Abstract
Background
Recent studies have reported the existence of a Trichuris species complex parasitizing primate. Nevertheless, the genetic and evolutionary relationship between Trichuris spp. parasitizing humans and Non-Human Primates (NHP) is poorly understood. The hypothesised existence of different species of Trichuris in primates opens the possibility to evaluate these primates as reservoir hosts of human trichuriasis and other putative new species of whipworms.
Results
In this paper, we carried out a morphological, biometrical and molecular study of Trichuris population parasitizing Macaca sylvanus from Spain based on traditional morpho-biometrical methods, PCA analysis and ribosomal (ITS2) and mitochondrial (cox1 and cob) DNA sequencing. Morphological results revealed that Trichuris sp. from M. sylvanus is Trichuris trichiura. Ribosomal datasets revealed that phylogenetic relationships of populations of Trichuris sp. from M. sylvanus were unresolved. The phylogeny inferred on mitochondrial datasets (partitioned and concatenated) revealed similar topologies; Thus, phylogenetic trees supported the existence of clear molecular differentiation between individuals of Trichuris sp. from M. sylvanus appearing in two different subclades.
Conclusions
Based on morphological parameters, biometrical measurements, and molecular sequence analysis, we conclude that the whipworms isolated from M. sylvanus were T. trichiura. Further, the evolutionary relationship showed that these worms belonged to two genotypes within the T. trichiura lineage. Since T. trichiura is of public health importance, it is important to carry out further studies to improve the understanding of its hosts range, evolution and phylogeography.
Keywords: Barbary macaque, Macaca sylvanus, Mitochondrial DNA, Trichuris trichiura, Zoonosis
Background
Trichuris species are nematodes belonging to Order Trichocephalida (Class Enoplea) and they parasitize the caecum of different hosts. For many years, Trichuris trichiura Linnaeus, 1771 was considered as the whipworm present in humans and Non-Human-Primates (NHP). Until now, it is known that several whipworm species are able to parasitize humans: T. trichiura (human whipworm), Trichuris suis Schrank, 1788 (pig whipworm) and Trichuris vulpis Froelich, 1789 (dog whipworm), but only T. trichiura has been considered for many years to be the specific whipworm of primates.
Whipworms’ genetic and evolutionary relationship between human and NHP is poorly understood. Moreover, given the phenotypic plasticity of these parasites themselves: host-induced variation, lack of morphological characteristics, and overlap between species in morphological characteristics, it is very difficult to distinguish among closely related Trichuris species [1–4].
Traditionally, the research on T. trichiura from humans and NHP had its main objective on differentiating this species from T. suis found in pigs [5–9]. Morphological studies of Trichuris isolated from primates and humans concluded that the species infecting these hosts is the same, despite slight morphological variations observed using scanning electron microscopy [6]. However, these studies were based on a few morphological features such as the total length or spicule length, but not on discriminative analysis of many morpho-biometrical significant parameters using statistical tests. Ravasi et al. [10] carried out a study to discriminate parasite species from human and NHP using exclusively molecular techniques. Thus, these authors [10] suggested the need for morphological analysis of Trichuris sp. adult collected from Papio ursinus (Chacma baboon) from South Africa to determine whether the genetic lineages corresponded with different morphological species. It seems to be a pattern of infection with different Trichuris species infecting host species, thus, some authors [11] concluded that it would be necessary to apply multiple genetic markers to Trichuris collected from humans and NHP from sympatric areas and worldwide locations. This would clarify parasite transmission routes between these primates allowing the implementation of appropriate control and prevention measures [11].
Hawash et al. [12] suggested the existence of a Trichuris species complex in primates and pigs based on complete mitochondrial genome analysis. Recently, Cutillas et al. [13] proposed a new species, Trichuris colobae, from Colobus guereza kikuyensis, which is distinguished from T. suis from pigs and T. trichiura from humans. Afterward, Callejón et al. [14] reported a morpho-biometric study showing the new species Trichuris ursinus from another NHP (P. ursinus) that differed significantly from T. trichiura (nine different characters) and T. colobae (six different characters). Furthermore, T. ursinus showed features close to T. suis (only three different characters). In all these studies, Trichuris specimens were measured according to parameters reported by Spakulová and Lýsek [15], Suriano and Navone [16] and Robles et al. [17], who summarized the morpho-biometric parameters used in recent years.
The Barbary macaque or Gibraltar macaque (Macaca sylvanus) is the only member of this genus found outside Asia, distributed in Africa, North of the Sahara Desert, and the only NHP to occur in Europe (Rock of Gibraltar) [18, 19]. In early times, it was widespread throughout North Africa from Libya to Morocco, but its current distribution is limited to patches of forest and scrub in Algeria and Morocco [19–22]. A long-established introduced Barbary macaque population lives in Gibraltar [23–25].
To date, morphological and molecular studies of the Trichuris populations of M. sylvanus have not been carried out. Hence, the main objective of this work is to determine the morphologic, biometric and molecular features of Trichuris sp. parasitizing M. sylvanus from Zoo Castellar (Spain) in order to: (i) identify at species level adult parasites of these specimens and (ii) to evaluate genetic variation and evolutionary relationships between Trichuris spp. from NHP and humans.
Results
Morphological and biometrical description
General
The anterior part of the body is long, narrow, tapered and whip-like; the posterior part of the body broad and handle-like. The cuticle has a fine transversal striation. The bacillary band is located laterally in the anterior portion of the body.
Male
The proximal cloacal tube is wide and continued with the distal cloacal tube that contains the spicule, which projected into the anterior portion of the body in a spicule tube (Figs. 1a and 2a). The spicule was observed to have two more chitinized extreme zones and a lighter central part (Figs. 1a and 2a). Spicule sheath was cylindrical without a distal bulb (Figs. 1c and 2a-b) and with triangular spines distributed from the proximal to distal portion (Figs. 1c and 2b). The testis ends near the union of the ejaculator duct and intestine. The cloaca with anus subterminal with one pair of paracloacal papillae not ornamented (Figs. 1b and 2c) was observed when the spicule sheath was invaginated. No cluster of papillae was observed.
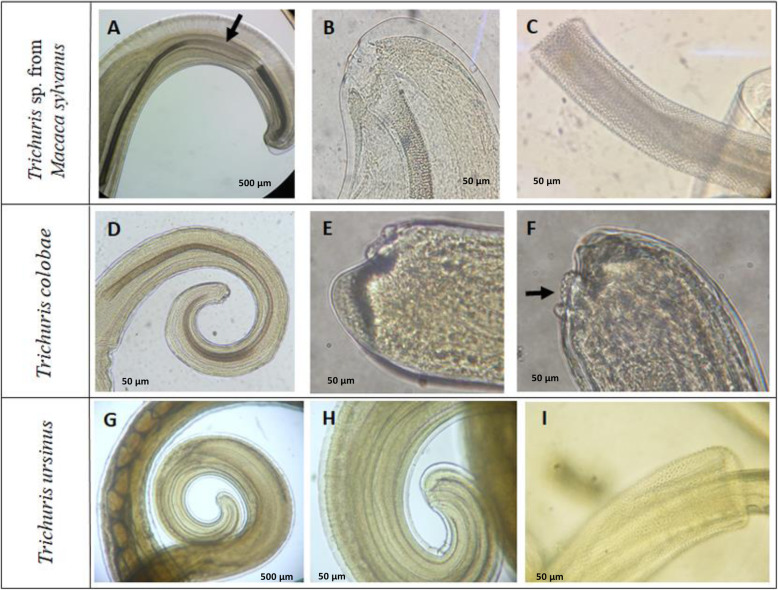
Fig. 1.
Morphology of males of Trichuris sp. from primates. Trichuris sp. from M. sylvanus (a-c), Trichuris colobae [13] (d-f) and Trichuris ursinus [14] (g-i). a-b Posterior end showing spicule (arrowed) and spicule sheath. c posterior end, spiny spicule sheath and cloaca. d Posterior end with spicule invaginated. e-f Posterior end showing cluster of papillae (arrowed). g-h: Posterior end with spicule invaginated. i spiny spicule sheath
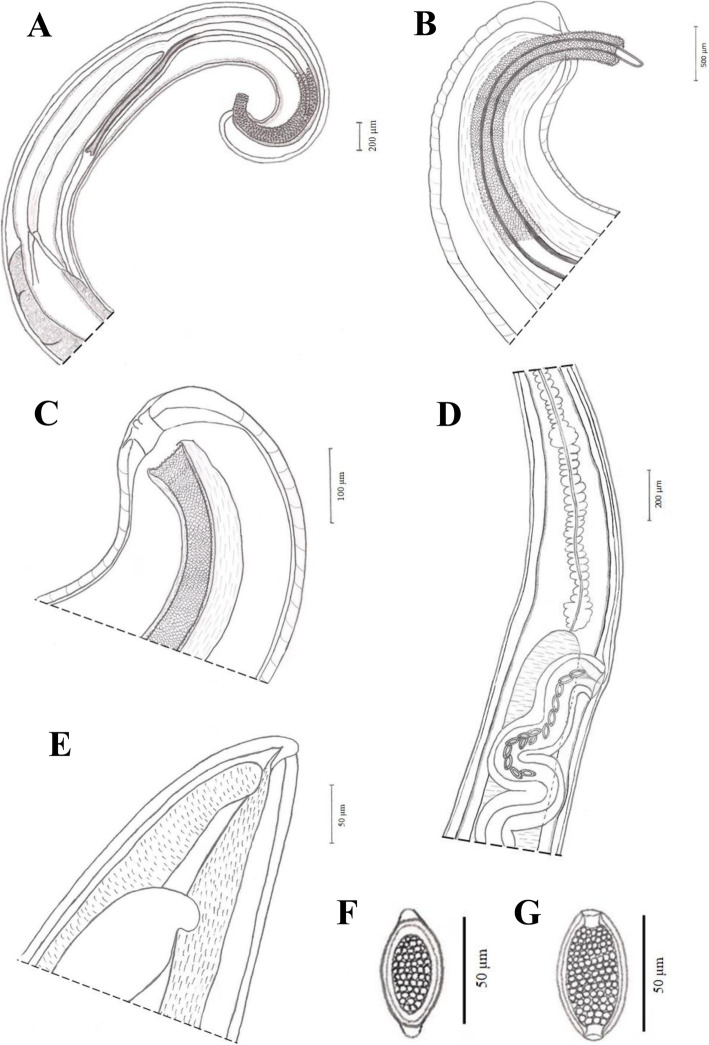
Fig. 2.
Drawings of Trichuris sp. from Macaca sylvanus. a Male, posterior end, spiny spicule sheath, spicule, spicule tube and proximal and distal cloacal tube, lateral view. b Male, detail of the posterior end, lateral view. c Male, cloaca subterminal with one pair of pericloacal papilla, lateral view. d Female, esophagus-intestine junction, vulva and vagina, lateral view. e Female, posterior end, lateral view. f and g Eggs
Female
A non-protrusive vulva without ornamentation was located at esophagus-intestinal junction level (Figs. 3a-b and 2d) and a short and straight vagina continuing with circumvolutions (Figs. 3a-b and2d) without papillae. The anus was subterminal (Figs. 3b and 2e). In addition, two different types of eggs were found in feces of macaque (Figs. 2f-g) which measurements ranged 60–50.6 × 38.4–23.9 μm.
Fig. 3.
Morphology of females of Trichuris sp. from primates. Morphology of females of Trichuris sp. from primates. Trichuris sp. from M. sylvanus (a-c), Trichuris colobae [13] (d-f) and Trichuris ursinus [14] (g-i). a Stychocites, esophagus-intestine junction and vagina. b Vulva and vagina. c Anus subterminal. d Vagina and like-crater vulva (arrowed). e Vulva. f Anus. g Vagina and vulva. h Vulva. i Anus
Morphological results revealed that the Trichuris populations of M. sylvanus in Spain correspond to T. trichiura.
The comparative morphological study with other males of Trichuris species from primates revealed a typical spicule in Trichuris sp. from macaque presenting a central clear part (Fig. 1a, arrowed, 2a) that was not present in that of T. colobae and T. ursinus (Fig. 1d and g-h). In addition, the cluster of typical papillae was only present in T. colobae (Figs. 1e-f). Females of these species appeared to have a non-protrusive vulva (Trichuris sp. and T. ursinus) (Fig. 3b, e and h) and a vulva like a crater with papillae in T. colobae (Figs. 3D-E). The vagina was very long and straight in T. ursinus (Fig. 3g) but appeared with circumvolutions in Trichuris sp. from macaque (Figs. 2d, 3a-b).
The preliminary biometrical study carried out in males and females of Trichuris sp. from M. sylvanus revealed data of 15 different parameters (see Additional files 1 and 2).
The Student’s t test was carried out considering the two different populations according to the two observed genetic lineages: TT2a and TT2c. P values for all measurements were higher than 0.05, thus, no any significant differences between both populations were detected (Tables 1 and 2).
Table 1.
Biometrical and morphological data of 33 males of Trichuris sp. isolated from M. sylvanus. TM = Trichuris male. EL/BL = Esophagus length/Body length. Б: standard deviation. X: arithmetic mean. Min: Minimum value. Max: Maximum value. All measurements are in millimetres
| Lineage TT2a | Total length | Esophagus length | Body length | Ratio EL/BL | Wide body | Spicule length | Spicular length sheath | Spicular tube | Proximal cloacal tube | Pericloacal papillae | Spicular sheath with spines | Distal bulb | Cluster with papillae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TM3 | 38 | 24 | 14 | 1.71 | 0.64 | 2.46 | 0.86 | Present | Present | Not visible | Present | Not present | Not present |
| TM5 | 39 | 24 | 15 | 1.60 | 0.75 | 0.84 | Present | Present | Not visible | Present | Not present | Not present | |
| TM8 | 41 | 28 | 13 | 2.15 | 0.74 | 2.87 | 0.91 | Present | Present | Not visible | Present | Not present | Not present |
| TM18 | 31 | 19 | 12 | 1.58 | 0.63 | 2.77 | 0.97 | Present | Present | Present | Present | Not present | Not present |
| MIN | 31 | 19 | 12 | 1.58 | 0.63 | 2.46 | 0.84 | ||||||
| MAX | 41 | 28 | 15 | 2.15 | 0.75 | 2.87 | 0.97 | ||||||
| X | 37.25 | 23.75 | 13.50 | 1.76 | 0.69 | 2.70 | 0.90 | ||||||
| Б | 4.35 | 3.69 | 1.29 | 0.27 | 0.06 | 0.21 | 0.06 | ||||||
| Lineage TT2c | |||||||||||||
| TM1 | 35 | 22 | 13 | 1.69 | 0.69 | 2.75 | 0.87 | Present | Present | Not visible | Present | Not present | Not present |
| TM2 | 34 | 23 | 11 | 2.09 | 0.67 | 2.56 | 0.90 | Present | Present | Not visible | Present | Not present | Not present |
| TM4 | 36 | 22 | 14 | 1.57 | 0.63 | 2.82 | 0.81 | Present | Present | Not visible | Present | Not present | Not present |
| TM6 | 32 | 21 | 11 | 1.91 | 0.66 | 2.46 | 0.73 | Present | Present | Not visible | Present | Not present | Not present |
| TM7 | 31 | 20 | 11 | 1.82 | 0.50 | 2.33 | 1.12 | Present | Present | Present | Present | Not present | Not present |
| TM9 | 34 | 22 | 12 | 1.83 | 0.54 | 2.55 | 1.01 | Present | Present | Present | Present | Not present | Not present |
| TM10 | 35 | 21 | 14 | 1.50 | 0.63 | 2.61 | 0.70 | Present | Present | Not visible | Present | Not present | Not present |
| TM11 | 35 | 22 | 13 | 1.69 | 0.59 | 2.51 | 0.47 | Present | Present | Present | Present | Not present | Not present |
| TM12 | 35.50 | 24.50 | 11 | 2.23 | 0.67 | 2.48 | 1.02 | Present | Present | Present | Present | Not present | Not present |
| TM13 | 37 | 24 | 13 | 1.85 | 0.7 | 2.73 | 0.96 | Present | Present | Not visible | Present | Not present | Not present |
| TM14 | 39 | 29 | 10 | 2.90 | 0.61 | 2.66 | 0.85 | Present | Present | Not visible | Present | Not present | Not present |
| TM15 | 27 | 16 | 11 | 1.45 | 0.47 | 2.16 | 0.91 | Present | Present | Not visible | Present | Not present | Not present |
| TM16 | 36 | 23 | 13 | 1.77 | 0.57 | 2.71 | 1.16 | Present | Present | Not visible | Present | Not present | Not present |
| TM17 | 32 | 21 | 11 | 1.91 | 0.65 | 2.53 | 0.77 | Present | Present | Not visible | Present | Not present | Not present |
| TM19 | 35 | 23 | 12 | 1.92 | 0.62 | 2.64 | 0.86 | Present | Present | Not visible | Present | Not present | Not present |
| TM20 | 33 | 21 | 12 | 1.75 | 0.65 | 2.63 | 0.95 | Present | Present | Not visible | Present | Not present | Not present |
| TM21 | 32 | 21 | 11 | 1.91 | 0.63 | 2.88 | 0.75 | Present | Present | Not visible | Present | Not present | Not present |
| TM22 | 35 | 21 | 14 | 1.50 | 0.65 | 2.68 | 0.64 | Present | Present | Not visible | Present | Not present | Not present |
| TM23 | 33 | 21 | 12 | 1.75 | 0.67 | 2 | 0.63 | Present | Present | Not visible | Present | Not present | Not present |
| TM24 | 31 | 20 | 11 | 1.82 | 0.69 | 2.55 | 0.79 | Present | Present | Not visible | Present | Not present | Not present |
| TM25 | 32 | 20.5 | 11.50 | 1.78 | 0.67 | 2.77 | 0.88 | Present | Present | Not visible | Present | Not present | Not present |
| TM26 | 34 | 23 | 11 | 2.09 | 0.66 | 2.39 | 1.10 | Present | Present | Present | Present | Not present | Not present |
| TM27 | 30 | 20 | 10 | 2.00 | 0.5 | 2.19 | 1.01 | Present | Present | Present | Present | Not present | Not present |
| TM28 | 11.50 | 0.57 | 2.81 | 0.88 | Present | Present | Not visible | Present | Not present | Not present | |||
| TM29 | 30 | 19 | 11 | 1.73 | 0.53 | 2.22 | 0.73 | Present | Present | Not visible | Present | Not present | Not present |
| TM30 | 34 | 23 | 11 | 2.09 | 0.62 | 2.53 | 0.57 | Present | Present | Not visible | Present | Not present | Not present |
| TM31 | 31 | 20 | 11 | 1.82 | 0.56 | 2.39 | 0.90 | Present | Present | Not visible | Present | Not present | Not present |
| TM32 | 34 | 22 | 12 | 1.83 | 0.7 | 2.71 | 0.94 | Present | Present | Present | Present | Not present | Not present |
| TM33 | 35 | 23 | 12 | 1.92 | 0.59 | 2.46 | 0.87 | Present | Present | Not visible | Present | Not present | Not present |
| MIN | 27 | 16 | 10 | 1.45 | 0.47 | 2 | 0.47 | ||||||
| MAX | 39 | 29 | 14 | 2.90 | 0.70 | 2.88 | 1.16 | ||||||
| X | 33.48 | 21.71 | 11.70 | 1.88 | 0.61 | 2.54 | 0.85 | ||||||
| Б | 2.50 | 2.22 | 1.12 | 0.28 | 0.06 | 0.21 | 0.16 | ||||||
Table 2.
Biometrical and morphological data of 32 females of Trichuris sp. isolated from M. sylvanus. TF = Trichuris female. Б: standard deviation. X: arithmetic mean. Min: Minimum value. Max: Maximum value. All measurements are in millimetres
| Lineage TT2a | Total length | Esophagus length | Body length | Ratio EL/BL | Wide body | Vulvar diameter | Vulva non protusive | Vulva with papillae or ornamentation | Vagina with circumvolutions | Anus subterminal |
|---|---|---|---|---|---|---|---|---|---|---|
| TF10 | 27 | 17 | 10 | 1.70 | 0.6 | 0.07 | Present | No | Present | Present |
| TF27 | 35 | 22 | 13 | 1.69 | 0.66 | 0.09 | Present | No | Present | Present |
| TF30 | 36 | 24 | 12 | 2.00 | 0.69 | 0.07 | Present | No | Present | Present |
| TF32 | 32 | 22 | 10 | 2.20 | 0.61 | 0.08 | Present | No | Present | Present |
| MIN | 27 | 17 | 10 | 1.69 | 0.60 | 0.07 | ||||
| MAX | 36 | 24 | 13 | 2.20 | 0.69 | 0.09 | ||||
| X | 32.50 | 21 | 11.33 | 1.91 | 0.64 | 0.08 | ||||
| Б | 4.04 | 2.99 | 1.50 | 0.25 | 0.04 | 0.01 | ||||
| Lineage TT2c | ||||||||||
| TF1 | 33 | 23 | 10 | 2.30 | 0.69 | 0.06 | Present | No | Present | Present |
| TF2 | 35 | 23 | 12 | 1.92 | 0.67 | 0.05 | Present | No | Present | Present |
| TF3 | 33 | 21 | 11 | 1.91 | 0.7 | 0.06 | Present | No | Present | Present |
| TF4 | 35 | 22 | 13 | 1.69 | 0.75 | 0.05 | Present | No | Present | Present |
| TF5 | 32 | 21 | 11 | 1.91 | 0.7 | 0.06 | Present | No | Present | Present |
| TF6 | 32.50 | 21 | 11.50 | 1.83 | 0.76 | 0.06 | Present | No | Present | Present |
| TF7 | 37 | 25.50 | 11.50 | 2.22 | 0.72 | 0.05 | Present | No | Present | Present |
| TF8 | 36 | 22 | 14 | 1.57 | 0.52 | 0.05 | Present | No | Present | Present |
| TF9 | 33 | 22 | 11 | 2.00 | 0.69 | 0.05 | Present | No | Present | Present |
| TF11 | 37 | 26 | 11 | 2.36 | 0.74 | 0.05 | Present | No | Present | Present |
| TF12 | 32 | 21 | 11 | 1.91 | 0.66 | 0.06 | Present | No | Present | Present |
| TF13 | 32 | 22 | 10 | 2.20 | 0.57 | 0.05 | Present | No | Present | Present |
| TF14 | 37 | 24 | 13 | 1.85 | 0.71 | 0.06 | Present | No | Present | Present |
| TF15 | 38 | 25 | 13 | 1.92 | 0.65 | 0.06 | Present | No | Present | Present |
| TF16 | 31 | 21 | 10 | 2.10 | 0.63 | 0.05 | Present | No | Present | Present |
| TF17 | 34 | 22 | 12 | 1.83 | 0.71 | 0.05 | Present | No | Present | Present |
| TF18 | 34 | 22 | 12 | 1.83 | 0.71 | 0.05 | Present | No | Present | Present |
| TF19 | 33 | 22 | 11 | 2.00 | 0.73 | Present | No | Present | Present | |
| TF20 | 34 | 22 | 12 | 1.83 | 0.59 | 0.06 | Present | No | Present | Present |
| TF21 | 33 | 23 | 10 | 2.30 | 0.83 | 0.05 | Present | No | Present | Present |
| TF22 | 34 | 24 | 10 | 2.40 | 0.69 | 0.06 | Present | No | Present | Present |
| TF23 | 30 | 20 | 10 | 2.00 | 0.62 | 0.07 | Present | No | Present | Present |
| TF24 | 36 | 23 | 13 | 1.77 | 0.69 | 0.05 | Present | No | Present | Present |
| TF25 | 32 | 21 | 11 | 1.91 | 0.77 | 0.05 | Present | No | Present | Present |
| TF26 | 37 | 26 | 11 | 2.36 | 0.71 | 0.06 | Present | No | Present | Present |
| TF28 | 34 | 23 | 11 | 2.09 | 0.7 | 0.06 | Present | No | Present | Present |
| TF29 | 37 | 26 | 11 | 2.36 | 0.79 | 0.06 | Present | No | Present | Present |
| TF31 | 35 | 24 | 11 | 2.18 | 0.61 | 0.07 | Present | No | Present | Present |
| MIN | 30 | 20 | 10 | 1.57 | 0.52 | 0.05 | ||||
| MAX | 38 | 26 | 14 | 2.40 | 0.83 | 0.07 | ||||
| X | 34.15 | 22.78 | 11.40 | 2.02 | 0.69 | 0.06 | ||||
| Б | 2.10 | 1.72 | 1.09 | 0.23 | 0.07 | 0.01 | ||||
The resulting factor maps for male and female populations of Trichuris sp. adults are represented in Fig. 4a and b, respectively. Both male populations slightly overlapped while the female communities appeared 100% overlapped. Males factor map was PC1: 57% and PC2: 35%, while females factor map was PC1: 81% and PC2: 18%.
Fig. 4.
Factor maps corresponding to adult Trichuris sp. Samples are projected onto the first (PC1) and second (PC2) principal components. Each group is represented by its perimeter. a Males factor map. PC1: 57% and PC2: 35%. b Females factor map. PC1: 81% and PC2: 18%
Annotation and features of ribosomal and mitochondrial genomes
The ITS2 region was amplified from the genomic DNA of 13 Trichuris sp. specimens from M. sylvanus from Spain revealing 7 haplotypes. These sequences were 587 base pairs (bp) in length and their G + C content ranged from 66.1–66.6%. Cox1 mitochondrial DNA (mtDNA) partial gene was sequenced from 43 individuals revealing the presence of 4 haplotypes. Cox1 sequences were 370 bp in length and their G + C content ranged from 38 to 39.2%. The cob partial gene was amplified from 13 specimens of Trichuris sp. revealing the presence of 4 haplotypes. The sequences (520 bp) revealed a G + C content ranging from 30.2 to 31.0%.
The datasets generated and analyzed during the current study are available in the GenBank™, EMBL and DDBJ repository, [Accession numbers: LR130781–4, LR132031–4, LR535742, LR535746–51] (Table 3).
Table 3.
Sequences of Trichuris spp. and outgroups species obtained from GenBank and used for phylogenetic analysis
| Species | Host species/Family | Geographical origin | Marker | Accession number |
|---|---|---|---|---|
| Trichuris colobae | Colobus guereza kikuyensis/ Cercopithecidae | Spain (Europe) | Cox1 | |
| Trichuris sp. | Colobus guereza kikuyensis/Cercopithecidae | Czech Republic (Europe) | JF690968 | |
| Homo sapiens/Hominidae | Czech Republic (Europe) | JF690962 | ||
| Macaca fuscata/ Cercopithecidae | Italy (Europe) | |||
| Macaca sylvanus/ Cercopithecidae | Spain (Europe) | |||
| Papio anubis/ Cercopithecidae | USA (North America) | KT449825 | ||
| Papio hamadryas/ Cercopithecidae | Czech Republic (Europe) | JF690963 | ||
| Papio hamadryas/ Cercopithecidae | Denmark (Europe) | KT449824 | ||
| Trichuris trichiura | Homo sapiens/Hominidae | China (Asia) | GU385218 | |
| Japan (Asia) | AP017704 | |||
| Uganda (Africa) | KT449826 | |||
| Trichuris suis | Sus scrofa domestica/Suidae | Spain (Europe) | HE653124 | |
| Denmark (Europe) | KT449822 | |||
| Uganda (Africa) | KT449823 | |||
| China (Asia) | ||||
| Sus scrofa scrofa/Suidae | Spain (Europe) | HE653127 | ||
| Trichuris ursinus | Papio ursinus/ Cercopithecidae | South Africa (Africa) | LT627353 | |
| Trichinella spiralis | USA (North America) | AF293969 | ||
| Trichinella pseudospiralis | Australia (Oceania) | KM357411 | ||
| Trichuris colobae | Colobus guereza kikuyensis/ Cercopithecidae | Spain (Europe) | Cob | LM994704 |
| Trichuris sp. | Macaca fuscata/ Cercopithecidae | Italy (Europe) | ||
| Macaca sylvanus/ Cercopithecidae | Spain (Europe) | |||
| Papio anubis/ Cercopithecidae | USA (North America) | KT449825 | ||
| Papio hamadryas/ Cercopithecidae | Denmark (Europe) | KT449824 | ||
| Papio sp. /Cercopithecidae | Spain (Europe) | LM994703 | ||
| Trichuris trichiura | Homo sapiens/Hominidae | China (Asia) | GU385218 | |
| Uganda (Africa) | KT449826 | |||
| Trichuris suis | Sus scrofa domestica/Suidae | China (Asia) | GU070737 | |
| Denmark (Europe) | KT449822 | |||
| Uganda (Africa) | KT449823 | |||
| Sus scrofa scrofa/Suidae | Spain (Europe) | LM994696 | ||
| Trichuris ursinus | Papio ursinus/ Cercopithecidae | South Africa (Africa) | ||
| Trichinella spiralis | USA (North America) | NC_002681 | ||
| Trichinella pseudospiralis | Australia (Oceania) | KM357411 | ||
| Trichuris colobae | Colobus guereza kikuyensis/ Cercopithecidae | Spain (Europe) | ITS2 | FM991956 |
| Trichuris trichiura | Macaca fuscata/ Cercopithecidae |
China (Asia) Japan (Asia) |
AM992987 | |
| AB586133 | ||||
| Homo sapiens | Cameroon (Africa) | GQ301555 | ||
| Trichuris sp. | Chlorocebus aethiops/ Cercopithecidae | Tanzania (Africa) | ||
| Homo sapiens/Hominidae | Czech Republic (Europe) | JF690940 | ||
| Macaca leonina/ Cercopithecidae | China (Asia) | MH390365 | ||
| KT344828 | ||||
| Macaca mulatta/ Cercopithecidae | China (Asia) | MH390367 | ||
| MH390369 | ||||
| Macaca silenus/ Cercopithecidae | Czech Republic (Europe) | JF690945 | ||
| Macaca sylvanus/ Cercopithecidae | Spain (Europe) | |||
| Papio anubis/ Cercopithecidae | Czech Republic (Europe) | JF690942 | ||
| Papio hamadryas/ Cercopithecidae | Czech Republic (Europe) | JF690941 | ||
| Papio hamadryas ursinus/ Cercopithecidae | South Africa (Africa) | |||
| Pan troglodytes/Hominidae | Netherlands (Europe) | JF690948 | ||
| Trichuris suis | Sus scrofa | Slovakia (Europe) | JF690951 | |
| Tanzania (Africa) | JN181811 | |||
| Sus scrofa domestica | Spain (Europe) | AJ249966 | ||
| Trichuris ursinus | Papio hamadryas ursinus/ Cercopithecidae | South Africa (Africa) | GQ301554 |
aAccess number of this study
Phylogenetic analysis
All trees (cox1, cob and ITS2) obtained by ML, MP and BI for Trichuris sp. revealed two highly supported phylogenetic clades (observed in a previous study) [26] that we named: “T. trichiura lineage” and “T. suis lineage” (Fig. 5 and Table 4). Clade 1 (“T. suis lineage”) included T. colobae, T. ursinus and T. suis and Clade 2 (“T. trichiura lineage”) included T. trichiura and Trichuris sp. from NHP corresponding to the genus Macaca, Papio and Chlorocebus.
Fig. 5.
Phylogenetic tree of Trichuris species based on ITS2 (rDNA) inferred using Bayesian Inference. Bayesian Posterior Probabilities of clades are listed first, followed by Maximum Parsimony and Maximum Likelihood bootstrap values, respectively, for clade frequencies exceeding 65%
Table 4.
Monophyly based on different markers (ITS2, cox1, and cob) of selected group based on different combinations of datasets and inference methods
| Nuclear region ITS2 | Cox1 mt gene | Cob mt gene | Mitochondrial genes (cox1 + cob) | Mitochondrial and nuclear markers (cox1, cob and ITS2) | |
|---|---|---|---|---|---|
| Clade 1 | 100/100/100 | 76/−/93 | 93/69/99 | 92/61/100 | 100/−/100 |
| Clade 2 | 100/−/100 | 73/98/80 | 92/−/100 | 90/−/100 | 100/100/100 |
| Subclade TT2a | – | 88/99/84 | 92/100/100 | 96/100/100 | 89/100/100 |
| Subclade TT2b | – | 98/100/93 | 92/99/99 | 98/100/96 | – |
| Subclade TT2c | – | 98/100/92 | 68/100/89 | 96/−/100 | 100/100/100 |
| Subclade TT2d | – | −/100/− | 92/100/99 | 100/100/100 | – |
| Subclade TT2a + TT2b | – | – | 100/96/99 | 70/96/84 | – |
| Subclade TT2a + TT2b + TT2c | – | – | 69/100/68 | – | – |
| Subclade TT2c + TT2d | – | 88/96/86 | – | 74/−/82 | – |
| Trichuris colobae | – | 88/100/100 | – | – | – |
| Trichuris ursinus | – | – | 100/100/100 | – | – |
| Trichuris suis | 100/100/100 | −/−/100 | 95/99/100 | 98/100/100 | – |
| Trichuris suis subclade 1a | – | 100/100/96 | 91/−/63 | 88/100/100 | – |
| Trichuris suis Subclade 1b | – | 87/98/82 | – | −/99/70 | – |
| Trichuris colobae + Trichuris ursinus | – | – | −/64/− | 76/−/86 | −/74/62 |
ML Maximum Likelihood bootstrap, MP Maximum Parsimony bootstrap, BPP Bayesian Posterior Probability. Clade 2: Trichuris trichiura lineage; Clade 1: Trichuris suis lineage; subclade TT2a: Trichuris sp. from M. sylvanus (haplotypes 2, 3 and 4), Trichuris sp. from H. sapiens from Czech Republic; subclade TT2b: Trichuris sp. from H. sapiens and P. anubis from Asia and USA; subclade TT2c: Trichuris sp. from M. sylvanus (haplotypes 1 and 2), H. sapiens from Uganda, P. hamadryas from Denmark and Czech Republic, subclade TT2d: Trichuris sp. from M. fuscata from Europe
The alignment of 29 ITS2 ribosomal DNA (rDNA) sequences of Trichuris species from human, swine and NHP from different geographic origins yielded a dataset of 584 characters. The phylogenetic tree inferred on DatasetITS2 (Fig. 5) placed Trichuris spp. from M. sylvanus within “T. trichiura lineage” (Clade 2) without any pattern of distribution according to the host species or geographical origin. Nevertheless, Trichuris sp. populations from genus Macaca from Asia (M. leonina, Macaca fuscata and M. mulatta) clustered together and separated from Trichuris sp. from M. sylvanus from Europe (Spain) (Fig. 5). In addition, populations of Trichuris sp. from M. sylvanus appeared in different groups, out of which the haplotypes H1, H2, H5 and H6 clustered together (95% ML BV, 95% BPP), H3 and H4 clustered to the rest of the populations of Trichuris spp. from humans and NHP and separated from H7 (Fig. 5). Exceptionally, one individual of T. trichiura from Homo sapiens from Cameroon and one individual of Trichuris sp. from M. silenus from the Czech Republic were included within “T. suis lineage” (Clade 1). The DatasetITS2 provided moderate phylogenetic resolution among most of Trichuris taxa regardless of inference method (Fig. 5).
The phylogeny inferred on mitochondrial datasets (partitioned and concatenated) revealed similar topologies; therefore, we assumed the concatenated tree based on mitochondrial datasets (cox1 and cob) to be the most representative (Fig. 6; Additional files 3 and 4). The concatenated dataset included 746 aligned positions and 22 taxa, including outgroups. ML, MP and BI methods showed congruence between each other revealing two main clades (corresponding with “T. suis lineage” and “T. trichiura lineage”) and respect to the sister-group relationships between Trichuris spp. from NHP, humans and pigs (Fig. 6 and Table 4). Four different highly supported subclades were observed within “T. trichiura lineage” (Clade 2): subclade TT2a including: Trichuris sp. from M. sylvanus from Spain (haplotypes 2 and 3); subclade TT2b: T. trichiura from H. sapiens from China and Trichuris sp. from Papio anubis from the USA; subclade TT2c: Trichuris sp. from M. sylvanus from Spain (majority haplotype 1), T. trichiura from H. sapiens from Uganda, Trichuris sp. from Papio hamadryas from Europe and two haplotypes of Trichuris sp. from M. fuscata from Europe; subclade TT2d: five haplotypes of Trichuris sp. from M. fuscata from Europe (Fig. 6). The phylogenetic topology revealed a sister relationship between subclades TT2a and TT2b and between subclades TT2c and TT2d with high bootstrap and BPP values (Fig. 6 and Table 4).
Fig. 6.
Phylogenetic tree of Trichuris species based on combined analysis of mitochondrial DNA (cox1 and cob) inferred using Bayesian Inference. Bayesian Posterior Probabilities of clades are listed first, followed by Maximum Parsimony and Maximum Likelihood bootstrap values, respectively, for clade frequencies exceeding 60%
The multiple alignments of 33 cox1 nucleotide sequences (including outgroups) yielded a dataset of 301 characters. Population from M. sylvanus was represented by 4 haplotypes, out of which, haplotypes 3 and 4 clustered together with Trichuris sp. from H. sapiens from the Czech Republic (subclade TT2a) and separated from the main haplotype 1 and haplotype 2 (subclade TT2c) (Additional file 3 and Table 4).
The multiple alignments of 27 cob nucleotide sequences (including outgroups) yielded a dataset of 520 characters. Within Clade 2, population from M. sylvanus was represented by 4 haplotypes, out of which, haplotypes 2, 3 and 4 clustered together within subclades TT2a whereas haplotype 1 appeared within subclade TT2c (see Additional file 4).
The concatenated mitochondrial and ribosomal sequences included 1463 aligned positions and 8 taxa (outgroups not included). Phylogenetic trees again supported the existence of the two main evolutionary lineages previously recognized and the existence of clear differentiation between individuals of Trichuris sp. from M. sylvanus separated in two different subclades (TT2a and TT2c) (see Additional file 5 and Table 4).
Comparative sequence analysis
The range of intra-population similarity of Trichuris sp. from M. sylvanus based on ITS2 rDNA sequences (seven haplotypes) was 99.6–100%. The similarity obtained within “T. trichiura lineage” ranged from 94.4 to 100% while within “T. suis lineage” this value ranged from 85.1 to 100%. When we compared ITS2 sequences corresponding with both lineages, the similarity observed ranged from 74 to 78.6%. Within “T. trichiura lineage”, the minimum similarity observed between Trichuris populations from different species of genus Macaca corresponded to Trichuris sp. from M. sylvanus from Spain and M. fuscata from Japan, and the maximum value of similarity was obtained when we compared Trichuris sp. from M. sylvanus and M. leonina from China (95.9 -99.4% respectively) (Fig. 5).
The cox1 sequences (four haplotypes) of Trichuris sp. obtained from M. sylvanus from Spain showed an intra-specific similarity of 83.6–100%. Thus, haplotype 1 was the main haplotype with 37 individuals showing the same cox1. In addition, haplotypes H1 and H2 showed a similarity percentage from 99.3 to 100% with respect to Trichuris sp. of M. fuscata from Europe and Trichuris sp. of P. hamadryas from the Czech Republic, respectively. The similarity observed within these subclades ranged from 95.6% (TT2b) to 100% (TT2a and TT2c). On the other hand, the similarity observed between these subclades ranged from 79.1% (when we compared TT2b with TT2c) to 87.0% (when we compared TT2c with TT2d). Furthermore, the similarity observed when we compared populations of Trichuris spp. from human and NHP with T. suis, T. colobae and T. ursinus, ranged from 73.4 to 80.6% (Table 5).
Table 5.
Intra-specific and inter-specific similarity observed in cox1 partial sequences in Trichuris species isolated from different hosts
| Cox1 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Trichuris spp. (Subclade TT2a) | Trichuris spp. (Subclade TT2b) | Trichuris spp. (Subclade TT2c) | Trichuris spp. (Subclade TT2d) Subclade. M. fuscata | T. suis (Subclade 1b) | T. suis (Subclade 1a) | T. colobae | T. ursinus | |
| Trichuris spp. (Subclade TT2a) | 98–100 | |||||||
| Trichuris spp. (Subclade TT2b) | 80.1–82.3 | 95.6–98.6 | ||||||
| Trichuris spp. (Subclade TT2c) | 83.4–84.1 | 79.1–82.1 | 99.3–100 | |||||
| Trichuris spp. (Subclade TT2d) Subclade. M. fuscata | 84.1–86.4 | 81.7–83.4 | 85.4–87.0 | 97.7–99 | ||||
| T. suis Spain (Subclade 1a) | 78.4–79.2 | 78.4–80.6 | 78.1–78.7 | 77.4–79.7 | 99.7–100 | |||
| T. suis China (Subclade 1b) | 77–78.1 | 76.7–79.5 | 77.1–78.7 | 75.4–78.4 | 90.9–92 | 95.3–99.5 | ||
| T. colobae | 75.6–76.1 | 76.2–78.9 | 76.4–77.4 | 76.7–78.7 | 78.4–78.9 | 77.8–79.2 | 99.7–100 | |
| T. ursinus | 74.8–75.6 | 73.4–74.2 | 75.1–76.1 | 74.8–75.4 | 81.2–81.4 | 81.4–83.9 | 80.4–80.9 | 100 |
Subclade TT2a: Macaca sylvanus from Spain, Homo sapiens from Czech Republic,. Subclade TT2b: H. sapiens from China and Japan, P. anubis from USA. Subclade TT2c: M. sylvanus from Spain, Macaca fuscata from Italy, H. sapiens from Uganda and P. hamadryas from Denmark and Czech Republic. Subclade TT2d: M. fuscata from Italy. Subclade 1a: Sus scrofa domestica from Denmark and Spain and S. s. scrofa from Spain. Subclade 1b: S. s. domestica from China
The 13 cob sequences of Trichuris sp. revealed the existence of four different haplotypes corresponding to two different lineages. The intra-specific similarity between those haplotypes ranged from 84.2 to 100%, corresponding to the lowest values observed when haplotype 1 was compared with haplotypes 2, 3 and 4. The cob sequences similarity observed within and between subclades revealed similar results that those obtained by cox1 sequences (Table 6).
Table 6.
Intra-specific and inter-specific similarity observed in cob partial sequences in Trichuris species isolated from different host
| cob | ||||||||
|---|---|---|---|---|---|---|---|---|
| Trichuris spp. (Subclade TT2a) | Trichuris spp. (Subclade TT2b) | Trichuris spp. (Subclade TT2c) | Trichuris spp. (subclade TT2d) Macaca fuscata from Europe | T. suis (Subclade 1b) | T. suis (Subclade 1a) | T. colobae | T. ursinus | |
| Trichuris spp. (Subclade TT2a) | 97.1–100 | |||||||
| Trichuris spp. (Subclade TT2b) | 81.7–88.1 | 93.9 | ||||||
| Trichuris spp. (Subclade TT2c) | 83.8–84.4 | 84–84.7 | 99.1–99.8 | |||||
| Trichuris spp. (subclade TT2d) Macaca fuscata from Europe | 84.7–86.7 | 86–87.6 | 86–88.2 | 98.7–100 | ||||
| T. suis (Subclade 1b) | 74.7–75.8 | 73.8–74.2 | 72.4–72.8 | 73–74.2 | 90–100 | |||
| T. suis (Subclade 1a) | 72.9–73.8 | 73.3–74.4 | 72.1–73.9 | 73.5–75.1 | 71.4–72.9 | – | ||
| T. colobae | 70–71.6 | 72.3 | 73.3–74.2 | 73.5–73.9 | 77.4 | 78.7 | – | |
| T. ursinus | 72.1–72.3 | 72.7–73.3 | 73.2–74.2 | 73.5–74.2 | 75.9–79.1 | 77.4–77.8 | 78.5–78-6 | 99.25–99.8 |
Subclade TT2a: Macaca sylvanus from Spain. Subclade TT2b: Homo sapiens from China, P. anubis from USA. Subclade TT2c: M. sylvanus from Spain, Macaca fuscata from Italy, Papio sp. from Spain and P. hamadryas from Denmark. Subclade TT2d: M. fuscata from Italy. Subclade 1a: Sus scrofa domestica from Uganda and Denmark and S.s. scrofa from Spain. Subclade 1b: S. s. domestica from China
Discussion
Morphological results revealed that the whipworm isolated from M. sylvanus is T. trichiura. Thus, in agreement with Cutillas et al. [7], Zaman [27] and Tenora et al. [28], the males of this species showed a pair of typical paracloacal papillae. Nevertheless, this is not in agreement with Ooi et al. [6] who reported the existence of a pair of paracloacal papillae associated to a cluster of small papillae not only in T. trichiura from human but in males of T. trichiura from M. fuscata and Papio papio and, furthermore, they reported females showing everted vagina covered with sharply pointed spines. We did not observe this type of vagina in T. trichiura from M. sylvanus. On the other hand, the comparative morphological study carried out on Trichuris species from other host primates (C. guereza and P. ursinus) revealed clear differences in respect to Trichuris sp. from M. sylvanus. Thus, this can be differentiated from T. colobae by the presence of a typical subterminal paracloacal papillae but not associated to a cluster of small papillae and a different spicule to that of T, colobae and T. ursinus, while females presented a non-everted vagina with a non-ornamented vulva. From a biometrical point of view, a preliminary study [29], based on modern morphometric approach, revealed that the analysis based on three measurements of males (maximum width of the posterior region of the body [thickness, M4], length of the spicule [M8], maximum length of the spicule sheath [M9], clearly illustrates globalized differences in the population of Trichuris sp. from M. sylvanus showing larger values of the males collected from the macaques with respect to T. trichiura from chimpanzees [7, 29]. The occurrence of different biometrical measurements in the same species was explained by Nissen et al. [9] as phenotypic adaptations [1]. This fact was also reported by Cutillas et al. [7, 13]. Furthermore, the existence of different types of eggs of Trichuris sp. in the same host has been previously reported [30].
On the other hand, molecular analyses based on mtDNA revealed the existence of two different genotypes corresponding to two different lineages within “T. trichiura lineage” that did not correlate with two different morphospecies. Nevertheless, we must be cautious since the number of individuals from one of the populations was very low. This fact would agree with Ghai et al. [31] who found that the host range of Trichuris sp. varies depending on the taxonomic group, with some groups showing host specificity and others showing host generality [31]. For this reason, these authors observed that one group was specific to humans, another one had an intermediate host range, and an additional group could infect all primates sampled, including humans. Furthermore, Ravasi et al. [10] found two different genotypes of Trichuris sp. from P. ursinus from two different geographical locations, but they did not carry out a morphological study to characterize different morphospecies. This morphological study was carried out by Callejón et al. [14] in one of these populations and they described the new species named T. ursinus related with T. suis lineage.
The combination of certain nuclear and mitochondrial markers could be considered as a useful taxonomic tool in order to infer phylogenetic relationships within Trichuris genus. The phylogeny of Trichuris spp. from humans and NHP inferred on ribosomal and mitochondrial datasets reported the existence of two main clades previously cited by different authors [10, 14, 32–34]. The similarity between different clades based on DatasetITS2 (“T. trichiura lineages” and “T. suis lineage”) showed clearly lower value (74–78.6% suggesting that Trichuris population of M. sylvanus could be considered as T. trichiura attending to the intra-population similarity observed) [32].
In addition, phylogenetic relationships within Clade 2 based on ribosomal datasets revealed that phylogenetic relationships of populations of Trichuris sp. from M. sylvanus were unresolved. Furthermore, Trichuris spp. isolated from genus Macaca (M. fuscata, M. leonina and M. mulatta) clustered within the same clade separated of Trichuris population from M. sylvanus. This fact could be explained since M. sylvanus is the unique macaque primate extant African representative, all other species being Asiatic suggesting a co-evolutionary process together with the host [35].
The phylogeny inferred on mitochondrial datasets revealed Trichuris sp. from M. sylvanus (Spain) is separated into two different subclades: TT2a (minority haplotype) and TT2c (majority haplotype). Subclade TT2c is considered the most frequent subclade observed in Trichuris spp. from NHP and humans. “T. trichiura lineage” included a species complex with hypothetical sibling/cryptic species. In this last lineage, and based on cox1 partial gene sequences, Trichuris sp. from M. sylvanus appeared distributed in two different subclades according to an African or European origin of T. trichiura from H. sapiens. This phylogenetic pattern of distribution could suggest that different populations are circulating, although samples were taken from the same host. Hawash et al. [36] found no difference between T. trichiura from humans and Trichuris from NHP in Uganda, and he indicated a specific African parasite origin, which would then has been transmitted to Asia and South America suggesting that Trichuris in humans represents an heirloom parasite. We observed that most individuals of Trichuris sp. from M. sylvanus clustered with T. trichiura from H. sapiens from Uganda (Africa) and only a few individuals clustered with Trichuris sp. of H. sapiens from the Czech Republic (Europe). Since only one reference from Africa is used, further molecular studies would be carried out to clarify if there are a specific African parasite origin and a posterior transmission to Europe and Asia.
In agreement with this study, similar results were observed on Trichuris sp. from M. fuscata [34]. Besides, this population showed two potentially distinct entities of Trichuris present in two different subclades: subclade TT2d (analogous to subclade MF reported by Cavallero et al. [34]) and subclade TT2c. These authors suggested the possibility of two different sources of infection for Japanese macaques corresponding with two Trichuris taxa. Within Clade 2, subclades TT2a, TT2b and TT2c correspond to taxonomic species able to infect primates and humans without strict host specificity. These results agree with Doležalová et al. [37] revealing the existence of Trichuris spp., which are shared by humans and several NHP (baboons and macaques).
Despite the fact that there seems to be a pattern of infection with different Trichuris species infecting particular host species, the existence of more species of Trichuris in primates opens up the possibility of studying the zoonotic potential of different hosts harboring T. trichiura and/or other putative new species of whipworms [31].
In addition, it would be necessary to carry out further morphological and molecular studies of Trichuris populations parasitizing NHP from different geographical origins to improve taxonomy and clarify different Trichuris species in primates, and to know if the diversity of Trichuris spp. parasitizing NHP is due to a host specific process, or if these species share different primate hosts, as well as, to evaluate these primates as reservoir hosts of human trichuriasis.
Conclusions
The morphological, biometrical, and molecular results showed that adults of Trichuris sp. from M. sylvanus were T. trichiura. Molecular analyses revealed the existence of two different genotypes corresponding to two different lineages within “T. trichiura lineage” that did not correspond to different morphospecies.
Methods
Isolation of material
Sixty-five adults (32 females and 33 males) of Trichuris sp. were collected from the caecum of a male Barbary macaque (M. sylvanus), which had died of natural causes, from the Zoo Castellar (Cádiz, Spain). This macaque male was 15-year-old, and it was born in the Zoo Castellar (Cádiz, Spain) and it was in captivity in contact with other individuals of the same species but without contact with others non-human primates’ species. It was in contact with the animal keepers and veterinary from the zoo. The parasitic evaluations revealed the presence of Trichuris eggs in the feces for many years and the anthelmintic treatment was mebendazole (10 mg/kg for 3–4 days). A pulmonary pathology was the cause of the natural death.
We previously received consent from the Zoo Castellar to collect these samples. Worms were washed extensively in 0.9% saline solution to remove remains of the host, then, frozen at − 20 °C or preserved in 70% ethanol for morphological, biometrical, and molecular analysis. Posteriorly, worms were cleared with glycerine/alcohol or acetic acid for morphological studies.
Morphological studies
Species identification was performed according to previous studies [7, 13, 14]. Morphological examinations were carried out as described by Oliveros et al. [38] and Skrjabin et al. [39]. A comparative study of morphological data of T. trichiura (present study), T. colobae [7, 13] and T. ursinus [14] was carried out.
Biometrical analysis of Trichuris specimens was carried out according to parameters reported by Spakulová and Lýsek [15], Suriano and Navone [16] and Robles et al. [17]. Subsequently, a biometrical study was carried out using those measurements that are significant in the differentiation of Trichuris species and reported previously by García-Sánchez et al. [29]. Descriptive univariate statistics (mean values, standard deviations, and range) for all parameters were determined for all individuals of Trichuris sp. from M. sylvanus.
We carried out many of the most common tests (including mean, standard deviation, Student’s t) using spreadsheet of Microsoft Excel. The Student’s t assess was used to test the equality of means for each variable in both lineages. The following non-redundant measurements (one measurement is not included in another) used for whipworm adults were: Total length, esophagus length (EL), body length (BL), ratio EL/BL, wide body, spicule length, spicule length sheath for male; total length, esophagus length, body length, ratio EL/BL, wide body and vulvar diameter for females [29]. It was considered a value statistically significant when P < 0.05. Biometric characters of Trichuris sp. from M. sylvanus were compared and assayed for a geometric morphometric analysis. Multivariate analyses were used to calculate the phenotypic variations between Trichuris specimens, using size-free canonical discriminant analyses on the covariance of log-transformed measurements. These analyses were applied to exclude the effect of within-group ontogenetic variations by reducing the effect of each character on the first-pooled, within-group, principal component (a multivariate size estimator) [39]. The principal component analysis (PCA) was used to summarize most of the variations in a multivariate dataset in a few dimensions. The multivariate analyses of the morphometric data were carried out by using BAC v.2 software [29, 40, 41].
DNA amplification and sequencing
Genomic DNA from 43 individual was extracted using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s protocol. Each nematode was placed in a sterile 1.5 ml Eppendorf tube and a pestle was used to facilitate the mechanic rupture of the cuticle. The genomic DNA was extracted from the complete body. Quality of extractions was assessed using 0.8% agarose gel electrophoresis infused with SYBR® Safe DNA gel stain.
All molecular markers sequenced in the present study (cox1 and cob, mtDNA and ITS2 rDNA) were amplified by polymerase chain reaction (PCR) using a thermal cycler (Eppendorf AG; Hamburg, Germany). PCR mix, PCR conditions and PCR primers are summarized in the Supporting information (see Additional file 6). The PCR products were checked on SYBR® Safe stained 2% Tris-Borate-EDTA (TBE) agarose gels and purified using the Wizard® SV Gel and PCR Clean-Up System (Promega). The purified PCR products were concentrated and sequenced in both directions using same primers used for PCR by Stab Vida (Portugal).
Phylogenetic analysis
rDNA (ITS2) and mtDNA (cox1 and cob) sequences were aligned using the MUSCLE alignment method [26] included in MEGA, version 7.0 [42]. For comparison, additional ribosomal and mitochondrial sequences from Trichuris infecting human, NHP and pigs from different geographical regions from the National Centre for Biotechnology Information (NCBI) GenBank™ database were included in the alignments. Trichinella spiralis and Trichinella pseudospiralis were used as outgroups for mitochondrial datasets (Table 3). No outgroups were used to infer phylogenetic trees based on ITS region because the sequences of nucleotides are not sufficiently conserved for there to be a reasonably unambiguous match. Nevertheless, sequences of Trichuris spp. from Macaca silenus and Macaca fascicularis from the Czech Republic were not included in the phylogenetic analysis due to errors found in these sequences. Moreover, cox1 sequences of Trichuris spp. from Macaca mulatta and Macaca leonina were not considered for phylogenetic analysis because they did not correspond with the partial fragment of cox1 gene analyzed in the present study; however, they were included in the phylogenetic analysis based on the ITS2 sequence (Table 3).
Phylogenetic analysis was performed by Maximum Parsimony (MP) algorithm using MEGA 7 [42], Maximum Likelihood (ML) using the PHYML package from Guindon and Gascuel [43] and Bayesian Inference (BI) using MrBayes, version 3.2.6 [44]. Each dataset was analyzed separately from each other, and both mitochondrial and ribosomal datasets were combined into a total evidence dataset. jModeltest was employed to compute the best partitioning scheme, as well as the best nucleotide substitution models for each partition [45]. Models of evolution were chosen for subsequent analysis according to the Akaike Information Criterion [46]. The concatenated dataset was partitioned by gene and models for individual genes within partitions were those selected by jModeltest. For ML inference, best-fit nucleotide substitution models included general time reversible (GTR) model with gamma-distributed rate variation and a proportion of invariable sites (GTR + I (cox1)), GTR model with gamma-distributed rate variation and a proportion of invariable sites (GTR + I + G (cob)) and GTR model with gamma-distributed rate (GTR + G (ITS2)). Support for the topology was examined using bootstrapping (heuristic option) [47] over 1000 replications to assess the relative reliability of clades. The Bayesian posterior probabilities (BPP) comprise the percentage converted for BI; the standard deviation of split frequencies used to determine whether the number of generations completed was enough. Models selected by jModeltest for BI were nst = 6 with inv. rates (cox1), nst = 6 with invgamma rates (cob) and nst = 6 with gamma rates (ITS2). BI analysis was run for ten million generations, and the tree was sampled every 500 generations. Trees from the first million generations were discarded based on an assessment of convergence. Burn-in was determined empirically by examination of the log likelihood values of the chains. After eliminating the first million trees as “burn-in”, we constructed a 50% majority-rule consensus tree, with nodal values representing the probability (posterior probability) that the recovered clades exist, given the aligned sequence data. We accepted a clade in the Bayesian tree at around 70% posterior probability.
The number of base differences per sequence with respect to the sequences under investigation was evaluated using the number of differences method of MEGA 7 to assess the similarity among all marker sequences of all specimens analyzed in the present study and other Trichuris species.
Since molecular analysis showed two different genetic lineages in Trichuris sp. from macaque, we carried out a posterior biometrical study based on those measurements and the method previously used considering the two different lineages observed in Trichuris sp. from M. sylvanus:
Lineage TT2a: Individuals showing the minority genetic lineage.
Lineage TT2c: Individuals showing the main genetic lineage.
Thus, the Student’s t test was used to test the equality of means for each variable in both lineages and biometric characters of Trichuris sp. from both lineages were compared and assayed for a geometric morphometric analysis.
Supplementary Information
Additional file 1. Biometrical data of 15 males of Trichuris sp. isolated from M. sylvanus.
Additional file 2. Biometrical data of 15 females of Trichuris sp. isolated from M. sylvanus.
Additional file 3. Phylogenetic tree of Trichuris species based on cox1 mtDNA sequences inferred using Bayesian method. Bayesian Posterior Probabilities of clades are listed first, followed by Maximum Parsimony and Maximum Likelihood bootstrap values, respectively, for clade frequencies exceeding 60%.
Additional file 4. Phylogenetic tree of Trichuris species based on cob mtDNA sequences inferred using Bayesian method. Bayesian Posterior Probabilities of clades are listed first, followed by Maximum Parsimony and Maximum Likelihood bootstrap values, respectively, for clade frequencies exceeding 60%.
Additional file 5. Phylogenetic tree of Trichuris species based on combined analysis of mitochondrial DNA (cox1 and cob) and nuclear ribosomal DNA (ITS2) inferred using Bayesian Inference. Bayesian Posterior Probabilities of clades are listed first, followed by Maximum Parsimony and Maximum Likelihood bootstrap values, respectively, for clade frequencies exceeding 65%.
Additional file 6. PCR mix, primers and conditions used for each molecular marker sequenced.
Acknowledgements
The authors thank Zoo Castellar (Cádiz, Spain) for providing samples of Trichuris sp. from M. sylvanus naturally died. We wish to thank Mrs. Anne Kendall for the language revision of the manuscript.
Abbreviations
- BI
Bayesian inference
- bp
base pairs
- BPP
Bayesian posterior probabilities
- BV
Bootstrap value
- cob
cytochrome b
- cox1
cytochrome c-oxidase subunit 1
- DNA
Deoxyribonucleic acid
- ITS
Internal transcribed spacer
- ML
Maximum likelihood
- mm
millimeters
- MP
Maximum parsimony
- mtDNA
mitochondrial DNA
- NHP
Non-human primates
- PCA
Principal component analysis
- PCR
Polymerase chain reaction
- rDNA
ribosomal DNA
Authors’ contributions
Analyzed the data: JRF RCF AGS. Conceived, sampled, and designed the experiments: CCB. Contributed reagents/materials/analysis tools: JRF RCF AZC CCB AGS. First drafted the paper: JRF RCF AZC CCB. Performed the experiments: JRF RCF AGS, AZC. Wrote the paper: JRF RCF AZC CCB. The author(s) read and approved the final manuscript.
Funding
This research has been funded by a grant from the Ministry of Economy, Industry and Competitiveness (CGL2017–83057), which included FEDER funds, the Junta de Andalucía (BIO-338) and a grant from the V and VI Plan Propio de Investigaciónof the University of Seville, Spain.
Availability of data and materials
The datasets generated and analyzed during the current study are available in the GenBank™, EMBL and DDBJ repository, [Accession numbers: LR130781–4, LR132031–4, LR535742, LR535746–51] (Table 3).
Ethics approval and consent to participate
Not applicable. This study does not require approval by an ethics committee. Macaca sylvanus, from which Trichuris specimens were collected from their caeca post-mortem, died of natural death. The specimen was handled and housed in a zoo in strict accordance with good animal practices.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12917-020-02661-4.
References
- 1.Knight RA. Morphological differences in Trichuris ovis associated with different host species. J Parasitol. 1984;70:842–843. doi: 10.2307/3281784. [DOI] [PubMed] [Google Scholar]
- 2.Spakulová M. Discriminant analysis as a method for the numerical evaluation of taxonomic characters in male trichurid nematodes. Syst Parasitol. 1994;29:113–119. doi: 10.1007/BF00009807. [DOI] [Google Scholar]
- 3.Robles MR, Navone GT. Redescription of Trichuris laevitestis (Nematoda: trichuridae) from Akodon azarae and Scapteromys aquaticus (Sigmodontinae: Cricetidae) in Buenos Aires province, Argentina. J Parasitol. 2006;92:1053–1057. doi: 10.1645/GE-827R.1. [DOI] [PubMed] [Google Scholar]
- 4.Robles MR. New species of Trichuris (Nematoda: Trichuridae) from Akodon montensis Thomas, 1913, of the Paranaense Forest in Argentina. J Parasitol. 2011;97:319–327. doi: 10.1645/GE-2434.1. [DOI] [PubMed] [Google Scholar]
- 5.Beer RJ. The relationship between Trichuris trichiura (Linnaeus 1758) of man and Trichuris suis (Schrank 1788) of the pig. Res Vet Sci. 1976;20:47–54. doi: 10.1016/S0034-5288(18)33478-7. [DOI] [PubMed] [Google Scholar]
- 6.Ooi HK, Tenora F, Itoh K, Kamiya M. Comparative study of Trichuris trichiura from nonhuman primates and form man, and their differences with Trichuris suis. J Vet Med Sci. 1993;55:363–366. doi: 10.1292/jvms.55.363. [DOI] [PubMed] [Google Scholar]
- 7.Cutillas C, Callejón R, De Rojas M, Tewes B, Úbeda JM, Ariza C, Guevara DC. Trichuris suis and Trichuris trichiura are different nematode species. Acta Trop. 2009;111:299–307. doi: 10.1016/j.actatropica.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Liu GH, Gasser RB, Su A, Nejsum P, Peng L, Lin RQ, Li M, Xu M, Zhu X. Clear genetic distinctiveness between human-and pig-derived Trichuris based on analysis of mitochondrial datasets. PLoS Negl Trop Dis. 2012;6:e1539. doi: 10.1371/journal.pntd.0001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nissen S, Al-Jubury A, Hansen TV, Olsen A, Christensen H, Thamsborg SM, Nejsum P. Genetic analysis of Trichuris suis and Trichuris trichiura recovered from humans and pigs in a sympatric setting in Uganda. Vet Parasitol. 2012;188:68–77. doi: 10.1016/j.vetpar.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Ravasi DF, O’Riain MJ, Davids F, Illing N. Phylogenetic evidence that two distinct Trichuris genotypes infect both humans and non-human primates. PLoS One. 2012;7:e44187. doi: 10.1371/journal.pone.0044187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betson M, Søe MJ, Nejsum P. Human trichuriasis: whipworm genetics, phylogeny, transmission and future research directions. Curr Trop Med Rep. 2015;2:209–217. doi: 10.1007/s40475-015-0062-y. [DOI] [Google Scholar]
- 12.Hawash MB, Andersen LO, Gasser RB, Stensvold C, Nejsum P. Mitochondrial genome analysis suggest multiple Trichuris species in humans, baboons, and pigs from different geographical regions. PLoS Negl Trop Dis. 2015;9:e0004059. doi: 10.1371/journal.pntd.0004059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cutillas C, De Rojas M, Zurita A, Oliveros R, Callejón R. Trichuris colobae n. sp. (Nematoda: Trichuridae), a new species of Trichuris from Colobus guereza kikuyensis. Parasitol Res. 2014;113:2725–2732. doi: 10.1007/s00436-014-3933-6. [DOI] [PubMed] [Google Scholar]
- 14.Callejón R, Halajian A, Cutillas C. Description of a new species, Trichuris ursinus n. sp. (Nematoda: Trichuridae) from Papio ursinus Keer, 1792 from South Africa. Infect Genet Evol. 2017;51:182–193. doi: 10.1016/j.meegid.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Spakulová M, Lýsek H. A biometric study of two populations of Trichocephalus suis Schrank, 1788 from swine and wild boars. Helminthologia. 1981;18:91–98. [Google Scholar]
- 16.Suriano DM, Navone GT. Three new species of the genus Trichuris Roederer, 1761 (Nematoda: Trichuridae) from Cricetidae and Octodontidae rodents in Argentina. Res Re Parasitol. 1994;54:39–46. [Google Scholar]
- 17.Robles MR, Navone GT, Notarnicola J. A new species of Trichuris (Nematoda: Trichuridae) from Phyllotini rodents in Argentina. J Parasitol. 2006;92:100–104. doi: 10.1645/GE-GE-552R.1. [DOI] [PubMed] [Google Scholar]
- 18.Delson E. Fossil macaques phyletic relationships and a scenario of development. In: Lindburg DG, editor. The macaques: studies in ecology behavior and evolution. New York: Van Nostrand Reinhold Co.; 1980. pp. 10–30. [Google Scholar]
- 19.Campeiro A. Intertroop agonistic behavior of a feral rhesus macaque troop ranging in town and forest areas in India. Aggress Behav. 1986;12:433–439. doi: 10.1002/1098-2337(1986)12:6<433::AID-AB2480120606>3.0.CO;2-C. [DOI] [Google Scholar]
- 20.Fa JE, editor. The Barbary macaque: a case study in conservation. New York: Plenum Press; 1984. [Google Scholar]
- 21.Menard N, Vallet D. Population dynamics of Macaca sylvanus in Algeria: an 8-year study. Am J Primatol. 1993;30:101–118. doi: 10.1002/ajp.1350300203. [DOI] [PubMed] [Google Scholar]
- 22.Scheffrahn W, Menard N, Vallet D, Gaci B. Ecology, demography, and population genetics of Barbary macaques in Algeria. Primates. 1993;34:381–394. doi: 10.1007/BF02382634. [DOI] [Google Scholar]
- 23.Fa JE. Apes on the rock. Oryx. 1981;16:73–76. doi: 10.1017/S003060530001680X. [DOI] [Google Scholar]
- 24.Von Starck D. Macaca sylvanus (Linnaeus, 1758) - Berberaffe, Magot. In: Niethammer J, Krapp F, editors. Handbuch der Saugetiere Europas, Band 3/I: Insektenfresser, Primaten. Wiesbaden: Akademische Verlagsgesellschaft; 1990. [Google Scholar]
- 25.Hodges JK, Cortes J, editors. The Barbary macaque: biology and conservation. Nottingham: Nottingham University Press; 2006. [Google Scholar]
- 26.Callejón R, Nadler S, De Rojas M, Zurita A, Petrášová J, Cutillas C. Molecular characterization and phylogeny of whipworm nematodes inferred from DNA sequences of cox1 mtDNA and 18S rDNA. Parasitol Res. 2013;112:3933–3949. doi: 10.1007/s00436-013-3584-z. [DOI] [PubMed] [Google Scholar]
- 27.Zaman V. Scanning electron microscopy of Trichuris trichiura (Nematoda) Acta Trop. 1984;41:287–292. [PubMed] [Google Scholar]
- 28.Tenora F, Hovorka I, Hejlkova D. A supplement to the scanning electron microscopy of some Trichocephalus spp. (Nematoda) Helminthologia. 1988;25:227–234. [Google Scholar]
- 29.García-Sánchez ÁM, Rivero J, Zurita A, Callejón R, Cutillas C. Differentiation of Trichuris species using a morphometric approach. Int J Parasitol Parasites Wild. 2019;9:218–223. doi: 10.1016/j.ijppaw.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinmann P, Rinaldi L, Cringoli G, Du ZW, Marti H, Jiang JY, Zhou H, Zhou XN, Utzinger J. Morphological diversity of Trichuris spp. eggs observed during an anthelminthic drug trial in Yunnan, China, and relative performance of parasitologic diagnostic tools. Acta Trop. 2015;141:184–189. doi: 10.1016/j.actatropica.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 31.Ghai RR, Chapman CA, Omeja PA, Davies TJ, Goldberg TL. Nodule worm infection in humans and wild primates in Uganda: cryptic species in a newly identified region of human transmission. PLoS Negl Trop Dis. 2014;8:e2641. doi: 10.1371/journal.pntd.0002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callejón R, Cutillas C, Nadler SA. Nuclear and mitochondrial genes for inferring Trichuris phylogeny. Parasitol Res. 2015;114:4591–4599. doi: 10.1007/s00436-015-4705-7. [DOI] [PubMed] [Google Scholar]
- 33.Cavallero S, De Liberato C, Friedrich KG, Di Cave D, Masella V, D’Amelio S, Berrilli F. Genetic heterogeneity and phylogeny of Trichuris spp. from captive non-human primates based on ribosomal DNA sequence data. Infect Genet Evol. 2015;34:450–456. doi: 10.1016/j.meegid.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Cavallero S, Nejsum P, Cutillas C, Callejón R, Doležalová J, Modrý D, D’Amelio S. Insights into the molecular systematics of Trichuris infecting captive primates based on mitochondrial DNA analysis. Vet Parasitol. 2019;272:23–30. doi: 10.1016/j.vetpar.2019.06.019. [DOI] [PubMed] [Google Scholar]
- 35.Modolo L, Salzburger W, Martin RD. Phylogeography of Barbary macaques (Macaca sylvanus) and the origin of the Gibraltar colony. Proc Natl Acad Sci. 2005;102:7392–7397. doi: 10.1073/pnas.0502186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hawash MB, Betson M, Al-Jubury A, Ketzis J, LeeWillingham A, Bertelsen MF, Cooper PJ, Littlewood DT, Zhu XQ, Nejsum P. Whipworms in humans and pigs: origins and demography. Parasit Vectors. 2016;9:37. doi: 10.1186/s13071-016-1325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doležalová J, Oborník M, Hajdušková E, Jirků M, Petrželková KJ, Bolechová P, Cutillas C, Callejon R, Jozef J, Berankova Z, Modry D. How many species of whipworms do we share? Whipworms from man and other primates form two phylogenetic lineages. Folia Parasitol (Praha) 2015;62:1–12. doi: 10.14411/fp.2015.063. [DOI] [PubMed] [Google Scholar]
- 38.Oliveros R, Cutillas C, Aris P, Guevara D. Morphologic, biometric and isoenzyme characterization of Trichuris suis. Parasitol Res. 1998;84:513–515. doi: 10.1007/s004360050438. [DOI] [PubMed] [Google Scholar]
- 39.Skrjabin KI, Shikhobalova NP, Orlow IV. Trichocephalidae and Capillariidae of animals and the man and the diseases caused by them. In: Greenberg D, editor. Translated by Birron, A. Israel: Essentials of Nematodology; 1957. [Google Scholar]
- 40.Dujardin JP. BAC software. France: Institut de Recherche pour le Développement, IRD; 2002. [Google Scholar]
- 41.Valero MA, Perez-Crespo I, Periago MV, Khoubbane M, Mas-Coma S. Fluke egg characteristics for the diagnosis of human and animal fascioliasis by Fasciola hepatica and F. gigantica. Acta Trop. 2009;111:150–159. doi: 10.1016/j.actatropica.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 44.Ronquist F, Huelsenbeck JP. MrBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 45.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 46.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 47.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Biometrical data of 15 males of Trichuris sp. isolated from M. sylvanus.
Additional file 2. Biometrical data of 15 females of Trichuris sp. isolated from M. sylvanus.
Additional file 3. Phylogenetic tree of Trichuris species based on cox1 mtDNA sequences inferred using Bayesian method. Bayesian Posterior Probabilities of clades are listed first, followed by Maximum Parsimony and Maximum Likelihood bootstrap values, respectively, for clade frequencies exceeding 60%.
Additional file 4. Phylogenetic tree of Trichuris species based on cob mtDNA sequences inferred using Bayesian method. Bayesian Posterior Probabilities of clades are listed first, followed by Maximum Parsimony and Maximum Likelihood bootstrap values, respectively, for clade frequencies exceeding 60%.
Additional file 5. Phylogenetic tree of Trichuris species based on combined analysis of mitochondrial DNA (cox1 and cob) and nuclear ribosomal DNA (ITS2) inferred using Bayesian Inference. Bayesian Posterior Probabilities of clades are listed first, followed by Maximum Parsimony and Maximum Likelihood bootstrap values, respectively, for clade frequencies exceeding 65%.
Additional file 6. PCR mix, primers and conditions used for each molecular marker sequenced.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the GenBank™, EMBL and DDBJ repository, [Accession numbers: LR130781–4, LR132031–4, LR535742, LR535746–51] (Table 3).