Abstract
Objectives
The purpose of the current study was to evaluate the analytical performance of seven kits for detecting IgM/IgG antibodies against coronavirus (SARS-CoV-2) by using four chemiluminescence immunoassay systems.
Methods
Fifty patients diagnosed with SARS-CoV-2 infection and 130 controls without coronavirus infection from the General Hospital of Chongqing were enrolled in the current retrospective study. Four chemiluminescence immunoassay systems, including seven IgM/IgG antibody detection kits for SARS-CoV-2 (A_IgM, A_IgG, B_IgM, B_IgG, C_IgM, C_IgG and D_Ab), were employed to detect antibody concentrations. The chi-square test, the receiver operating characteristic (ROC) curve and Youden’s index were determined to verify the cut-off value of each detection system.
Results
The repeatability verification results of the A, B, C and D systems are all qualified. D_Ab performed best (92% sensitivity and 99.23% specificity), and B_IgM performed worse than the other systems. Except for the A_IgM and C_IgG systems, the optimal diagnostic thresholds and cut-off values of the other kits and their recommendations are inconsistent with each other. B_IgM had the worst AUC, and C_IgG had the best diagnostic accuracy. More importantly, the B_IgG system had the highest false-positive rate for testing patients with AIDS, tumours and pregnancies. The A_IgM system test showed the highest false-positive rates among elderly individuals over 90 years old. COVID-2019 IgM/IgG antibody test systems exhibit performance differences.
Conclusions
The Innodx Biotech Total Antibody serum diagnosis kit is the most reliable detection system for anti-SARS-CoV-2 antibodies, which can be used together with nucleic acid tests as an alternative method for SARS-CoV-2 detecting.
Keywords: COVID-19, SARS-CoV-2, antibody, chemiluminescence immunoassay, performance verification
Introduction
Coronavirus pneumonia (coronavirus disease 2019, COVID-19) is an acute respiratory infection caused by severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2).1 The epidemic of the disease has not ended since the winter of 2019, and it is still raging worldwide. SARS-CoV-2 is highly contagious through aerosols, droplets and contact.2 Generally, the incubation period of SARS-CoV-2 is three to seven days, but the longest incubation period can reach 14 days.3 It has caused more than 7,250,000 human infections and nearly 410,000 deaths worldwide as of the end of 9 June. Therefore, the early diagnosis of SARS-CoV-2 infection is crucial. Previous studies have shown that the SARS-CoV-2 antigen stimulates the immune system to produce an immune response and that specific IgM and IgG antibodies appear in the serum of patients after infection.4 The SARS-CoV-2-specific IgM and IgG antibody tests have been involved in the diagnosis criteria for suspected patients whose COVID-19 viral nucleic acid test appears false negative, according to the recently published guidelines of Novel Coronavirus Pneumonia Diagnosis and Treatment (Trial Version 7), which were advocated by the National Health Committee.5
Current popular detection methods for anti-SARS-CoV-2 antibodies include colloidal gold and chemiluminescence immunoassays.6 Chemiluminescence immunoassays are a laboratory technology that combines a luminescence system with an immune response. It not only uses the specificity of the immune response but also has the high sensitivity of the luminescence reaction and is widely used in immunoassays.7 Our laboratory currently has four automatic chemiluminescence immunoassay systems, A, B, C and D, of which the three detection systems A, B and C detect SARS-CoV-2-specific IgM and IgG antibodies, and the D system detects total IgM/IgG antibodies. The current investigation intends to evaluate the repeatability, clinical sensitivity and specificity of seven antibody detection kits for four detection systems, as well as the false-positive rate in special populations. Youden’s index verifies the best diagnostic threshold (cut-off value) of each detection system to understand the analytical detection performance of each system and ensure the detection results.
Material and methods
Sample collection
Fifty serum samples from patients with SARS-CoV-2 infection diagnosed in 26 January to 6 February 2020 and 130 serum samples from patients with other conditions, including 20 late-term pregnant women, 20 patients with solid tumours, 20 patients with AIDS, 21 patients over 90 years old and 49 normal controls, were enrolled from the Immunology Department of the Laboratory Department of Chongqing General Hospital (three hospitals) from late February to March 2020. Control populations are selected based on common false-positive populations (interfering factors, such as rheumatoid factor, heterophilic antibody, complement, acquired animal Ig antibody, lysozyme, etc.) reported in the daily work and literature reports. All patients with SARS-CoV-2 infection were confirmed by nucleic acid testing (NAT) and computed tomography (CT) scan. All collected serum specimens were inactivated in a water bath at 56°C for 1 h and then stored in a freezer at –80°C.8,9
Reagents and instruments
The automatic immunochemiluminescence analyser A was called detection system A (Bioscience Diagnostic Technology Co., Ltd). Reagents included the anti-coronavirus (SARS-CoV-2) IgM antibody detection kit (referred to as A_IgM, batch number: G202002415), and S/CO (sample cut-off value) ≥1.0 was designated positive. For the SARS-CoV-2 IgG antibody detection kit (referred to as A_IgG, batch number: G202002414), S/CO ≥ 1.0 was designated positive. The fully automatic immunochemiluminescence analyser B was called detection system B (Shenzhen New Industries Biomedical Engineering Co., Ltd). Reagents included the SARS-CoV-2 IgM antibody detection kit (referred to as B_IgM, batch number: 271200201), for which S/CO ≥ 1.0 AU/ml was designated positive. For the SARS-CoV-2 IgG antibody detection kit (referred to as B_IgG, batch number: 2722000101), S/CO ≥ 1.0 AU/ml was designated positive. The automatic immunochemiluminescence analyser C was called detection system C (Shenzhen YHLO Biotech Co., Ltd). Reagents included the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) IgM antibody detection kit (referred to as C_IgM, batch number: 20200206), for which S/CO ≥ 10 AU/ml was designated positive. For the SARS-CoV-2 IgG antibody detection kit (referred to as C_IgG, batch number: 20200202), S/CO ≥ 10 AU/ml was designated positive. The fully automatic immunochemiluminescence analyser D was called detection system D (Xiamen Innodx Biotech Co., Ltd). Reagents included the SARS-CoV-2 antibody detection kit (referred to as D_Ab, batch number: 20200309), for which S/CO ≥ 1.0 was designated positive.
Precision verification
Under the conditions of calibration and quality control of the detection systems, all of them are qualified, and the following experiments are carried out. The cut-off value is 1.0, 1.0, 1.10 AU/ml, 1.10 AU/ml, 10 AU/ml, 10 AU/ml and 1.0 in the A_IgM, A_IgG, B_IgM, B_IgG, C_IgM, C_IgG and D_Ab detection systems, respectively. Among 50 specimens of patients infected with SARS-CoV-2, one case of a weakly positive specimen with an S/CO value less than three times the cut-off value (level 1, L1) and one case with an S/CO value greater than three times the cut-off value (level 2, L2) were selected. Within-run precision was determined first. All detection systems were used to analyse the corresponding L1 and L2 specimens, conducting 20 consecutive tests. All tests were completed within one day, 20 S/CO values were observed, the results were judged and the standard deviation and coefficient of variation were calculated. The result was judged to be 100% in line, and the coefficient of variation was less than 10%. Between-run precision was examined second. The detection system analyses the corresponding L1 and L2 specimens, which were performed once per day, continuously detecting for 20 days, observing the S/CO value 20 times, judging the result and calculating the standard deviation and coefficient of variation. The result was judged to be 100% in line, and the coefficient of variation was less than 15%.
Statistical analysis
All statistical analyses were conducted using R software (http://www.R-project.org/).
Evaluations of sensitivity with the 95% CI, specificity with the 95% CI and false positives in specific populations were conducted separately. The ROC curve (R package pROC) and Youden’s index were used to calculate the optimal diagnostic threshold (cut-off value) of the detection system.
Results
Clinical characteristics of all samples are present in Table 1. Sex distribution between two groups shows no significant difference. Age in case group was younger than control groups. About 32% patients endure severe respiratory syndrome.
Table 1.
Clinical characteristic of all samples.
| Characteristics | Case (n = 50) | Control (n = 130) |
|---|---|---|
| Sex(male/female) | 22/28 | 70/60 |
| Age (mean ± SD) | 47.44 ± 17.0 | 61.9 ± 23.0 |
| Symptomatic | ||
| Severe | 16 (32%) | |
| Mild | 34 (68%) | |
| Complications (none/Yes) | ||
| Cough | 15/35 | |
| Fever | 15/35 | |
| Fatigue | 39/11 | |
| Dyspnoea | 40/10 |
To test the precision of each kit, we performed within-run and between-run detections. As seen from Table 2, the repeatability verification results of the A, B, C and D systems are all qualified. Among them, system D performed best and system B performed worst in the weak-positive specimens. More importantly, the B_IgM and B_IgG systems were nearly twice as precise as C_IgM and D_Ab.
Table 2.
Diagnosis precision within different kits.
| Kits | Within-run |
Between-run |
||
|---|---|---|---|---|
| L1 (CV%) | L2 (CV%) | L1 (CV%) | L2 (CV%) | |
| A_IgM | 2.9 ± 0.17 (5.71) | 357.4 ± 11.95 (3.34) | 2.85 ± 0.22 (7.72) | 349.9 ± 12.76 (3.64) |
| A_IgG | 1.51 ± 0.06 (4.19) | 205.3 ± 7.59 (3.7) | 1.66 ± 0.10 (6.02) | 208.6 ± 8.22 (3.94) |
| B_IgM | 1.32 ± 0.04 (3.18) | 3.74 ± 0.34 (8.98) | 1.45 ± 0.07 (4.83) | 3.88 ± 0.45 (11.6) |
| B_IgG | 1.94 ± 0.16 (8.04) | 22.75 ± 0.61 (2.7) | 2.11 ± 0.19 (9) | 21.09 ± 0.63 (3) |
| C_IgM | 32.7 ± 0.92 (2.8) | 163.9 ± 4.27 (2.6) | 35.7 ± 1.13 (3.12) | 160.5 ± 4.87 (3.03) |
| C_IgG | 16.11 ± 0.69 (4.25) | 117.43 ± 1.50 (1.28) | 17.0 ± 0.74 (4.35) | 115.1 ± 1.55 (1.35) |
| D_Ab | 2.74 ± 0.06 (2.15) | 23.05 ± 0.57 (2.46) | 2.88 ± 0.09 (3.13) | 23.67 ± 0.59 (2.49) |
L1: level 1; L2: level 2.
A total of 50 patients were considered to have COVID-19 because a viral nucleic acid test appeared positive, and the other 130 controls had negative viral nucleic acid results. Overall, 180 subjects were tested with the COVID-19-specific serological assay. The results showed varying sensitivity and specificity among different kits. D_Ab performed best (92% sensitivity and 99.23% specificity), and B_IgM performed worse than the others (Table 3).
Table 3.
Diagnosis sensitivity and specificity within different kits.
| Kits | Positive in case | Positive in control | Sensitivity (95%CI) | Specificity (95%CI) |
|---|---|---|---|---|
| A_IgM | 41 (9) | 8 (122) | 82.00% (69.20%,90.23%) | 93.85% (88.33%,96.85%) |
| A_IgG | 43 (7) | 4 (126) | 86.00% (73.81%,93.05%) | 96.92% (92.36%,98.80%) |
| B_IgM | 13 (37) | 8 (122) | 26.00% (15.87%,39.55%) | 93.85% (88.33%,96.85%) |
| B_IgG | 43 (7) | 28 (102) | 86.00% (73.81%,93.05%) | 78.46% (70.63%,84.66%) |
| C_IgM | 31 (19) | 3 (127) | 62.00% (48.15%,74.14%) | 97.69% (93.44%,99.21%) |
| C_IgG | 44 (6) | 3 (127) | 88.00% (76.20%,94.38%) | 97.69% (93.44%,99.21%) |
| D_Ab | 46 (4) | 1 (129) | 92.00% (81.16%,96.85%) | 99.23% (95.77%,99.86%) |
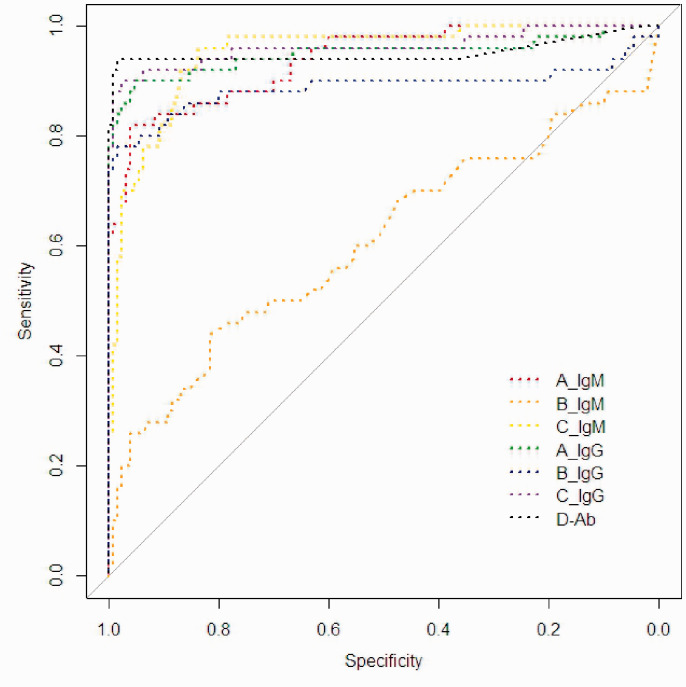
The ROC curve was depicted by using the original S/CO value (Figure 1). According to the ROC curve, we obtained the optimal operating point of the different kits (Table 3). It can be concluded that, except for the optimal operating thresholds of A_IgM and C_IgG, the optimal diagnostic thresholds of the other kits and the cut-off values from the recommendations are inconsistent with each other. The results showed that the AUC of D_Ab reached 0.95 and that Youden’s index was 0.93 (Table 4). The optimal cut-off value was 0.54, with sensitivity and specificity values of 99% and 94%, respectively. According to the optimal operating threshold, there were only three patients who had a negative result, and two controls had a positive result. Additionally, B_IgM had the worst AUC, and C_IgG had the best diagnostic accuracy.
Figure 1.
ROC curve for different kits.
Table 4.
Cut-off value and ROC-related parameters within different kits.
| Kit | Cut-off value | Optimal operating point | Specificity | Sensitivity | AUC | Youden’s index |
|---|---|---|---|---|---|---|
| A_IgM | 1 | 0.9 | 0.96 | 0.82 | 0.94 | 0.78 |
| A_IgG | 1 | 0.49 | 0.95 | 0.9 | 0.95 | 0.85 |
| B_IgM | 0.9 | 0.56 | 0.82 | 0.44 | 0.61 | 0.26 |
| B_IgG | 0.9 | 3.17 | 0.99 | 0.78 | 0.89 | 0.77 |
| C_IgM | 10 | 1.85 | 0.84 | 0.96 | 0.95 | 0.8 |
| C_IgG | 10 | 9.61 | 0.98 | 0.9 | 0.96 | 0.88 |
| D_Ab | 1 | 0.54 | 0.99 | 0.94 | 0.95 | 0.93 |
Considering that endogenous and exogenous factors exist in the process of antibody assays, subgroups of controls, including patients with acquired immune deficiency syndrome (AIDS), tumours, or pregnancies or those older than 90 years old, were involved in the current analysis. Each system had false-positive results in the selected subgroup of controls (Table 5). It is worth noting that the B_IgG system had the highest false-positive rate for testing patients with AIDS, tumours and pregnancies. The A_IgM system test showed the highest false-positive rates among elderly individuals over 90 years old.
Table 5.
The false-positive rate (%) in specific patients.
| Subgroup | A_IgM | A_IgG | B_IgM | B_IgG | C_IgM | C_IgG | D_Ab |
|---|---|---|---|---|---|---|---|
| HIVs (n = 20) | 5 | 0 | 5 | 55 | 5 | 5 | 5 |
| Tumours (n = 20) | 10 | 15 | 10 | 40 | 5 | 0 | 0 |
| Pregnant (n = 20) | 5 | 0 | 10 | 15 | 0 | 0 | 0 |
| Elder (≥90, n = 21) | 9.5 | 0 | 4.8 | 0 | 4.8 | 4.8 | 0 |
Discussion
SARS-CoV-2 belongs to the β genus and is the seventh most well-known coronavirus that infects humans. Its nucleocapsid protein (NP) stimulates the human immune system to cause chemical reactions. Specific IgM antibodies emerge on the seventh day of infection and appear to peak after 28 days. Specific IgG antibodies emerge around the 10th day of infection and reach a peak after 49 days, which can be maintained for a long time in the blood. The median time for total plasma antibodies appears on the 12th day after infection.10,11 In the current investigation, the average time of serum collection in all subjects was 13 days after diagnosis; therefore, specific IgM and IgG antibodies should already exist in the specimens.
With the published guidelines of the Novel Coronavirus Pneumonia Diagnosis and Treatment Program (Trial Version 7),5 the suspected cases are positive for serum SARS-CoV-2-specific IgM/IgG antibodies, are positive for SARS-CoV-2-specific IgG antibodies, or have SARS-CoV-2 IgG antibody concentrations that are four times greater in the recovery period than in the acute period, which can confirm the diagnosis of COVID-19.12,13 The diagnosis standard of COVID-19 is a changing situation. There is substantial market demand for SARS-CoV-2 antibody detection reagents worldwide. Manufacturers domestically produce antibody detection reagents that are used in the clinical laboratory. Previous investigations have shown that the clinical specificity and sensitivity of some anti-SARS-CoV-2 IgM antibodies are 96.2% and 70.24%, respectively. The clinical specificity and sensitivity of anti-SARS-CoV-2 IgG antibodies are 92.4% and 96.1%, respectively. Therefore, false-negative and false-positive results will appear, which will cause confusion in clinical judgment. Thus, the laboratory needs to pay close attention to the performance indicators of the reagents used.
Seven detection kits from four chemiluminescence systems were used in the current study. All of the kits have been permitted for use via the emergency approval of the China National Drug Administration or the EU CE sales and have been applied to clinical detection. According to the requirements of the People's Republic of China Health Industry Standard WS/T 505–2017 ‘Qualitative Measurement Performance Evaluation Guidelines’,13 the performance indicators of qualitative kits should focus on repeatability, clinical accuracy (including clinical sensitivity and specificity) and verification of the cut-off value. The results showed that the repeatability of all detection systems is in line with the manufacturer's statement, but the variance among them is relatively large. Specifically, the coefficients of variation regarding B-IgM and B-IgG are larger than those of the others.
According to the WS/T 494–2017 guideline, the sensitivity and specificity of qualitative items for different occasions are also regulated.14,15 In the use of preliminary screening tests, the sensitivity should be greater than 95%. In the case of diagnosis, both the sensitivity and specificity should be greater than 95%. In a confirmed diagnostic test, the specificity should be greater than 98%.12 According to the results of the current study, the clinical sensitivity and specificity of all detection systems do not meet the requirements of screening, diagnosis and confirmation of diagnosis experiments. Therefore, all detection systems cannot be used independently for the diagnosis of SARS-CoV-2 infections and need to be used together with nucleic acid tests and clinical symptoms.
Regarding the confounding factors influencing detection results, we divided controls into subgroups that included patients with AIDS, tumours, or pregnancy and older people over 90 years old.6,16,17 The results of the current investigation showed that B_IgM has the lowest sensitivity, indicating the possibility of more false negatives that can occur during a period of viral infection or in patients with low immunity.
B_IgG has the lowest specificity, which indicates a higher false-positive rate; this situation is prone to occur in special patients, such as those with AIDS, solid tumours, or pregnancy and elderly individuals.18 The reason for false positives may be due to some interfering substances (such as rheumatoid factor, which is homologous to the kit antibodies) present in the specimens. Simultaneously, according to the area under the ROC curve of each detection system, the diagnostic accuracy of B_IgM and B_IgG was also the worst, and the diagnostic accuracy of the other systems was better. In addition, according to the ROC curve and Youden’s index, the best diagnostic thresholds were exhibited by A-IgM and C-IgG, and those of the others were inconsistent with the manufacturer's declaration. The optimal thresholds of A_IgG, B_IgM, C_IgM and D_Ab are less than the cut-off value, indicating increased false-positive results. The optimal threshold of C_IgM is greater than the cut-off value, indicating additional false-negative cases.
Therefore, the laboratory should conduct the necessary performance evaluation of the selected anti-novel coronavirus antibody, carefully interpret the results of the anti-novel coronavirus antibody, perform necessary further testing requirements and reduce missed diagnoses and misdiagnoses.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The project was supported partly by grants from National Natural Science Foundation of China (81572089), and partly by grants from Yuzhong District Scientific Research Project (20180129).
Ethical approval: The study was approved by Chongqing General Hospital Ethics Committee. Informed consent was obtained from all individuals included in this study.
Guarantor: PL.
Contributorship: YFW, PL performed the sequencing and statistical analyses and article preparing. ZJL, KW, and TL were responsible for sample collection. YFW, PL devised such research and also revised article for publishing. YFW and PL gained ethical approval.
ORCID iD: Yafang Wan https://orcid.org/0000-0001-7786-1341
References
- 1.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (London, England) 2020; 395: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren SY, Wang WB, Hao YG, et al. Stability and infectivity of coronaviruses in inanimate environments. World J Clin Cases 2020; 8: 1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet (London, England) 2020; 395: 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L, Lu L, Cao W, et al. Hypothesis for potential pathogenesis of Sars-Cov-2 infection-a review of immune changes in patients with viral pneumonia. Emerg Microbes Infect 2020; 9: 727–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.P.R.China NHCoNovel corona virus pneumonia diagnosis and treatment guideline (trial version 7). [EB/OL], www.nhc.gov.cn/xcs/zhengcwj/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml (2020, accessed 24 September 2020).
- 6.Zhong L, Chuan J, Gong BO, et al. Detection of serum IgM and IgG for covid-19 diagnosis. Sci China Life Sci 2020; 63: 777–780. [DOI] [PMC free article] [PubMed]
- 7.Wang Q, Du Q, Guo B, et al. A method to prevent SARS-CoV-2 IgM false positives in gold immunochromatography and enzyme-linked immunosorbent assays. J Clin Microbiol 2020; 58: e00375–20. [DOI] [PMC free article] [PubMed]
- 8.Hu X, An T, Situ B, et al. Heat inactivation of serum interferes with the immunoanalysis of antibodies to SARS-CoV-2. J Clin Lab Anal 2020; 34: e23411. [DOI] [PMC free article] [PubMed]
- 9.Hu X, Zhang R, An T, et al. Impact of heat-inactivation on the detection of SARS-CoV-2 IgM and IgG antibody by ELISA. Clin Chim Acta 2020; 509: 288–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis 2020; ciaa344. [DOI] [PMC free article] [PubMed]
- 11.Tan W, Lu Y, Zhang J, et al. Viral kinetics and antibody responses in patients with covid-19. medRxiv. 2020. doi: 10.1101/2020.03.24.20042382. [DOI]
- 12.Pan Y, Li X, Yang G, et al. Serological immunochromatographic approach in diagnosis with sars-cov-2 infected covid-19 patients. J Infect 2020; 81: e28–e32. [DOI] [PMC free article] [PubMed]
- 13.Ting-Jun Z, Chang-Hai Z, Long-Qi X, et al. interpretation of detection of intestinal helminthes – the Kato-Katz method (ws/t 570-2017). Zhongguo Xue xi Chong Bing Fang Zhi za Zhi 2018; 30: 575–577. [DOI] [PubMed] [Google Scholar]
- 14.Schnurra C, Reiners N, Biemann R, et al. Comparison of the diagnostic sensitivity of SARS-CoV-2 nucleoprotein and glycoprotein-based antibody tests. J Clin Virol 2020; 129: 104544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Zhou L, Liu H, et al. Establishing a high sensitivity detection method for SARS-CoV-2 IgM/IgG and developing a clinical application of this method. Emerging Microbes Infect 2020; 9: 2020–2029. [DOI] [PMC free article] [PubMed]
- 16.Padoan A, Cosma C and Sciacovelli L. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin Chem Lab Med 2020; 58: 1081–1088. [DOI] [PubMed]
- 17.Li Z, Yi Y, Luo X, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol 2020; 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed]
- 18.Parazzini F, Bortolus R, Mauri PA, et al. Delivery in pregnant women infected with sars-cov-2: a fast review. Int J Gynaecol Obstet 2020; 150: 41–46. [DOI] [PMC free article] [PubMed]