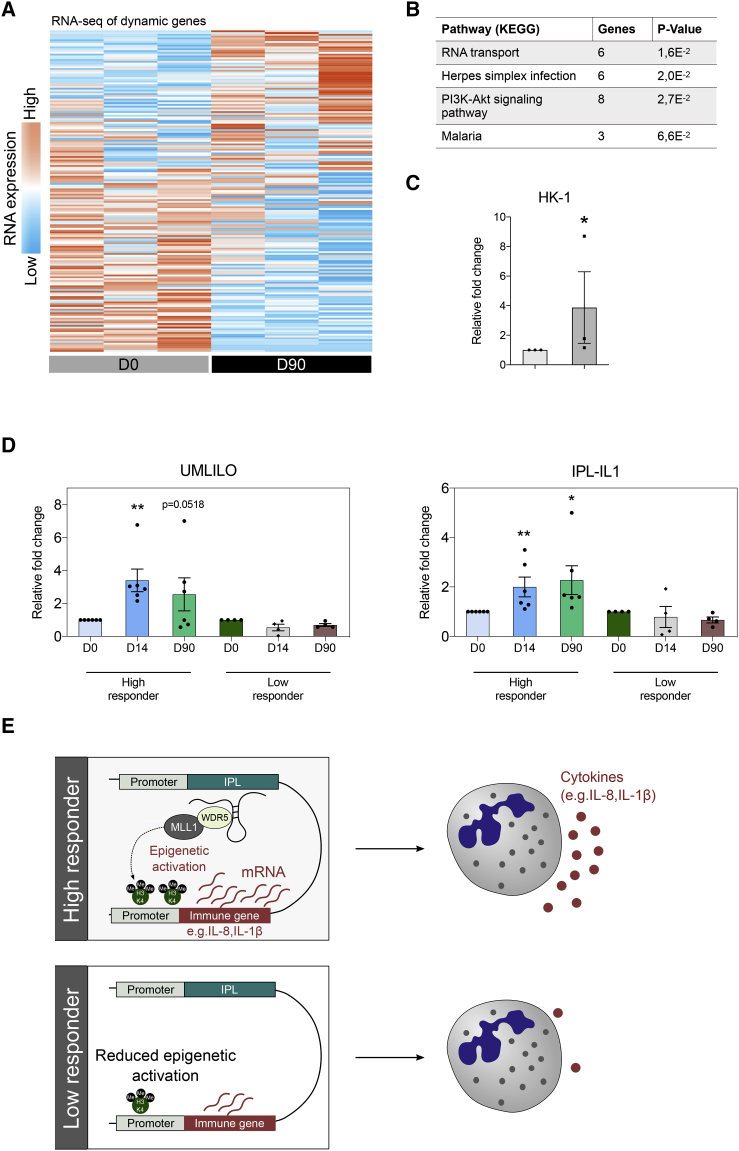
In line with previous studies on trained monocytes, analysis of the transcription profiles of neutrophils in their resting state revealed fewer changes upon BCG vaccination as compared to the changes in epigenetic profile (Figure 6A). This indicates that BCG reprograms neutrophils epigenetically to respond differently to secondary stimuli, leading to pronounced differences in gene expression after secondary stimulation, rather than at the resting state. However, analysis revealed enhanced expression of genes involved in the PI3K-Akt signaling pathway (Figure 6B). This pathway plays a crucial role in neutrophil effector functions and has been shown to mediate a metabolic shift that is essential for the induction of trained immunity in monocytes (Arts et al., 2016b; Cheng et al., 2014). Furthermore, the transcription of hexokinase 1 (HK1) was increased upon BCG vaccination (Figure 6C). As HK1 is a rate-limiting enzyme of glycolysis, this finding may further support our observation of long-term metabolic changes in neutrophils upon BCG vaccination.
Figure 6.
Transcriptomic Analysis of Neutrophils Trained by BCG
(A) Heatmap showing genes that are differentially expressed in human neutrophils upon BCG vaccination (p < 0.2, fold change > 1.5, n = 3).
(B) Pathways associated with genes that are differentially expressed 3 months after BCG vaccination as compared to baseline.
(C) Fold change in the expression of HK1 (3 months upon BCG as compared to baseline, mean ± SEM, n = 3).
(D) Fold change in the expression of IPLs UMLILO and IPL-IL1 after BCG vaccination as compared to levels before vaccination in high responders and low responders (mean ± SEM, n = 6 high responders, n = 4 low responders, ∗p < 0.05, ∗∗p < 0.01, Wilcoxon signed-rank test).
(E) In contrast to low responders, high responders display increased IPL expression upon BCG vaccination, leading to higher levels of H3K4me3 on target genes and enhanced gene transcription upon secondary exposure.
A long-standing question in the field of trained immunity has been how the H3K4me3 modification is directed only to the promoters of specific genes. Recently, it was demonstrated that a novel class of long non-coding RNAs (lncRNAs), termed immune priming lncRNAs (IPLs), coordinates the H3K4me3 accumulation at inflammatory gene promoters, including IL-8 and IL-1β, during trained immunity responses in monocytes (Fanucchi et al., 2019). UMLILO, a prototypical IPL, specifically regulates H3K4me3 deposition at the promoter site of IL-8, whereas IPL-IL1 regulates IL-1β. This study revealed that transcription of IPLs is increased in neutrophils after BCG vaccination as compared to levels before vaccination. Importantly, when we stratified volunteers into high responders (fold increase in killing capacity ≥2) and low responders (fold increase in killing capacity <2), we observed that the increase in IPL expression upon BCG vaccination was strongly associated with responder status (Figure 6D). Together, these findings might suggest that BCG vaccination increases IPL expression in neutrophils and, as a result, H3K4me3 levels on target genes, resulting in enhanced gene transcription and antimicrobial function in trained neutrophils (Figure 6E).
Discussion
Various reports have demonstrated that BCG vaccination can train innate immune cells to exhibit an enhanced and long-lasting activation upon challenge with mycobacterial or unrelated heterologous microbial pathogens. Since the large majority of studies on trained immunity has been performed on monocytes, macrophages, and NK cells, very little is known about the long-term impact of BCG on other innate immune cell populations. Neutrophils are the most abundant innate immune cells in humans and play a crucial role in the front line of immune defense (Nicolás-Avila et al., 2017). Intriguingly, various recent reports studying the effect of trained immunity on progenitors of the hematopoietic system revealed a strong long-lasting bias toward granulopoiesis, indicating a potential role for neutrophil activation in the process of innate immune memory (Christ et al., 2018; Cirovic et al., 2020; Kaufmann et al., 2018; Mitroulis et al., 2018). Here, we have shown that the induction of trained immunity by BCG induces long-term functional reprogramming of human circulating neutrophils in vivo via chromatin remodeling.
Specifically, we have shown that BCG vaccination induces a long-term switch in the phenotype of unstimulated (circulating) neutrophils, which is characterized by enhanced expression levels of activation markers and decreased expression of markers associated with immunosuppression. Higher expression of CD11b (the αM subunit of the integrin receptor αMβ2) and CD66b (CECAM8) indicates activation of the various functions of neutrophils, such as migration (Li, 1999), degranulation (Borregaard et al., 2007), and respiratory burst priming (McLeish et al., 2013), all important for pathogen clearance. Various recent studies have revealed that neutrophils are heterogeneous in phenotype and function. Once released into the blood, fresh neutrophils undergo phenotypic changes until they disappear from the circulation (aged neutrophils) by a process called neutrophil aging (Adrover et al., 2019). Aging has been described to positively correlate with proinflammatory activity, and aged neutrophils show lower expression of CD62L and higher levels of CD11b as compared to freshly released neutrophils (Adrover et al., 2019; Casanova-Acebes et al., 2013; Zhang et al., 2015). These alterations in aged neutrophils are similar to those observed upon BCG vaccination. However, future studies are necessary to better understand the neutrophil phenotype upon BCG vaccination, to assess similarities and differences with previously described phenotypes, and to investigate if the life span/aging of neutrophils is affected by BCG-induced training.
Interestingly, large differences were found in the expression of programmed death-ligand 1 (PD-L1), which was significantly decreased 3 months after BCG vaccination. PD-L1 is a cell membrane protein that binds to programmed cell death protein 1 (PD-1) and plays a major role in suppressing immune responses. Expression of PD-L1 on cancer cells serves as an effective mechanism for tumors to escape from host immune responses, and the development of PD-1 and PD-L1 inhibitors proved a breakthrough in the treatment of cancer (Iwai et al., 2002). Neutrophils are one of the most abundant immune cells in the tumor microenvironment of solid tumors (Coffelt et al., 2016), and in patients with gastric cancer, the presence of neutrophils with an immunosuppressive phenotype is associated with disease progression and a decrease in patient survival (Wang et al., 2017). Based on our findings, modulation of neutrophil phenotype by the induction of trained immunity might be a strategy for cancer treatment (Netea et al., 2017), and future studies investigating the effect of trained neutrophils on T cell function are warranted.
Since BCG vaccination results in functional changes in monocytes, we explored whether BCG would also impact neutrophil antimicrobial function. Interestingly, BCG vaccination significantly increased the production of the chemokine IL-8 and the serine protease elastase upon ex vivo exposure to secondary pathogenic stimuli. Moreover, neutrophils demonstrated a significant increase in killing of C. albicans upon BCG vaccination, increased ROS production, and enhanced expression of degranulation markers. These changes in neutrophil effector function are in line with an earlier study showing increased ROS production and killing of C. albicans in BCG-trained macrophages in mice (van ’t Wout et al., 1992) and our findings of enhanced S. aureus phagocytosis and ROS production in BCG-vaccinated mice as compared to control mice. Whether an increase in phagocytosis also occurs in human neutrophils upon BCG vaccination needs to be assessed in future clinical trials. Together, these results suggest that the enhanced antimicrobial function of neutrophils might contribute to the observed nonspecific protective effects of BCG vaccination against unrelated subsequent infections. Promoting neutrophil effector function via the induction of trained immunity may also offer avenues to counteract the adverse effects of (chemotherapy-induced) neutropenia. On the other hand, suppression of neutrophil activity by inhibition of trained immunity mechanisms may be beneficial in patients suffering from diseases characterized by neutrophil-mediated inflammation.
As epigenetic reprogramming mediates the functional ability of monocytes to respond more strongly to secondary infections (Arts et al., 2018), the involvement of chromatin remodeling in neutrophils was assessed before and after BCG vaccination. Indeed, our findings indicate that the functional changes observed in neutrophils after vaccination are most likely mediated by epigenetic modifications at the promoter sites of genes essential for antimicrobial function, as indicated by increased levels of H3K4me3 upon BCG vaccination. However, the short duration of life of neutrophils in the circulation, corroborated with the identification of functional and epigenetic changes in neutrophils months after the vaccination, can only lead to the conclusion that these effects are mediated through epigenetic and functional changes in neutrophils precursors. This hypothesis is supported by recent experimental studies in mice (Christ et al., 2018; Mitroulis et al., 2018), as well as studies in humans in our laboratory (Cirovic et al., 2020). One important question is whether the BCG-induced effects can be solely attributed to chromatin remodeling or if persistent low-grade BCG infection also contributes to these effects. A previous study found that viable BCG was present at the vaccination site in only half of the individuals 1 month after BCG vaccination (Minassian et al., 2012). For this reason, we have chosen to study neutrophils 3 months after BCG vaccination, when it is likely that microorganisms are no longer present. Indeed, a separate study reported that all volunteers tested negative for M. bovis in the bone marrow by qPCR 3 months after BCG vaccination (Cirovic et al., 2020).
Previous work has demonstrated a strong link between epigenetic and metabolic changes in trained cells (Arts et al., 2016a). More specifically, a strong increase in glycolysis in BCG-trained monocytes has been observed, and this was found to be crucial for the induction of trained immunity. Neutrophils rely almost exclusively on glycolysis for their energy demands, as they have a very small number of mitochondria (Borregaard and Herlin, 1982). Interestingly, we observed an increase in glycolytic rate in ex-vivo-stimulated neutrophils upon BCG vaccination. Moreover, levels of lactate correlated with neutrophil killing capacity, supporting the importance of glycolysis for effective trained immunity responses in neutrophils. In line with these findings, we found significantly increased levels of H3K4me3 at promoters of genes encoding key regulators of glycolysis, such as phosphofructokinase and mTOR, and increased gene transcription of the rate-limiting glycolysis enzyme HK1.
This study also opens new questions for future studies. One important task for future research will be to decipher the exact molecular mechanisms that modulate HSPCs once individuals have received the BCG vaccine. In addition, the impact of trained neutrophils on other cell types needs further attention. Evidence accumulated in recent years has indicated an important role for neutrophils in the modulation of neighboring cells, including macrophages (Rosales, 2018). In this context, it would be particularly interesting to study whether an altered neutrophil phenotype upon BCG vaccination could induce long-term effects on immune responses displayed by other cell subpopulations, such as lymphocytes and macrophages. Intriguingly, it has been shown that neutrophils upon helminth infection are indeed able to train macrophages to acquire a long-lasting enhanced protective phenotype (Chen et al., 2014). Whether a similar effect exists upon BCG vaccination and if this represents a strategy for vaccine development remains to be explored. Additionally, RNA-seq studies and epigenomic analysis at the single-cell level are needed to investigate whether the trained immunity profile is present in only a subset of neutrophils or if it involves the entire neutrophil population. Interestingly, a new field of investigation may be opened by the intriguing findings of changes in regulation of immune activators in eosinophils. More studies are warranted to study and eventually validate this observation, as well as the implications of BCG vaccination in allergic diseases. Indeed, several epidemiological studies have suggested effects of BCG vaccination on the incidence and severity of allergic diseases, but results are conflicting (Aaby et al., 2000; Alm et al., 1997; Grüber et al., 2001).
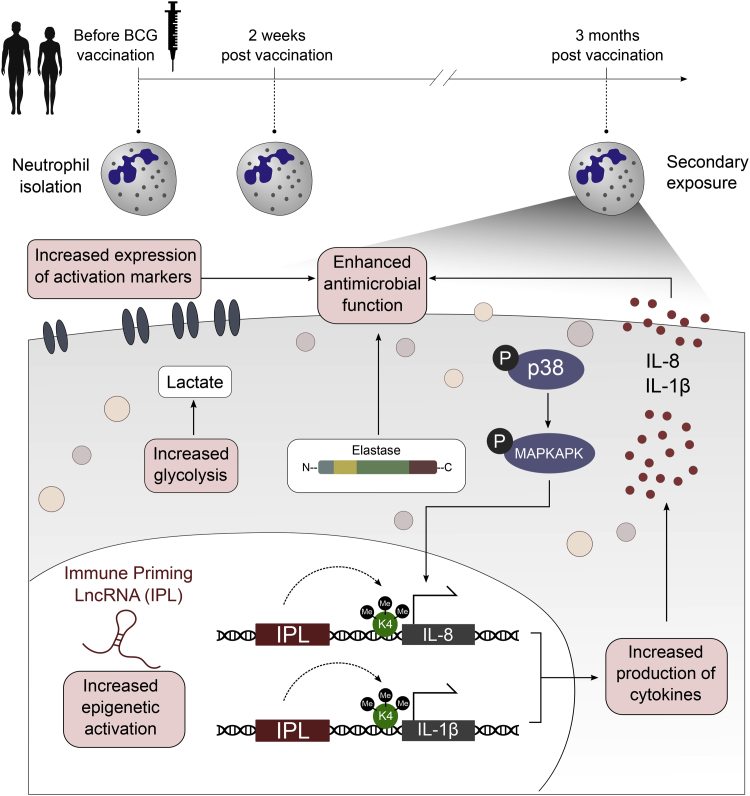
In conclusion, we show that neutrophils can be functionally reprogrammed to exhibit a propensity to respond more efficiently to microbial stimulation after vaccination with BCG. Circulating neutrophils not only show an increased expression in activation markers, but also display increased antimicrobial function upon ex vivo stimulation with various pathogens. An increase in transcription of lncRNAs and epigenetic changes at the level of histone methylation might be the mechanism through which BCG enhances innate immune responses in neutrophils (Figure 7). Considering the role of neutrophils in various disease states, our findings suggest the potential for the modulation of trained immunity as a therapeutic strategy to modulate neutrophil effector function.
Figure 7.
Schematic Overview of the Mechanisms That May Underlie Enhanced Neutrophil Effector Function upon BCG Vaccination
BCG-induced trained immunity results in an increased responsiveness of neutrophils, with effector functions such as cytokine production and killing capacity being increased upon secondary stimulation with non-related pathogens. Changes in the inflammatory profile of neutrophils upon BCG are associated with changes in epigenetic profile and cellular metabolism. Increased expression of lncRNAs, termed IPLs, might lead to elevated H3K4me3 accumulation at genes coding for antimicrobial molecules. Together with the upregulation of signaling pathways and glycolysis, epigenetic changes at the level of histone methylation result in enhanced transcription of antimicrobial proteins upon exposure to a secondary stimulus.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-H3K4me3 | Diagenode | pab-003-050; RRID: AB_2616052 |
| anti-CD16 FITC (clone 3G8) | Beckman Coulter | Cat#B49215; RRID: AB_2848116 |
| anti-CD10 PE (HI10A) | BioLegend | Cat#312203; RRID: AB_314914 |
| anti-CD11b PE-Dazzle (ICRF44) | BioLegend | Cat#301347; RRID: AB_2564080 |
| anti-CD62L PE-Cy7 (clone DREG-56) | BioLegend | Cat#304821; RRID: AB_830800 |
| anti-PD-L1 APC (clone MIH1) | ThermoFisher | Cat#17598342; RRID: AB_10597586 |
| anti-CD66b AF700 (G10F5) | BioLegend | Cat#305113; RRID: AB_2566037 |
| anti-CD15 Brilliant Violet 421 (clone W6D3) | BioLegend | Cat#323039; RRID: AB_2566519 |
| anti-CD45 Krome Orange (clone J33) | Beckman Coulter | Cat#A96416; RRID: AB_2833027 |
| biotin anti-mouse Ly6g (clone 1A8) | BioLegend | Cat#127603; RRID: AB_1036096 |
| anti-CD11b (clone M1/70) | BioLegend | Cat#101207; RRID: AB_10702027 |
| anti-myeloperoxidase (clone CLB-MPO-1) | Beckman Coulter | Cat#M3455U; RRID: AB_131041 |
| anti-CD14 PE-Cy5.5 (clone M5E2) | BioLegend | Cat#301847; RRID: AB_2564058 |
| Bacterial and Virus Strains | ||
| Heat-killed Mycobacterium tuberculosis | Gift | H37Rv |
| Candida albicans | Gift | ATCCMYA-3573, UC820 |
| Staphylococcus aureus (Wood strain without protein A) BioParticles | Thermo Scientific | Cat#S23371 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Bacille Calmette-Guérin Vaccine | Intervax | Bulgaria strain |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat#16561-29-8 |
| Lipopolysaccharide | Sigma-Aldrich | From E. coli serotype 055:B5, L2880 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#P8465 |
| BSA | Wisent | Cat#800-195-EG |
| 5′-Deoxy-5′-(methylthio) adenosine (MTA) | Sigma-Aldrich | Cat#D5011 |
| Percoll | Sigma-Aldrich | Cat#P1644 |
| Ficoll-Paque | GE Healthcare | Cat#17-1440-03 |
| Roswell Park Memorial Institute medium (RPMI) | Invitrogen | Cat#22406031 |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | Cat#A7030 |
| iScript reverse transcriptase | Bio-Rad | Cat#1708840 |
| TRIzol reagent | Life Technologies | Cat#15596018 |
| SYBR Green | Applied Biosciences | Cat#4368708 |
| 16% Formaldehyde | Fisher Scientific | Cat#28908 |
| A23187 | Sigma Aldrich | Cat#C7522 |
| MNase | Boehringer Mannheim | Cat#10107921001 |
| R848 | Invivogen | Cat#tlrl-r848 |
| CpG | Invivogen | Cat#tlrl-2336 |
| Smart Tube stabilizer | Fisher Scientific | Cat#501351692 |
| Saponin | Sigma-Aldrich | Cat# 8047-15-2 |
| Fixation/Permeabilization Concentrate | Thermo Fisher | Cat#00-5123-43 |
| Anti-biotin microbeads | Miltenyi Biotec | Cat#130-090-485 |
| Critical Commercial Assays | ||
| Human IL-1β ELISA | R&D systems | Cat#DY201 |
| Human TNFα ELISA | R&D systems | Cat#DY210 |
| Human IL-6 ELISA | Sanquin | Cat#M1916 |
| Human IL-8 ELISA | Sanquin | Cat#M1918 |
| iScript cDNA Synthesis Kit | Bio-Rad | Cat#1708891 |
| MinElute PCR purification Kit | QIAGEN | Cat#28006 |
| DNase I | QIAGEN | Cat#79254 |
| Rneasy Mini Kit | QIAGEN | Cat#74106 |
| Lactate Fluorometric Assay Kit | Biovision | Cat#K607 |
| Human Neutrophil Elastase DuoSet ELISA Kit | R&D | Cat# DY9167-05 |
| KAPA library preparation kit | Kapa Biosystems | KK8400 |
| riboZero gold rRNA removal kit | Illumina | MRZG12324 |
| Nextera DNA Library Prep Kit | Illumina | FC-121-1031 |
| TruSeq SBS KIT v3 - HS (50 cycles) | Illumina | FC-401-3002 |
| NextSeq 500/550 High Output v2 kit (75 cycles) | Illumina | FC-404-2005 |
| NEBNext High-Fidelity 2 × PCR Master Mix | New England Biolabs | Cat#M0541 |
| iQ SYBR® Green Supermix | Bio-Rad | Cat#1708880 |
| 100 × SYBR Green I Nucleic Acid Gel Stain | Thermo Fisher Scientific | Cat#S7563 |
| SPRIselect reagent kit | Beckman Coulter | Cat#B23218 |
| E-Gel® SizeSelect Agarose Gels, 2% | Thermo Fisher Scientific | Cat#G661002 |
| dNTP set 100 mM | Life Technologies | Cat#10297-018 |
| dUTP 100 mM | Promega | Cat#U119A |
| Glycogen (20 mg/ml) | Life Technologies | Cat#10814-010 |
| Random Hexamer primers | Sigma-Aldrich | Cat#11034731001 |
| Second Strand Buffer | Life Technologies | Cat#10812-014 |
| Superscript III Reverse Transcriptase | Life Technologies | Cat#18080-044 |
| DNA polymerase I, E. coli | New England Biolabs | Cat#M0209S |
| USER enzyme | New England Biolabs | Cat#M5505L |
| E.Coli Ligase | New England Biolabs | Cat#M0205L |
| Rnasin Plus Rnase Inhibitor | Promega | Cat#N2615 |
| Ribonuclease H | Life Technologies | Cat#AM2293 |
| T4 DNA polymerase | New England Biolabs | Cat#M0203L |
| Sodium Acetate (3M) | Life Technologies | Cat#AM9740 |
| Qubit RNA HS assay kit | Life Technologies | Cat#Q32852 |
| Ribozero Gold Kit | Illumina | Cat#MRZG12324 |
| Bradford Concentration Assay | Thermo Fisher | Cat#23246 |
| Cell Rox Green flow cytometry assay kit | Thermo Fisher | Cat#C10492 |
| Deposited Data | ||
| RNA-seq and ChIP-seq data of neutrophils before and three months after BCG vaccination | This paper | GEO: GSE153729 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 mice | Jackson Laboratories | NA |
| Oligonucleotides | ||
| Primers for qPCR, see Table S1 | This paper | NA |
| Software and Algorithms | ||
| CyTOF software | Fluidigm | https://www.fluidigm.com |
| FACSDiva Software | BD Biosciences | SCR_001456 |
| FlowJo software v.10 | Tree Star | SCR_000410 |
| GraphPad Prism v. 5.03 | Graphpad Software | https://www.graphpad.com |
| MMSEQ | (Turro et al., 2011) | https://github.com/eturro/mmseq |
| DEseq | (Anders and Huber, 2010) | http://bioconductor.org/packages/release/bioc/html/DESeq.html |
| Bwa | (Li and Durbin, 2009) | http://bio-bwa.sourceforge.net/ |
| Samtools and Bamtools | (Barnett et al., 2011; Li et al., 2009) | http://code.google.com/p/phantompeakqual-tools/http://samtools.sourceforge.net/https://github.com/pezmaster31/bamtools |
| MACS2 | (Zhang et al., 2008) | https://github.com/taoliu/MACS/ |
| HOMER | (Heinz et al., 2010) | http://homer.ucsd.edu/homer/motif/ |
| DAVID | (Huang et al., 2009) | https://david.ncifcrf.gov/ |
| Deeptools | (Ramírez et al., 2014) | https://deeptools.readthedocs.io/en/develop/ |
Resource availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mihai G. Netea at the Radboud University Medical Center, Nijmegen, the Netherlands (mihai.netea@radboudumc.nl).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The RNA and ChIP sequencing data of neutrophils before and 3 months after BCG vaccination as reported in this paper can been found in the NCBI Gene Expression Omnibus under accession number GEO: GSE153729.
Experimental model and Subject Details
A total of 25 healthy individuals of Western-European descent (10 males and 15 females, age range 20-70 years) were included between February 2018 and June 2018 at the Radboud University Medical Center, the Netherlands. All volunteers were BCG naive and had not lived in a tuberculosis endemic country. Exclusion criteria were chronic or acute disease at the time of sampling, pregnancy or breastfeeding, any vaccination other than BCG within three months before start of the study, and use of medication other than paracetamol and oral anti-contraceptive drugs in the last month before samples of venous blood were drawn. Throughout the study period, subjects were not allowed to take any drugs, except paracetamol and oral ant-contraceptive drugs. All volunteers received BCG (InterVax, Canada, strain Bulgaria). Blood was drawn before BCG vaccination and 2 weeks and 3 months after vaccination. 2 Volunteers did not show up at their 3-months appointment, therefore the time point 3 months after vaccination contains data of 23 volunteers. All individuals were taking part in the 300BCG study, which was approved by the Ethical Committee of Radboud University Medical Center (no. NL58553.091.16). Experiments were conducted according to the principles expressed in the Declaration of Helsinki. All individuals gave written informed consent to donate venous blood for research.
All blood collections were performed in the morning and isolation started within two hours after blood draw. There were no significant differences in time between blood collection and neutrophil isolation or start of functional assays at the different time points.
Method Details
Stimuli
The Candida albicans strain ATCC MYA-3573, UC820 was grown for 24 hours at 37°C in liquid Sabouraud medium. Growth was monitored during the entire growth period. After the growth period, C. albicans was washed three times in cold PBS and counted before stimulation experiments. For some experiments, C. albicans was heat killed by incubation for 45 minutes at 98°C, followed by resuspension in RPMI 1640 to an inoculum size corresponding with 1 × 106 / mL yeast cells. Cultures of H37Rv M. tuberculosis were grown to mid-log phase in Middlebrook 7H9 liquid medium (Difco, Becton Dickinson, East-Rutherford) supplemented with oleic acid/albumin/dextrose/catalase (OADC) (BBL, Becton Dickinson), washed three times in sterile saline solution, heat-killed and then disrupted using a bead beater. Concentration was measured using a bicinchoninic acid (BCA) assay (Pierce, Thermo Scientific, Rockville). Escherichia coli lipopolysaccharide (LPS; serotype 055:B5; Sigma) was further purified as described previously (Hirschfeld et al., 2000).
Peripheral blood mononuclear cells and neutrophil isolation
Venous blood of human healthy volunteers was collected into 10 mL EDTA tubes (Monoject). Peripheral blood mononuclear cell (PBMC) isolation was performed by density centrifugation of blood diluted 1:1 in pyrogen-free PBS over Ficoll-Paque (GE healthcare, UK). Upon density gradient centrifugation and removal of the PBMC fraction, PMNs were isolated by hypotonic lysis. In order to remove erythrocytes, the cells were treated twice with hypotonic lysis buffer (155 mM NH4Cl, 10 mM KHCO3) during incubation on ice for 15 minutes. Next, PMNs were washed twice in PBS and resuspended in RPMI culture medium (Roswell Park Memorial Institute medium, Invitrogen, CA, USA) supplemented with 50 μg/mL gentamicin (Centrafarm), 2 mM glutamax (GIBCO), and 1 mM pyruvate (GIBCO). The PMN fraction consisted of > 95% neutrophils (Figure S4). Viability of the cells was measured by annexin-V and propidium iodide (PI) staining and showed 11.04 ± 4.2% annexin-V+PI- cells and 6.75 ± 1.99% annexin-V+ PI+ cells (Figure S4).
Neutrophil stimulation assays
5 × 105 neutrophils in a 100 μL volume were added to flat-bottom 96-well plates (Corning, NY, USA). Cells were incubated either with culture medium only as a negative control, 1 μg/mL sonicated M. tuberculosis H37Rv, 10 ng/mL LPS, 500 ng/mL PMA or 5 × 105 CFU heat-killed C. albicans for 17 hr at 37°C and 5% CO2 (all in the presence of 10% pooled human serum). After incubation, supernatants were collected and stored at −20°C until assayed. Viability of the cells after 17 hr incubation was measured by annexin-V and propidium iodide (PI) staining and showed 83.2 ± 1.7% of annexin-V+ PI- cells and 4.2 ± 0.6% annexin-V+ PI+ cells (Figure S4).
Flow Cytometry
Total leukocytes were obtained after erythrocyte lysis in isotonic NH4CL buffer and washed twice with PBS. White blood cell counts were determined by a cell counter (Coulter Ac-T Diff® cell counter; Beckman Coulter, Fullerton, CA, USA) and used to calculate the absolute numbers of CD45+ leukocytes identified by flow cytometry as described in detail previously (Aguirre-Gamboa et al., 2016).
Briefly, half a million of total leukocytes were used to analyze surface markers with the NaviosTM flow cytometer (Beckman Coulter, Fullerton, CA, USA). The cells were transferred to a V-bottom plate and washed twice with PBS + 0.2% bovine serum albumin (BSA; Sigma-Aldrich, St. Louis, USA), stained for 20 min at room temperature in the dark with the fluorochrome-conjugated monoclonal antibodies (mAbs) of interest and washed twice with PBS + 0.2% BSA. For surface staining of granulocytes in total leukocytes, the following conjugated mAbs specific for human cells were used: anti-CD16 FITC (Beckman Coulter; Suarlée, Belgium, clone 3G8), anti-CD10 PE (BioLegend, San Diego, CA, USA HI10A), anti-CD11b PE-Dazzle (BioLegend, San Diego, CA, USA ICRF44), anti-CD14 PE-Cy5.5 (BioLegend, San Diego, CA, USA, clone M5E2), anti-CD62L PE-Cy7 (BioLegend, San Diego, CA, USA, clone DREG-56), anti-PD-L1 APC (ThermoFisher, Waltham, MA USA, clone MIH1), anti-CD66b AF700 (BioLegend, San Diego, CA, USA, clone G10F5), anti-CD15 Brilliant Violet 421 (BioLegend, San Diego, CA, USA, clone W6D3) and anti-CD45 Krome Orange (Beckman Coulter, Suarlée, Belgium, clone J33).
After stimulation of isolated neutrophils for 17 hours, cells were stained for surface markers as described above, permeabilized and fixed according to manufacturer’s instructions (Thermo Fisher), and followed by intracellular staining with anti-myeloperoxidase (Beckman Coulter, Suarlée, Belgium, clone CLB-MPO-1).
viSNE algorithm settings
Manually gated singlets, viable granulocytes (as shown in Figure S1) were imported into Cytobank (Kotecha et al., 2010) and subjected to viSNE analysis, 9 of 15 channels were used to perform the analysis: CD16, CD10, CD11b, CD14, CD62L, PD-L1, CD66b, CD15 and CD45. Equal event sampling was done using 52268 events per individual.
viSNE analysis quality check
To check for batch effects due to slight alterations in staining reagents or measurement parameters, we organized the data files by date of measurement and visually look for similar patterns in the 2D maps. Once we excluded the data (15 of the 75 original files) in which batch effect was apparent, we concatenated the files obtained after the viSNE run, to have one file per time-point measured, and displayed a global viSNE 2D map as the one in Figure 1D.
CITRUS algorithm settings
Manually gated singlets, viable granulocytes (as shown in Figure S1) were imported into Cytobank and subjected to CITRUS analysis. We ran this analysis using the following settings: granulocytes were chosen as input population, 9 of 15 channels were selected (same as for viSNE analysis). The files were assigned to the appropriate experimental group (before, two weeks and three months after BCG vaccination). We chose the Nearest Shrunken Centroid (PAMR) as association model, abundance was chosen as cluster of characterization, and equal event sampling (545 of a total of 29785) with a minimum cluster size of 1% were applied. We used the default Cytobank values for Cross Validation Folds and False Discovery Rate (1%).
CITRUS analysis quality check
To assess the impact of the stochastic nature of CITRUS in choosing features to build the model, we ran CITRUS seven times and checked that the Model Error Rate, the number of features selected and the characteristics of these features (abundance and marker expression) were consistent throughout the runs. Here we show the hierarchical plots and significant clusters for one of the seven analyses (Figure 1; Data S1).
ROS assay
The production of ROS was determined by a luminol (5-amino-2,3, dihydro-1,4-phtalazinedione)-enhanced luminescence assay. 2 × 105 neutrophils were added to white 96-well plates (Corning) in a volume of 200 μL. Next, neutrophils were left either unstimulated or exposed to serum-opsonized 2 × 105 C. albicans (UC820, heat-killed). Chemiluminescence was measured at 37°C for every 142 s during 1 hour in a BioTek Synergy HTreader. The integral of relative luminescence units per second (RLU/sec) was measured. All measurements were performed in quadruplicate. Opsonized C. albicans was prepared by incubation of C. albicans in human pooled serum for 1 hour at 37°C. The fold change was calculated by dividing the level of ROS production after vaccination by the amount of ROS production before vaccination.
Killing of C. albicans by neutrophils
Freshly isolated neutrophils (5 × 105) were incubated with C. albicans (1 × 106 CFU) in the presence of 10% pooled human serum in 96-well plates in a final volume of 200 μL. After 17 hr at 37°C and 5% CO2, the cells were washed five times with water and plated in serial dilution on Sabouroud agar plates in triplicate. CFUs were counted after 24 hr at 37°C. The average number of colonies of three plates was calculated.
NETosis assay
Neutrophils were seeded in 96-well plates (0.1 million cells in 50 μl per well) and allowed to adhere to the plates for 30 minutes at 37°C. Next, cells were stimulated with 10 μg/mL LPS (Sigma Aldrich), 5 nM PMA (Sigma Aldrich), 4 μM A23187 (Sigma Aldrich) or multiplicity of infection (MOI) 5 heat-killed C. albicans. Neutrophils were incubated for three hours at 37°C to form NETs. The supernatant was then discarded and replaced by 100 μl U/mL MNase (Boehringer Mannheim) in medium. After incubation of ten minutes at 37°C, the supernatant was carefully collected as NET harvest. The DNA content of the harvest was quantified by adding 5 mM Sytox green and measure the fluorescence on a Clariostar device at 480/520 nm excitation/emission. In each experiment we included seven technical duplicates, the mean of these duplicates was used for further calculations. All data was first normalized to mock treated cells before calculation of fold changes between the different visits. Wilcoxon Signed Rank Test was used to determine statistic differences.
Cytokine, lactate and elastase measurements
Cytokine production was assessed in supernatants using commercial ELISA kits for human IL-1β, IL-8 and TNF-α (R&D systems, MN, USA), in accordance with the manufacturer’s instructions. Levels of lactate were determined using a Lactate Fluorometric Assay Kit (Biovision, CA, USA). Levels of elastase were assessed using a Human Neutrophil Elastase DuoSet ELISA Kit (R&D systems, MN, USA), according to manufacturer’s instructions.
Mass cytometry phospho-signaling analysis
Fresh blood aliquots (90 μl per stimulation) from 11 donors were incubated with the indicated TLR agonist (LPS, 100 ng/ml (Escherichia coli, serotype 055:B5, Sigma); R848, 1μg/ml (Invivogen); CpG 10ug/ml (Invivogen)) or left unstimulated as a control for 15 minutes at 37°C. Next, blood aliquots were fixed with Smart Tube stabilizer (Fisher Scientific) and frozen according to the manufacturer’s protocol. In preparation for mass cytometry cell labeling, all frozen samples from an individual donor were thawed in parallel with the cells re-suspended in complete RPMI medium (GIBCO, Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Institut de Biotechnologies Jacques Boy), and then washed in CSM-S buffer (PBS supplemented with 5 mg/mL BSA and 0.3% saponin (Sigma-Aldrich)). Barcoding of donor samples by visit was performed by incubating cells with one of six different metal-conjugated anti-CD45 antibodies for 30 min at room temperature (RT). Cells were washed in PBS and 0.3% saponin (Sigma-Aldrich), then labeled with one of five different palladium isotopes chelated to isothiocyano-benzyl-EDTA (final concentration of 200 nM) (Zunder et al., 2015). Following 30 minutes incubation at 4°C, cells were re-suspended in PBS supplemented with 10% FBS, washed with CSM-S then all 30 samples from an individual donor where pooled. Next, cells were washed and incubated for 30 min at RT with a 100 μL cocktail of metal-conjugated antibodies. Cells were washed and fixed for 10 min at RT with 2.4% paraformaldehyde (Sigma-Aldrich). Total cells were identified by DNA intercalation (1 μM Cell-ID intercalator; Fluidigm/DVS Science) in 2% PFA at 4°C overnight. Labeled samples were assessed by using a Helios mass cytometer instrument (Fluidigm), using a flow rate of 0.030 ml/min and an event rate of ~300 cells/second. Flow cytometry standard (FCS) files were normalized to EQ Four Element calibration beads using CyTOF software. FCS files were imported into Cytobank data analysis software for conventional cytometric analysis of immune cell populations. Phosphosignaling activity was then simultaneously quantified per cell for the following functional markers: p38, pMAPKAPK2, pNFKb and pCREB. Endogenous phosphosignaling immune features were derived from the analysis of unstimulated blood. Stimulation response features were calculated as the difference in values between stimulated and unstimulated conditions. Co-expression features were derived from the proportion of cells expressing both functional markers. For both endogenous and stimulation response features, the arcsinh transform of the mean values of the neutrophil population was used.
Mouse experiments and analysis
C57BL/6 mice were purchased from Jackson Laboratories and were maintained in the animal facility of BRFAA. Mice were housed under specific pathogen-free conditions on a standard 12/12h light/dark cycle and were used at the age of 8-10 weeks. Procedures were approved by the Institutional Committee of Protocol Evaluation in conjunction with the related veterinary authority of the Region of Attika, Greece. BCG (or PBS as control) was administered to the mice by retro-orbital injection. Each mouse received a single dose of around 1x10ˆ6 CFU. 7 days after injections, single-cell suspensions from spleens were generated by passing them through a 40-μm cell strainer. To determine intracellular levels of ROS by flow cytometry, isolated splenic single cell suspensions were stained with antibodies against neutrophil surface markers (anti-CD11b, clone M1/70 and anti-Ly6g, clone 1A8) and were then stained with Cell Rox Green flow cytometry assay kit (Thermo Fisher).
To isolate neutrophils, splenocytes were incubated with a biotin anti-mouse Ly6g antibody (clone 1A8; BioLegend) followed by anti-biotin microbeads (Miltenyi Biotec). Neutrophils were then positively selected on a magnetic field according to the manufacturer’s instructions (MACS separation columns, Miltenyi Biotec). For the phagocytosis assay, isolated neutrophils were incubated with fluorescein conjugated Staphylococcus aureus (Wood strain without protein A) BioParticles (Thermo Scientific) in a ratio of 1/30 (neutrophil/particles) for 30 minutes at 37°C. In order to measure only the particles that were phagocytosed and discriminate from the ones that were not, trypan blue was used as a quenching agent. The particles that were phagocytosed were quantified by mean fluorescent intensity of FITC in CD11b+ Ly6g+ cells.
RNA isolation and Chromatin immunoprecipitation (ChIP)
Neutrophils isolated from volunteers were lysed in Trizol reagent and stored at −20°C. RNA was extracted from cells using the QIAGEN RNeasy RNA extraction kit (QIAGEN, Netherlands), using on-column DNaseI treatment. For ChIP experiments, purified neutrophils were isolated from human volunteers before and after vaccination as described above and fixed in methanol-free 1% formaldehyde. Cells were sonicated using a Diagenode Bioruptor Pica sonicator and immunoprecipitation was performed using antibodies against H3K4me3 (Diagenode, Seraing, Belgium). DNA was purified using QIAGEN MinElute PCR purification Kit and processed further for qPCR analysis using the SYBR green method or ChIP-sequencing as described previously (Bekkering et al., 2018). For qPCR analysis, samples were analyzed by a comparative Ct method according to the manufacturer’s instructions. Myoglobulin was used as a negative control and H2B as a positive control in experiments assessing H3K4me3. Primer sequences can be found in Table S1.
Upstream RNA-seq and ChIP-seq processing
ChIP-sequencing reads were aligned to human genome assembly hg38 (NCBI version 38) using bwa. BAM files were first filtered to remove the reads with mapping quality less than 15, followed by fragment size modeling. MACS2 (https://github.com/macs3-project/MACS/) was used to call the peaks. Data (H3K4me3 reads/peak) were normalized using the R package DESeq and then pairwise comparisons were performed. To infer gene expression levels, RNA-seq reads were aligned to hg19 human transcriptome using Bowtie. Quantification of gene expression was performed using MMSEQ.
Quantification and Statistical Analysis
Statistics human experiments
Statistical parameters including the exact value of n, the definition of center, dispersion and precision measures (mean ± SEM) and statistical significance are reported in the Figures and Figure Legends. Results were analyzed using Wilcoxon signed-rank test where applicable. Calculations were performed in Graphpad prism 5 (CA, USA) and in figures, asterisks indicate statistical significance (∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001). Data are shown as mean ± SEM.
Statistics ChIP-seq and RNA-seq
Dynamic genes and H3K4me3 peaks were identified using DEseq with change in signal determined as fold change > 1.5 and padj < 0.05, with a mean RPKM > 1 for gene expression or mean reads/peak > 20 for histone modifications (Novakovic et al., 2016). Genes of interest were downloaded from Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000). H3K4me3 peaks were assigned to the nearest transcription start site (TSS) within 1Mb using the GREAT tool (McLean et al., 2010).
Acknowledgments
The authors thank all volunteers for their participation in this study. In addition, we would like to thank Helga Dijkstra, Heidi Lemmers, and Trees Jansen for their help. M.G.N. was supported by an ERC Advanced Grant (833247) and a Spinoza grant from the Netherlands Organisation for Scientific Research (NWO). Y.A.R.-R. was supported by a CONACyT scholarship for graduate studies abroad (410825) from the Mexican Council for Science and Technology (CONACyT). C.M.d.B. and G.J.M.P. were supported in part by the Netherlands Organisation for Scientific Research (NWO) (05188). B.N. was supported by an NHMRC (Australia) New Investigator grant (1157556). T.C. was supported by an ERC grant (DEMETINL). S.F., Y.N., and M.M.M. received grant support from the Bill and Melinda Gates Foundation (BMGF).
Author Contributions
Conceptualization, S.J.C.F.M.M. and M.G.N.; Methodology, S.J.C.F.M.M., Y.A.R.-R., K.T., L.K., J.G., P.V., B.N., C.M.d.B., E.T.F., S.F., Y.N., C.F., V.P.M., V.A.C.M.K., and L.C.J.d.B.; Writing – Original Draft, S.J.C.F.M.M.; Writing – Review & Editing, Y.A.R.-R., J.G., M.G.N., I.J., H.K., R.v.C., G.J.M.P., and T.C.; Supervision, M.G.N., I.J., H.K., L.A.B.J., G.J.M.P., T.C., M.M.M., and D.A.D.
Declaration of Interests
The authors declare no competing interests.
Published: November 17, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108387.
Supplemental Information
References
- Aaby P., Shaheen S.O., Heyes C.B., Goudiaby A., Hall A.J., Shiell A.W., Jensen H., Marchant A. Early BCG vaccination and reduction in atopy in Guinea-Bissau. Clin. Exp. Allergy. 2000;30:644–650. doi: 10.1046/j.1365-2222.2000.00803.x. [DOI] [PubMed] [Google Scholar]
- Aaby P., Roth A., Ravn H., Napirna B.M., Rodrigues A., Lisse I.M., Stensballe L., Diness B.R., Lausch K.R., Lund N. Randomized trial of BCG vaccination at birth to low-birth-weight children: beneficial nonspecific effects in the neonatal period? J. Infect. Dis. 2011;204:245–252. doi: 10.1093/infdis/jir240. [DOI] [PubMed] [Google Scholar]
- Adrover J.M., Del Fresno C., Crainiciuc G., Cuartero M.I., Casanova-Acebes M., Weiss L.A., Huerga-Encabo H., Silvestre-Roig C., Rossaint J., Cossío I. A Neutrophil Timer Coordinates Immune Defense and Vascular Protection. Immunity. 2019;50:390–402.e10. doi: 10.1016/j.immuni.2019.01.002. [DOI] [PubMed] [Google Scholar]
- Aguirre-Gamboa R., Joosten I., Urbano P.C.M., van der Molen R.G., van Rijssen E., van Cranenbroek B., Oosting M., Smeekens S., Jaeger M., Zorro M. Differential Effects of Environmental and Genetic Factors on T and B Cell Immune Traits. Cell Rep. 2016;17:2474–2487. doi: 10.1016/j.celrep.2016.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi K., Traver D., Miyamoto T., Weissman I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Alm J.S., Lilja G., Pershagen G., Scheynius A. Early BCG vaccination and development of atopy. Lancet. 1997;350:400–403. doi: 10.1016/S0140-6736(97)02207-1. [DOI] [PubMed] [Google Scholar]
- Amir A.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., Pe’er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur J.S., Ley S.C. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- Arts R.J., Novakovic B., Ter Horst R., Carvalho A., Bekkering S., Lachmandas E., Rodrigues F., Silvestre R., Cheng S.C., Wang S.Y. Glutaminolysis and Fumarate Accumulation Integrate Immunometabolic and Epigenetic Programs in Trained Immunity. Cell Metab. 2016;24:807–819. doi: 10.1016/j.cmet.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J.W., Carvalho A., La Rocca C., Palma C., Rodrigues F., Silvestre R., Kleinnijenhuis J., Lachmandas E., Gonçalves L.G., Belinha A. Immunometabolic Pathways in BCG-Induced Trained Immunity. Cell Rep. 2016;17:2562–2571. doi: 10.1016/j.celrep.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J.W., Moorlag S.J.C.F.M., Novakovic B., Li Y., Wang S.Y., Oosting M., Kumar V., Xavier R.J., Wijmenga C., Joosten L.A.B. BCG Vaccination Protects against Experimental Viral Infection in Humans through the Induction of Cytokines Associated with Trained Immunity. Cell Host Microbe. 2018;23:89–100.e5. doi: 10.1016/j.chom.2017.12.010. [DOI] [PubMed] [Google Scholar]
- Barnett D.W., Garrison E.K., Quinlan A.R., Strömberg M.P., Marth G.T. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkering S., Arts R.J.W., Novakovic B., Kourtzelis I., van der Heijden C., Li Y., Popa C.D., Ter Horst R., van Tuijl J., Netea-Maier R.T. Metabolic Induction of Trained Immunity through the Mevalonate Pathway. Cell. 2018;172:135–146. doi: 10.1016/j.cell.2017.11.025. [DOI] [PubMed] [Google Scholar]
- Bekkering S., Blok B.A., Joosten L.A., Riksen N.P., van Crevel R., Netea M.G. In Vitro Experimental Model of Trained Innate Immunity in Human Primary Monocytes. Clin. Vaccine Immunol. 2016;23:926–933. doi: 10.1128/CVI.00349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biering-Sørensen S., Aaby P., Napirna B.M., Roth A., Ravn H., Rodrigues A., Whittle H., Benn C.S. Small randomized trial among low-birth-weight children receiving bacillus Calmette-Guérin vaccination at first health center contact. Pediatr. Infect. Dis. J. 2012;31:306–308. doi: 10.1097/INF.0b013e3182458289. [DOI] [PubMed] [Google Scholar]
- Borregaard N., Herlin T. Energy metabolism of human neutrophils during phagocytosis. J. Clin. Invest. 1982;70:550–557. doi: 10.1172/JCI110647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borregaard N., Sørensen O.E., Theilgaard-Mönch K. Neutrophil granules: a library of innate immunity proteins. Trends Immunol. 2007;28:340–345. doi: 10.1016/j.it.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Bruggner R.V., Bodenmiller B., Dill D.L., Tibshirani R.J., Nolan G.P. Automated identification of stratifying signatures in cellular subpopulations. Proc. Natl. Acad. Sci. USA. 2014;111:E2770–E2777. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova-Acebes M., Pitaval C., Weiss L.A., Nombela-Arrieta C., Chèvre R., A-González N., Kunisaki Y., Zhang D., van Rooijen N., Silberstein L.E. Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell. 2013;153:1025–1035. doi: 10.1016/j.cell.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavakis T., Mitroulis I., Hajishengallis G. Hematopoietic progenitor cells as integrative hubs for adaptation to and fine-tuning of inflammation. Nat. Immunol. 2019;20:802–811. doi: 10.1038/s41590-019-0402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Wu W., Millman A., Craft J.F., Chen E., Patel N., Boucher J.L., Urban J.F., Jr., Kim C.C., Gause W.C. Neutrophils prime a long-lived effector macrophage phenotype that mediates accelerated helminth expulsion. Nat. Immunol. 2014;15:938–946. doi: 10.1038/ni.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell. 2016;167:1398–1414.e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H., Rao N.A., Aghajanirefah A. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ A., Günther P., Lauterbach M.A.R., Duewell P., Biswas D., Pelka K., Scholz C.J., Oosting M., Haendler K., Baßler K. Western Diet Triggers NLRP3-Dependent Innate Immune Reprogramming. Cell. 2018;172:162–175.e14. doi: 10.1016/j.cell.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirovic B., de Bree L.C.J., Groh L., Blok B.A., Chan J., van der Velden W.J.F.M., Bremmers M.E.J., van Crevel R., Händler K., Picelli S. BCG Vaccination in Humans Elicits Trained Immunity via the Hematopoietic Progenitor Compartment. Cell Host Microbe. 2020;28:322–334.e5. doi: 10.1016/j.chom.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffelt S.B., Wellenstein M.D., de Visser K.E. Neutrophils in cancer: neutral no more. Nat. Rev. Cancer. 2016;16:431–446. doi: 10.1038/nrc.2016.52. [DOI] [PubMed] [Google Scholar]
- Dos Santos J.C., Barroso de Figueiredo A.M., Teodoro Silva M.V., Cirovic B., de Bree L.C.J., Damen M.S.M.A., Moorlag S.J.C.F.M., Gomes R.S., Helsen M.M., Oosting M. β-Glucan-Induced Trained Immunity Protects against Leishmania braziliensis Infection: a Crucial Role for IL-32. Cell Rep. 2019;28:2659–2672.e6. doi: 10.1016/j.celrep.2019.08.004. [DOI] [PubMed] [Google Scholar]
- Ecker S., Chen L., Pancaldi V., Bagger F.O., Fernández J.M., Carrillo de Santa Pau E., Juan D., Mann A.L., Watt S., Casale F.P., BLUEPRINT Consortium Genome-wide analysis of differential transcriptional and epigenetic variability across human immune cell types. Genome Biol. 2017;18:18. doi: 10.1186/s13059-017-1156-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanucchi S., Fok E.T., Dalla E., Shibayama Y., Börner K., Chang E.Y., Stoychev S., Imakaev M., Grimm D., Wang K.C. Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments. Nat. Genet. 2019;51:138–150. doi: 10.1038/s41588-018-0298-2. [DOI] [PubMed] [Google Scholar]
- Fulurija A., Ashman R.B., Papadimitriou J.M. Neutrophil depletion increases susceptibility to systemic and vaginal candidiasis in mice, and reveals differences between brain and kidney in mechanisms of host resistance. Microbiology. 1996;142:3487–3496. doi: 10.1099/13500872-142-12-3487. [DOI] [PubMed] [Google Scholar]
- Furze R.C., Rankin S.M. Neutrophil mobilization and clearance in the bone marrow. Immunology. 2008;125:281–288. doi: 10.1111/j.1365-2567.2008.02950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüber C., Kulig M., Bergmann R., Guggenmoos-Holzmann I., Wahn U., MAS-90 Study Group Delayed hypersensitivity to tuberculin, total immunoglobulin E, specific sensitization, and atopic manifestation in longitudinally followed early Bacille Calmette-Guérin-vaccinated and nonvaccinated children. Pediatrics. 2001;107:E36. doi: 10.1542/peds.107.3.e36. [DOI] [PubMed] [Google Scholar]
- Hayashi F., Means T.K., Luster A.D. Toll-like receptors stimulate human neutrophil function. Blood. 2003;102:2660–2669. doi: 10.1182/blood-2003-04-1078. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld M., Ma Y., Weis J.H., Vogel S.N., Weis J.J. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J. Immunol. 2000;165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Iwai Y., Ishida M., Tanaka Y., Okazaki T., Honjo T., Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA. 2002;99:12293–12297. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann E., Sanz J., Dunn J.L., Khan N., Mendonça L.E., Pacis A., Tzelepis F., Pernet E., Dumaine A., Grenier J.C. BCG Educates Hematopoietic Stem Cells to Generate Protective Innate Immunity against Tuberculosis. Cell. 2018;172:176–190.e19. doi: 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- Keeter W.C., Moriarty A., Butcher M.J., Ma K.W., Nadler J.L., Galkina E. IL-12 induced STAT4 activation plays a role in pro-inflammatory neutrophil functions. J. Immunol. 2018;200:166.159. [Google Scholar]
- Keeter W.C., Moriarty A., Mehrpouya-Baharami P., Melo P., Nadler J., Serezani C.H., Kaplan M.H., Galkina E. STAT4 promotes critical neutrophil functions and is required for antimicrobial immunity in mice. J. Immunol. 2020;204:148.122. [Google Scholar]
- Kleinnijenhuis J., Quintin J., Preijers F., Joosten L.A., Ifrim D.C., Saeed S., Jacobs C., van Loenhout J., de Jong D., Stunnenberg H.G. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. USA. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinnijenhuis J., Quintin J., Preijers F., Joosten L.A.B., Jacobs C., Xavier R.J., van der Meer J.W.M., van Crevel R., Netea M.G. BCG-induced trained immunity in NK cells: Role for non-specific protection to infection. Clin. Immunol. 2014;155:213–219. doi: 10.1016/j.clim.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecha N., Krutzik P.O., Irish J.M. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 2010;Chapter 10 doi: 10.1002/0471142956.cy1017s53. Unit10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. The alphaMbeta2 integrin and its role in neutrophil function. Cell Res. 1999;9:171–178. doi: 10.1038/sj.cr.7290015. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini O., Costa S., Bevilacqua D., Calzetti F., Tamassia N., Spina C., De Sabata D., Tinazzi E., Lunardi C., Scupoli M.T. Mature CD10+ and immature CD10- neutrophils present in G-CSF-treated donors display opposite effects on T cells. Blood. 2017;129:1343–1356. doi: 10.1182/blood-2016-04-713206. [DOI] [PubMed] [Google Scholar]
- McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeish K.R., Uriarte S.M., Tandon S., Creed T.M., Le J., Ward R.A. Exocytosis of neutrophil granule subsets and activation of prolyl isomerase 1 are required for respiratory burst priming. J. Innate Immun. 2013;5:277–289. doi: 10.1159/000345992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minassian A.M., Satti I., Poulton I.D., Meyer J., Hill A.V., McShane H. A human challenge model for Mycobacterium tuberculosis using Mycobacterium bovis bacille Calmette-Guerin. J. Infect. Dis. 2012;205:1035–1042. doi: 10.1093/infdis/jis012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitroulis I., Ruppova K., Wang B., Chen L.S., Grzybek M., Grinenko T., Eugster A., Troullinaki M., Palladini A., Kourtzelis I. Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell. 2018;172:147–161.e12. doi: 10.1016/j.cell.2017.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal M., Siddiqui M.R., Tran K., Reddy S.P., Malik A.B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 2014;20:1126–1167. doi: 10.1089/ars.2012.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Joosten L.A., Latz E., Mills K.H., Natoli G., Stunnenberg H.G., O’Neill L.A., Xavier R.J. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Joosten L.A.B., van der Meer J.W.M. Hypothesis: stimulation of trained immunity as adjunctive immunotherapy in cancer. J. Leukoc. Biol. 2017;102:1323–1332. doi: 10.1189/jlb.5RI0217-064RR. [DOI] [PubMed] [Google Scholar]
- Ng L.G., Ostuni R., Hidalgo A. Heterogeneity of neutrophils. Nat. Rev. Immunol. 2019;19:255–265. doi: 10.1038/s41577-019-0141-8. [DOI] [PubMed] [Google Scholar]
- Nicolás-Ávila J.A., Adrover J.M., Hidalgo A. Neutrophils in Homeostasis, Immunity, and Cancer. Immunity. 2017;46:15–28. doi: 10.1016/j.immuni.2016.12.012. [DOI] [PubMed] [Google Scholar]
- Novakovic B., Habibi E., Wang S.Y., Arts R.J.W., Davar R., Megchelenbrink W., Kim B., Kuznetsova T., Kox M., Zwaag J. β-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance. Cell. 2016;167:1354–1368.e14. doi: 10.1016/j.cell.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay J., den Braber I., Vrisekoop N., Kwast L.M., de Boer R.J., Borghans J.A., Tesselaar K., Koenderman L. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood. 2010;116:625–627. doi: 10.1182/blood-2010-01-259028. [DOI] [PubMed] [Google Scholar]
- Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187-91. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales C. Neutrophil: A Cell with Many Roles in Inflammation or Several Cell Types? Front. Physiol. 2018;9:113. doi: 10.3389/fphys.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A., Gustafson P., Nhaga A., Djana Q., Poulsen A., Garly M.L., Jensen H., Sodemann M., Rodriques A., Aaby P. BCG vaccination scar associated with better childhood survival in Guinea-Bissau. Int. J. Epidemiol. 2005;34:540–547. doi: 10.1093/ije/dyh392. [DOI] [PubMed] [Google Scholar]
- Turro E., Su S.Y., Gonçalves Â., Coin L.J., Richardson S., Lewin A. Haplotype and isoform specific expression estimation using multi-mapping RNA-seq reads. Genome Biol. 2011;12:R13. doi: 10.1186/gb-2011-12-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ’t Wout J.W., Poell R., van Furth R. The role of BCG/PPD-activated macrophages in resistance against systemic candidiasis in mice. Scand. J. Immunol. 1992;36:713–719. doi: 10.1111/j.1365-3083.1992.tb03132.x. [DOI] [PubMed] [Google Scholar]
- Walk J., de Bree L.C.J., Graumans W., Stoter R., van Gemert G.J., van de Vegte-Bolmer M., Teelen K., Hermsen C.C., Arts R.J.W., Behet M.C. Outcomes of controlled human malaria infection after BCG vaccination. Nat. Commun. 2019;10:874. doi: 10.1038/s41467-019-08659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.T., Zhao Y.L., Peng L.S., Chen N., Chen W., Lv Y.P., Mao F.Y., Zhang J.Y., Cheng P., Teng Y.S. Tumour-activated neutrophils in gastric cancer foster immune suppression and disease progression through GM-CSF-PD-L1 pathway. Gut. 2017;66:1900–1911. doi: 10.1136/gutjnl-2016-313075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Chen G., Manwani D., Mortha A., Xu C., Faith J.J., Burk R.D., Kunisaki Y., Jang J.E., Scheiermann C. Neutrophil ageing is regulated by the microbiome. Nature. 2015;525:528–532. doi: 10.1038/nature15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder E.R., Finck R., Behbehani G.K., Amir A.D., Krishnaswamy S., Gonzalez V.D., Lorang C.G., Bjornson Z., Spitzer M.H., Bodenmiller B. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA and ChIP sequencing data of neutrophils before and 3 months after BCG vaccination as reported in this paper can been found in the NCBI Gene Expression Omnibus under accession number GEO: GSE153729.