Abstract
Polymerase eta (Polη) is a translesion synthesis DNA polymerase directly linked to cancer development. It can bypass several DNA lesions thereby rescuing DNA damage-stalled replication complexes. We previously presented evidence implicating Saccharomyces cerevisiae Polη in transcription elongation, and identified its specific RNA extension and translesion RNA synthetic activities. However, RNA synthesis by Polη proved rather inefficient under conditions optimal for DNA synthesis. Searching for factors that could enhance its RNA synthetic activity, we have identified the divalent cation of manganese. Here, we show that manganese triggers drastic changes in the activity of Polη. Kinetics experiments indicate that manganese increases the efficiency of ribonucleoside incorporation into RNA by ~400–2000-fold opposite undamaged DNA, and ~3000 and ~6000-fold opposite TT dimer and 8oxoG, respectively. Importantly, preference for the correct base is maintained with manganese during RNA synthesis. In contrast, activity is strongly impaired, and base discrimination is almost lost during DNA synthesis by Polη with manganese. Moreover, Polη shows strong preference for manganese during RNA synthesis even at a 25-fold excess magnesium concentration. Based on this, we suggest that a new regulatory mechanism, selective metal cofactor utilization, modulates the specificity of Polη helping it to perform distinct activities needed for its separate functions during replication and transcription.
Keywords: polymerase η, enzyme kinetics, yeast, manganese
1. Introduction
DNA polymerases possess catalytic activity to synthesize DNA in a template-dependent fashion using deoxy-ribonucleoside-triphosphates (dNTPs). However, the attributes of their activities differ considerably reflecting their diverse cellular functions [1,2]. Replicative DNA polymerases are responsible for faithful duplication of the genome and because of that they have a highly selective and restrictive active center ensuring that the correct complementary deoxy-ribonucleoside-monophosphates (dNMPs) are inserted into the growing DNA strand [3,4]. Due to their high selectivity, modifications or lesions in the template strand hinder the movement of replicative DNA polymerases during replication, leading to cell death if unattended. However, translesion synthesis (TLS) DNA polymerases evolved that are capable of synthesizing through DNA lesions [5,6]. These polymerases can take over synthesis from stalled replicative DNA polymerases and carry out synthesis across lesion sites, maintaining the continuity of replication. Contrary to replicative DNA polymerases, the active centers of TLS DNA polymerases are more spacious and less selective, enabling them to accommodate damaged, modified nucleotides. As a result of this, TLS DNA polymerases are error-prone on undamaged DNA, frequently inserting non-complementary nucleosides, which can lead to mutagenesis. Therefore, strict regulatory mechanisms restricting their activities to DNA damage sites are visualized. DNA polymerase η (Polη) is a TLS DNA polymerase that is uniquely able to carry out an efficient and error-free bypass of the most frequent ultraviolet (UV) light-induced DNA lesions, cyclobutane pyrimidine dimers [7]. The importance of this activity is well emphasized by the fact that inactivity of Polη in humans causes the cancer-prone xeroderma pigmentosum variant (XP-V) disorder [8,9]. Polη carries out a mostly error-free bypass of one of the most frequent spontaneous oxidative DNA lesions, 8-oxoguanine (8-oxoG), as well, and it was shown to bypass several other DNA lesions with varying fidelity [5,6,10].
Because of their high fidelity, it was of surprise that replicative DNA polymerases could incorporate ribonucleoside-monophosphates (rNMPs) with relatively high frequency into DNA due to incomplete exclusion of ribonucleoside-triphosphates (rNTPs) from their active centers [11]. Though rNMP incorporation occurs with a much reduced efficiency compared to dNMP, it has been estimated that replicative DNA polymerases incorporate ~10,000 rNMPs into the genome of a yeast cell during a single round of replication, putting rNMPs among the most abundant DNA lesions. The excess presence of rNMPs in DNA went undetected for a long time because they are efficiently removed by ribonucleotide excision repair [12,13]. Beside replicative polymerases, almost all DNA polymerases have been shown to be able to utilize rNMPs during DNA synthesis, though most of them do so with very low efficiencies [14,15,16,17]. However, there are a few exceptions, such as E. coli PolV, mycobacterial DinB2, and Polµ, that can utilize rNTPs and dNTPs with comparable efficiencies [18,19,20]. We and others have recently identified the ability of Polη to use rNTPs during synthesis [21,22,23,24]. Akin to other DNA polymerases, Polη incorporates rNMPs during DNA synthesis with a very low efficiency. Even so, experiments with both human and yeast Polη showed that they could contribute to the accumulation of ribonucleosides in the genomes of human and yeast cells [25,26]. Unexpectedly, our results indicated that the RNA synthetic activity of yeast Polη was specific as it inserted rNMPs at least 10-fold more efficiently into RNA over DNA [21]. During RNA extension it could even perform TLS opposite a TT dimer and 8-oxoG in an error-free manner. Moreover, we found that the lack of Polη impaired transcription elongation and caused transcriptional inhibition of several genes. These findings suggested a role for Polη during transcription elongation, and possibly in TLS during transcription. However, we also found that Polη utilized dNTPs with a much higher efficiency than rNTPs during RNA extension. Even though in yeast cells rNTP concentrations are in the millimolar, whereas dNTP concentrations are in the micromolar range, Polη would still synthesize a mixed strand consisting of ribo- and deoxyribonucleosides, which would have detrimental effects on cells. Hence, we surmised that certain cellular factors could improve the specific RNA extension activity of Polη.
DNA polymerases apply a mechanism based on two divalent metal cations during catalysis [27]. The catalytic and nucleotide metal-binding sites at their active center coordinate two metal ions that facilitate the nucleophilic attack by the 3′-OH group of the primer on the α-phosphate of the incoming nucleotide. The presence of a third metal ion at the active center has been recently discovered which is probably needed to reduce the product release barrier [28,29,30]. Mg2+ is presumed to be the physiological metal cofactor for DNA polymerases due to its widespread occurrence in nature, its much higher cellular concentration compared with other divalent metal cations, and that in vitro it universally activates DNA polymerases. However, other metal ions such as Mn2+, Co2+, and Ni2+ can be substituted for Mg2+ in in-vitro polymerization reactions, but the replacement usually significantly diminishes either the efficiency or the fidelity of the enzyme, or both [31,32]. Notwithstanding, Polβ, Polλ, Polµ, Polι, and PrimPol represent the growing group of exemptions where the activity is improved with the replacement metal. For example, human Polβ exhibited higher reactivity in the presence of Mn2+ as compared with Mg2+ so that it could even extend a blunt-ended double-stranded DNA template [33]. Kinetic and thermodynamic analysis suggested that Polλ evolved as a Mn2+-specific enzyme [34]. Mn2+ had positive effects on both the efficiency and the fidelity of gap-filling synthesis by Polµ [35]. Similarly, Polι was more active with Mn2+ compared to Mg2+ [36]. PrimPol was also found to be a Mn2+-dependent enzyme as its DNA primase and polymerase activities, as well its DNA primer/template-binding affinity, significantly improved upon Mn2+-binding [37]. Interestingly, the optimal Mn2+ concentrations for the above DNA polymerases spanned from the micromolar to the millimolar range. For example, Polι was most active at 75–250 µM Mn2+ concentrations on both undamaged and on damage-containing templates [36]. The optimal Mn2+ concentrations for Polµ were between 10 and 100 µM during gap-filling and non-homologous end joining [35]. However, Primpol and Polλ were most active around 1, and 1–5 mM Mn2+ concentrations, respectively [34,38]
In the present study, we report that substitution of manganese for the metal cofactor magnesium implements drastic changes in the activity of Polη. It greatly impairs its activity and sharply decreases its fidelity during DNA synthesis, whereas RNA synthesis becomes 400–2000-fold more efficient with manganese concomitantly maintaining the base selectivity of the enzyme. Moreover, the weak damage bypass activity of Polη observed during RNA synthesis with magnesium is augmented by 3000–6000-fold with manganese opposite TT dimer and 8-oxoG, respectively. Additionally, of note, manganese is preferred by Polη over magnesium even at a 25-fold lower concentration during RNA synthesis. Based on these findings, we propose a model with a new regulatory mechanism contributing to a shift between the DNA and RNA synthetic activities of Polη.
2. Results
2.1. Effect of Divalent Metal Ions on the Synthetic Activity of Polη
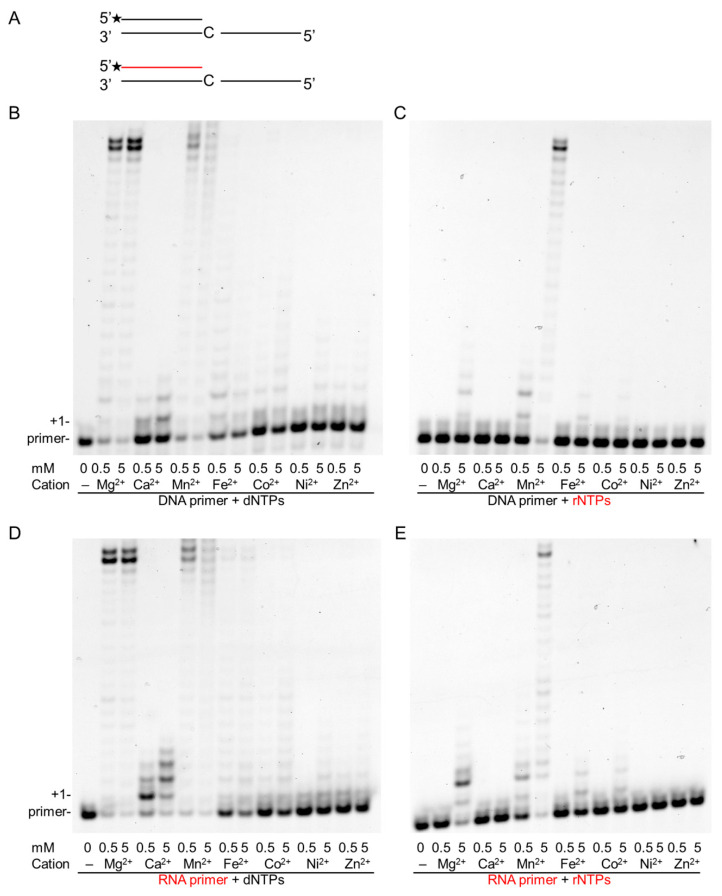
In our attempt to unravel conditions that could enhance the RNA synthetic activity of Polη, we tested the metal ion dependence of both of its DNA and RNA synthetic activities. Besides magnesium, we compared activities in the presence of six other divalent metal cations that had been implicated in enzymatic activation. In in-vitro primer extension assays, each tested cation supported dNMP insertion into both DNA and RNA primers to a varying extent, although Polη exhibited the highest activity in the presence of Mg2+ and Mn2+, lengthening almost all the primers and synthesizing until the end of the template (Figure 1B,D). In the case of each metal cofactor, activation could be detected even at low 0.5-mM metal concentrations, whereas a high 5-mM metal concentration resulted in higher activity. Surprisingly, when the reactions were supplemented with rNTPs instead of dNTPs, activity was observed only with Mn2+ at a 0.5-mM metal ion concentration using either a DNA or an RNA primer (Figure 1C,E). Moreover, Polη exhibited strong activity only in the presence of Mn2+ even at a 5-mM metal concentration, and only a weak activity could be detected with Mg2+, Fe2+, and Co2+, in agreement with our previous results, whereas no activity was detected with the other metals—Ca2+, Ni2+, and Zn2+ [21]. These data suggest that Mn2+ could be the proper cation needed for the activation of the RNA synthetic activity of Polη.
Figure 1.
Effect of various divalent cations on the primer extension activities of Polη. In-vitro primer extension reactions were performed with 40-nM Polη for 5 min in the presence of two different concentrations of the indicated divalent cations. (A) The structures of the primer/template used in the experiments are shown. The RNA primer is depicted in red. Asterisks (*) indicate fluorescently labeled primer ends. Reactions were carried out in the presence of (B) DNA primer and deoxy-ribonucleoside-triphosphates (dNTPs), (C) DNA primer with ribonucleoside-triphosphates (rNTPs), (D) RNA primer with dNTPs, and (E) RNA primer and rNTPs. Reactions in (B–E) contained 30 nM of the hybridized primer/template, and close to physiological concentrations of nucleotides, either 50 µM of dNTPs or 1 mM of rNTPs. The positions of the primer and its extension by one nucleotide are indicated. RNA primers and rNTPs are highlighted in red.
2.2. Mg2+ and Mn2+ Concentration-Dependent Synthesis by Polη
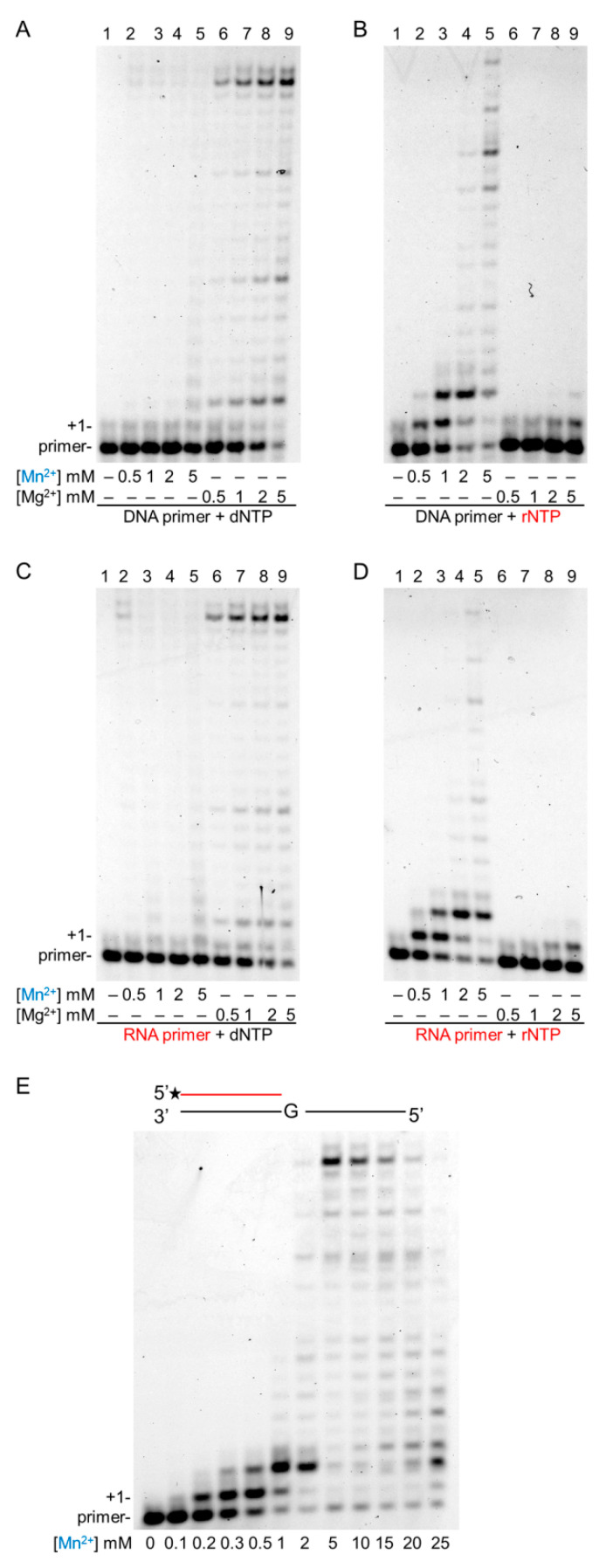
To compare the effect of Mg2+ and Mn2+ on the synthetic activities of Polη, we applied increasing concentrations of the metal ions and carried out synthesis reactions using a DNA or an RNA primer with dNTPs or rNTPs, in all four combinations. In these experiments, the highest synthetic activities were observed at the highest applied (5 mM) metal ion concentrations in all primer–substrate combinations with both Mg2+ and Mn2+ (Figure 2). As expected, in the presence of a DNA primer and Mg2+, Polη exhibited high activity with dNTPs, which sharply increased with increasing Mg2+ concentrations (Figure 2A, lanes 6–9). At the highest 5 mM Mg2+ concentration, Polη extended ~90% of the primers and synthesis reached the end of the template. Interestingly, a similarly strong activity was observed when the DNA primer was replaced with an RNA primer (Figure 2C, lanes 6–9). On the other hand, substituting Mn2+ for Mg2+ resulted in greatly diminished dNMP insertions by Polη on both DNA and RNA primers (Figure 2A,C, lanes 1–5). Though a higher Mn2+ concentration resulted in a somewhat higher activity, it did not become considerably stronger even at the highest Mn2+ concentration, extending only ~30% of the primers. Contrary to the low activation of DNA synthesis, Mn2+ dramatically enhanced rNMP insertion by Polη utilizing either a DNA or an RNA primer. Although rNMP incorporation was inefficient with Mg2+ extending ~10% of the primers with only 1–2 nucleotides at the highest Mg2+ concentration (Figure 2B,D, lanes 6–9), it sharply increased with increasing Mn2+ concentration lengthening ~90% of the primers and resulting in fully extended primers at a 5-mM Mn2+ concentration (Figure 2B,D, lanes 1–5). The same Mn2+ concentration-dependent activation could be detected using individual rNTPs in the assays (Figure S1). In summary, these results showed that although Mn2+ strongly reduced the DNA synthetic activity of Polη compared to Mg2+, it dramatically elevated its RNA synthetic activity in a concentration-dependent manner.
Figure 2.
Effect of increasing concentrations of magnesium and manganese ions on the primer extension activity of Polη in the presence of dNTPs or rNTPs. (A–D) Reactions were performed for 5 min with 10-nM Polη in the presence of 20-nM primer/template, and close to physiological concentrations of nucleotides, either 50-µM dNTPs, or 1-mM rNTPs. (A) DNA primer (S3) and dNTPs, (B) DNA primer (S3) with rNTPs, (C) RNA primer (S7) with dNTPs, and (D) RNA primer (S7) and rNTPs. The concentrations of Mn2+ and Mg2+ are indicated below each panel. Lanes are numbered at the top. (E) Optimal manganese concentration needed for the highest activation of Polη during RNA synthesis. Primer extension reactions containing the indicated concentration of Mn2+ were performed for 5 min with 20-nM Polη and 1-mM rNTP. The positions of the primer and its extension by one nucleotide are indicated. RNA primers and rNTPs are highlighted in red, and manganese in blue.
In order to determine the proper concentration of Mn2+ needed for the highest activation of RNA synthesis, we tested reactions containing Mn2+ in a broad range of 0.1–25 mM. As Figure 2E shows, Polη was activated at all Mn2+ concentrations, and the highest activity was detected with 5-mM Mn2+. Lower or higher concentrations resulted in a gradually decreasing activity, though the changes were more moderate at the higher range.
2.3. Kinetics of Correct rNMP Incorporation into RNA in the Presence of Mn2+
Previously, we determined the kinetic parameters of the RNA synthetic activity of Polη in the presence of Mg2+ and found that it incorporated single rNMPs into RNA with an efficiency of ~10−3–10−4 min−1µM−1 [21]. To quantitate the enhancement of its RNA synthetic activity observed in the presence of Mn2+, we carried out similar steady-state kinetic studies using 5-mM Mn2+ instead of 5-mM Mg2+ in the reactions (Figure S2). Remarkably, as Table 1 shows, Polη inserted rNMPs into RNA ~1000-fold more efficiently when utilizing Mn2+. The smallest ~400-fold increase was detected during rCMP insertion, whereas the highest ~2000-fold difference was measured during rAMP insertion. The overall enhancement was due to a ~100-fold decrease in the Michaelis–Menten constant (KM) indicating stronger rNTP-binding of Polη, and to a ~10-fold increase in the catalytic constant (kcat) values reflecting the velocity of the reactions. These observed changes in the Km and kcat values indicated a greatly improved specificity of the RNA extension reactions achieved in the presence of Mn2+.
Table 1.
Comparison of the kinetic parameters of rNTP incorporation into RNA by Polη using Mg2+ or Mn2+ as cofactors.
| Templating Nucleotide | Incoming Nucleotide | Cation | kcat (min−1) | KM (µM) | kcat/KM (min−1µM−1) | Relative Efficiency a |
|---|---|---|---|---|---|---|
| T | rATP | Mg2+ | 0.24 ± 0.01 b | 466 ± 47.3 b | 5.15 × 10−4 b | |
| Mn2+ | 2.61 ± 0.14 | 2.51 ± 0.64 | 1.04 | 2019 | ||
| G | rCTP | Mg2+ | 2.76 ± 0.06 b | 438 ± 37.5 b | 6.30 × 10−3 b | |
| Mn2+ | 4.68 ± 0.22 | 1.89 ± 0.42 | 2.48 | 394 | ||
| C | rGTP | Mg2+ | 0.45 ± 0.01 b | 394 ± 52 b | 1.14 × 10−3 b | |
| Mn2+ | 5.07 ± 0.27 | 2.55 ± 0.63 | 1.99 | 1746 | ||
| A | rUTP | Mg2+ | 0.10 ± 0.01 b | 423 ± 90.4 b | 2.36 × 10−4 b | |
| Mn2+ | 3.51 ± 0.19 | 12.8 ± 2.25 | 2.74 × 10−1 | 1161 | ||
| 8-oxo-G | rCTP | Mg2+ | 0.034 ± 0.004b | 974 ± 270 b | 3.52 × 10−5 | |
| Mn2+ | 0.275 ± 0.01 | 1.25 ± 0.28 | 2.20 × 10−1 | 6286 | ||
| TT dimer | rATP | Mg2+ | 0.0083 ± 0.001 b | 1678 ± 445 b | 4.94 × 10−6 | |
| Mn2+ | 0.174 ± 0.005 | 11.3 ± 1.35 | 1.54 × 10−2 | 3117 |
a Relative efficiency was calculated using the following equation: frel = (kcat/KM)Mn2+/(kcat/KM)Mg2+. b Published in [21].
2.4. The Effect of Mn2+ on the Base Selectivity of Polη
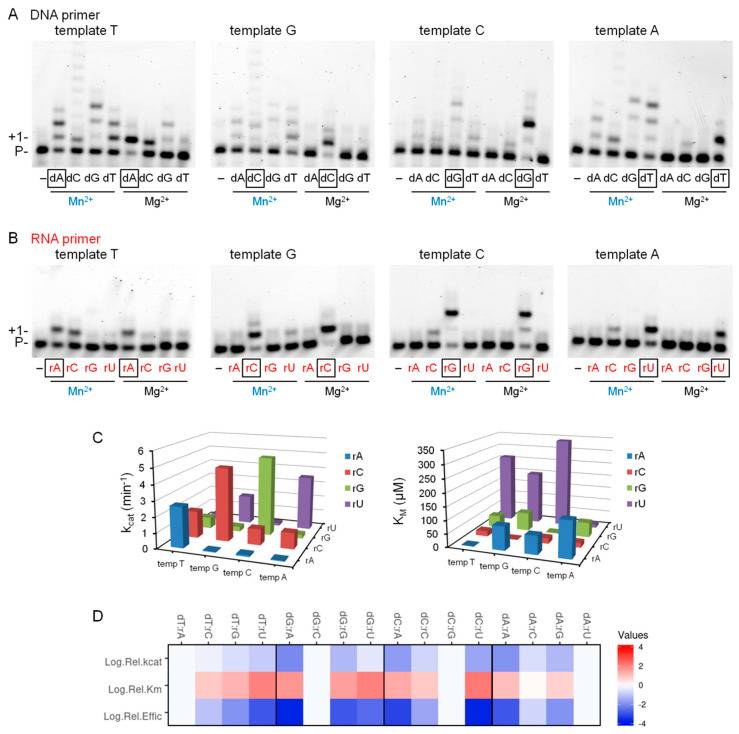
In-vitro Mn2+ can be substituted for Mg2+ in the activation of several DNA polymerases. However, in most cases the accuracy of DNA synthesis supported by Mn2+ drastically decreases, spoiling the activity of the polymerase. Hence, we investigated the effects of Mn2+ on the base selectivity of Polη by testing the incorporation of all four dNMPs and rNMPs individually into DNA and RNA primers, respectively, opposite each of the four possible DNA template residues. When DNA synthesis was assayed using Mg2+, Polη showed preference for the correct dNTP in accordance with its reported 10−2–10−4 fidelity (Figure 3A). However, base selectivity was almost completely lost with Mn2+ and the correct and incorrect dNMPs were inserted with comparably weak efficiencies. During RNA synthesis with Mg2+, Polη discriminated against the incorrect bases catalyzing only weak misinsertions as opposed to robust correct rNMP insertions (Figure 3B). Surprisingly, although Mn2+ resulted in a somewhat decreased base selectivity indicated by the stronger intensity of the bands representing misinsertions, a clear preference for the correct rNTPs was maintained. To obtain a more accurate insight on the effect of Mn2+ on base selection, we quantitated the fidelity of RNA synthesis in the presence of Mn2+ in steady-state kinetic experiments (Figure S2). The results of these studies showed that Polη had a lower affinity to incorrect rNTPs than to correct ones as seen by a ~100-fold increased Km and it incorporated them slower, as seen by a ~10-fold decreased kcat (Table 2 and Figure 3C). Overall, Polη exhibited base selectivity in the 10−2–10−4 range during RNA synthesis with Mn2+ (Table 2 and Figure 3D), which corresponded well with its reported accuracy during DNA synthesis with Mg2+ [39]. Interestingly, rAMP misincorporations were the weakest (~10−3–10−4), whereas rCMP was misinserted with the highest (10−1–10−2) relative efficiencies opposite all three non-complementary template residues. In summary, the above data indicate that Polη maintained its fidelity during RNA synthesis in the presence of Mn2+.
Figure 3.
Fidelity of Polη during DNA and RNA synthesis in the presence of magnesium or manganese. (A,B) Reactions contained 5-mM Mn2+ or Mg2+ as indicated, 6-nM Polη, 20-nM (A) DNA/DNA (S1–4) or (B) RNA/DNA (S5–8) primer/template, and (A) 0.1-mM individual dNTPs or (B) 4 mM individual rNTP, as indicated. Reaction times were 1 min except for the 15-min reactions with Mg2+ in (B). The templating base in the incoming position is denoted. RNA primer and rNTPs are highlighted in red, and manganese is in blue. The nucleotides representing correct insertions are boxed. (C) Measured kcat (left) and KM (right) values of various ribunucleotide incorporations opposite the indicated templating bases. (D) Heat map showing relative catalytic constants of incorporation of incorrect versus correct rNTPs. Log.Rel.kcat = Log10(kcat incorrect/kcat correct), Log.Rel.Km = Log10(KM incorrect/KM correct) and Log.Rel.Effic = Log10 [(kcat/KM)incorrect/(kcat/KM)correct]. Heat map was generated using http://www.heatmapper.ca [40].
Table 2.
Kinetic parameters of rNTP incorporation and misincorporation into RNA by Polη using Mn2+ as cofactor.
| Templating Nucleotide | Incoming Nucleotide | kcat (min−1) | KM (µM) | kcat/KM (min−1µM−1) | Relative Efficiency a | Discrimination1/f b |
|---|---|---|---|---|---|---|
| T | rATP | 2.61 ± 0.14 | 2.51 ± 0.64 | 1.04 | ||
| rCTP | 1.72 ± 0.06 | 19.4 ± 3.14 | 0.09 | 8.7 × 10−2 | 12 | |
| rGTP | 0.72 ± 0.03 | 44.6 ± 7.03 | 0.016 | 1.5 × 10−2 | 67 | |
| rUTP | 0.36 ± 0.02 | 261 ± 50.6 | 0.0014 | 1.3 × 10−3 | 769 | |
| G | rATP | 0.06 ± 0.00 | 89.3 ± 17.7 | 0.0007 | 2.8 × 10−4 | 3571 |
| rCTP | 4.68 ± 0.22 | 1.89 ± 0.42 | 2.48 | |||
| rGTP | 0.31 ± 0.02 | 68.9 ± 14.9 | 0.0045 | 1.8 × 10−3 | 555 | |
| rUTP | 1.86 ± 0.04 | 199 ± 14.5 | 0.0093 | 3.8 × 10−3 | 263 | |
| C | rATP | 0.10 ± 0.00 | 69.2 ± 11.9 | 0.0014 | 7.0 × 10−4 | 1429 |
| rCTP | 1.03 ± 0.04 | 19.9 ± 3.74 | 0.052 | 2.6 × 10−2 | 38 | |
| rGTP | 5.07 ± 0.27 | 2.55 ± 0.63 | 1.99 | |||
| rUTP | 0.16 ± 0.01 | 339 ± 49.4 | 0.0005 | 2.5 × 10−4 | 4000 | |
| A | rATP | 0.05 ± 0.00 | 138 ± 26.2 | 0.0004 | 1.48 × 10−3 | 676 |
| rCTP | 1.03 ± 0.07 | 17.5 ± 4.92 | 0.059 | 2.2 × 10−1 | 5 | |
| rGTP | 0.24 ± 0.01 | 55.9 ± 8.64 | 0.0043 | 1.6 ×10−2 | 63 | |
| rUTP | 3.51 ± 0.19 | 12.8 ± 2.25 | 0.27 |
a Relative efficiency was calculated using the following equation: frel = (kcat/KM)incorrect/(kcat/KM)correct. b Inverse relative efficiency: 1/frel = (kcat/KM)correct/(kcat/KM)incorrect.
2.5. DNA Damage Bypass Activity of Polη with Mn2+
Next, we examined the effect of Mn2+ on the TLS activity of Polη during RNA extension (Figure 4A,B). Our previous results obtained in the presence of Mg2+ revealed a very inefficient bypass of 8-oxoG and TT dimer during RNA extension [21]. Importantly, Mn2+ had a profound effect on insertion that was opposite to both of these damages (Figure S3). Table 1 shows that a ~6000-fold and a ~3000-fold enhancement in TLS efficiency was measured in steady-state kinetic experiments opposite 8-oxoG and TT dimer, respectively, compared to data obtained in the presence of Mg2+. As in the case of undamaged templates and correct incoming nucleotides, the apparent Km values decreased by two orders of magnitude, whereas the Kcat values increased by an order of magnitude with Mn2+. Moreover, Polη kept its fidelity during the bypass reactions as it preferably inserted the correct rNMPs opposite the damage sites and no significant insertions of the incorrect nucleotides were observed (Figure 4C,D). Based on these results, we concluded that Mn2+ was a specific activator of the RNA synthetic activity of Polη both on undamaged templates and opposite DNA damages.
Figure 4.
DNA damage bypass by Polη during RNA synthesis in the presence of manganese. (A,B) The template in S12 contained 8-oxoG in the incoming position. Reactions were performed for 3 min with 5-mM Mn2+, 1.6-nM Polη, 8-nM RNA/DNA primer/template and the indicated amount of individual rNTPs. (C,D) The template in S16 contained TT dimer in the incoming position. Reactions were performed for 15 min with 5-mM Mn2+, 1.6-nM Polη, 20-nM RNA/DNA primer/template and the indicated amount of individual rNTPs. RNA primers and rNTPs are highlighted in red.
2.6. Metal Preference of Polη during RNA Synthesis
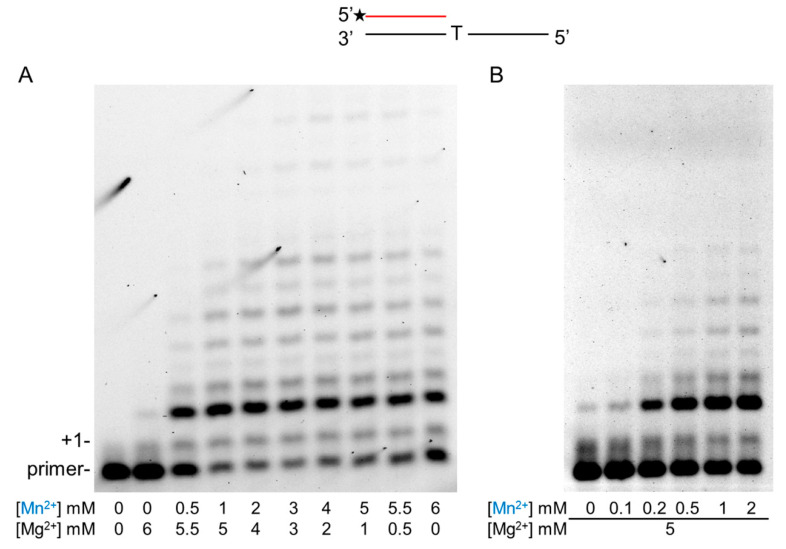
Since Mn2+ exerted a dramatic effect on Polη activity, it was important to examine which metal cation was preferred by Polη during RNA synthesis. For this reason, we carried out RNA extension experiments with rNTPs in the joint presence of Mg2+ and Mn2+. In the first set of reactions, the concentration of Mg2+ gradually decreased from 6 to 0 mM, whereas the concentration of Mn2+ increased from 0 to 6 mM in parallel, maintaining the total metal cation concentration at 6 mM. The pattern of reaction products contrasted strikingly in the sole presence of Mg2+ or Mn2+ (Figure 5A, compare the first and last lanes), enabling an easy detection of cation utilization. The results showed that, even at eleven-fold excess of Mg2+, reaction products specific to Mn2+ appeared (Figure 5A second lane). In the next set of reactions, the Mg2+ concentration was kept at 5 mM and Mn2+ concentration was gradually increased. In this setup, the reaction products showed a Mn2+-specific pattern already at a 0.2-mM Mn2+ concentration despite the 25-fold higher Mg2+ level indicating that Polη preferred Mn2+ over Mg2+ in the reactions (Figure 5B, lane 3).
Figure 5.
Metal ion preference of Polη during RNA synthesis. Primer extension reactions were performed with 10-nM Polη, 20-nM S6 RNA/DNA primer/template and 1-mM rNTP mix for 5 min. (A) Reactions contained both Mn2+ and Mg2+ in the indicated concentrations. (B) Reactions contained 5-mM Mg and the indicated concentrations of Mn2+. Manganese is highlighted in blue. The positions of the primer and its one nucleotide extension (+1) are indicated.
3. Discussion
The aim of the present study was to identify cellular factors that could improve the RNA synthetic activity of yeast Polη. The conception was based on our previous results showing that Polη has a specific RNA synthetic activity inserting rNMPs at least ten times more efficiently into an RNA primer as opposed to a DNA primer [21]. Despite its specificity, the observed efficiency of RNA synthesis was rather weak raising the possibilities that either the applied reaction conditions were not appropriate or Polη required accessory proteins for efficient RNA synthesis, or both.
Hence, first we tried to optimize the reaction by replacing the generally used metal cofactor Mg2+ with other divalent metal cations. We tested several metal cations and all could activate DNA synthesis to varying degrees, but Mg2+ and Mn2+ achieved the highest activity. On the other hand, during RNA synthesis Ca2+, Ni2+, and Zn2+ were inactive, Mg2+, Fe2+, and Co2+ conferred very limited activity, and only Mn2+ supported an efficient reaction. These results suggest that Mn2+ is the proper metal cofactor of Polη during RNA synthesis and that other metal cations cannot be substituted for it. Steady-state kinetic experiments revealed that in comparison with Mg2+, Mn2+ caused a 400–2000-fold increase in efficiency during RNA synthesis on undamaged templates, and a 6000- and 3000-fold increase opposite 8-oxoG and TT dimer, respectively. The specificity of the activation is underpinned by the fact that the enzyme maintained its base selectivity in the 10−2–10−4 range with Mn2+, similarly to its base discrimination during DNA synthesis with Mg2+ [39]. Moreover, Polη preferentially utilized Mn2+ even in a 25-fold excess of Mg2+ during RNA synthesis. Taken together these data reinforce our previous finding that the RNA synthetic activity of Polη is specific, and identify Mn2+ as its apposite metal cofactor. Most importantly, our experiments also demonstrate that selective utilization of the two metal cations Mg2+ and Mn2+ results in a strong difference in specificity. When utilizing Mg2+, Polη is proficient in DNA synthesis but very inefficient during RNA synthesis, and preference for the correct base is sustained in both cases. On the other hand, contrary to the remarkable enhancement of RNA synthesis by Mn2+, it adversely affected the DNA synthetic activity of Polη by strikingly decreasing both the efficiency and the fidelity of the reaction. This differential effect of Mn2+ on the DNA and RNA synthetic activities of Polη sharply contrasts with the effect it had on the reported Mn2+-dependent polymerases, Pols λ, ι, µ, and Primpol, in which case Mn2+ exerted an overall positive effect on synthesis with either dNTPs or rNTPs [34,35,36,37]. The advantage of enhanced dNMP incorporation is obvious during DNA synthesis. Nevertheless, as it was suggested, increased rNTP utilization could also be advantageous during the repair of DNA double-strand breaks by non-homologous end joining given the easy accessibility of rNTPs. During transcription, however, the DNA synthetic activity of Polη has to be repressed and the RNA synthetic activity has to be elevated to avoid excess dNMP insertion into RNA, which could hinder elongation, lead to miscoding, or otherwise could alter key steps of transcription and translation [41,42,43,44,45,46].
Yet, we have to consider that Mg2+ is the most abundant divalent metal cation in the cell. The intracellular concentration of Mg2+ is in the millimolar range, which is much higher than the concentration of Mn2+ which is in the micromolar range [47]. Therefore, Mg2+ could be readily acquired by a plethora of enzymes including Polη during DNA replication. In turn, though our results indicate that Mn2+ is preferred over Mg2+ by Polη during RNA synthesis even at a ~25-fold lower concentration, given the huge difference between the intracellular concentrations of the two metal ions the involvement of additional factors assisting Mn2+-binding has to be presumed. We hypothesize that direct interactions with the transcription machinery could have an effect on the metal utilization of Polη so that Mn2+ would be preferred over other cations. One possible way to achieve this could be through direct metal delivery by a metal chaperone, which has been described for many enzymes [48,49]. For example, the metal chaperone copper chaperone for Sod1 (Ccs1) activates the superoxide dismutase Sod1 by directly transferring copper to Sod1, and similarly Cox17 conveys copper to Cox11 for eventual transfer to cytochrome C oxidase for activation [50,51]. Further experiments are needed to unravel the identity of factors that can influence the metal selectivity of yeast Polη and the mechanism of their action.
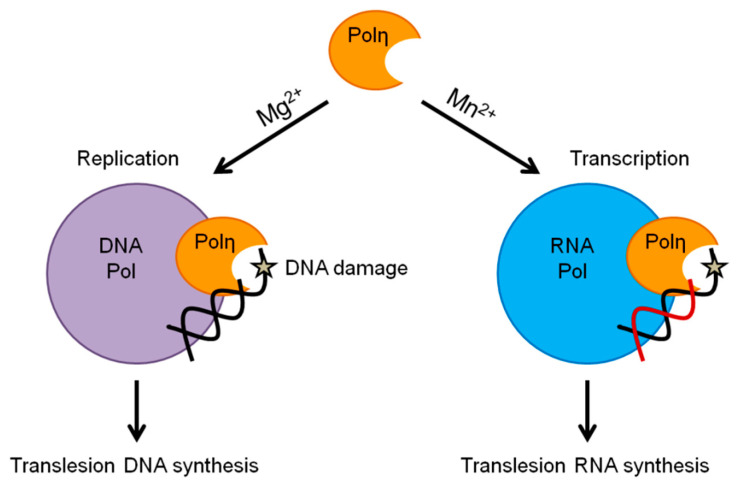
In conclusion, we propose that preferential activation of the DNA or RNA synthetic activity with concomitant impairment of the other through selective metal utilization constitutes a new regulatory mechanism that, with the contribution of other yet unidentified factors, enables Polη to take part in synthesis and DNA damage bypass during replication and also during transcription (Figure 6). Future studies with other polymerases and other enzymes would be necessary to determine the generality of the mechanism. However, since yeast and human Polη exhibit very similar biochemical characteristics during DNA synthesis including processivity, fidelity, damage bypass ability, and human Polη was also shown to be able to utilize rNTPs during DNA extension [23,24,25], therefore it would be of high significance to investigate whether human Polη also has Mn2+-activated specific RNA synthetic and translesion RNA synthesis activities, adding an additional layer to its contribution to genome stability.
Figure 6.
Proposed function of the selective metal cation-dependent activities of yeast Polη. The arrows next to Mg2+ and Mn2+ symbolize specific strong enhancement of the DNA or RNA synthetic activities of Polη, respectively.
4. Materials and Methods
4.1. Protein Purification
Saccharomyces cerevisiae Polη was overexpressed in yeast in N-terminal fusion with GST and affinity purified on glutathione–Sepharose beads (GE Healthcare, Uppsala Sweden). as described previously [21]. The GST-tag was removed in the last step of the purification by incubating the beads with PreScission protease (Merck KGaA, Darmstadt, Germany). Efficiency of the purification was verified by polyacrylamide gel electrophoresis and Coomassie staining (Merck KGaA, Darmstadt, Germany).
4.2. Oligonucleotides and Primer Extension Assays
Sequences of DNA/DNA and RNA/DNA primer/template substrates used in this study are shown in Table S1. Oligonucleotides used as primers contained a fluorophore indocarbocyanine (Cy3) label at the 5′-ends. Oligonucleotides used in these experiments were purchased from Integrated DNA Technologies, Coralville, Iowa, USA, except for the 8-oxoG-containing primer which was from Midland Certified Reagent Co., Midland, Texas, USA and the TT dimer-containing oligonucleotide was from Trilink Biotechnologies, San Diego, California, USA. Results were also verified with DNA and RNA primers purchased from Sigma-Aldrich Merck KGaA, Darmstadt, Germany. Standard primer extension reactions (5 μL) contained 25-mM Tris/HCl pH 7.5, 1-mM dithiothreitol, 100-μg/mL bovine serum albumin, 10% glycerol, the specified divalent cation as chloride salt, and substrate and enzyme as described in the figure legends. Reactions were initiated by the addition of the cation at the indicated concentrations, incubated at 30 °C and quenched by the addition of 15-μL loading buffer containing 95% formamide, 18-mM EDTA, 0.025% SDS, 0.025% bromophenol blue and 0.025% xylene cyanol. The reaction products were resolved on 10–14% polyacrylamide gels containing 7-M urea and analyzed with a Typhoon TRIO Phosphorimager (GE Healthcare, Little Chalfont, Buckinghamshire, UK).
4.3. Determination of Steady-State Kinetic Parameters
Primer extension reactions were performed as described above with the following modifications. On undamaged templates, 1-nM Polη was incubated with 20-nM of primer substrate in standard buffer containing 5-mM Mn. Reactions were initiated by adding the corresponding single rNTP (varied from 0.05 to 500 µM) and incubated at 30 °C from 30 s to 2 min. For kinetic analysis of 8-oxoG or TT dimer bypass, 1-nM Polη was incubated with 8- or 16-nM primer substrate, respectively, in standard buffer containing 5-mM Mn. Reactions were initiated by adding rCTP (0.05 to 500 µM) or rATP (0.05 to 500 µM), and incubated at 30 °C for 3 and 10 min, respectively. The intensity of the gel bands corresponding to the substrate and the product were quantitated with Typhoon TRIO Phosphorimager (GE Healthcare, Little Chalfont, Buckinghamshire, UK) using ImageQuant TL software (version 7.0, GE Healthcare, Little Chalfont, Buckinghamshire, UK) and the observed rates of nucleotide incorporation were plotted as a function of rNTP concentration. The data were fit by non-linear regression using the SigmaPlot program (version 12.5 Systat Software, San Jose, CA, USA) to the Michaelis–Menten equation describing a hyperbola, v = Vmax×[rNTP]/(Km + [rNTP]. The kcat and Km steady-state parameters were obtained from the fit and were used to calculate the efficiency (kcat/Km) and the relative efficiency (activation by Mn2+ versus Mg2+) using the formula frel = (kcat/Km)Mn2+/(kcat/Km)Mg2+. A heat map was generated using http://www.heatmapper.ca [40].
Acknowledgments
We thank Szilvia Minorits and Aniko Bozo-Toth for technical assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/21/8248/s1, Figure S1, Figure S2, Figure S3, Table S1.
Author Contributions
Conceptualization, E.B. and I.U.; formal analysis, E.B., I.U.; investigation, E.B.; writing—original draft preparation, I.U.; writing—review and editing, E.B., I.U.; visualization, E.B.; supervision, I.U.; funding acquisition, I.U. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Research, Development and Innovation Office (grant numbers GINOP-2.3.2-15-2016-00001, GINOP-2.3.2-15-2016-00024).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Garcia-Diaz M., Bebenek K. Multiple functions of DNA polymerases. CRC Crit. Rev. Plant Sci. 2007;26:105–122. doi: 10.1080/07352680701252817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acharya N., Khandagale P., Thakur S., Sahu J.K., Utkalaja B.G. Quaternary structural diversity in eukaryotic DNA polymerases: Monomeric to multimeric form. Curr. Genet. 2020;66:635–655. doi: 10.1007/s00294-020-01071-1. [DOI] [PubMed] [Google Scholar]
- 3.Echols H., Goodman M.F. Fidelity Mechanisms In DNA Replication. Annu. Rev. Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 4.Johansson E., Dixon N. Replicative DNA polymerases. Cold Spring Harb. Perspect. Biol. 2013;5:a012799. doi: 10.1101/cshperspect.a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prakash S., Johnson R.E., Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 6.Vaisman A., Woodgate R. Translesion DNA polymerases in eukaryotes: What makes them tick? Crit. Rev. Biochem. Mol. Biol. 2017;52:274–303. doi: 10.1080/10409238.2017.1291576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson R.E., Prakash S., Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 8.Johnson R.E., Kondratick C.M., Prakash S., Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 9.Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K., Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 10.Haracska L., Yu S.L., Johnson R.E., Prakash L., Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 11.Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Lundström E.B., Burgers P.M.J., Johansson E., Chabes A., Kunkel T.A. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eder P.S., Walder R.Y., Walder J.A. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-O. [DOI] [PubMed] [Google Scholar]
- 13.Rydberg B., Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl. Acad. Sci. USA. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joyce C.M. Choosing the right sugar: How polymerases select a nucleotide substrate. Proc. Natl. Acad. Sci. USA. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaisman A., Woodgate R. Ribonucleotide discrimination by translesion synthesis DNA polymerases. Crit. Rev. Biochem. Mol. Biol. 2018;53:382–402. doi: 10.1080/10409238.2018.1483889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova A.V., McElhinny S.A.N., Watts B.E., Kunkel T.A., Burgers P.M. Ribonucleotide incorporation by yeast DNA polymerase ζ. DNA Repair. 2014;18:63–67. doi: 10.1016/j.dnarep.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sassa A., Çaǧlayan M., Rodriguez Y., Beard W.A., Wilson S.H., Nohmi T., Honma M., Yasui M. Impact of ribonucleotide backbone on translesion synthesis and repair of 7,8-Dihydro-8-oxoguanine. J. Biol. Chem. 2016;291:24314–24323. doi: 10.1074/jbc.M116.738732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaisman A., Kuban W., McDonald J.P., Karata K., Yang W., Goodman M.F., Woodgate R. Critical amino acids in Escherichia coli UmuC responsible for sugar discrimination and base-substitution fidelity. Nucleic Acids Res. 2012;40:6144–6157. doi: 10.1093/nar/gks233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ordonez H., Uson M.L., Shuman S. Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation. Nucleic Acids Res. 2014;42:11056–11070. doi: 10.1093/nar/gku752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nick McElhinny S.A., Ramsden D.A. Polymerase Mu Is a DNA-Directed DNA/RNA Polymerase. Mol. Cell. Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gali V.K., Balint E., Serbyn N., Frittmann O., Stutz F., Unk I. Translesion synthesis DNA polymerase η exhibits a specific RNA extension activity and a transcription-associated function. Sci. Rep. 2017;7:13055. doi: 10.1038/s41598-017-12915-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donigan K.A., Cerritelli S.M., McDonald J.P., Vaisman A., Crouch R.J., Woodgate R. Unlocking the steric gate of DNA polymerase η leads to increased genomic instability in Saccharomyces cerevisiae. DNA Repair. 2015;35:1–12. doi: 10.1016/j.dnarep.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su Y., Egli M., Guengerich F.P. Human DNA polymerase η accommodates RNA for strand extension. J. Biol. Chem. 2017;292:18044–18051. doi: 10.1074/jbc.M117.809723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su Y., Egli M., Guengerich F.P. Mechanism of ribonucleotide incorporation by human DNA polymerase η. J. Biol. Chem. 2016;291:3747–3756. doi: 10.1074/jbc.M115.706226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mentegari E., Crespan E., Bavagnoli L., Kissova M., Bertoletti F., Sabbioneda S., Imhof R., Sturla S.J., Nilforoushan A., Hübscher U., et al. Ribonucleotide incorporation by human DNA polymerase η impacts translesion synthesis and RNase H2 activity. Nucleic Acids Res. 2017;45:2600–2614. doi: 10.1093/nar/gkw1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meroni A., Nava G.M., Bianco E., Grasso L., Galati E., Bosio M.C., Delmastro D., Muzi-Falconi M., Lazzaro F. RNase H activities counteract a toxic effect of Polymerase η in cells replicating with depleted dNTP pools. Nucleic Acids Res. 2019;47:4612–4623. doi: 10.1093/nar/gkz165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beese L.S., Steitz T.A. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: A two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y., Yang W. Capture of a third Mg2+ is essential for catalyzing DNA synthesis. Science. 2016;352:1334–1337. doi: 10.1126/science.aad9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon H., Warshel A. Simulating the Fidelity And The Three Mg Mechanism of Pol η and clarifying the validity of transition state theory in enzyme catalysis. Proteins. 2017;85:1446–1453. doi: 10.1002/prot.25305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens D.R., Hammes-Schiffer S. Exploring the Role of the Third Active Site Metal Ion in DNA Polymerase η with QM/MM Free Energy Simulations. J. Am. Chem. Soc. 2018;140:8965–8969. doi: 10.1021/jacs.8b05177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sirover M.A., Loeb L.A. Metal activation of DNA synthesis. Biochem. Biophys. Res. Commun. 1976;70:812–817. doi: 10.1016/0006-291X(76)90664-1. [DOI] [PubMed] [Google Scholar]
- 32.Sirover M.A., Dube D.K., Loeb L.A. On the fidelity of DNA replication. Metal activation of Escherichia coli DNA polymerase I. J. Biol. Chem. 1979;254:107–111. [PubMed] [Google Scholar]
- 33.Pelletier H., Sawaya M.R., Wolfle W., Wilson S.H., Kraut J. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase β. Biochemistry. 1996;35:12762–12777. doi: 10.1021/bi9529566. [DOI] [PubMed] [Google Scholar]
- 34.Blanca G., Shevelev I., Ramadan K., Villani G., Spadari S., Hübscher U., Maga G. Human DNA polymerase λ diverged in evolution from DNA polymerase β toward specific Mn++ dependence: A kinetic and thermodynamic study. Biochemistry. 2003;42:7467–7476. doi: 10.1021/bi034198m. [DOI] [PubMed] [Google Scholar]
- 35.Martin M.J., Garcia-Ortiz M.V., Esteban V., Blanco L. Ribonucleotides and manganese ions improve non-homologous end joining by human Polm No Title. EMBO J. 2013;41:2428–2436. doi: 10.1093/nar/gks1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank E.G., Woodgate R. Increased catalytic activity and altered fidelity of human DNA polymerase iota in the presence of manganese. J. Biol. Chem. 2007;282:24689–24696. doi: 10.1074/jbc.M702159200. [DOI] [PubMed] [Google Scholar]
- 37.Zafar M.K., Ketkar A., Lodeiro M.F., Cameron C.E., Eoff R.L. Kinetic analysis of human PrimPol DNA polymerase activity reveals a generally error-prone enzyme capable of accurately bypassing 7,8-dihydro-8-oxo-2′-deoxyguanosine. Biochemistry. 2014;53:6584–6594. doi: 10.1021/bi501024u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.García-Gómez S., Reyes A., Martínez-Jiménez M.I., Chocrón E.S., Mourón S., Terrados G., Powell C., Salido E., Méndez J., Holt I.J., et al. PrimPol, an Archaic Primase/Polymerase Operating in Human Cells. Mol. Cell. 2013;52:541–553. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Washington M.T., Johnson R.E., Prakash S., Prakash L. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J. Biol. Chem. 1999;274:36835–36838. doi: 10.1074/jbc.274.52.36835. [DOI] [PubMed] [Google Scholar]
- 40.Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., Wishart D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan T., Loria A., Zhong K. Probing of tertiary interactions in RNA: 2′-hydroxyl-base contacts between the RNase P RNA and pre-tRNA. Proc. Natl. Acad. Sci. USA. 1995;92:12510–12514. doi: 10.1073/pnas.92.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindqvist M., Sarkar M., Winqvist A., Rozners E., Strömberg R., Gräslund A. Optical spectroscopic study of the effects of a single deoxyribose substitution in a ribose backbone: Implications in RNA-RNA interaction. Biochemistry. 2000;39:1693–1701. doi: 10.1021/bi992055n. [DOI] [PubMed] [Google Scholar]
- 43.Fahlman R.P., Olejniczak M., Uhlenbeck O.C. Quantitative analysis of deoxynucleotide substitutions in the codon-anticodon helix. J. Mol. Biol. 2006;355:887–892. doi: 10.1016/j.jmb.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 44.Morin B., Whelan S.P.J. Sensitivity of the polymerase of vesicular stomatitis virus to 2′ substitutions in the template and nucleotide triphosphate during initiation and elongation. J. Biol. Chem. 2014;289:9961–9969. doi: 10.1074/jbc.M113.542761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D., Bushnell D.A., Westover K.D., Kaplan C.D., Kornberg R.D. Structural Basis of Transcription: Role of the Trigger Loop in Substrate Specificity and Catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brunelle J.L., Shaw J.J., Youngman E.M., Green R. Peptide release on the ribosome depends critically on the 2′ OH of the peptidyl tRNA substrate. RNA. 2008;14:1526–1531. doi: 10.1261/rna.1057908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cyert M.S., Philpott C.C. Regulation of cation balance in Saccharomyces cerevisiae. Genetics. 2013;193:677–713. doi: 10.1534/genetics.112.147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tottey S., Harvie D.R., Robinson N.J. Understanding how cells allocate metals using metal sensors and metallochaperones. Acc. Chem. Res. 2005;38:775–783. doi: 10.1021/ar0300118. [DOI] [PubMed] [Google Scholar]
- 49.Capdevila D.A., Edmonds K.A., Giedroc D.P. Metallochaperones and metalloregulation in bacteria. Essays Biochem. 2017;61:177–200. doi: 10.1042/EBC20160076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Culotta V.C., Klomp L.W.J., Strain J., Casareno R.L.B., Krems B., Gitlin J.D. The copper chaperone for superoxide dismutase. J. Biol. Chem. 1997;272:23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 51.Moira Glerum D., Shtanko A., Tzagoloff A. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 1996;271:14504–14509. doi: 10.1074/jbc.271.24.14504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.