Abstract
Anopheles gambiae and An. arabiensis are major malaria vectors in sub-Saharan Africa. Knowledge of how geographical factors drive the dispersal and gene flow of malaria vectors can help in combatting insecticide resistance spread and planning new vector control interventions. Here, we used a landscape genetics approach to investigate population relatedness and genetic connectivity of An. gambiae and An. arabiensis across Kenya and determined the changes in mosquito population genetic diversity after 20 years of intensive malaria control efforts. We found a significant reduction in genetic diversity in An. gambiae, but not in An. arabiensis as compared to prior to the 20-year period in western Kenya. Significant population structure among populations was found for both species. The most important ecological driver for dispersal and gene flow of An. gambiae and An. arabiensis was tree cover and cropland, respectively. These findings highlight that human induced environmental modifications may enhance genetic connectivity of malaria vectors.
Subject terms: Ecological genetics, Malaria
Introduction
Anopheles gambiae s.l. are the primary vectors of human malaria in sub-Saharan Africa, a disease responsible for 405,000 deaths worldwide annually, with around 90% occurring in Africa1. While the species commonly occupy similar ecological niches, An. gambiae s.s. (hereafter referred to as An. gambiae) are generally associated with more humid environments, whereas An. arabiensis have a higher tolerance for drier environments2,3. Another notable difference between the two species is that An. gambiae are highly anthropophagic4,5, whereas An. arabiensis are more catholic in their feeding behavior6. Since mosquitoes primarily disperse to seek blood meals and oviposit7, these differences in habitat and feeding preferences may result in ecological variables differentially driving the dispersal patterns of these two malaria vectors. These differences potentially lead to a complex system influencing malaria parasite spread. Knowledge of how geographical factors influence the dispersal of malaria vectors can help in efforts to contain insecticide resistance, planning effective vector control interventions, and identifying potential areas susceptible to parasite re-introduction from infected mosquitoes following antimalarial interventions8.
Organism dispersal patterns and population connectedness can be inferred from measuring genetic relatedness among populations or gene flow. While studies have reported genetic differentiation between An. arabiensis populations, neither physical barriers nor geographic distance have been identified as factors responsible for An. arabiensis population structuring9–12. Likewise, geographical distance alone does not appear to be a barrier to gene flow among populations of An. gambiae, as Lehmann et al.13 found high gene flow between populations in Kenya (East Africa) and Senegal (West Africa). However, An. gambiae populations were found to be highly differentiated between western Kenya and eastern Kenya12. The eastern arm of the Great Rift Valley, which bisects Kenya, has been speculated to be the cause of genetic differentiation in An. gambiae populations due to its surrounding low temperatures and arid conditions making it inhospitable to agriculture, and as such, lacks human settlements12,14. Since the eastern arm of the Great Rift Valley is characterized by several attributes, such as low temperatures, low precipitation, as well as low human population density12, these factors cannot be disentangled for driving population structure of An. gambiae using traditional population genetics.
However, using a landscape genetics approach, we can test the impacts of ecological variables on organism dispersal to parse out the importance of key variables influencing malaria vector dispersal8,15. Broadly, the approach involves inferring population movement from the distribution of genetic markers, quantifying the distribution of ecological factors hypothesized to drive dispersal, and statistically testing the relationships between genetic variation and landscape heterogeneity15–17. These inferences enable us to identify potential hotspot areas of disease movement for targeted public health interventions and containment of disease and drug and insecticide resistance8. For example, this approach has been used to identify dispersal corridors for Ae. albopictus mosquitoes in the United States, which were found to be primarily highways and non-forested areas18. Moreover, landscape genetics has been used to study infectious diseases, including chronic wasting disease19, rabies20,21, hantavirus22, H5N1 avian influenza23, and malaria24,25. The objective of this study is to use a landscape genetics approach to test the hypothesis that genetic connectivity of An. gambiae and An. arabiensis is influenced by environmental, landscape, or social factors, as opposed to geographic distance alone. By testing relationships between population genetic structure of malaria vectors and ecological factors, we can parse out confounding factors and determine the importance of key variables influencing malaria vector dispersal8,15.
Results
Anopheles gambiae s.l. larvae were collected in 2014–2015 from thirteen sites across Kenya, which fall within four distinct geographical areas: western Kenya lowlands, western Kenya highlands, Great Rift Valley, and coastal Indian Ocean (Supplementary Table 1; Supplementary Fig. 1). An. arabiensis specimens included in analyses originated from ten populations in western Kenya, Great Rift Valley and coastal Indian Ocean (total individuals = 357); whereas, An. gambiae specimens included in analyses originated from six populations in western Kenya only (total individuals = 254). To assess genetic diversity and structure of vector populations, six microsatellite loci were genotyped in An. arabiensis and five microsatellites were genotyped in An. gambiae specimens (Supplementary Table 2).
Genetic diversity analysis
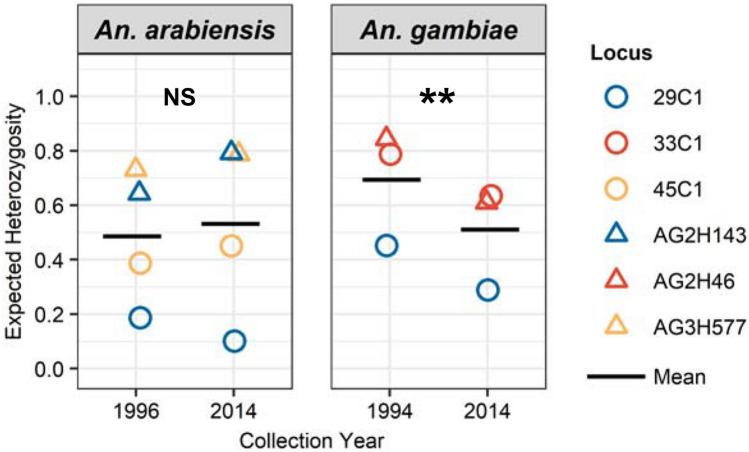
For An. arabiensis, a measure of genetic diversity, expected heterozygosity (HE), ranged from 0.461 in an Indian Ocean coastal site to 0.723 in an Indian Ocean coastal study site (Supplementary Table 3). HE did not vary significantly among regions (Supplementary Fig. 2). Mean allelic richness (AR) ranged from 3.46 in an Indian Ocean coastal site to 6.24 in a western lowland site (Supplementary Table 3). For An. gambiae, HE and AR were generally higher in the highland sites (HE:0.588–0.653; AR:3.74–5.87) compared to the western Kenya lowland sites (HE:0.503–0.584; AR:3.57–4.90) (Supplementary Fig. 2; Supplementary Table 3). An overall heterozygote deficiency was detected across all populations (P < 0.05) for both species. There was no evidence indicating linkage disequilibrium at any locus across all population for An. arabiensis (P > 0.05). By population, there was significant linkage disequilibrium in An. arabiensis detected at two study sites in western Kenya (HB: 45C1/29C1 and AG2H143/AG3H577; KB: 29C1/AG3H577), one study site in the Great Rift Valley (MT: AG3H249/AG2H143), and two study sites in Indian Ocean coastal Kenya (JU: AG2H46/AG2H143; KK: AG2H46/29C1). For An. gambiae, linkage disequilibrium was detected in two locus pairs across all populations (AG2H143/29C1 and AG2H143/AG3H577; P < 0.05). By population, there was significant linkage disequilibrium in An. gambiae at three study sites in the western lowlands (PB: AG2H143/AG3H577, AG2H46/AG2H143, and AG2H46/AG3H577; KN: AG2H143/29C1; HB: AG2H143/29C1 and AG2H143/AG3H577) and one study site in the western highlands (EE: AG2H46/29C1). Since all of the locus pairs observed to be in linkage disequilibrium for both species do not occur in the same chromosomal region (Supplementary Table 2), this result suggests the possibility that random drift or non-random mating are leading to the observed linkage disequilibrium in An. arabiensis and An. gambiae. Compared to previously published HE values for An. arabiensis samples collected from nearby study sites within the Lake Victoria basin area of Western Kenya in 199612, we observed no significant change in overall HE between 1996 and 2014 (Fig. 1; P = 0.417; paired t-test). In comparison to previously published HE values for An. gambiae samples collected in 199426, overall HE was significantly lower in 2014 (Fig. 1; P = 0.002, paired t-test).
Figure 1.
Comparison of genetic diversity (expected heterozygosity) between 1994/1996 and 2014 from nearby study sites within the Lake Victoria basin area of western Kenya for Anopheles arabiensis and An. gambiae. **P < 0.01; NS indicates P > 0.05 by paired t-tests. Data from 1996 was obtained from Kamau et al.12. Data from 1994 was obtained from Lehmann et al.26.
Population structure and migration rates
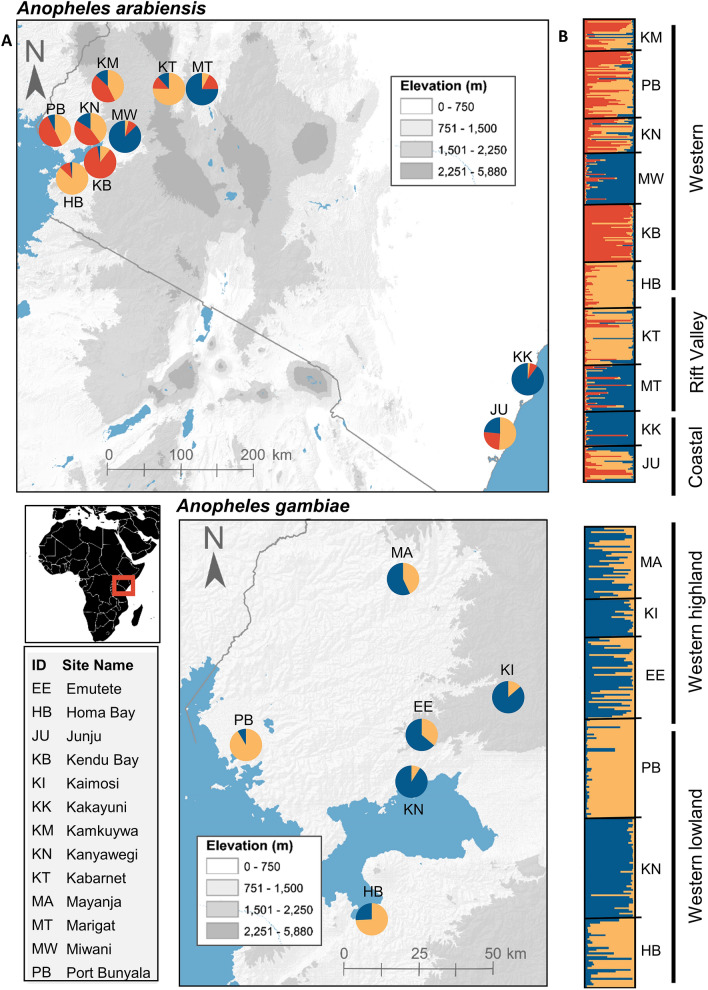
For An. arabiensis, we identified three clusters consistently across ten runs in Structure (Fig. 2). Predominant cluster membership varied between coastal Kenya and the other two regions. The AMOVA indicated that 12.77% of variation occurred among groups, which were populations grouped by their predominant cluster (FCT = 0.128; P < 0.001). Variation within populations (79.7%; FST = 0.203; P < 0.001) and variation occurred among populations within groups (7.49% FSC = 0.086; P < 0.001) were also significant. For An. gambiae, we identified two clusters consistently across ten runs in Structure (Fig. 2). Predominant cluster membership varied between the western lowland and western highland regions. The AMOVA indicated that 6.22% of variation occurred among groups, which were populations grouped by their predominant cluster (FCT = 0.062; P = 0.058). Variation within populations (86.9%; FST = 0.131; P < 0.001) and variation occurred among populations within groups (6.84% FSC = 0.073; P < 0.001) were significant. Based on a linear mixed effects model with maximum likelihood population effects (MLPE), populations did not conform to an isolation-by-distance model for either species (P > 0.05; Supplementary Fig. 3).
Figure 2.
Inferred population structure estimated by STRUCTURE among Anopheles arabiensis (K = 3) and An. gambiae (K = 2). (A) Map of mean ancestral coefficients by study location. Pie chart color indicates proportion of population assigned to each ancestral cluster. (B) Individual level ancestral coefficients. Individuals are represented as rows. Horizontal black lines indicate division in sampling location and vertical black bars indicates region of sampling location. The most probable cluster is indicated by color. Within each individual, the extent of the component colors indicates the magnitude of the membership coefficient corresponding to each cluster. The background elevation maps were created from Shuttle Radar Topography Mission (SRTM) data in ArcMap 10.6.1 (https://desktop.arcgis.com/en/arcmap/).
Historical migration rates between vector populations were evaluated by analysis in Migrate-N, and the highest directional migration rates (migration rate [M] > 18) were visualized (Fig. 3).Analysis of past migration rates for An. arabiensis revealed that frequent migrations (M > 18) occurred between populations across regions, as well as between populations within regions (Fig. 3). The Great Rift Valley was a source of migration for four sites in western Kenya. Frequent migrations were common among sites within western Kenya. For An. gambiae, lowland populations were more often sources for migration than highland populations. Frequent migrations occurred most often from a lowland population to a highland population. Migration rates for all pairwise population comparisons are available in Supplementary Table 4.
Figure 3.
Migration directionality and intensity among Anopheles arabiensis and An. gambiae populations in Kenya. The intensity of gene flow is indicated by the width of migration arrows. Only migration rates with median M > 18 are indicated on the map. The background elevation maps were created from Shuttle Radar Topography Mission (SRTM) data in ArcMap 10.6.1 (https://desktop.arcgis.com/en/arcmap/).
Landscape genetic analysis
We used landscape genetics to test for associations between landscape factors and gene flow between populations. We created landscape resistance raster files for the following variables, which we hypothesized to potentially influence gene flow of the studied malaria vector species: annual mean temperature, annual precipitation, percent tree cover data, presence or absence of croplands or cropland/natural vegetation mosaics, human population density, and road proximity (Table 1). Landscape genetic analysis was conducted using two measures of genetic distance, FST and DPS. Since the results from both analyses did not vary substantially, FST analyses are presented here, and DPS analyses are presented in the supplementary material (Supplementary Table 5; Supplementary Fig. 4). Notably, the genetic distance metrics were highly correlated (P < 0.0001 for both species). For An. arabiensis, cropland was the top single-surface model in explaining population genetic structure of An. arabiensis (Table 2). The cropland model had the lowest ΔAIC (0.000), third lowest average rank (2.335), and highest percentage for being the top model across 1000 bootstrap iterations (34.8%). For An. gambiae, the tree cover model was the top model in explaining population genetic structure, which had the lowest ΔAIC (0.000), second lowest average rank (1.891), and highest percentage for being the top model across bootstrap iterations (40.3%).
Table 1.
Predictor variables used for landscape genetic analysis.
| Variable | Source | |
|---|---|---|
| Environmental | Average temperature | WorldClim BIO1 |
| Annual precipitation | WorldClim BIO12 | |
| Landscape | Percent tree cover | NASA MOD44B |
| Cropland | NASA MOD12Q | |
| Social | Human population density | Worldpop |
| Road proximity | OpenStreetMap |
Table 2.
Model selection results of for linear mixed-effects models optimized on pairwise genetic differentiation (FST) in ResistanceGA.
| Model | K | Avg. ΔAIC | Avg. rank | Top model (%) |
|---|---|---|---|---|
| Anopheles arabiensis | ||||
| (1) Cropland | 2 | 0 | 2.335 | 34.8 |
| (2) Temperature | 2 | 1.865 | 1.878 | 30.0 |
| (3) Precipitation | 2 | 2.257 | 2.220 | 26.3 |
| (4) Geographic distance | 1 | 3.526 | 3.567 | 8.9 |
| Anopheles gambiae | ||||
| (1) Tree cover | 2 | 0 | 1.891 | 40.3 |
| (2) Temperature | 2 | 1.520 | 1.778 | 35.2 |
| (3) Precipitation | 2 | 3.428 | 2.635 | 19.3 |
| (4) Geographic distance | 1 | 6.271 | 3.696 | 5.2 |
K, number of parameters in the mixed effects model; Avg., averaged over 1000 bootstrap iterations; ΔAICc, Difference in Akaike information criterion from the lowest AIC model; rank, model ranking; top model, percentage of the bootstrap iterations that a model was the top model.
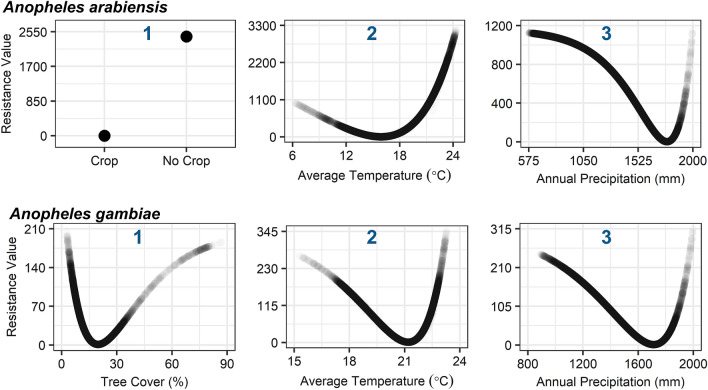
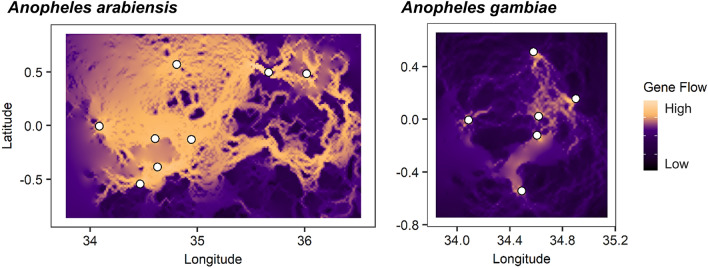
To determine the relationship between model factors and landscape resistance to gene flow, response curves were generated (Fig. 4). In the highest performing model for An. arabiensis, cropland, areas with croplands or cropland/natural vegetation mosaics had a low resistance to gene flow. Average temperature was the next highest performing model. An average temperature of 15.9 °C had the lowest landscape resistance to gene flow. Areas with an average temperature exceeding 23.3 °C had the highest landscape resistance to gene flow (highest 30% resistance values). For the third highest performing model, landscape resistance to gene flow was lowest in areas with annual precipitation 1530.8–1914.8 mm (lowest 30% resistance values) and highest in areas with annual precipitation less than 1230.8 mm and greater than 1970.8 mm. In the highest performing model for An. gambiae, the lowest landscape resistance values occurred in areas with 9.2–38.2% tree cover and the highest landscape resistance values occurred in areas with less than 4.9% tree cover and greater than 61.2% tree cover. For the second highest performing model, the lowest landscape resistance occurred in areas with an average annual temperature between 18.9 and 22.6 °C and the highest landscape resistance values occurred in areas with an average temperature less than 16.1 °C and greater than 23.0 °C. For the third highest performing model, the lowest landscape resistance to gene flow occurred in areas with 1401.5–1890.8 mm annual precipitation and the highest landscape resistance to gene flow occurred in areas with less than 1017.5 mm and greater than 1963.0 mm annual precipitation. Our visualization of gene flow between study sites based on the highest performing models, cropland for An. arabiensis and tree cover for An. gambiae, identifies likely pathways for gene flow between study sites (Fig. 5).
Figure 4.
Response curves signifying the relationship between ecological variables and landscape resistance to gene flow in the three highest performing single-surface models for Anopheles gambiae and An. arabiensis. Blue number in plot indicates model ranking by Avg. ΔAIC value.
Figure 5.
Gene flow map based on highest performing landscape resistance model for Anopheles gambiae and An. Arabiensis. Black circles indicate sampling locations. Yellow color indicates areas of high hypothesized gene flow between sampling locations. For An. arabiensis, map is based on the cropland model. For An. gambiae, map is based on the tree cover model. The maps were created in R 3.6.0 (https://www.r-project.org/rdata) with output currents from Circuitscape 4.0 (https://circuitscape.org/) based on the highest performing landscape resistance models.
Discussion
Using a landscape genetics framework to test hypotheses related to whether environmental, landscape, or social factors predominantly influence the population structures of An. gambiae and An. arabiensis, we found that landscape and environmental factors primarily influence the population structures of these species in western Kenya. For An. gambiae, tree cover was the most important factor shaping population structure. While, for An. arabiensis, presence of cropland was the highest performing explanatory factor. These ecological drivers of gene flow may help to explain broad-scale population structuring patterns previously observed.
In this study, we observed a significant reduction in diversity (HE) in An. gambiae from 199426 to 2014 (this study), which were collected from neighboring populations within the Lake Victoria basin area of western Kenya. For An. arabiensis, we did not observe a comparable reduction in HE in between similar time points, 199612 to 2014 (this study). Notably, we do not report on diversity from within the 18 to 20-year period, so we cannot describe how diversity may have fluctuated within this time frame. With that said, these findings of reduced HE in An. gambiae are consistent with a population decline, which has been observed throughout much of East Africa in response to mass distribution and increased coverage of insecticide-treated bed nets (ITNs)27–32. Conversely, the maintenance of HE in An. arabiensis suggests that An. arabiensis populations have remained relatively more stable despite the high coverage of ITNs in Kenya. Anopheles gambiae are known to be more susceptible to malaria interventions than An. arabiensis due to their preference for feeding on humans and resting indoors4,5. Whereas An. arabiensis are more catholic in their feeding behavior, taking blood meals from both human and non-human hosts, especially cattle33–35, as well as more commonly feed and rest outdoors27,28,36,37. These findings highlight the role that An. arabiensis may play in maintaining residual malaria transmission in sub-Saharan Africa28,38 and the need for alternative intervention strategies to target this species. Moreover, understanding how ecological features influence An. arabiensis dispersal will likely become increasingly important to interrupting residual malaria transmission.
Neither An. arabiensis nor An. gambiae conformed to an isolation-by-distance model in this study. These findings are consistent with previous studies9–13, which suggests the possibility that factors aside from geographic distance alone drive gene flow, such as climate, landscape, or social features. Additionally, for An. arabiensis, we found frequent past migration across regions of Kenya. This finding is also consistent with previous studies and may be in part due to a past range expansion, facilitated by the expansion of human settlement and agriculture, rather than large-scale, contemporary cross-country migrations39–41. For An. gambiae, most frequent migrations occurred from lowland to highland populations. Nonetheless, we did find significant population structuring, indicating the presence of gene flow barriers for An. arabiensis and An. gambiae in Kenya. Notably, FST values for both species were on average higher than those observed in the 1990s12,26, which suggests that population connectivity has reduced since then. This pattern of reduced connectivity may be potentially confounded by the recent history of vector control. With that said, these results together suggest the possibility that factors aside from distance alone drive gene flow patterns of malaria vectors in Kenya.
We tested the hypothesis that low human population densities and scant agriculture between An. gambiae populations provide a barrier to gene flow12,14. This notion was postulated since populations of An. gambiae between western Kenya and eastern Kenya were found to be more distinct than between western Kenya and Senegal in western Africa, despite that the two countries are separated by more than 5000 km12, whereas western Kenya and coastal Kenya populations are only 700 km apart12. The lack of human settlement on the high plateaus of the eastern arm of the Great Rift Valley, which bisects Kenya, was thought to explain the population structuring pattern, since unlike the eastern arm of the rift, human agricultural activity occurs in a broad band across the area between Senegal and western Kenya12. However, we provide evidence that tree cover primarily explains gene flow patterns in western Kenya rather than human population densities or agriculture. Both low (< 5%) and high (> 61%) tree cover were found to be gene flow barriers for An. gambiae in western Kenya. Whereas an intermediate tree cover of 9–38% was associated with enhanced gene flow for An. gambiae. This finding is consistent with a previous study in Kenya, which found that a canopy cover of 26% was associated with the presence of the An. gambiae complex, while a canopy cover of 44% was associated with the absence of the An. gambiae complex42. Moreover, this finding may also be consistent with the broad-scale population structuring observed across Africa12,14, as unlike with the western arm of the Great Rift Valley, a wide swath of land to the East of the eastern arm of the Great Rift Valley is characterized by short vegetation and a tree cover < 5% (Supplementary Fig. 5).
While populations of An. gambiae across the eastern arm of the rift were previously found to be highly genetically differentiated, lower differentiation was found between populations of An. arabiensis in this and other studies12. This result suggested that different factors drive gene flow patterns for An. arabiensis than for An. gambiae despite that the species commonly occupy similar ecological niches2,3. We provide evidence in support of this conjecture, as unlike for An. gambiae, a low percent tree cover was not a barrier to gene flow for An. arabiensis, but rather, the absence of cropland was the primary barrier to gene flow. Notably, however, the presence and absence of cropland does not explain the lack of population structuring between western and eastern Kenya observed by Kamau et al. in the late 1990s12, as western and eastern Kenya are not connected by cropland areas (Supplementary Fig. 5). That this cropland model does not explain population structuring patterns outside of western Kenya may be explained by previous observations that An. arabiensis is more of a climate generalist than An. gambiae38. Anopheles arabiensis are found across a very diverse environmental range, which ultimately allows the survival of the species in many locations in Africa38. Moreover, An. arabiensis are known to be relatively resilient to drastic environmental changes through both natural causes and control measures43. Lastly, since An. arabiensis are able to withstand more arid conditions than An. gambiae44, it is possible that other environmental indicators which were not evaluated in this study explain gene flow patterns, such as maximum temperature. Since An. arabiensis may not be as constrained by environmental heterogeneity, as well as are less susceptible to traditional malaria interventions, controlling residual malaria transmission may prove more challenging and require additional, alternative interventions.
The importance of tree cover in shaping population structure of An. gambiae and cropland for An. arabiensis suggests the significant role that land cover and land use plays in providing suitable microhabitats for larvae to develop. This notion is underscored by the fact that the tree cover model outperformed models based on environmental and social factors in An. gambiae, and that cropland was the top model for An. arabiensis. Too much tree cover may lead to a lack of sunlight and lower temperatures, which slows or inhibits the growth of larvae42. Whereas a lack of tree cover may lead to or be indicative of conditions that are too dry, potentially leaving adult An. gambiae susceptible to desiccation44,45. Whereas An. arabiensis are more desiccation tolerant44, and as such, can thrive in drier environments. These findings of high tree cover hindering gene flow of An. gambiae highlight the effect that deforestation may have in promoting gene flow of malaria vectors. Further, deforestation for agricultural purposes may promote gene flow of An. arabiensis, as cropland areas may enhance gene flow. Thus, deforestation may increase invasion risk of novel malaria vector and parasite genotypes to surrounding areas, potentially also enhancing the spread of insecticide resistant vectors and drug resistant parasites.
This study has several limitations. First, the landscape genetic analysis included genetic data from a single time point. It is not clear whether these spatial trends would be consistent across time. Second, we only examined relationships between genetic distance and ecological variables at one spatial scale. Patterns driving gene flow for these species may vary across spatial scales, i.e. one factor may be more important at a finer scale than at a coarser scale8. Third, the genetic data used for this study was based on five to six microsatellite loci per species, which may provide a limited sample of the genome. Future studies capturing a larger proportion of the genome may improve the resolution of population structure assignment. Finally, this landscape genetics analysis only examined populations in western Kenya. It is likely that different factors drive gene flow patterns depending on the ecological setting, and thus the patterns identified here may differ from those in another region. Thus, the generalizability of these findings to different scales and regions is unknown and requires further investigation. With that said, this study has sought to extend our understanding of local malaria transmission settings in Kenya. Further, this approach may be broadly applicable for investigating factors driving gene flow of malaria vectors in other geographic regions, as well as for additional studies in Kenya.
To conclude, we found that corridors for An. gambiae in western Kenya are most likely to be areas of moderate tree cover (9–38%). Potential corridors for An. arabiensis are areas with cropland or cropland/natural vegetation mosaics. These findings underscore that human induced land cover and land use modifications may enhance connectivity of these species. Specifically, agricultural deforestation may promote dispersal and gene flow for both species. Understanding the factors that limit and promote movement (gene flow) of malaria vectors can help us to more effectively deploy interventions to maintain vector control by identifying areas susceptible to invasion from neighboring mosquito populations. Moreover, this knowledge can be leveraged to help limit the spread of malaria parasites by mosquitoes from nearby areas, as well as contain insecticide resistance8. Finally, knowledge of factors influencing malaria vector dispersal are likely to become increasingly important as malaria transmission becomes increasingly heterogeneous46.
Methods
Sample collection
An. gambiae s.l. larvae were collected between May 2014 and January 2015 from thirteen sites across Kenya (Supplementary Fig. 1). These sites fall within four distinct geographical areas: western Kenya lowlands, western Kenya highlands, Great Rift Valley, and coastal Indian Ocean (Supplementary Table 1. Larvae were collected using a standard mosquito dipper. No more than five larvae were collected per habitat (pool of water) to reduce potential bias from collecting mosquito full siblings47. Larvae in a given study site were collected from between 14 and 53 habitats within a 1.5 km diameter area. Collected larvae were stored in 100% ethanol until DNA purification. Genomic DNA was extracted using standard ethanol extraction procedures with phenol:chloroform48. DNA was eluted into 20 µl of TE buffer. Then, DNA was quantified using a NanoDrop 8000 Spectrophotomer and diluted to a concentration of 1 µg/1 µl sterile water. We identified An. arabiensis and An. gambiae species within the An. gambiae s.l. complex using a ribosomal DNA polymerase chain reaction (PCR) assay49.
Microsatellite genotyping
Six microsatellite loci were selected for genotyping An. gambiae and five loci were selected for An. arabiensis based on evidence of polymorphism in previous studies, reliable amplification, and distribution across chromosomes (Supplementary Table 2)50. We used the M13 tailed primer method to fluorescently label our primers51. Amplification was conducted in a total volume of 10 µl with 5 µl of 2 × DreamTaq Green PCR Master Mix (Thermo Fisher, USA), 0.5 µl of 10 µM primer (forward primer with M13 tail), and 1 µl of DNA template. Thermocycling conditions for An. gambiae and An. arabiensis were as follows: initial denature of 94 °C for 3 min, followed by 35 amplification cycles of 94 °C for 30 s, annealing temperature (Supplementary Table 3) for 30 s, and 72 °C for 45 s, and then a final extension of 72 °C for 6 min. PCR products were analyzed on an automated 4300 DNA analyzer (Li-Cor, Lincoln, NE), and alleles were quantified with the use of Gene ImagIR 4.33 software (Li-Cor).
Population genetic analysis
To test whether population genetic diversity has been altered over the past two decades in the face of intense malaria vector control campaigns, namely the increase in use of insecticide-treated bednets (ITNs)27–32, we assessed measures of heterozygosity in Arlequin52. We then compared expected heterozygosity, a measure of genetic diversity, from western Kenya lowland specimens collected in this study to previously published expected heterozygosity values in nearby lowland populations of An. arabiensis and An. gambiae, which were collected in 199612 and 199426, respectively, prior to the scale-up in use of ITNs. Expected heterozygosity was averaged across nearby populations within the Lake Victoria basin area of western Kenya (n = 2 for An. arabiensis-1996 and An. gambiae-1994; n = 5 for An. arabiensis-2014; n = 3 for An. gambiae-2014) for each loci and then compared. We used paired t-tests to assess statistical significance between expected heterozygosity in pre-ITN scale-up populations (1994/1996) and post-ITN scale-up (2014) populations at matching loci. In addition, for populations from this study (2014), we tested for deviation from Hardy–Weinberg equilibrium and linkage disequilibrium (LD), independence of microsatellite loci, as well as estimated the inbreeding coefficient in Genepop 4.253,54. We also estimated allelic richness at each study site in the R package diveRsity55.
Population structure and migration rates
We estimated population structure using STRUCTURE v. 2.3.4, which uses a Bayesian algorithm to group samples into genetically distinct clusters56. We tested K = 1–7 for An. gambiae and K = 1–10 for An. arabiensis, with ten replicates for each K-level, an initial burn-in of 100,000, and then 500,000 Monte Carlo Markov Chain iterations. The program was run using an admixture model. ∆K was used to detect the number of K (clusters). The output data for the best estimate of K were analyzed using CLUMMP to calculate the mean cluster membership coefficients across multiple runs57. An analysis of molecular variance (AMOVA) was conducted in Arlequin to test for significance of genetic differentiation52. To test for isolation-by-distance, we used linear mixed effects models with the maximum likelihood population effects58 to fit pairwise geographic distance to FST measured in Genepop 4.253,54. Gene flow frequency among populations was estimated for each species in Migrate-N v3.7.2 with the Brownian motion model and Bayesian inference search strategy59. Bidirectional gene flow between all pairwise population comparisons was considered. Four independent runs were conducted with a burn-in of 104 steps, sampling increment of 100 steps, and 5,000 recorded steps in each chain for a total of 2 × 106 visited parameter values.
Landscape genetic analysis
To test for hypothesized associations between landscape factors and gene flow between populations, we used a landscape genetic analysis approach. When testing the effects of environmental factors on gene flow between populations, between-site characteristics are of the greatest concern60. Hence, landscape resistance surfaces were created based on factors hypothesized to prevent or promote gene flow between sites. Since mosquitoes primarily disperse to seek blood meals and oviposit, as well as have physiological constraints regarding development and desiccation, we selected variables which influence these phenomena, i.e., the availability of blood meals and oviposition sites (larval habitats) and environmental suitability. Further, we selected variables that did not closely correlate with each other, which can lead to erroneous model selection results. All pairs of variables had a Pearson correlation coefficient <| 0.6 | (Supplementary Table 6). We created landscape resistance raster files in ArcGIS 10 using climate data (annual average temperature and annual precipitation) from WorldClim (BIO1 and BIO12)61, percent tree cover data from NASA (MOD44B)62,63, cropland from NASA (MOD12Q)62,63, human population density from WorldPop64, and road proximity from OpenStreetMap (Table 1). Specifically, tree cover and cropland variables were chosen because these factors may alter the local ambient temperature and humidity as well as the availability and nutrient content of larval habitats. The cropland raster file was created by reclassifying croplands and cropland/natural vegetation mosaics (IGBP land cover classification system) to “presence of cropland” and all other land cover types to “absence of cropland.” The cropland raster was categorized into two classes to limit the number of parameters estimated and to ensure classes were adequately represented in the landscape. The road proximity raster file was created using the proximity analysis tool in ArcGIS 10. All raster files were resampled to a grain size (resolution) of 2 km to balance computational efficiency. Landscape resistance distance among all pairs of sites was measured using the commuteDistance function in PopGenReport, which is based upon electrical circuit theory65. Resistance distance, defined as the effective resistance between a pair of nodes where all edges are replaced by analogous resistors, reflects the movement cost, as well as availability of alternative pathways, providing an advantage over the commonly used least cost path method65.
Genetic distance between populations was calculated as FST measured in Genepop 4.253,54 and DPS measured in PopGenReport66 (Supplementary Table 7; Supplementary Table 8). FST is a commonly used metric in population and landscape genetic studies, but relies on equilibrium assumptions, which may not be met. Whereas DPS does not rely on any such theoretical assumptions. Evaluation of locus-specific effects to overall genetic distance (FST) values indicated that FST values obtained from each individual locus positively correlates with overall FST for both species, but loci contribution to overall FST was not even, particularly with regard to the 29C1 locus for An. arabiensis (Supplementary Fig. 6). Since the 29C1 locus for An. arabiensis was an outlier in contributing to overall FST, pairwise FST was calculated both with and without the 29C1 locus. Notably, the omission of 29C1 in calculating overall FST did not substantially alter the isolation-by-distance analysis (Supplemental Fig. 6). Therefore, we used pairwise FST and DPS values inclusive of 29C1 for landscape genetic analyses.
We used the R package ResistanceGA to unbiasedly optimize resistance surfaces to our genetic data67. This method has been demonstrated to have a low type 1 error rate for continuous and categorical surfaces, as well as a high correlation between true and optimized resistance surfaces in simulations68. Prior to optimizing our terrestrial resistance surfaces of interest (Table 1), we optimized a surface with lakes and land (no lakes) only to assign a resistance value to lakes. We then masked lakes in subsequent optimizations with the optimized resistance assignment value from the prior analysis, so that continuous land factors were optimized independent of lake features. Linear mixed effects models with the maximum likelihood population effects (MLPE) were used to fit optimized resistance surfaces to genetic data58. The three optimized resistance surfaces with the highest MLPE were then bootstrapped over 1000 iterations by subsampling 80% of the sample locations and refitting the MLPE model. Akaike information criterion (AIC) was used to compare model fitness to genetic data. We excluded eastern Kenya sites from landscape genetic analysis for Anopheles arabiensis due to the biases uneven sampling can introduce in analysis, as population sampling was not evenly distributed across the country69.
Supplementary information
Acknowledgements
We thank our field team of the ICEMR project in Kenya for collecting field samples. This work is supported by grants from the National Institutes of Health (NIH) (U19AI129326, R01AI050243, D43TW001505, and F32AI147460).
Author contributions
E.H.S., D.Z., T.L., and G.Y. contributed to the conception of the study. E.H.S., T.L., M.M., H.N., S.T., S.K., C.M., H.A., and A.G. contributed to the acquisition and analysis of data. E.H.S., J.W.K., and G.Y. contributed to the data interpretation. E.H.S., H.N., and S.T. drafted the work. E.H.S., D.Z., J.W.K., and G.Y. substantially revised it.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files (Supplementary File S1; Supplementary File S2).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-76248-2.
References
- 1.World Health Organization . World malaria report 2019. Geneva: WHO; 2019. [Google Scholar]
- 2.Wirtz RA, Burkot TR. Detection of malarial parasites in mosquitoes. In: Maudlin I, Sinha RC, editors. Advances in Disease Vector Research. New York: Sprinter; 1991. [Google Scholar]
- 3.Trape JF, Rogier C. Combating malaria morbidity and mortality by reducing transmission. Parasitol. Today. 1996;12:236–240. doi: 10.1016/0169-4758(96)10015-6. [DOI] [PubMed] [Google Scholar]
- 4.Mala AO, et al. Plasmodium falciparum transmission and aridity: a Kenyan experience from the dry lands of Baringo and its implications for Anopheles arabiensis control. Malar. J. 2011;10:121. doi: 10.1186/1475-2875-10-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macdonald G. The Epidemiology and Control of Malaria. London: Oxford Univ. Press; 1957. [Google Scholar]
- 6.Gillies M, de Meillon B. The Anophelini of Africa South of the Sahara (Ethiopian Zoogeographical Region) Johannesburg: South African Institute of Medical Research; 1968. [Google Scholar]
- 7.Service, M. W Mosquito (Diptera: Culicidae) dispersal—the long and short of it. J. Med. Entomol. 1997;34:579–588. doi: 10.1093/jmedent/34.6.579. [DOI] [PubMed] [Google Scholar]
- 8.Hemming-Schroeder E, Lo E, Salazar C, Puente S, Yan G. Landscape genetics: a toolbox for studying vector-borne diseases. Front. Ecol. Evol. 2018;6:21. doi: 10.3389/fevo.2018.00021. [DOI] [Google Scholar]
- 9.Ramsdale CD, Fontaine RE. Ecological Investigations of Anopheles gambiae and Anopheles funestus. Geneva: World Health Organization; 1970. [Google Scholar]
- 10.Charlwood JD, Vij R, Billingsley PF. Dry season refugia of malaria-transmitting mosquitoes in a dry savannah zone of east Africa. Am. J. Trop. Med. Hyg. 2000;62:726–732. doi: 10.4269/ajtmh.2000.62.726. [DOI] [PubMed] [Google Scholar]
- 11.Aniedu I. Dynamics of malaria transmission near two permanent breeding sites in Baringo district, Kenya. Indian J. Med. Res. 1997;105:206–211. [PubMed] [Google Scholar]
- 12.Kamau L, et al. Analysis of genetic variability in Anopheles arabiensis and Anopheles gambiae using microsatellite loci. Insect Mol. Biol. 1999;8:287–297. doi: 10.1046/j.1365-2583.1999.820287.x. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann T, et al. Genetic differentiation of Anopheles gambiae populations from East and West Africa: comparison of microsatellite and allozyme loci. Heredity. 1996;77:192–200. doi: 10.1038/hdy.1996.124. [DOI] [PubMed] [Google Scholar]
- 14.Kamau L, Lehmann T, Hawley WA, Orago AS, Collins FH. Microgeographic genetic differentiation of Anopheles gambiae mosquitoes from Asembo Bay, western Kenya: a comparison with Kilifi in coastal Kenya. Am. J. Trop. Med. Hyg. 1998;58:64–66. doi: 10.4269/ajtmh.1998.58.64. [DOI] [PubMed] [Google Scholar]
- 15.Storfer A, et al. Putting the ‘landscape’ in landscape genetics. Heredity. 2007;98:128–142. doi: 10.1038/sj.hdy.6800917. [DOI] [PubMed] [Google Scholar]
- 16.Biek R, Real LA. The landscape genetics of infectious disease emergence and spread. Mol. Ecol. 2010;19:3515–3531. doi: 10.1111/j.1365-294X.2010.04679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storfer A, Murphy MA, Spear SF, Holderegger R, Waits LP. Landscape genetics: Where are we now? Mol. Ecol. 2010;19:3496–3514. doi: 10.1111/j.1365-294X.2010.04691.x. [DOI] [PubMed] [Google Scholar]
- 18.Medley KA, Jenkins DG, Hoffman EA. Human-aided and natural dispersal drive gene flow across the range of an invasive mosquito. Mol. Ecol. 2015;24:284–295. doi: 10.1111/mec.12925. [DOI] [PubMed] [Google Scholar]
- 19.Blanchong JA, et al. Landscape genetics and the spatial distribution of chronic wasting disease. Biol. Lett. 2008;4:130–133. doi: 10.1098/rsbl.2007.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cullingham CI, Kyle CJ, Pond BA, Rees EE, White BN. Differential permeability of rivers to raccoon gene flow corresponds to rabies incidence in Ontario, Canada. Mol. Ecol. 2009;18:43–53. doi: 10.1111/j.1365-294X.2008.03989.x. [DOI] [PubMed] [Google Scholar]
- 21.Côté H, Garant D, Robert K, Mainguy J, Pelletier F. Genetic structure and rabies spread potential in raccoons: the role of landscape barriers and sex-biased dispersal. Evol. Appl. 2012;5:393–404. doi: 10.1111/j.1752-4571.2012.00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guivier E, et al. Landscape genetics highlights the role of bank vole metapopulation dynamics in the epidemiology of Puumala hantavirus. Mol. Ecol. 2011;20:3569–3583. doi: 10.1111/j.1365-294X.2011.05199.x. [DOI] [PubMed] [Google Scholar]
- 23.Carrel M, Wan XF, Nguyen T, Emch M. Genetic variation of highly pathogenic H5N1 avian influenza viruses in Vietnam shows both species-specific and spatiotemporal associations. Avian Dis. 2011;55:659–666. doi: 10.1637/9785-051811-Reg.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo E, et al. Transmission dynamics of co-endemic Plasmodium vivax and P. falciparum in Ethiopia and prevalence of antimalarial resistant genotypes. PLoS Negl. Trop. Dis. 2017;11:e0005806. doi: 10.1371/journal.pntd.0005806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo E, et al. Frequent spread of Plasmodium vivax malaria maintains high genetic diversity at the Myanmar–China Border, without distance and landscape barriers. J. Infect. Dis. 2017;216:1254–1263. doi: 10.1093/infdis/jix106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehmann T, et al. Microgeographic structure of Anopheles gambiae in western Kenya based on mtDNA and microsatellite loci. Mol. Ecol. 1997;6:243–253. doi: 10.1046/j.1365-294X.1997.00177.x. [DOI] [PubMed] [Google Scholar]
- 27.Bayoh MN, et al. Anopheles gambiae: historical population decline associated with regional distribution of insecticide-treated bed nets in western Nyanza Province, Kenya. Malar. J. 2010;9:62. doi: 10.1186/1475-2875-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitau J, et al. Species shifts in the Anopheles gambiae complex: do LLINs successfully control Anopheles arabiensis? PLoS ONE. 2012;7:e31481. doi: 10.1371/journal.pone.0031481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mwangangi JM, et al. The role of Anopheles arabiensis and Anopheles coustani in indoor and outdoor malaria transmission in Taveta District, Kenya. Parasit. Vectors. 2013;6:114. doi: 10.1186/1756-3305-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ototo EN, et al. Surveillance of malaria vector population density and biting behaviour in western Kenya. Malar. J. 2015;14:244. doi: 10.1186/s12936-015-0763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sougoufara S, Harry M, Doucouré S, Sembène PM, Sokhna C. Shift in species composition in the Anopheles gambiae complex after implementation of long-lasting insecticidal nets in Dielmo, Senegal. Med. Vet. Entomol. 2016;30:365–368. doi: 10.1111/mve.12171. [DOI] [PubMed] [Google Scholar]
- 32.Hemming-Schroeder E, et al. Emerging pyrethroid resistance among Anopheles arabiensis in Kenya. Am. J. Trop. Med. Hyg. 2018;98:704–709. doi: 10.4269/ajtmh.17-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Githeko AK, et al. Some observations on the biting behavior of Anopheles gambiae ss, Anopheles arabiensis, and Anopheles funestus and their implications for malaria control. Exp. Parasitol. 1996;82:306–315. doi: 10.1006/expr.1996.0038. [DOI] [PubMed] [Google Scholar]
- 34.Massebo F, Balkew M, Gebre-Michael T, Lindtjørn B. Blood meal origins and insecticide susceptibility of Anopheles arabiensis from Chano in South-West Ethiopia. Parasit. Vectors. 2013;6:44. doi: 10.1186/1756-3305-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tirados I, Costantini C, Gibson G, Torr SJ. Blood-feeding behaviour of the malarial mosquito Anopheles arabiensis: implications for vector control. Med. Vet. Entomol. 2006;20:425–437. doi: 10.1111/j.1365-2915.2006.652.x. [DOI] [PubMed] [Google Scholar]
- 36.Sinka ME, et al. The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic précis. Parasit. Vectors. 2010;3:117. doi: 10.1186/1756-3305-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charlwood JD, et al. The rise and fall of Anopheles arabiensis (Diptera: Culicidae) in a Tanzanian village. Bull. Entomol. Res. 1995;85:37–44. doi: 10.1017/S0007485300051993. [DOI] [Google Scholar]
- 38.Drake JM, Beier JC. Ecological niche and potential distribution of Anopheles arabiensis in Africa in 2050. Malar. J. 2014;13:213. doi: 10.1186/1475-2875-13-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donnelly MJ, Cuamba N, Charlwood JD, Collins FH, Townson H. Population structure in the malaria vector, Anopheles arabiensis Patton, in East Africa. Heredity. 1999;83:408–417. doi: 10.1038/sj.hdy.6885930. [DOI] [PubMed] [Google Scholar]
- 40.Donnelly MJ, Townson H. Evidence for extensive genetic differentiation among populations of the malaria vector Anopheles arabiensis in Eastern Africa. Insect Mol. Biol. 2000;9:357–367. doi: 10.1046/j.1365-2583.2000.00197.x. [DOI] [PubMed] [Google Scholar]
- 41.Donnelly MJ, Licht MC, Lehmann T. Evidence for recent population expansion in the evolutionary history of the malaria vectors Anopheles arabiensis and Anopheles gambiae. Mol. Biol. Evol. 2001;18:1353–1364. doi: 10.1093/oxfordjournals.molbev.a003919. [DOI] [PubMed] [Google Scholar]
- 42.Minakawa N, et al. Spatial distribution of anopheline larval habitats in Western Kenyan highlands: effects of land cover types and topography. Am. J. Trop Med. Hyg. 2005;73:157–165. doi: 10.4269/ajtmh.2005.73.157. [DOI] [PubMed] [Google Scholar]
- 43.Muturi EJ, et al. Population genetic structure of Anopheles arabiensis (Diptera: Culicidae) in a rice growing area of central Kenya. J. Med. Entomol. 2014;47:144–151. doi: 10.1093/jmedent/47.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray EM, Bradley TJ. Physiology of desiccation resistance in Anopheles gambiae and Anopheles arabiensis. Am. J. Trop Med. Hyg. 2005;73:553–559. doi: 10.4269/ajtmh.2005.73.553. [DOI] [PubMed] [Google Scholar]
- 45.Yamana TK, Eltahir EA. Incorporating the effects of humidity in a mechanistic model of Anopheles gambiae mosquito population dynamics in the Sahel region of Africa. Parasit. Vectors. 2013;6:235. doi: 10.1186/1756-3305-6-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nkumama IN, O’Meara WP, Osier FH. Changes in malaria epidemiology in Africa and new challenges for elimination. Trends Parasitol. 2017;33:128–140. doi: 10.1016/j.pt.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H, et al. Monooxygenase levels and knockdown resistance (kdr) allele frequencies in Anopheles gambiae and Anopheles arabiensis in Kenya. J. Med. Entomol. 2014;45:242–250. doi: 10.1093/jmedent/45.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Severson DW. RFLP analysis of insect genomes. In: Crampton JM, Beard CB, Louis C, editors. The Molecular Biology of Insect Disease Vectors. Dordrecht: Springer; 1997. [Google Scholar]
- 49.Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 50.Zheng L, Benedict MQ, Cornel AJ, Collins FH, Kafatos FC. An integrated genetic map of the African human malaria vector mosquito, Anopheles gambiae. Genetics. 1996;143:941–952. doi: 10.1093/genetics/143.2.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oetting WS, et al. Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics. 1995;30:450–458. doi: 10.1006/geno.1995.1264. [DOI] [PubMed] [Google Scholar]
- 52.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 53.Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. doi: 10.1093/oxfordjournals.jhered.a111573. [DOI] [Google Scholar]
- 54.Rousset F. Genepop'007: a complete reimplementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 55.Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013;4:782–788. doi: 10.1111/2041-210X.12067. [DOI] [Google Scholar]
- 56.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bates D, et al. Package ‘lme4’. Convergence. 2015;12:2. [Google Scholar]
- 59.Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl. Acad. Sci. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cushman S, Storfer A, Waits L. Landscape Genetics: Concepts, Methods, Applications. West Sussex: Wiley; 2015. [Google Scholar]
- 61.Roy J. Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. J. R. Meteor. Soc. 2005;25:1965–1978. [Google Scholar]
- 62.Channan S, Collins K, Emanuel WR. Global Mosaics of the Standard MODIS Land Cover Type Data. College Park: University of Maryland and the Pacific Northwest National Laboratory; 2014. [Google Scholar]
- 63.Friedl MA, et al. MODIS Collection 5 global land cover: algorithm refinements and characterization of new datasets. Remote Sens. Environ. 2010;114:168–182. doi: 10.1016/j.rse.2009.08.016. [DOI] [Google Scholar]
- 64.Tatem AJ. WorldPop, open data for spatial demography. Sci. Data. 2017;4:1–4. doi: 10.1038/sdata.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McRae BH, Dickson BG, Keitt TH, Shah VB. Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology. 2008;89:2712–2724. doi: 10.1890/07-1861.1. [DOI] [PubMed] [Google Scholar]
- 66.Adamack AT, Gruber B. PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol. Evol. 2014;5:384–387. doi: 10.1111/2041-210X.12158. [DOI] [Google Scholar]
- 67.Peterman WE. ResistanceGA: An R package for the optimization of resistance surfaces using genetic algorithms. Methods Ecol. Evol. 2018;9:1638–1647. doi: 10.1111/2041-210X.12984. [DOI] [Google Scholar]
- 68.Peterman WE, et al. A comparison of popular approaches to optimize landscape resistance surfaces. Landsc. Ecol. 2019;34:2197–2208. doi: 10.1007/s10980-019-00870-3. [DOI] [Google Scholar]
- 69.Oyler-McCance SJ, Fedy BC, Landguth EL. Sample design effects in landscape genetics. Conserv. Genet. 2013;14:275–285. doi: 10.1007/s10592-012-0415-1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files (Supplementary File S1; Supplementary File S2).