Abstract
A novel coronavirus strain, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in China. This study aims to characterize key attributes of SARS-CoV-2 epidemiology as the infection emerged in China. An age-stratified mathematical model was constructed to describe transmission dynamics and estimate age-specific differences in biological susceptibility to infection, age-assortativeness in transmission mixing, and transition in rate of infectious contacts (and reproduction number R0) following introduction of mass interventions. The model estimated the infectious contact rate in early epidemic at 0.59 contacts/day (95% uncertainty interval-UI = 0.48–0.71). Relative to those 60–69 years, susceptibility was 0.06 in those ≤19 years, 0.34 in 20–29 years, 0.57 in 30–39 years, 0.69 in 40–49 years, 0.79 in 50–59 years, 0.94 in 70–79 years, and 0.88 in ≥80 years. Assortativeness in transmission mixing by age was limited at 0.004 (95% UI = 0.002–0.008). R0 rapidly declined from 2.1 (95% UI = 1.8–2.4) to 0.06 (95% UI = 0.05–0.07) following interventions' onset. Age appears to be a principal factor in explaining the transmission patterns in China. The biological susceptibility to infection seems limited among children but high among those >50 years. There was no evidence for differential contact mixing by age.
Keywords: SARS-CoV-2, COVID-19, Coronavirus, Epidemiology, China, Mathematical model
Introduction
An outbreak of a novel coronavirus strain, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was identified in Wuhan, Hubei province, China, in late December 2019 [1,2]. The outbreak started with identification of four cases of severe pneumonia of unknown etiology, but with symptoms similar to those of Severe Acute Respiratory Syndrome (SARS) and the Middle East Respiratory Syndrome (MERS) [1,3]. Initial cases were linked to exposure at the Huanan Seafood Market, but subsequent infections resulted from rapid community transmission [[1], [2], [3]]. Within about two months, over 80,000 cases and 3000 deaths occurred across China [2,4], amid extreme preventive measures and outstanding healthcare mobilization [2,3]. The resulting disease was named Coronavirus Disease 2019 (COVID-2019) by the World Health Organization (WHO) [5], and has been declared a pandemic [6] after affecting tens of countries and territories [4]. For simplicity, we will thereafter refer to this virus as “COVID-19”, though it is the name of the disease form, given its prevalent current use in the public sphere and to avoid confusion between SARS-CoV-2 and SARS-CoV.
The aims of this study are to investigate and characterize key attributes of COVID-19 epidemiology as the infection emerged in China including 1) age-specific differences in the biological susceptibility to infection, 2) age-assortativeness in infection transmission, and 3) transition in the rate of infectious contacts (and reproduction number) following the introduction of mass interventions.
Materials and methods
Mathematical model
A deterministic compartmental mathematical model was constructed to describe COVID-19 transmission dynamics in a given population, and was applied here to the population of China (S1 Fig of Supplementary Information (SI)). The model was designed based on current understanding of the infection's natural history and epidemiology. Nine age groups were considered, each representing a ten-year age band except for the last category (0–9, 10–19, …, ≥80 years). For each age group, eight coupled nonlinear differential equations were used to describe population flow across compartments based on infection status, infection stage, and disease stage. The model consisted in total of 72 nonlinear differential equations. Analyses were performed in MATLAB R2019a [7].
Susceptible individuals in each age group are at risk of being exposed to the infection at varying hazard rates, which are age- and time- dependent, to capture the variability in the risk of exposure and the impact of public health interventions. Following a latency period, infected individuals are stratified to develop mild infection followed by recovery, or severe infection followed by severe disease then recovery, or critical infection followed by critical disease and either recovery or disease mortality.
The model parameterized the variation in the rate of infectious contacts through a Woods-Saxon function [[8], [9], [10], [11]] to characterize the transition after China's robust public health response in terms of its scale or strength, smoothness or abruptness, duration, and the turning point. The model also incorporated an age mixing matrix that allows a range of contact mixing between individuals varying from fully assortative (mixing only with individuals in the same age group) to fully proportionate (mixing with individuals with no preferential bias for a specific age group). The degree of assortativeness in infection transmission mixing by age is a parameter defined through the age mixing matrix that describes the mixing between the different age groups. The latter parameter was estimated through model fitting. Relevant equations pertaining to the age mixing matrix and its components can be found in Section 1 of SI.
Further details on model structure can be found in Section 1 of SI.
Model parameterization
The model was parameterized using current data on COVID-19 natural history and epidemiology. The duration of latent infection was set at 3.69 days based on an existing estimate [12] and based on a median incubation period across confirmed cases of 5.1 days [1], adjusted for the observed viral load among infected persons following exposure [13] and reported infection transmission prior to onset of symptoms [14]. The age-stratified proportions of infected individuals that will eventually progress to develop mild, severe, or critical infections were based on the observed distribution of cases across these infection stages in China [3,15,16]. The duration of infectiousness was assumed to last for 3.48 days based on an existing estimate [12] and based on the observed time to recovery in persons with mild infection [3,12] and the observed viral load among infected persons [13,14].
Individuals with severe (or critical) infections develop severe (or critical) disease over a period of 28 days prior to recovery, as informed by the observed duration from onset of severe (or critical) disease to recovery [3]. Individuals with critical disease had the additional risk of disease mortality [17]. The age-specific disease mortality rate was fitted factoring the observed crude case fatality rate in each age group in China as of February 11, 2020 [2,17].
The population size, demographic structure (age distribution), and life expectancy of the population of China, as of 2020, were obtained from the United Nations World Population Prospects database [18].
Further details on model parameters, values, and justifications can be found in S1–S2 Tables and Section 2 of SI.
Model fitting
The model was fitted to the following sources of data: 1) time series of diagnosed COVID-19 cases and of the cumulative number of diagnosed COVID-19 cases and of recovered individuals [3], 2) time series of reported COVID-19 deaths and of the cumulative number of COVID-19 deaths [3], 3) crude case fatality rate in each age group [2,17], 4) relative attack rate by age, that is the proportion of the population that has already been infected by February 11, 2020 stratified by age [17], 5) proportion of infections in each age group that will progress to be mild infections [3,15,16], 6) proportion of infections in each age group that will progress to be severe infections [3,15,16], 7) proportion of infections in each age group that will progress to be critical infections [3,15,16]. China's reported cases and deaths were adjusted to reflect the change in coronavirus case definition to include, in addition to the laboratory-confirmed cases, those who are clinically-diagnosed [19,20] (Section 2 of SI).
Model fitting was used to estimate the infectious contact rate, nine parameters for the age-stratified susceptibility to the infection, degree of assortativeness in the age group mixing, overall attack rate, overall disease mortality rate, time delay between onset of actual infection and case notification, and between actual death and reported death, and transition in the basic reproduction number R 0 (Section 3 of SI). A nonlinear least-square data fitting method, based on the Nelder-Mead simplex algorithm, was used to minimize the sum of squares between data points and model predictions [21]. The model was further used to estimate the susceptibility effect sizes (relative susceptibility) for each age group while accounting for the infection transmission dynamics and the effects of assortativeness in mixing in the population.
Uncertainty analyses
A multivariable uncertainty analysis was conducted to determine the range of uncertainty around model predictions. Five-hundred simulation runs were performed, applying at each run, Latin Hypercube sampling from a multidimensional distribution of the model parameters, where parameter values are selected from ranges specified by assuming ±30% uncertainty around parameters' point estimates. These parameters included the duration of latent infection, the duration of infectiousness, the duration of severe disease following onset of severe disease, and the duration of hospitalization for critical infection. The model was then refitted to the input data, and the resulting distributions of estimates, across all 500 runs, were used to calculate the model predictions' means and 95% uncertainty intervals (UIs).
Results
The model fitted the different COVID-19 empirical data such as time-series of diagnosed cases (Fig. 1A), time-series of reported deaths (Fig. 1B), and age-stratified attack rate as of February 11, 2020 (Fig. 1C). The model estimated the epidemic emergence at ~49 days (95% UI: 48–50) prior to January 17, 2020, that is towards the end of November 2019. At the beginning of the epidemic, the predicted infectious contact rate was 0.59 contacts per day (95% UI: 0.48–0.71; S2D Fig of SI). The predicted (average) time delay was 5.4 days (95% UI: 5.2–5.6) between onset of actual infection and reported infection, and 1.6 days (95% UI: 1.5–1.7) between actual death and reported death.
Fig. 1.
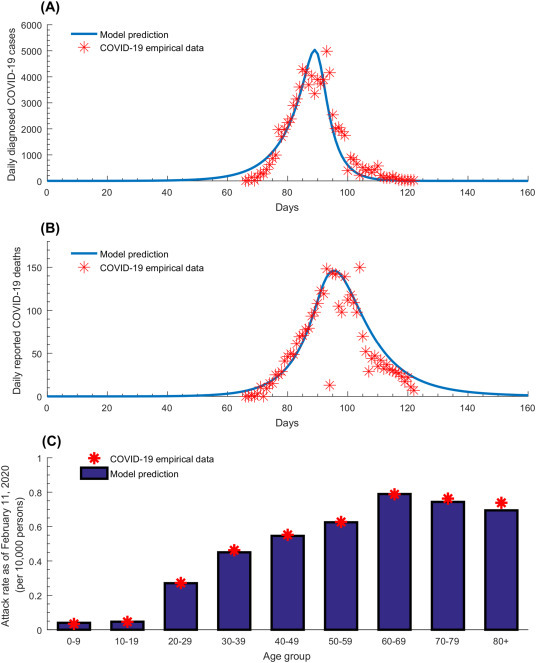
Model fitting of COVID-19 empirical data. Model fits to (A) the time-series of daily diagnosed cases, (B) the time-series of daily reported deaths, and (C) the age-stratified attack rate, that is the proportion of the population that has already been infected by February 11, 2020 stratified by age.
Fig. 2 shows the predicted time evolution of COVID-19 crude case fatality rate (CFR). In the early phase of the epidemic, the crude CFR increased rapidly following the rise in incidence, but plateaued shortly after (towards the end of the first month) and remained so till incidence reached its peak. When incidence started declining (~90 days), the crude CFR grew rapidly, eventually saturating at ~150 days. End of outbreak CFR, estimated through the 500 simulation runs, was 5.1% (95% UI = 4.8–5.4%; S2A Fig of SI).
Fig. 2.
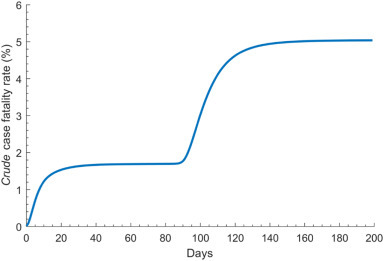
Model predictions for the time evolution of the COVID-19 crude case fatality rate defined as the cumulative number of deaths out of the cumulative number of diagnosed infections.
Fig. 3 features the estimated age-stratified susceptibility profile to COVID-19 infection. Susceptibility was lowest in individuals 0–9 years of age and highest in the 60–69 years age group. Relative to those 60–69 years of age, susceptibility to the infection was only 0.05 in those 0–9 years of age and 0.06 in those 10–19 years of age, but 0.34 in those 20–29 years of age, 0.57 in those 30–39 years of age, 0.69 in those 40–49 years of age, 0.79 in those 50–59 years of age, 0.94 in those 70–79 years of age, and 0.88 in those ≥80 years of age. The uncertainty analysis affirmed these results with narrow uncertainty intervals (S2B Fig of SI).
Fig. 3.
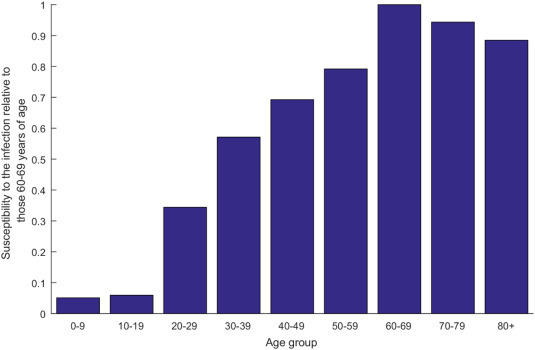
Model predictions for the age-stratified susceptibility profile to COVID-19 infection, relative to those 60–69 years of age.
Fig. 4 illustrates the predicted degree of assortativeness in the age group mixing for each of the 500 simulation runs and model fitting of the uncertainty analysis. The mean degree of assortativeness (mean of the parameter e Age as described in SI) was estimated at 0.004 (95% UI = 0.002–0.008)—there was virtually no assortativeness in infection transmission mixing by age.
Fig. 4.
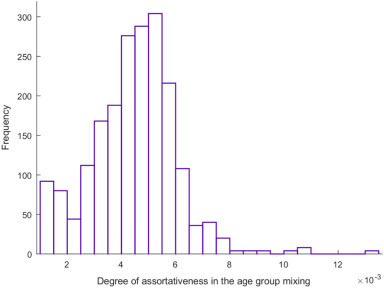
Model predictions for the degree of assortativeness in infection transmission mixing by age across the 500 uncertainty analysis simulation runs. This parameter, defined through the age mixing matrix, describes the mixing between the different age groups. Relevant equations pertaining to the age mixing matrix and its components can be found in Section 1 of Supporting Information.
Fig. 5 and S2C Fig of SI show the time evolution of R 0, and its predicted mean and 95% UI across the 500 uncertainty runs, respectively. In the early phase of the epidemic, R 0 was estimated at 2.1 (95% UI: 1.8–2.4), but rapidly declined to 0.06 (95% UI: 0.05–0.07) following the onset of interventions. The sharp transition duration for R 0 was estimated at 11.5 days (95% UI: 9.5–13.0).
Fig. 5.
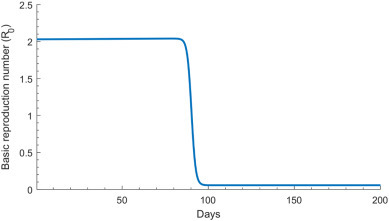
Model predictions for the time evolution of the basic reproduction number R0 before and after onset of the interventions in China.
Discussion
Several key attributes of the epidemiology of COVID-19 have been investigated and estimated. A finding is that the biological susceptibility to the infection appears to vary by age (Fig. 3). Susceptibility to COVID-19 was substantially higher among those >50 years of age compared to those in the younger age groups. For instance, compared to those 60–69 years of age, those ≤19 years of age, 20–29 years of age, and 30–39 years of age were, respectively, 94%, 68%, and 43% less susceptible to being infected. Notably, this age-dependence in the susceptibility to the infection could not be explained by differences in mixing between age groups, as the results indicated limited assortativeness in infection transmission mixing by age (Fig. 4).
These findings support an important role for age in the epidemiology of this infection and affirm other studies suggesting lower susceptibility to the infection at younger age [3,[22], [23], [24], [25], [26], [27], [28], [29]]. Remarkably, the observed attack rate pattern for COVID-19 by age (Fig. 1C) is the inverse (or better complement) of the age-specific cumulative incidence pattern of the 2009 influenza A (H1N1) pandemic (H1N1pdm) infection (S3 Fig of SI) [30]. Presumably, prior recent exposure to other common cold coronaviruses (which are believed to have a similar attack rate pattern to that of H1N1) could be potentially acting as a protective factor (cross immunity) against COVID-19 acquisition (or rapid clearance) at young age [31]. Growing evidence indicates that exposure to other common cold coronaviruses may induce cross-reactive T cell responses and this development of T cell immune memory may be protective against COVID-19 or its severe forms in unexposed individuals [[32], [33], [34]].
An alternative hypothesis has suggested immune imprinting to a similar virus among adults [22]. This being said, the underlying immunological and/or epidemiological factors driving this age effect remain to be investigated with several alternative mechanisms potentially explaining this pattern. For instance, children and young adults may have subclinical infection with low viral load and rapid clearance with or with no transmission potential to others [35]. Of note that the contact tracing data from China suggest that children did not appear to play a significant role in the transmission [3,24,25]. Differences in social behavior and contact patterns by age, which can be very complex [36,37], may also play a role in explaining the variation in the susceptibility to the infection. This being said, additional studies among children exploring viral load, immune response, and social behavior factors are needed before the role of children in community transmission can be ascertained to satisfaction.
Our results indicated that the crude CFR observed in the first three months of the outbreak underestimated the end of outbreak CFR by about 50% (Fig. 2). This is because most infections were still recent infections and have not yet progressed to critical disease or death—death is a late outcome that was estimated to occur 2–8 weeks after onset of symptoms [3].
Another finding of this study is the limited assortativeness in infection transmission mixing by age (Fig. 4), that is equal mixing between the different age groups, which is rather uncommon for respiratory infections [36,37]. This finding is possibly explained by the fact that, with the rapid implementation of stringent lockdown, most transmissions occurred in the context of households rather than of schools, workplaces, or other settings, as supported by existing evidence [3]. Indeed, analysis of 344 clusters in Guangdong and Sichuan provinces indicated that 78–85% of clusters pertained to families [3]. This finding is also supported by analysis of the contacts stratified by age in China [23]. It is, however, important to highlight that while this finding may apply to the China epidemic (given the lockdown measures that may have limited all other transmission pathways such as in schools, workplaces or other settings), it may not necessarily be generalizable to other settings, as existing evidence suggests strong age assortativeness in the transmission dynamics of respiratory infections [36,37]. Indeed, we recently observed strong age assortativeness when we modeled the COVID-19 epidemic in Qatar [38].
The present study affirmed the impact and success of the drastic preventive measures in curtailing infection transmission. R 0 was sharply reduced by 97% over a short duration (Fig. 5). At the beginning of the epidemic, on average, each infected person had 0.6 infectious contacts per day, that is each person passed the infection to 0.6 persons per day (S2D Fig of SI). The rate of infectious contacts can be expressed roughly as c × p, where c is the number of “social” contacts conducive to COVID-19 transmission per day and p is the transmission probability of the virus in a single contact. While p is unknown, it is possibly in the order of 1–5%, as suggested by contact tracing data from China—1-5% of contacts were subsequently laboratory-confirmed as COVID-19 cases [3]. This implies that, on average, each infected person had somewhere between 10 and 60 contacts per day at the beginning of the epidemic, but very few contacts after the lockdown. The latter further affirms the role of the lockdown in severely cutting the contact rate, making sustainable infection transmission very difficult. This finding demonstrates how strong action by policymakers can have an immense impact on limiting infection spread and sequelae.
This study has limitations. Model projections are contingent on the quality and representativeness of the input data. For instance, we assumed infection levels to be as officially documented, but evidence suggested that many infections may have been undocumented [12]. Case ascertainment could also be age dependent. The natural history of this infection is not yet firmly established, and the case management protocols have evolved over time [19,20]. Mortality data seem to suggest that the standard of care improved over time especially in recent weeks when the healthcare sector was no longer overwhelmed with a large case load. We conducted this study using early epidemic data for one specific country, but our knowledge of the epidemic has been rapidly expanding, and thus an alternative broader view of the epidemiology may emerge with accumulation of data. For example, differences in susceptibility among adults may not be as large as those estimated based on China's early epidemic data. We used a deterministic compartmental model, but this type of model may not be representative of the stochastic transmission dynamics when the number of infections is small, such as in the very early phase of the epidemic, thereby adding uncertainty to our estimate for the day of outbreak emergence. Despite these limitations, our parsimonious model, tailored to the nature of available data, was able to reproduce the COVID-19 epidemic as observed in China, and provided insights about infection transmission and disease progression in the population.
In conclusion, age appears to be a principal factor in explaining the patterns of COVID-19 transmission dynamics in China. The biological susceptibility to the infection seems limited among children, intermediate among young adults and those mid-age, but high among those >50 years of age. There was no evidence for considerable differential contact mixing by age, consistent with most transmission occurring in households rather than in schools or workplaces. Further mathematical modeling research is needed to understand the COVID-19 epidemics in other countries, and to draw further inferences about the global epidemiology of this infection.
Data availability
All data generated or analysed during this study are included in this article and its Supplementary Material.
Funding
The modeling infrastructure was made possible by NPRP grant number 9-040-3-008 from the Qatar National Research Fund (a member of Qatar Foundation). GM acknowledges support by UK Research and Innovation as part of the Global Challenges Research Fund, grant number ES/P010873/1. The statements made herein are solely the responsibility of the authors. The authors are also grateful for support provided by the Biostatistics, Epidemiology, and Biomathematics Research Core at Weill Cornell Medicine-Qatar. The publication of this article was funded by Qatar National Library.
Author contributions
Conceptualization, Laith J. Abu-Raddad; Data curation, Houssein Ayoub, Hiam Chemaitelly and Laith J. Abu-Raddad; Formal analysis, Houssein Ayoub and Laith J. Abu-Raddad; Funding acquisition, Laith J. Abu-Raddad; Investigation, Houssein Ayoub; Methodology, Houssein Ayoub and Laith J. Abu-Raddad; Project administration, Laith J. Abu-Raddad; Supervision, Laith J. Abu-Raddad; Writing – original draft, Hiam Chemaitelly; Writing – review & editing, Houssein Ayoub, Hiam Chemaitelly, Ghina Mumtaz, Shaheen Seedat, Susanne Awad, Monia Makhoul and Laith J. Abu-Raddad.
Declaration of Competing Interest
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gloepi.2020.100042.
Appendix A. Supplementary data
Supplementary material
References
- 1.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172:577–582. doi: 10.7326/M20-0504. (PubMed PMID: 32150748) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. (PubMed PMID: 32091533) [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization Report of the WHO-China joint mission on coronavirus disease 2019 (COVID-19) 2020. https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf Available from:
- 4.COVID-19 outbreak live update. 2020. https://www.worldometers.info/coronavirus/ Available from: (Accessed: March 14, 2020)
- 5.World Health Organization (WHO) Naming the coronavirus disease (COVID-19) and the virus that causes it. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it Available from: (Accessed: March 11 2020)
- 6.World Health Organization (WHO) WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020. 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 Available from: (Accessed: March 14, 2020)
- 7.MATLAB® . The MathWorks, Inc; 2019. The language of technical computing. [Google Scholar]
- 8.Woods R.D., Saxon D.S. Diffuse surface optical model for nucleon-nuclei scattering. Phys Rev. 1954;95:577. [Google Scholar]
- 9.Velicia F.F. On the moments of a (WS) β distribution. J Phys A Math Gen. 1987;20:2293. [Google Scholar]
- 10.Awad S.F., Abu-Raddad L.J. Could there have been substantial declines in sexual risk behavior across sub-Saharan Africa in the mid-1990s? Epidemics. 2014;8:9–17. doi: 10.1016/j.epidem.2014.06.001. (PubMed PMID: 25240899) [DOI] [PubMed] [Google Scholar]
- 11.Ayoub H.H., Abu-Raddad L.J. Impact of treatment on hepatitis C virus transmission and incidence in Egypt: a case for treatment as prevention. J Viral Hepat. 2017;24:486–495. doi: 10.1111/jvh.12671. 28039923 [DOI] [PubMed] [Google Scholar]
- 12.Li R., Pei S., Chen B., Song Y., Zhang T., Yang W. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. (PubMed PMID: 32179701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. (PubMed PMID: 32074444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. 32003551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. (PubMed PMID: 32109013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. (PubMed PMID: 31986264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novel Coronavirus Pneumonia Emergency Response Epidemiology Team The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. (PubMed PMID: 32064853) [DOI] [PubMed] [Google Scholar]
- 18.United Nations Department of Economic and Social Affairs Population Dynamics The 2019 revision of world population prospects. 2020. https://population.un.org/wpp/ Available from. (Accessed on March 1st, 2020)
- 19.BBC Coronavirus: sharp increase in deaths and cases in Hubei. 2020. https://www.bbc.com/news/world-asia-china-51482994 Available from: (Accessed on: March 16, 2020)
- 20.Gunia A.Z.M. China reported a huge increase in New COVID-19 cases. Here's why it's actually a step in the right direction. https://time.com/5783401/covid19-hubei-cases-classification/ Available from:
- 21.Lagarias J.C., Reeds J.A., Wright M.H., Wright P.E. Convergence properties of the Nelder--Mead simplex method in low dimensions. SIAM J Optimiz. 1998;9:112–147. [Google Scholar]
- 22.Mizumoto K., Omori R., Nishiura H. Age specificity of cases and attack rate of novel coronavirus disease (COVID-19) medRxiv. 2020 doi: 10.1101/2020.03.09.20033142. 2020.03.09.20033142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J., Litvinova M., Liang Y., Wang Y., Wang W., Zhao S. Changes in contact patterns shape the dynamics of the COVID-19 outbreak in China. Science. 2020;368:1481–1486. doi: 10.1126/science.abb8001. (PubMed PMID: 32350060; PubMed Central PMCID: PMCPMC7199529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies N.G., Klepac P., Liu Y., Prem K., Jit M., Group CC-w Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat Med. 2020 doi: 10.1038/s41591-020-0962-9. (PubMed PMID: 32546824) [DOI] [PubMed] [Google Scholar]
- 25.Zhu Y., Bloxham C.J., Hulme K.D., Sinclair J.E., Tong Z.W.M., Steele L.E. Children are unlikely to have been the primary source of household SARS-CoV-2 infections. medRxiv. 2020 doi: 10.1101/2020.03.26.20044826. 2020.03.26.20044826. [DOI] [Google Scholar]
- 26.Colson P., Tissot-Dupont H., Morand A., Boschi C., Ninove L., Esteves-Vieira V. Children account for a small proportion of diagnoses of SARS-CoV-2 infection and do not exhibit greater viral loads than adults. Eur J Clin Microbiol Infect Dis. 2020;39:1983–1987. doi: 10.1007/s10096-020-03900-0. (PubMed PMID: 32845413; PubMed Central PMCID: PMCPMC7447969) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macartney K., Quinn H.E., Pillsbury A.J., Koirala A., Deng L., Winkler N. Transmission of SARS-CoV-2 in Australian educational settings: a prospective cohort study. Lancet Child Adolesc Health. 2020 doi: 10.1016/S2352-4642:30251-0. (PubMed PMID: 32758454; PubMed Central PMCID: PMCPMC7398658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstein E., Lipsitch M., Cevik M. On the effect of age on the transmission of SARS-CoV-2 in households, schools and the community. medRxiv. 2020 doi: 10.1101/2020.07.19.20157362. (PubMed PMID: 32743609; PubMed Central PMCID: PMCPMC7386533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merckx J., Labrecque J.A., Kaufman J.S. Transmission of SARS-CoV-2 by children. Dtsch Arztebl Int. 2020;117:553–560. doi: 10.3238/arztebl.2020.0553. (PubMed PMID: 32705983; PubMed Central PMCID: PMCPMC7376445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Kerkhove M.D., Hirve S., Koukounari A., Mounts A.W., Group HNpsw Estimating age-specific cumulative incidence for the 2009 influenza pandemic: a meta-analysis of A(H1N1)pdm09 serological studies from 19 countries. Influenza Other Respir Viruses. 2013;7:872–886. doi: 10.1111/irv.12074. (PubMed PMID: 23331969; PubMed Central PMCID: PMCPMC5781221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020 doi: 10.1126/science.abb5793. (PubMed PMID: 32291278; PubMed Central PMCID: PMCPMC7164482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sette A., Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat Rev Immunol. 2020;20(8):457–458. doi: 10.1038/s41577-020-0389-z. (PubMed PMID: 32636479; PubMed Central PMCID: PMCPMC7339790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. (PubMed PMID: 32473127; PubMed Central PMCID: PMCPMC7237901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd2071. (PubMed PMID: 32591408; PubMed Central PMCID: PMCPMC7319493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maltezou H.C., Vorou R., Papadima K., Kossyvakis A., Spanakis N., Gioula G. Transmission dynamics of SARS-CoV-2 within families with children in Greece: a study of 23 clusters. J Med Virol. 2020 doi: 10.1002/jmv.26394. (PubMed PMID: 32767703; PubMed Central PMCID: PMCPMC7441283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prem K., Cook A.R., Jit M. Projecting social contact matrices in 152 countries using contact surveys and demographic data. Plos Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005697. (PubMed PMID: WOS:000411981000011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mossong J., Hens N., Jit M., Beutels P., Auranen K., Mikolajczyk R. Social contacts and mixing patterns relevant to the spread of infectious diseases. Plos Med. 2008;5:381–391. doi: 10.1371/journal.pmed.0050074. (PubMed PMID: WOS:000254928900013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abu-Raddad L.J., Chemaitelly H., Ayoub H.H., Al Kanaani Z., Al Khal A., Al Kuwari E. Characterizing the Qatar advanced-phase SARS-CoV-2 epidemic. medRxiv. 2020 doi: 10.1101/2020.07.16.20155317. 2020.07.16.20155317v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
All data generated or analysed during this study are included in this article and its Supplementary Material.


