Abstract
Intestinal microbiota dysbiosis has been described in inflammatory bowel disease (IBD), but data from China are limited. In this study, we performed molecular analysis of the fecal microbial community from 20 healthy Chinese subjects and 25 patients with Crohn’s disease (CD), and evaluated associations with bacterial and fungal compositions. Decreased richness and diversity of bacterial composition was observed in the CD group compared with healthy (H) subjects. Significant structural differences in bacterial (but not fungal) composition among healthy controls and CD patients were found. A reduction in Firmicutes and Actinobacteria abundance, and overrepresentation of Proteobacteria were observed in the CD patients compared with the H group. The Escherichia-Shigella genus was overrepresented in the CD group, whereas Faecalibacterium, Gemmiger, Bifidobacterium, Romboutsia, Ruminococcus, Roseburia, and Fusicatenibacter abundance were decreased in the CD group compared with H subjects. Differences in fungal microbiota between the H and CD groups were observed at the genus rather than at the phylum level. The Candida genus was overrepresented in the CD (active disease) group compared with the H group, whereas no difference between CD (remission) and H groups was observed. Aspergillus, unclassified_Sordariomycetes, and Penicillium genera had greater representation in the H subjects compared with the CD group. Bacterial and fungal intra- and inter-kingdom correlations were observed between the H and CD groups. Therefore, fecal bacterial and fungal microbiome communities differed considerably between H and CD patients, and between Chinese and Western populations. The role of gut microbiota in homeostasis and in gastrointestinal disorders should be investigated further.
Keywords: bacteria, Crohn’s disease, fungi, inflammatory bowel disease, intestinal microbiota
Introduction
Inflammatory bowel disease (IBD), generally comprising Crohn’s disease (CD) and ulcerative colitis (UC), is an inflammatory disease with a global prevalence. IBD is an important health problem in developed countries, with its incidence currently increasing in developing countries.1 The burden of IBD is rising globally. In Europe and North America, more than 2 million and 1.5 million people suffer from the disease, respectively.2 In China, the incidence of IBD is growing rapidly, with substantial variation in levels and disease trends in different regions, and is associated positively with gross domestic product.1,3,4
It is widely acknowledged that IBD is usually caused by an inappropriate immune reaction toward imbalanced gut microbiota in genetically predisposed patients under the influence of environmental factors.5 The intestinal microbiota, which comprises many microorganisms, including bacteria, fungi, viruses, and eukaryotic parasites, has been shown to be involved in many physiological functions, including maturation of the immune system, stimulation of the epithelial barrier, regulation of metabolism, and resistance to infection.6 Bacteria are the most abundant group in the gut microbiota; their number is approximately 1014 in the human body, and their function in intestinal inflammation is the most studied. Dysbiosis of gut bacteria is related strongly to the development of IBD.7 Previous studies have indicated that patients with IBD have decreased intestinal microbial diversity, reduced levels of beneficial bacteria, and increased proportions of harmful bacteria.8 In CD-associated dysbiosis, reduced Firmicutes and increased Proteobacteria numbers have been reported at the phylum level, while decreased Faecalibacterium prausnitzii, Erysipelotrichaceae, and Bifidobacteriaceae, and increased Fusobacterium and Escherichia numbers have been reported at the family, genus, and species levels.9–11 In UC-associated dysbiosis, decreased Prevotella, Coprococcus, F. prausnitzii, and Roseburia hominis,12,13 and increased Blautia and Veillonella numbers have been reported at the genus and species levels.13
Gut fungi make up only approximately 0.1% of the total gut microbiota, but the body of each fungal cell is approximately 100-fold larger than a typical bacterial cell.14 The role of gut fungal microbiota in IBD pathogenesis is attracting growing attention. In our previous research, we found that the fungal microbiota is markedly changed in IBD mice models, and that fungi-depleted mice exhibit aggravated acute dextran sulfate sodium (DSS)-induced colitis.15 Sokol et al. reported an increased Basidiomycota/Ascomycota ratio, overrepresentation of Candida albicans, and a reduced Saccharomyces cerevisiae proportion in patients with CD compared with healthy (H) subjects.16 Takayuki Imai et al. reported decreased Saccharomyces and Sarocladium and increased Candida numbers in patients with UC compared with H subjects.17 The Malassezia was reported to be associated with CD and can exacerbate colitis in mouse models.18
Many fungi and bacteria are in close relationship with each other,19,20 and studies relevant to the role of fungi in IBD patients are limited. Moreover, the recruitment domain family member 9 (CARD9) and nucleotide oligomerization domain 2 (NOD2) are important IBD-susceptible genes involved in immune responses to fungi and bacteria, respectively.5 Patients with IBD and with CARD9 or NOD2 mutations have an imbalanced microbiota, which can cause a more aggressive disease status.5
The intestinal microbiota is affected by many factors such as diet, age, ethnicity, and gender.21 Among these, age and diet represent major intrinsic and extrinsic factors that affect the composition of the resident intestinal microbiota.22 The incidence of IBD in the Western population reached a plateau in the twentieth century.23 Meanwhile, the incidence of this disease has been rising rapidly in China, the largest developing country in the world, over the past few decades.3,4,24 However, relevant knowledge about the dysbiosis of gut microbiota (e.g., bacteria and fungi) in Chinese IBD patients is limited.25–27 Therefore, in this study, we performed a molecular analysis of the bacterial and fungal microbiome of Chinese patients with CD and healthy Chinese subjects to explore the relative composition of the intestinal microbiota (bacteria and fungi) in the gut and their inter- and intra-kingdom interactions with intestinal inflammation. To avoid interference from the age factor, we studied only young adult subjects aged between 18 and 39 years (patients with CD and H subjects). Moreover, the bacterial and fungal composition in patients with CD in active disease (CD-act) and remission (CD-rem) stages were compared and the inter-kingdom interactions between bacteria and fungi were evaluated.
Material and methods
Study population
This study was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (2018-SR-154) and written informed consent was obtained before study enrolment. In total, 25 CD patients (19 in active stage and 6 in remission) and 20 healthy subjects (HS) were recruited from February 2019 to January 2020. All patients were diagnosed with CD according to established clinical, endoscopic, radiologic, and histologic criteria.28 None of the study participants had taken probiotics, prebiotics, antibiotics, anti-fungal agents, or colon-cleansing regimens for at least 8 weeks prior to enrolment.22 None of the subjects had a prior history of metabolic disease or gastroenterology surgery. Female subjects who were lactating or pregnant were excluded. CD patients received mesalazine, corticosteroids, or azathioprine before study enrolment. Control group was constituted by HS who had normal colonoscopy performance and did not have a history of intestinal disorders. Subjects characteristics are described in Table 1. The average CD Activity Index (CDAI) was evaluated when the patients were enrolled in the study.
Table 1.
Demographic and basic characteristics of CD patients and HS.
| HS | CD-act | CD-rem | |
|---|---|---|---|
| N | 20 | 19 | 6 |
| Female/male | 12/8 | 11/9 | 4/2 |
| Age (years) mean (range) | 30.3 (20–37) | 32.1 (19–39) | 33.4 (20–39) |
| CDAI mean (range) | 202.56 (167–304) | 59.56 (34–132) | |
| Montreal classification n (%) | |||
| A1 | 0 | 0 | |
| A2 | 19 | 6 | |
| A3 | 0 | 0 | |
| L1 | 6 | 1 | |
| L2 | 1 | 0 | |
| L3 | 12 | 5 | |
| L4 | 0 | 0 | |
| B1 | 6 | 6 | |
| B2 | 10 | 0 | |
| B3 | 3 | 0 | |
| p | 6 | 4 | |
| Current treatment n (%) | |||
| 5-aminosalicylic acid | 4 | 1 | |
| Corticosteroids | 1 | 2 | |
| Azathioprine | 4 | 3 | |
| None | 10 | 0 | |
CD, Crohn’s disease; ; CD-act, CD patients in active stage; CDAI, CD activity index; CD-rem, CD patients in remission; HS, healthy subjects.
Sample collection
Fresh fecal samples from patients with CD or healthy subjects were collected and were stored immediately in liquid nitrogen until further processing. CD patients with a CDAI > 220 were considered to be in an active disease stage, and CD patients with a CDAI < 150 were considered to be in clinical remission.
DNA isolation and library construction
DNA was extracted from the fecal samples using the FastDNA® SPIN Kit for Feces (MP Biomedicals, Santa Ana, CA, USA) according to the method described before.20 The extracted DNA was quantified on a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The 16S V3-V4 and ITS 1-2 represent the bacteria and fungi, respectively. The sequences for the universal primers of the V3–V4 region of 16S rRNA (341F&534R) (forward primer: 5′-NNNNNNNNCCTACGGGAGGCAGCAG-3′, reverse primer: 5′-NNNNNNNNATTACCGCGGCTGCTGG-3′) and internal transcribed spacer regions 1 and 2 (ITS1-2) (forward primer: 5′-NNNNNNNNCTTGGTCATTTAGAGGAAGTAA-3′, reverse primer: 5′-NNNNNNNNGCTGCGTTCTTCATCGATGC-3′) were amplified for sequence analysis as previously described.15,20 The NNNNNNNN stretches represent the sample-unique 8-bp barcode that can tag each polymerase chain reaction (PCR) amplicon. PCR was carried out using the HotStarTaq® Plus DNA Polymerase (Qiagen, Valencia, CA, USA) with a reaction volume of 50 µl containing 100–200 ng DNA template, 1 µl HotStar DNA Polymerase, 1 µl dNTP, 5 µl 10 × Buffer, 2.5 µl each primer (10 M), and ddH2O (to 50 µl). Amplifications were performed under the following conditions: initial denaturation at 95°C for 5 min, followed by 28 cycles (for 16S rDNA) and 38 cycles (for ITS1-2) of denaturation at 95°C for 15 s, primer annealing at 55°C for 15 sec and extension at 72°C for 60 sec, followed by final elongation at 72°C for 10 min. The cDNA products were checked by agarose [1.5% (w/v)] gel electrophoresis in 0.5 mg/ml ethidium bromide, and purified with the Qiaquick gel extraction kit (Qiagen). Before sequencing, the DNA concentration of each PCR product was determined using a Qubit® 2.0 Green double-stranded DNA assay and it was quality controlled using a bioanalyzer (Agilent 2100, Santa Clara, CA, USA). Depending on coverage needs, all libraries can be pooled for one run. The amplicons from each reaction mixture were pooled in equimolar ratios based on their concentration. DNA sequencing with specific tags were performed on the Illumina MiSeq system (Illumina MiSeq, San Diego, CA, USA), according to the manufacturer’s instructions.
Library construction, quantification, and sequencing processing
The AMPure XP beads were used to purify the free primers and primer dimer species in the amplicon product. the samples were soon shipped to Sangon BioTech (Shanghai) for library construction using a universal lighting adapter and index. The DNA concentration of each PCR product was determined by Qubit® 2.0 green double-stranded DNA analysis, and quality control was performed using a bioanalyzer (Agilent 2100). After all of the libraries achieved a fast run, the amplification products in each reaction mixture were pooled together in equimolar ratio according to their concentration. The Illumina MiSeq system (Illumina MiSeq) was used for sequencing according to the manufacturer’s instructions.
After assembling the short Illumina readings, deleting or removing inappropriate sequences, merging all the same sequences into one sequence, comparing the sequences according to the customized reference database, deleting all indexes and adaptors, we submitted the valid 16s RNA and ITS 1-2 sequences of each sample to the Ribosomal Database Project (RDP, version 16) classifier and Unite (version 8.0), respectively, to identify the bacterial and fungal sequences. Operational taxonomic units (OTUs) were defined using a 97% similarity cut-off value. OTUs present in 50% or more of the fecal samples in CD (or H groups) were identified as core OTUs. Partial least-squares discriminant analysis (PLS-DA) scores plot based on the core OTUs was generated with Simca-P version 12 (Umetrics), and heat maps based on core OTUs (with the specific taxa are listed) was performed using Multi-Experiment Viewer (MeV) software to visualize and cluster the Microbiota community into different groups. Species richness and diversity statistics including Chao1, ACE, Simpson, and Shannon indexes were calculated using the mothur website; the modified pipeline is described on the website (https://mothur.org/). In addition, all valid bacterial sequences without primers were submitted for downstream analysis
Statistical analyses
As the majority of the datasets did not meet the assumptions of normal distribution, we estimated the data by Kruskal–Wallis one-way analysis of variance (ANOVA) using the Statistical Package for Social Sciences version 23.0 (SPSS Inc., Chicago, IL, USA) to compare median values of microbiota data between CD-act, CD-rem, and H groups. Spearman correlation analysis was used to analyze the correlation between intestinal microbiota (bacteria and fungi) composition. Unless otherwise indicated, data were expressed as the mean ± standard deviation (SD); p < 0.05 was considered to be a statistically significant difference.
Results
Bacterial microbiota diversity in H subjects and CD patients
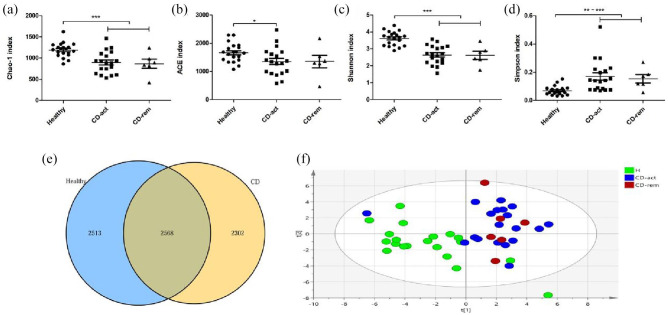
For each sample, a rarefaction curve was obtained with the observed number of bacterial OTUs on sequence counts at different sequencing depths. As shown in Supplemental Figure S1, all rarefaction curves were saturated. After comparing the four different indicators (Chao1, ACE, Shannon, and Simpson indices), we observed a decline in the gut microbiota biodiversity (alpha diversity) in samples from patients with CD compared with H subjects. The Chao1, ACE, and Shannon indices of H subjects were significantly higher than those of the CD (CD-act and/or CD-rem) groups, whereas the Simpson index of H subjects was significantly lower than that of CD patients, thus reflecting a decreased richness and diversity of bacterial composition in the CD group compared with the H subjects (Figure 1A–D). We obtained 2,274,075 trimmed sequences (~50,535 sequences/sample), and defined 5081 OTUs using a 97% similarity cut-off value in H subjects and 4870 OTUs in the CD group to profile the overall structural changes of gut fungi as partial least squares-discriminant analysis (PLS-DA) plots (Figure 1E–F) and heat maps (Figure 2). The PLS-DA plots and heat maps exhibit a remarkable difference in bacterial composition between H subjects and CD patients.
Figure 1.
Comparative analyses of the bacterial microbial communities of healthy controls and CD patients. Four different indexes were used to measure alpha diversity: Chao1 (A), ACE (B), Shannon (C), and Simpson (D) were compared between the healthy controls, CD-act and CD-rem patients. The bacterial OTU attribution in healthy and CD patients is illustrated. (E) The blue circle indicates the OTUs present in healthy samples, and the yellow circle indicates the OTUs present in the CD patients; overlap indicates the OTU shared by samples from both groups. PLS-DA scores plot based on the relative abundance of bacterial OTUs (97% similarity level) in H, CD-act, and CD-rem groups were analyzed.
*p < 0.05, **p < 0.01, ***p < 0.001.
CD, Crohn’s disease; CD-act, CD patients in active stage; CD-rem, CD patients in remission; H, healthy subjects; OTU, operational taxonomic unit; PLS-DA, partial least-squares discriminant analysis.
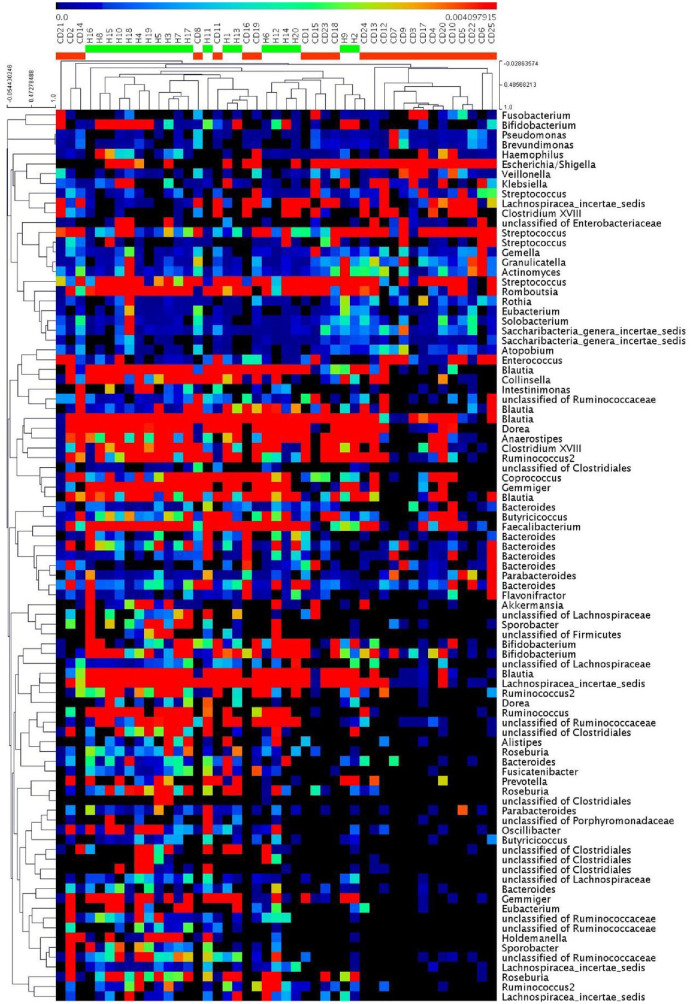
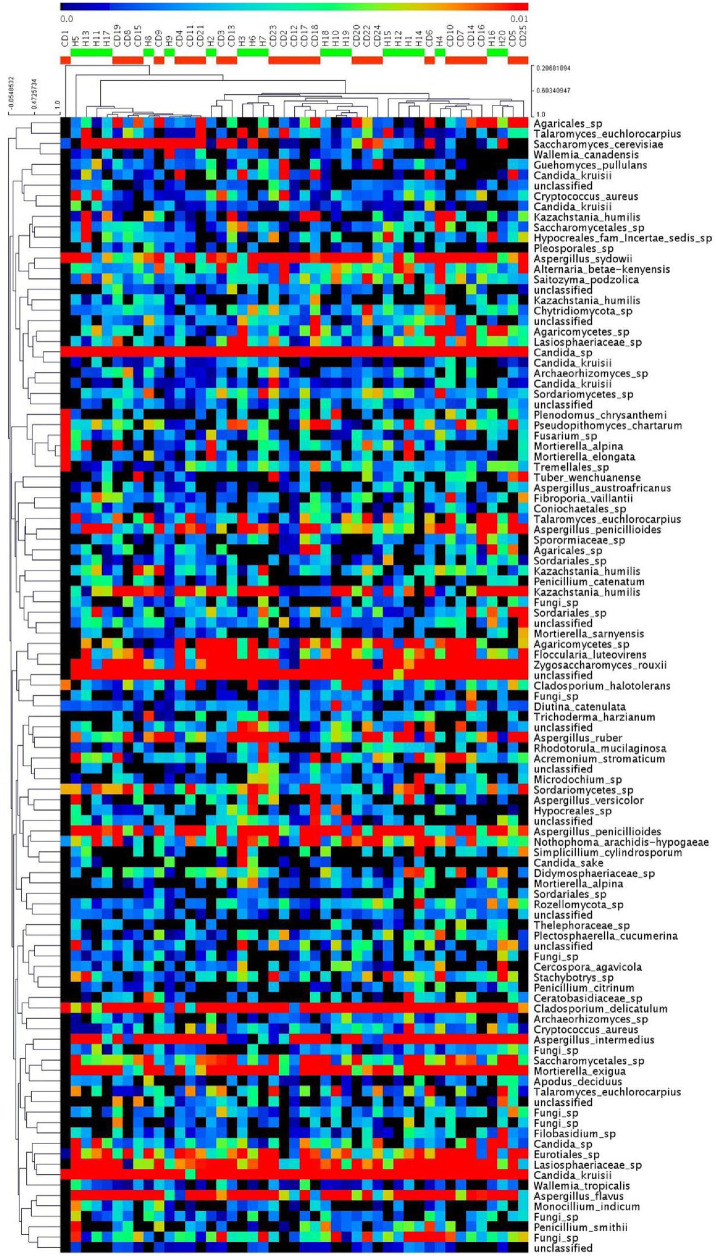
Figure 2.
Heat map of the core OTUs of bacterial communities inferred from fecal 16S rRNA V3–V4 region sequences, with each sample individually. The colored squares in each row indicate the relative abundance of the OTU among H and CD subjects.
CD, Crohn’s disease; H, healthy subjects; OTU, operational taxonomic unit.
Bacterial microbiota composition in H subjects and CD patients
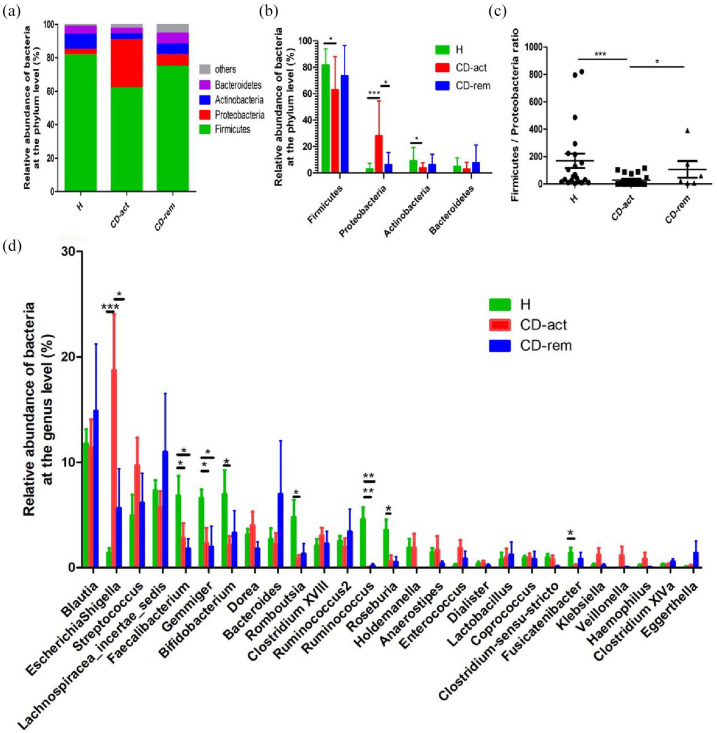
Four major bacterial phyla were observed in both H subjects and CD patients: Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes. Firmicutes and Proteobacteria were the most abundant in all three subject groups, followed by Actinobacteria and Bacteroidetes (Figure 3A). Importantly, up to 0.01 ± 0.035% of the sequences were assigned to unidentified bacteria in H subjects and 0.07 ± 0.033% in CD patients, thus reflecting the relatively sufficient annotation of the current bacterial database. In this study, a reduction of Firmicutes and Actinobacteria numbers and an overrepresentation of Proteobacteria were observed in CD patients compared with the H group, while Bacteroidetes did not show a significant difference between the groups (Figure 3A–B). In addition, we compared the ratio of Firmicutes with that of Proteobacteria, Actinobacteria, and Bacteroidetes, and found increased ratios in H subjects compared with the CD-act and/or CD-rem groups (Figure 3C and Supplemental Figure S2A and S2B). A total of 27 major genera, namely Blautia, Escherichia-Shigella, Streptococcus, Lachnospiracea incertae sedis, Faecalibacterium, Gemmiger, Bifidobacterium, Dorea, Bacteroides, Romboutsia, Clostridium, Ruminococcus2, Ruminococcus, Roseburia, Holdemanella, Anaerostipes, Enterococcus, Dialister, Lactobacillus, Coprococcus, Clostridium sensu stricto, Fusicatenibacter, Klebsiella, Veillonella, Haemophilus, Clostridium XlVa, and Eggerthella, were present in both H subjects and CD patients (Figure 3D). The Escherichia-Shigella genus was overrepresented in the CD group, while the Faecalibacterium, Gemmiger, Bifidobacterium, Romboutsia, Ruminococcus, Roseburia, and Fusicatenibacter genera numbers were decreased in the CD group compared with H subjects (Figure 3D).
Figure 3.
The taxonomic composition of the bacterial community of healthy controls and CD patients at the phylum and genus level. (A, B) Phylum level. (C) Firmicutes/Proteobacteria ratio. (D) Genus level.
*p < 0.05, **p < 0.01, ***p < 0.001.
CD, Crohn’s disease; CD-act, CD patients in active stage; CD-rem, CD patients in remission; H, healthy subjects.
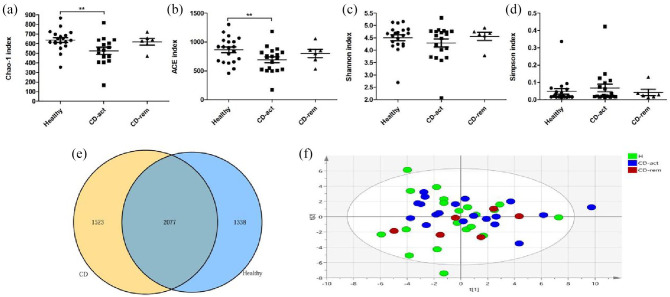
Fungal microbiota diversity in H subjects and CD patients
The Chao1 and ACE indices showed a decreasing trend in CD-act patients compared with the H group, reflecting a decreased fungal richness in the former, while both indices in CD-rem patients also exhibited a decreasing trend, but did not reveal a statistically significant difference compared with the H group, probably due to the limited sample size (Figure 4A–D). On the other hand, the Shannon and Simpson indices in H, CD-act, and CD-rem groups did not show significant differences. We obtained 1,684,238 trimmed sequences (~37,427 sequences/sample) and defined 4415 OTUs using a 97% similarity cut-off value in H subjects, and 4600 OTUs in CD group to profile the overall structural changes of gut fungi as PLS-DA plots (Figure 4E–F) and heat maps (Figure 5). However, the PLS-DA plots and heat maps do not exhibit a remarkable difference in fungal composition between H subjects and CD patients (Figures 4F and 5).
Figure 4.
Comparative analyses of the fungal microbial communities of healthy controls and CD patients. Four different indexes were used to measure alpha diversity: Chao1 (A), ACE (B), Shannon (C), and Simpson (D) were compared between healthy controls, CD-act and CD-rem patients. (E) The fungal OTU attribution in healthy and CD patients are illustrated. The blue circle indicates the OTUs present in healthy samples, and the yellow circle indicates the OTUs present in the CD patients; overlap indicates the OTU shared by samples from both groups. PLS-DA scores plot based on the relative abundance of fungal OTUs (97% similarity level) in H, CD-act, and CD-rem groups were analyzed.
*p < 0.05, **p < 0.01, ***p < 0.001.
CD, Crohn’s disease; CD-act, CD patients in active stage; CD-rem, CD patients in remission; H, healthy subjects; OTU, operational taxonomic unit; PLS-DA, partial least-squares discriminant analysis.
Figure 5.
Heat map of the core OTUs of fungal communities inferred from fecal ITS 1-2 region sequences, with each sample individually. The colored squares in each row indicate the relative abundance of the OTU among the H and CD subjects.
CD, Crohn’s disease; H, healthy subjects; OTU, operational taxonomic unit.
Fungal microbiota composition in H subjects and CD patients
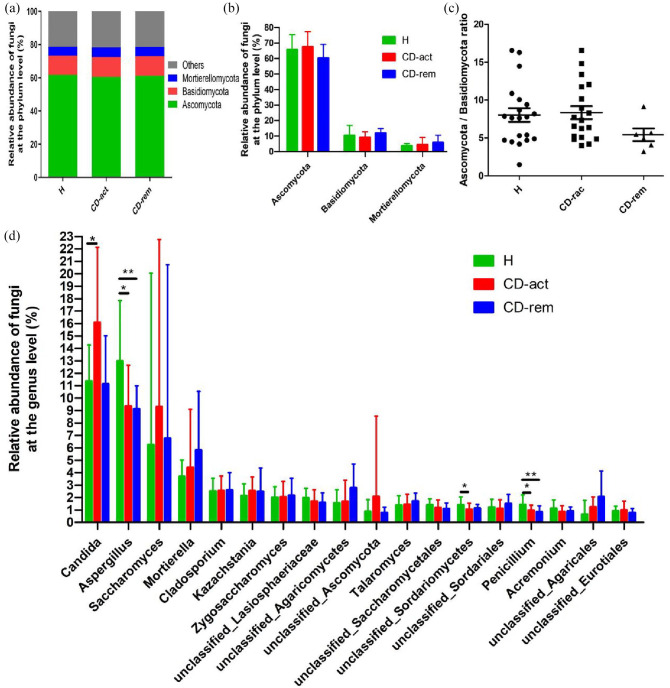
A total of 12 fungal phyla, namely Ascomycota, Basidiomycota, Mortierellomycota, Rozellomycota, Chytridiomycota, Mucoromycota, Cercozoa, Glomeromycota, Neocallimastigomycota, Kickxellomycota, Blastocladiomycota, and Olpidiomycota, were identified in all samples. Three major fungal phyla were observed in both H subjects and CD patients: Ascomycota, Basidiomycota, and Mortierellomycota (Figure 6A). Ascomycota was the most abundant phylum in all three subject groups, followed by Basidiomycota and Mortierellomycota (Figure 6A–B). Importantly, 20.09 ± 8.24% of the sequences were assigned to unidentified fungi in H subjects and 19.63 ± 6.90% in CD patients, thus reflecting the poor annotation of the current fungi database (Figure 6A). In this study, we did not find significant differences in the three major fungal phyla between the three groups. Moreover, the ratio of Ascomycota to either Basidiomycota or Mortierellomycota was not significantly different between the H, CD-act, and CD-rem groups (Figure 6C and Supplemental Figure S3). A total of 18 major genera were identified, namely Candida, Aspergillus, Saccharomyces, Mortierella, Cladosporium, Kazachstania, Zygosaccharomyces, unclassified_Lasiosphaeriaceae, unclassified_Agaricomycetes, unclassified_Ascomycota, Talaromyces, unclassified_Saccharomycetales, unclassified_Sordariomycetes, unclassified_Sordariales, Penicillium, Acremonium, unclassified_Agaricales, and unclassified_Eurotiales. Among these, the Candida genus was overrepresented in the CD-act group compared with the H group, while the difference between CD-rem and H groups was not significant (Figure 6E). The Aspergillus, unclassified_Sordariomycetes, and Penicillium genera had greater representation in the H subjects compared with the CD group (Figure 6E).
Figure 6.
The taxonomic composition of the fungal community of healthy controls and CD patients at the phylum and genus level. (A, B) Phylum level. (C) Ascomycota/Basidiomycota ratio. (D) Genus level.
*p < 0.05, **p < 0.01, ***p < 0.001.
CD, Crohn’s disease; CD-act, CD patients in active stage; CD-rem, CD patients in remission; H, healthy subjects.
Correlation between bacteria and fungi in the gut of CD patients
We observed correlations between the major bacteria and fungi at the genus level. Interestingly, most of the correlations were not strong and did not show statistical significance (Supplemental Figure S4). We next assessed whether bacterial and fungi microbiota genera that showed evident differences between the H and CD groups were correlated with each other. To address this, the Spearman’s correlation within the abundance of bacterial and fungal taxa was calculated and is presented in Tables 2 and 3.
Table 2.
Distance correlation plots of the relative abundance of fungi and bacteria genera.
| Escherichia Shigella | Faecalibacterium | Gemmiger | Bifido bacterium | Romboutsia | Ruminococcus | Roseburia | Fusicatenibacter | Candida | Aspergillus | Unclassified Sordariomycetes | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Faecali-bacterium | −0.214 | ||||||||||
| Gemmiger | −0.547** | 0.400* | |||||||||
| Bifido-bacterium | −0.212 | 0.402* | 0.284 | ||||||||
| Romboutsia | −0.347 | 0.377 | 0.05 | 0.095 | |||||||
| Ruminococcus | −0.262 | 0.269 | 0.369 | −0.025 | −0.036 | ||||||
| Roseburia | −0.325 | 0.062 | 0.415* | −0.068 | 0.316 | 0.652** | |||||
| Fusicateni bacter | −0.552** | 0.403* | 0.649** | 0.18 | 0.224 | 0.378 | 0.441* | ||||
| Candida | −0.038 | −0.264 | −0.101 | −0.421* | −0.191 | −0.156 | −0.076 | −0.06 | |||
| Aspergillus | 0.102 | 0.078 | −0.11 | −0.109 | 0.254 | −0.298 | −0.127 | −0.175 | 0.209 | ||
| Unclassified Sordariomy cetes | −0.017 | 0.093 | 0.363 | 0.386 | 0.008 | 0.067 | 0.211 | 0.408* | −0.028 | 0.024 | |
| Penicillium | 0.118 | −0.141 | −0.078 | −0.224 | 0.077 | −0.026 | 0.094 | 0.185 | 0.358 | 0.344 | 0.048 |
p < 0.05; **p < 0.01.
CD, Crohn’s disease.
Table 3.
Correlation between bacteria and fungi in healthy subjects. Distance correlation plots of the relative abundance of fungi and bacteria genera.
| EscherichiaShigella | Faecalibacterium | Gemmiger | Bifidobacterium | Romboutsia | Ruminococcus | Roseburia | Fusicatenibacter | Candida | Aspergillus | unclassified_Sordariomycetes | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Faecalibacterium | −0.358 | ||||||||||
| Gemmiger | 0.173 | −0.074 | |||||||||
| Bifidobacterium | 0.099 | −0.317 | −0.192 | ||||||||
| Romboutsia | 0.377 | −0.459* | 0.474* | 0.015 | |||||||
| Ruminococcus | −0.133 | 0.589** | 0.25 | −0.206 | −0.186 | ||||||
| Roseburia | −0.463* | 0.598** | 0.011 | −0.436 | −0.34 | 0.284 | |||||
| Fusicatenibacter | −0.098 | 0.233 | −0.331 | −0.027 | −0.214 | 0.326 | 0.325 | ||||
| Candida | −0.397 | 0.301 | −0.098 | −0.385 | −0.194 | 0.464* | 0.086 | 0.239 | |||
| Aspergillus | −0.259 | 0.341 | −0.179 | −0.359 | −0.263 | 0.44 | 0.162 | −0.002 | 0.614** | ||
| unclassified_Sordariomycetes | −0.047 | 0.022 | 0.032 | 0.117 | −0.1 | 0.554* | 0.115 | 0.304 | 0.267 | 0.297 | |
| Penicillium | −0.035 | 0.144 | −0.373 | −0.052 | −0.569** | 0.029 | 0.143 | 0.271 | 0.26 | 0.254 | 0.139 |
p < 0.05; **p < 0.01.
We observed quite different correlations between bacterial and fungal compositions in H subjects and CD patients. However, the number and strength of correlations were similar in both groups (Supplemental Figure S4). Interestingly, in patients with CD, there was a negative correlation between the Escherichia-Shigella genus and Gemmiger and Fusicatenibacter genera; a positive correlation between the abundance of Faecalibacterium and Gemmiger, Bifidobacterium, and Fusicatenibacter; a positive correlation between Gemmiger and Roseburia as well as Fusicatenibacter; a negative correlation between the abundance of Bifidobacterium and Candida; and a positive correlation between Roseburia and Ruminococcus as well as Fusicatenibacter (Table 2). In addition, Fusicatenibacter abundance was positively correlated with that of a genus belonging to unclassified_Sordariomycetes (Table 2).
On the other hand, in the H group, we found that Escherichia-Shigella showed a negative correlation with Roseburia; Faecalibacterium showed a negative correlation with Romboutsia, but a positive correlation with Ruminococcus and Roseburia; and Romboutsia showed a positive correlation with Gemmiger, but a negative correlation with Penicillium (Table 3). Ruminococcus showed a positive correlation with unclassified_Sordariomycetes; Candida showed a positive correlation with Ruminococcus and Aspergillus (Table 3). Taken together, these results suggest a complex relationship between the bacteria and fungi present in the gut microbiota and that specific alterations are present in CD and H subjects.
Discussion
The intestinal microbiota plays pivotal roles in the pathology of gastrointestinal tract inflammation. Bacteria and fungi coexist in the gut and may interact with each other: antibiotic treatment in mice leads to an increase in fungal numbers, suggesting that a balance exists between bacterial and fungal microbiota. The role of intestinal microbiota in IBD has been studied to some extent in the Western population. However, the Chinese population has a different genetic background and environmental influences (such as diet) and might thus harbor a distinct fecal microbiome (bacteria and fungi) composition. In this study, we investigated the fecal bacterial and fungal microbiomes in Chinese patients with CD and healthy controls using the high-throughput sequencing method, since most intestinal bacteria and fungi cannot be identified with the limited culture-based methods. We also analyzed the correlations between the bacterial and fungal microbiomes in CD and H groups.
We first investigated bacterial dysbiosis between the fecal samples, and confirmed the decreased richness (Chao1 and ACE indices) and diversity (Shannon and Simpson indices), and the altered bacterial community structure in the CD groups, which was consistent with the results of previous studies. Furthermore, we found remarkable structural differences in the bacterial communities between CD and H groups based on the PLS-DA analysis and the heat map based on the core OTU. We further explored the differences at the phylum and genus levels and found decreased numbers of Firmicutes and Actinobacteria and an increased number of Proteobacteria at the phylum level in the CD group compared with those in the H group. In addition, the ratios of Firmicutes to Proteobacteria, Actinobacteria, and Bacteroidetes were decreased significantly in CD-act patients compared with those in the H group. We next compared the bacterial differences at the genus level and identified 27 major genera (⩾ 0.01) in both groups. Interestingly, we found that the Escherichia-Shigella genus was overrepresented in the CD group. We also identified a reduced number of Bifidobacterium, Faecalibacterium, Gemmiger, Bifidobacterium, Romboutsia, Ruminococcus, Roseburia, and Fusicatenibacter in the CD group compared with that in the H group. However, in a previous Chinese study, Chen et al.25 found that Streptococcus and Enterococcus were overrepresented, while Coprococcus, Roseburia, Faecalibacterium, and Ruminococcus were less represented in fecal samples from patients with CD compared with those from healthy controls. However, they reported that, for unknown reasons, some microbial DNA could not be amplified from fecal samples using the barcoded 454 primers, and no single genus exceeded 1% of the total bacteria. The differences between their findings and ours could be due to the different DNA extraction methods, high-throughput sequencing methods, and the relatively incomplete database that they used 4 years ago. In another Chinese study, Ma et al. reported the outgrowth of Bacteroidetes and reduced Firmicutes in patients with CD compared with that in the control group.27 In their study, among the 10 most abundant genera found in the CD group (>1% of the total bacteria), the levels of Escherichia and Prevotella were significantly increased and those of Haemophilus were remarkably decreased in the feces of the CD group compared with that of the control. Although gut bacteria can be affected easily by diet, environment, age, and disease status, some of their findings are consistent with ours.
In addition to studies from Eastern countries,13,25,27 previous studies conducted in Western countries also showed a high prevalence of Escherichia coli isolated from biopsy or fecal specimens of patients with IBD,29,30 which suggests that E. coli plays a role in the initiation and promotion of the inflammatory processes in CD. This is in agreement with our findings of Escherichia-Shigella’s overrepresentation in the CD group. Meanwhile, the genus Bifidobacterium is known to represent typical gut microorganisms with presumed beneficial effects on human health, and treatment with these bacteria can improve the disease condition. Andoh et al. have also reported a reduction in Bifidobacterium numbers in patients with active CD, which is consistent with our findings.31 However, in relation to the depletion of beneficial bacteria in IBD, short-chain fatty acid (SCFA)-producing bacteria should be mentioned. SCFAs are small organic acids that can be produced by fermentation of undigested and unabsorbed components of food in the gut by several gut microorganisms32. SCFAs can provide energy to the colonic enterocytes and protect mucosal integrity by increasing mucus secretion and sealing the mucosa against bacterial adhesins.32 The number of SCFA-producing bacteria is usually reduced in IBD patients.7 In this study, we observed that the numbers of F. prausnitzii, Gemmiger, and Roseburia, three SCFA-producing bacteria,7,33 were decreased in the CD group compared with the H group, which could explain the pathogenesis of IBD. Lloyd-Price et al. reported the depletion of Roseburia hominis in patients with CD.34 However, the exact Roseburia species depleted in the patients with CD in our study is unknown.
The presence of Romboutsia – a recently identified bacterial genus commonly residing in the human gut – is associated with a healthy status of the bacterial population.33 Mangifesta et al. reported that the Romboutsia genus was depleted in cancerous mucosa as well as in adenomatous polyps,33 which indicated that these bacteria could represent novel microbial biomarkers associated with early tumor generation. In this study, we also found a reduction in the prevalence of the Romboutsia genus in patients with CD. Meanwhile, the function of the Ruminococcus genus is controversial. Franzosa et al.7 reported that Ruminococcus obeum numbers were reduced in patients with IBD; however, the Ruminococcus gnavus clade and Ruminococcus torques were reported to be overrepresented.35,36 Owing to the limitations of the current bacterial database, we could not annotate the bacteria at the species level.
There is very little research on the relationship between Fusicatenibacter and intestinal inflammation. To the best of our knowledge, only one study has reported that Fusicatenibacter saccharivorans numbers decreased with UC activity, and that it could suppress murine chemical colitis.17 Our study is the first to report the depletion of Fusicatenibacter in patients with CD. We then analyzed the fecal fungal microbiome composition. Contrary to bacterial communities, we found decreased fungal richness (Chao1 and ACE indices) only in CD-act patients compared with controls, while fungal diversity (Shannon and Simpson indices) did not show significant differences between both groups. Moreover, the differences in fungal community structure were not as significant as those in the bacterial community, as shown in PLS-DA and the heat-map based on the core OTU. We further explored the differences in the fungal community at the phylum and genus levels and found that three major phyla (Ascomycota, Basidiomycota, and Mortierellomycota) were dominant in the human gut. However, we did not find statistical differences in the three phyla among the H, CD-act, and CD-rem groups. In addition, we did not find significant differences in the ratio of Ascomycota to either Basidiomycota or Mortierellomycota among the three groups. We further investigated differences in fungal composition at the genus level, and found the Candida genus to be overrepresented in the CD-act (but not in the CD-rem) group, which was consistent with a Japanese study,37 but not with a Western study of a patient with CD.16
Candida and Aspergillus were the two most abundant genera identified in the gut in our study; however, in the Western population study mentioned before, Saccharomyces and Debaryomyces were the two most abundant genera.16 This may result from basic differences in fungal composition between the Eastern and Western populations. Mukhopadhya et al. reported an increase in the levels of Basidiomycota and a reduction in the levels of Ascomycota in de novo pediatric patients with IBD compared with those in the control.38 However, this latter study amplified fungal PCR products from only 8 out of 37 subjects, which could be due to inappropriate DNA extraction38; therefore, the results should be treated with caution. In our study, Candida numbers were clearly increased in the CD-act patients compared with those in the H group; additionally, the Aspergillus, unclassified_Sordariomycetes, and Penicillium genera numbers were reduced in the CD group compared with those in the H group. Candida was the most abundant genus in the human gut in this study, which is consistent with previous reports.5 This genus has been implicated in IBD pathogenesis. Candida tropicalis and Candida albicans have been demonstrated to initiate intestinal inflammation in IBD.16,19 Within the Candida genus, the most abundant member in healthy individuals and IBD patients is C. albicans, which has the greatest clinical relevance in humans.39
Aspergillus spores are ubiquitous and many Aspergillus species, such as Aspergillus flavus, Aspergillus fumigatus, and Aspergillus niger, can cause major invasive diseases.40 The gastrointestinal tract may represent a portal of entry for Aspergillus species in immunocompromised patients.40 Previous studies reported that specific Aspergillus species (e.g. Aspergillus amstelodami and Aspergillus fumigatus) could exacerbate colitis.18,41 Therefore, it is interesting that the Aspergillus genus, which has been reported to be harmful to the human body, was depleted in the gut of patients with CD. Penicillium numbers were also reduced in the gut of patients with CD, which was in concordance with a Western study.16 However, in a Japanese study, Penicillium species were not detected in samples of a healthy Japanese population.37 Aspergillus and Penicillium are most likely environmental or food-borne fungi that do not inhabit the gastrointestinal tract.42 The different results in our observations could be due to different dietary habits among the different countries. In addition, the Aspergillus and Penicillium genera belong to the Sordariomycetes class, and other unclassified genera belonging to this class also showed decreased numbers in the gut of patients with CD; however, the exact genera are not clear because of the limited fungal database. Growing data suggest that changes in gut fungi may be associated with the pathogenesis of IBD. Gut commensal fungi act synergistically with other members of the fungal and/or bacterial microbiota; however, their role in IBD is unclear.43 This may be due to the large number of intestinal microbiota and the complex interactions between the microorganisms. We have illustrated the correlations of bacteria and fungi that show statistical differences in Tables 2 and 3. Strikingly, these correlations in the CD and H groups were dramatically different, while the number and intensity of correlations between fungi and bacteria were similar between the groups. To investigate the global intra-kingdom and inter-kingdom equilibrium in patients with CD and H controls, we used heat-maps to illustrate the correlations at the genus level involving both bacteria and fungi (Supplemental Figure S4). However, most of the correlations were neither very strong nor significant but were dramatically different between the CD and H groups, which may be due to the different intestinal inflammatory conditions in the gut. Moreover, the correlations were complex and most of them were only moderate, which may be due to the existence of too many influencing factors in the gut, a considerably large population of gut microbiota, and intricate connections among them. Some correlations in patients with CD were very interesting and may provide a theoretical basis for future microecological treatment. For example, Escherichia-Shigella and Candida are conditioned pathogens that can aggravate intestinal inflammation in immunodeficient patients. However, Gemmiger and Fusicatenibacter showed a negative correlation with Escherichia-Shigella, and Bifidobacterium showed a negative correlation with Candida. Therefore, the administration of a supplement containing Gemmiger, Fusicatenibacter, and Bifidobacterium may help prevent pathogen invasion in the intestine. However, this hypothesis needs further investigation, and the role of Gemmiger and Fusicatenibacter in intestinal inflammation needs to be confirmed by further experiments. Furthermore, a positive correlation between the abundance of Faecalibacterium and Gemmiger, Bifidobacterium, and Fusicatenibacter was observed, thereby revealing that probiotic bacteria can help promote each other’s growth and protect the equilibrium of the intestinal microbial community. However, correlations in H subjects were observed to be quite different from those in patients with CD. Therefore, more studies are needed to uncover the intrinsic links between gut microbiota in inflammatory and healthy intestinal conditions.
In conclusion, with the application of high-throughput sequencing technology, associations between the gut microbiota structure and gastroenterological diseases were uncovered. However, there are some limitations in this study. First, although we have eliminated the interference of probiotics, prebiotics, antibiotics, anti-fungal agents, or colon-cleansing regimens on the intestinal microbiology of the patients, the medications (5-aminosalicylic acid, corticosteroids, and azathioprine) that the patients used could still affect the intestinal microbiota to some extent – a condition that cannot be fully avoided. Second, patients with CD in active status usually have a more severe B phenotype (e.g., B2 and B3). As a result, in this study, 13 out of the 19 CD-act patients were of the B2/B3 phenotype. The different B phenotypes could affect the distribution of microbiota. However, with respect to the limitation of the number of patients with CD, we did not find any significant difference in the bacterial and fungal distribution between the CD-act and CD-rem groups. Third, the luminal microbiota in the stools and the mucosal microbiota present in the intestinal epithelium are quite different ecosystems. Mouzan et al. reported different fungal microbiota communities present in the mucosa and feces of the patients with CD.44 As several of the subjects in this study refused to undergo colonoscopy, we could not evaluate the disease activity of the patients with CD by colonoscopy and obtain biopsy samples to study the mucosal microbiota communities; instead, only the fecal microbiota was evaluated. Moreover, due to the limitation of the gut microbiota database, there were many unknown bacteria and/or fungi identified in the gut. Nonetheless, in this study, we characterized the fecal bacterial and fungal microbiome of a healthy Chinese young population and identified bacterial and fungal dysbiosis in patients with CD and showed that the basic composition of the microbiota in the Chinese population differed considerably from that in the Western population, which could be explained partly by the influence of genetics and environmental factors such as diet.
In conclusion, the “chicken-and-egg” problem of whether intestinal microbiota alteration or inflammation occurs first in the development of CD remains unsolved. However, exploring the microbial distribution and the inter- and intra-kingdom interactions in the gut will help elucidate the pathophysiology of CD. The precise role of bacteria and fungi in maintaining gut homeostasis and in the pathophysiology of gastrointestinal disorders such as CD should be further investigated in the future. Moreover, functional changes in the gut microbiota (e.g., metabolomics) are proven to be more important than modifications in the bacterial composition45; thus, studies on the effect of the gut microbiota metabolites on intestinal inflammation could shed new light for the treatment of IBD.
Supplemental Material
Supplemental material, Supple_Figures for Characterization of fungal and bacterial dysbiosis in young adult Chinese patients with Crohn’s disease by Xinyun Qiu, Xiaojing Zhao, Xiufang Cui, Xiaqiong Mao, Nana Tang, Chunhua Jiao, Di Wang, Yue Zhang, Ziping Ye and Hongjie Zhang in Therapeutic Advances in Gastroenterology
Footnotes
Author contributions: XY.Q. designed the research; XY.Q., XJ.Z., XF.C., XQ.M, NN.T., CH.J., D.W., Y.Z., and ZP.Y. performed the research; XQ.M and XJ.Z. analyzed the data; XY.Q. wrote the manuscript; HJ.Z. helped edit the manuscript.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Natural Science Foundation of China (grant number: 81600416 and 81470827) and the Natural Science Foundation of Jiangsu Province (grant number: BK20161065).
ORCID iD: Xinyun Qiu  https://orcid.org/0000-0001-6409-2408
https://orcid.org/0000-0001-6409-2408
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Xinyun Qiu, Department of Gastroenterology, First Affiliated Hospital of Nanjing Medical University, Nanjing, China.
Xiaojing Zhao, Department of Gastroenterology, First Affiliated Hospital of Nanjing Medical University, Nanjing, China.
Xiufang Cui, Department of Gastroenterology, First Affiliated Hospital of Nanjing Medical University, Nanjing, China.
Xiaqiong Mao, Department of Gastroenterology, First Affiliated Hospital of Nanjing Medical University, Nanjing, China.
Nana Tang, Department of Gastroenterology, First Affiliated Hospital of Nanjing Medical University, Nanjing, China.
Chunhua Jiao, Department of Gastroenterology, First Affiliated Hospital of Nanjing Medical University, Nanjing, China.
Di Wang, Department of Gastroenterology, First Affiliated Hospital of Nanjing Medical University, Nanjing, China.
Yue Zhang, Department of Gastroenterology, First Affiliated Hospital of Nanjing Medical University, Nanjing, China.
Ziping Ye, Department of Gastroenterology, First Affiliated Hospital of Nanjing Medical University, Nanjing, China.
Hongjie Zhang, Department of Gastroenterology, First Affiliated Hospital of Nanjing Medical University, Guangzhou Road 300#, Nanjing, 210029, China.
References
- 1. Ng SC, Kaplan GG, Tang W, et al. Population density and risk of inflammatory bowel disease: a prospective population-based study in 13 countries or regions in Asia-Pacific. Am J Gastroenterol 2019; 114: 107–115. [DOI] [PubMed] [Google Scholar]
- 2. Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2018; 390: 2769–2778. [DOI] [PubMed] [Google Scholar]
- 3. Mehrmal S, Uppal P, Nedley N, et al. The global, regional, and national burden of psoriasis in 195 countries and territories, 1990 to 2017: a systematic analysis from the Global Burden of Disease Study 2017. J Am Acad Dermatol. Epub ahead 4 May 2020. DOI: 10.1016/j.jaad.2020.04.139. [DOI] [PubMed] [Google Scholar]
- 4. Su HJ, Chiu YT, Chiu CT, et al. Inflammatory bowel disease and its treatment in 2018: global and Taiwanese status updates. J Formos Med Assoc 2019; 118: 1083–1092. [DOI] [PubMed] [Google Scholar]
- 5. Glassner KL, Abraham BP, Quigley EMM. The microbiome and inflammatory bowel disease. J Allergy Clin Immunol 2020; 145: 16–27. [DOI] [PubMed] [Google Scholar]
- 6. Gao J, Xu K, Liu H, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol 2018; 8: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 2019; 4: 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khan I, Ullah N, Zha L, et al. Alteration of gut microbiota in Inflammatory Bowel Disease (IBD): cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 2019; 8: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008; 105: 16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pascal V, Pozuelo M, Borruel N, et al. A microbial signature for Crohn’s disease. Gut 2017; 66: 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014; 15: 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Knoll RL, Forslund K, Kultima JR, et al. Gut microbiota differs between children with inflammatory bowel disease and healthy siblings in taxonomic and functional composition: a metagenomic analysis; 2017; 312: G327–G339. [DOI] [PubMed] [Google Scholar]
- 13. Nishino K, Nishida A, Inoue R, et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol 2018; 53: 95–106. [DOI] [PubMed] [Google Scholar]
- 14. Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 2014; 14: 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qiu XY, Zhang F, Yang X, et al. Changes in the composition of intestinal fungi and their role in mice with dextran sulfate sodium-induced colitis. Sci Rep 2015; 5: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sokol H, Leducq V, Aschard H, et al. Fungal microbiota dysbiosis in IBD. Gut 2017; 66: 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takeshita K, Mizuno S, Mikami Y, et al. A single species of clostridium subcluster XIVa decreased in ulcerative colitis patients. Inflamm Bowel Dis 2016; 22: 2802–2810. [DOI] [PubMed] [Google Scholar]
- 18. Limon JJ, Tang J, Li D, et al. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe 2019; 25: 377–388 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Musis C, Granata L, Dallio M, et al. Inflammatory bowel diseases: the role of gut microbiota. Curr Pharm Des 2020; 26: 2951–2961. [DOI] [PubMed] [Google Scholar]
- 20. Qiu XY, Li X, Wu Z, et al. Fungal-bacterial interactions in mice with dextran sulfate sodium (DSS)-induced acute and chronic colitis. Rsc Advances 2016; 6: 65995–66006. [Google Scholar]
- 21. Neuman H, Debelius JW, Knight R, et al. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol Rev 2015; 39: 509–521. [DOI] [PubMed] [Google Scholar]
- 22. Clements SJ, Carding SR. Diet, the intestinal microbiota, and immune health in aging. Crit Rev Food Sci Nutr 2018; 58: 651–661. [DOI] [PubMed] [Google Scholar]
- 23. Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol 2015; 12: 720–727. [DOI] [PubMed] [Google Scholar]
- 24. Hu D, Ren J, Wang G, et al. Geographic mapping of Crohn’s disease and its relation to affluence in jiangsu province, an eastern coastal province of china. Gastroenterol Res Pract 2014; 2014: 590467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen L, Wang W, Zhou R, et al. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine (Baltimore) 2014; 93: e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Q, Wang C, Tang C, et al. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J Clinical Gastrol 2014; 48: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma HQ, Yu TT, Zhao XJ, et al. Fecal microbial dysbiosis in Chinese patients with inflammatory bowel disease. World J Gastroenterol 2018; 24: 1464–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maaser C, Sturm A, Vavricka SR, et al. ECCO-ESGAR guideline for diagnostic assessment in IBD part 1: initial diagnosis, monitoring of known IBD, detection of complications. J Crohns Colitis 2019; 13: 144–164. [DOI] [PubMed] [Google Scholar]
- 29. Darfeuille-Michaud A, Neut C, Barnich N, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology 1998; 115: 1405–1413. [DOI] [PubMed] [Google Scholar]
- 30. Mirsepasi-Lauridsen HC, Vallance BA, Krogfelt KA, et al. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin Microbiol Rev 2019; 32: e00060–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andoh A, Kuzuoka H, Tsujikawa T, et al. Multicenter analysis of fecal microbiota profiles in Japanese patients with Crohn’s disease. J Gastroenterol 2012; 47: 1298–1307. [DOI] [PubMed] [Google Scholar]
- 32. Parada Venegas D, De la Fuente MK, Landskron G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol 2019; 10: 277–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mangifesta M, Mancabelli L, Milani C, et al. Mucosal microbiota of intestinal polyps reveals putative biomarkers of colorectal cancer. Sci Rep 2018; 8: 13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019; 569: 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hall AB, Yassour M, Sauk J, et al. A novel ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 2017; 9:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schirmer M, Garner A, Vlamakis H, et al. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol 2019; 17: 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Imai T, Inoue R, Kawada Y, et al. Characterization of fungal dysbiosis in Japanese patients with inflammatory bowel disease. J Gastroenterol 2019; 54: 149–159. [DOI] [PubMed] [Google Scholar]
- 38. Mukhopadhya I, Hansen R, Meharg C, et al. The fungal microbiota of de-novo paediatric inflammatory bowel disease. Microbes Infect 2015; 17: 304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mao X, Qiu X, Jiao C, et al. Candida albicans SC5314 inhibits NLRP3/NLRP6 inflammasome expression and dampens human intestinal barrier activity in Caco-2 cell monolayer model. Cytokine 2020; 126: 154882. [DOI] [PubMed] [Google Scholar]
- 40. Eggimann P, Chevrolet JC, Starobinski M, et al. Primary invasive aspergillosis of the digestive tract: report of two cases and review of the literature. Infection 2006; 34: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wheeler ML, Limon JJ, Bar AS, et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe 2016; 19: 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Piovani D, Danese S, Peyrin-Biroulet L, et al. Inflammatory bowel disease: estimates from the global burden of disease 2017 study. Aliment Pharmacol Ther 2020; 51: 261–270. [DOI] [PubMed] [Google Scholar]
- 43. Lam S, Zuo T, Ho M, et al. Review article: fungal alterations in inflammatory bowel diseases. Aliment Pharmacol Ther 2019; 50: 1159–1171. [DOI] [PubMed] [Google Scholar]
- 44. El Mouzan M, Wang F, Al Mofarreh M, et al. Fungal microbiota profile in newly diagnosed treatment-naive children with Crohn’s disease. J Crohns Colitis 2017; 11: 586–592. [DOI] [PubMed] [Google Scholar]
- 45. Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012; 13: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supple_Figures for Characterization of fungal and bacterial dysbiosis in young adult Chinese patients with Crohn’s disease by Xinyun Qiu, Xiaojing Zhao, Xiufang Cui, Xiaqiong Mao, Nana Tang, Chunhua Jiao, Di Wang, Yue Zhang, Ziping Ye and Hongjie Zhang in Therapeutic Advances in Gastroenterology