Abstract
The whole world is still suffering substantially from the coronavirus disease 2019 (COVID-19) outbreak. Several protein-based molecules that are associated with the Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which are essential for its functionality, survival, and pathogenesis have been identified and are considered as potential therapeutic targets. These protein-based molecules are either structural/non-structural components of SARS-CoV-2 or host factors, which play a crucial role in this infection. Developing drug molecules against these essential functional molecules to hinder their regular functioning and associated physiological pathways could be promising for successful clinical management of this novel coronavirus infection. The review aims to highlight the functional molecules that play crucial roles in SARS-CoV-2 pathogenesis. We have emphasized how these potential druggable targets could be beneficial in tackling the COVID-19 crisis.
Abbreviations: SARS-CoV, Severe Acute Respiratory Syndrome Coronavirus; COVID-19, Coronavirus Disease – 2019; PHEIC, Public Health Emergency of International Concern; ACE2, Angiotensin-Converting Enzyme – 2; RME, Receptor-Mediated Endocytosis; RTC, Replication-Transcription Complex; NSP, Non-Structural Protein; RNP, Ribonuceloprotein; TMPRSS2, Transmembrane Protease Serine 2; ORF, Open Reading Frame
Keywords: Therapeutic target, COVID-19, Pathogenesis, Spike proteins, TMPRSS2
1. Introduction
The 21st century has witnessed severe pneumonic pandemics caused by various coronaviruses (CoVs). In 2003, Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) broke out, and the incidence of respiratory ailments caused by the virus with a lethal rate of approximately 10% was reported from five different countries (Cheng, 2007, Lee, 2003). During the year 2012, the Middle East Respiratory Coronavirus (MERS-CoV) caused a similar pneumonic outbreak in the Arabian Peninsula. The virus had a case fatality rate (CFR) of approximately 35% (Zaki, 2012, de Groot, 2013). More recently, a new coronavirus emerged at the end of the year 2019, in December in the city of Wuhan, situated in the Hubei province of China. This novel coronavirus was named SARS-CoV-2 or Severe Acute Respiratory Syndrome Coronavirus-2 (Piyush et al., 2020). This novel coronavirus was encountered for the first time and was reported to cause respiratory disorders in infected individuals.
After the outbreak in China, SARS-CoV-2 infections are promptly reported in countries like the USA, India, Brazil, Russia, Colombia, Peru, Spain, Mexico, Argentina, etc. (Zhu, 2020). At the beginning of the outbreak, the symptoms of the novel coronavirus infection were found to be somewhat similar to that of the regular flu, caused by the seasonal influenza virus, and hence were confused with the same disease (Rajarshi et al., 2020). But as soon as the situation got worsened in the following days, on 31st January 2020, WHO (World Health Organization) officially declared the SARS-CoV-2 caused coronavirus disease 2019 (COVID-19) as PHEIC (Public Health Emergency of International Concern). Keeping in view the severity of the situation, the World Health Organization decided to declare COVID-19 as a pneumonia pandemic on 11th March 2020. Till date (20th October 2020), the total numbers of SARS-CoV-2 infected cases reported are 40. 4 Million and the total number of death due to SARS-CoV-2 reported are 1,12 Million across the globe.
The mortality rate of COVID-19 was reported to be approximately 3.4%. This rate is relatively lower than that of the previously encountered SARS-CoV and MERS-CoV that had a mortality rate of around 9.6% and 35%, respectively (Fani et al., 2020). COVID-19 poses a grave threat to individuals who have a history of severe health issues or are still suffering from health disorders like diabetes, hypertension, CVD (cardiovascular disease), or respiratory diseases (Yang, 2020, Fang et al., 2020). Apart from pre-existing diseases, other factors like gender and age also contribute crucially to the death rate (World Health Organization, 2020). Depending upon the intensity of the disease and the immunity of the individual, COVID-19 may last up to 6 weeks. For the infection to establish completely, a varying period of incubation has been reported. The incubation time may be reduced by a second exposure to the viral inoculum (Munster, 2020). Several sources have reported an average incubation period to be of 14 days, which is relatively longer than that in the case of SARS and MERS (World Health Organization, 2020, Bai, 2020, Lauer, 2020, Lessler, 2009, Backer et al., 2020).
Usually, the transmission of SARS-CoV-2 occurs via contact of contaminated hands with mouth, nose, or eyes, or the virus may get airborne in the form of aerosols, which may spread in the surrounding air as a result of sneezing or coughing by an infected individual. The fine aerosols contain the viral particles which can be inhaled directly by the healthy individuals nearby (Yang, 2020, Liu, 2020). The mechanism of pathogenesis of SARS-CoV and SARS-CoV-2 similar to each other to some extent and involves an interaction of spike glycoprotein of the virus with the ACE2 (angiotensin-converting enzyme 2) receptor expressed on the host cells (Zhao, 2020, Prabakaran et al., 2004). Apart from the structural proteins of the virus, several non-structural proteins (NSPs), such as Nsp12 (having RNA-dependent RNA polymerase activity) and Nsp5 (3-Chymotrypsin-like protease), etc., and some host proteins like TMPRSS2 as well as ACE2 receptors are also known to play a significant role in proliferation, pathogenesis, and survival of the virus in the host body. These essential functional molecules associated with the SARS-CoV-2 can be targeted, and consequently, similar sets of therapeutic targets need to be investigated further to find effective therapeutic strategies for COVID-19.
2. SARS-CoV-2: structural analysis and essential functional molecules
The current pneumonic crisis, COVID-19, caused by SARS-CoV-2, is similar to the pandemic of 2003 caused by SARS-CoV. Both the strains, i.e., SARS-CoV and SARS-CoV-2, are associated with the B lineage of the genera of β-coronaviruses. It has been reported that, to date, these coronaviruses are the largest RNA viruses. SARS-CoV-2 has a positive-sense single-stranded RNA genome (Rane, et al., 2020), which is arranged into a beads-on-string fashion and are complexed with the capsid protein, thus forming virus particles protein-ribonucleic helical core (Llanes, et al., 2020). The structural proteins of the virus particle are anchored to a lipid bilayer, which enwraps the genetic core. The most abundant triple spanning protein essentially required for assembly of the virus is the membrane (M) protein. The spike (S) protein provides a crown-like structural look, forming the trimeric spike that extends from the lipid membrane. Due to this crown-like appearance, these viruses are termed as ‘corona’ viruses. The spike protein plays a pivotal role in infectivity and transmissibility of the virus, internalization of the virus into the host cells, and tissue tropism. The smallest among all other structural proteins, the envelope (E) protein plays a major role in budding and maturation and is primarily confined with vesicle trafficking organelles such as Golgi and Endoplasmic Reticulum/Golgi interface. To make the viral replication possible inside the host cells, the structural proteins of the virus work concomitantly (Bárcena, 2009, Tseng, 2013, Schoeman and Fielding, 2019). In the recently conducted studies, it was found that, for infusion and entry inside the host cell, the SARS-CoV-2 makes use of ACE2 receptors expressed on the host cell surface. The virus is considered to be internalized via the process of RME, i.e., receptor-mediated endocytosis, after its binding to the ACE2 receptor. The viral genome, i.e., the viral RNA is released into the cytoplasm of the host cell after the internalization of the virus. In the cytoplasm, the translation of the viral genome takes place for the formation of the non-structural proteins, which establishes the replication transcription complex (RTC). The sub-genomic mRNAs required for encoding accessory and structural proteins are formed by RTC. The accessory viral proteins present in the form of viral buds or virions in the Golgi and endoplasmic reticulum along with the new RNA genome of the virus, which is bound to the nucleocapsid (N) protein, are all transported through vesicle cargoes for fusion with the plasma membrane and are ultimately released out of the host cell (Tripp and Tompkins, 2018, Hussain, 2005, Perrier, 2019, Enjuanes, 2004, Xia, 2020, Yu, 2020, Zhang et al., 2014).
2.1. Viral RNA genome
An approximately 30 kB long positive-sense single-stranded RNA constitutes the viral genome of the SARS-CoV-2. After the complete phylogenetic analysis of the RNA genome of the virus, it was revealed that 29,903 nucleotides were present in the viral genome, and it shared 89.1% similarity with the previously encountered SARS-CoV (Rajarshi et al., 2020a, Rajarshi et al., 2020b). The viral genome consisted of 5́ and 3́ UTRs (untranslated regions) at either end. The open reading frames ORF1a and ORF1b, which encodes for a polyprotein, are situated at the 5́ proximal RNA genome. This polyprotein, after its proteolytic cleavage, leads to the formation of 16 different NSPs (non-structural proteins), including RNA-dependent RNA polymerases (RdRp) like Nsp12, viral proteases like Nsp3, Nsp5 and cysteine protease helicase like Nsp13, and several other NSPs which are anticipated to be involved in the transcription and replication of the viral genome. 12 nested ORFs which are essentially required for encoding the main structural proteins, i.e., envelope (E), spike (S), nucleocapsid (N), membrane (M), and several other accessory proteins, are situated at the distal portion of the genome of the virus, toward 3́ end. The analysis of the viral genome has significantly assisted in acquiring more understanding about the SARS-CoV-2. Previously reported recombination hotspots, i.e., spike, orf3b, orf8, regions, were remarkably differentiated via the aid of sequence analysis (Chan, 2020). Various frame shift elements and transcriptional regulatory elements like stem-loop structures situated at 5́ and 3́ UTRS contribute towards the complex translational and transcriptional properties of the RNA of the virus (Sola, 2015, Irigoyen, 2016, Rangan et al., 2020, Gordon, 2020). Comprehending the sub-genomic mRNA sequences and more understanding regarding the secondary genomic RNA structures might help in the establishment and development of genome targeted therapeutics, apart from just knowing about genetic annotations.
2.2. Spike (S) protein of SARS-CoV-2
Spike (S) proteins are class I fusion proteins expressed on the viral surface. These proteins are densely glycosylated and consist of a large ectodomain, i.e., a single-pass transmembrane domain which provides anchorage for the proteins to the lipid bilayer as well as to a small intracellular segment. S1 and S2 are the two subunits comprised of the ectodomain, which forms homotrimers. The S1 subunit consists of a C-terminal functional domain that is involved in binding with the receptor. The S2 subunit comprises a cytoplasmic domain that assists in the fusion of viral envelope with the membrane of the host cell through the endosomal pathway, a transmembrane domain, and a fusion peptide called heptad repeat 1 and 2 (HR1 and HR2) (Rane, et al., 2020). The S protein is present in a pre-fusion form on the surface of a virus particle (Li, 2016). After the contact of the virus with the host cell, the host cell membrane proteases like transmembrane protease serine-2 (TMPRSS2) are responsible for the priming of S protein, to carry out internalization efficiently via the process membrane wrapping (Hoffmann, 2020, Walls, 2020). The structural elucidation of SARS-CoV-2′s receptor-binding domain (RBD) reported that its binding affinity with the ACE2 receptor is approximately ten times stronger than the previously encountered SARS-CoV. Also, the S2 domain of the SARS-CoV-2 was reported to be relatively more flexible than the SARS-CoV (Wrapp, 2020). Another major difference between the structure of spike proteins of SARS-CoV and SARS-CoV-2 is the position of RBDs in their respective down conformations. In the case of SARS-CoV, the RBD packs tightly against the NTD (N-terminal domain) of the neighbouring protomer, in the down protomer, whereas in the case of SARS-CoV-2, RBD is angled closer to the trimer’s central cavity in the down conformation (Wrapp, 2020). The spike protein of SARS-CoV-2 bears proteolytic sites known as the S1/S furin-like rift spot, which is not present in SARS-CoV and is reported to enhance the pathogenicity of the virus, thus distinguishing both the viruses. It has also been reported that the furin-like rift spot also results in the enhancement of the tissue tropism of the viruses (Cheng, 2019, Coutard, 2020).
The spike protein is exposed to an immense evolutionary pressure as it is the first contact site between viruses and the host cells. The transmission and infectivity of the viruses are greatly influenced by the changes in the spike protein. In the case of the SARS-CoV-2, the spike protein underwent several changes like a furin-like cleavage site, and changes at the binding sites of the receptors are being considered as the reason behind species jumping and transmission among humans efficiently. Additionally, it has also been reported that the SARS-CoV-2 forms syncytium, which allows the spreading of viruses via cell–cell fusion and might also contribute towards its rapid infectivity (Xia, 2020).
2.3. Main protease of SARS-CoV-2
The 3C-like protease, encoded by Nsp5, is also known as the main protease (Mpro). The Mpro is known to be the first protein to get auto-cleaved and then further leads to the cleavage of polyprotein into discrete members of NSPs at the LeuGln↓ (Ser, Ala, Gly) cleavage site. The stable active form of the Mpro is achieved in the form of an octamer whilst its dimer and monomer subsist in equilibrium (Xia and Kang, 2011). The structural arrangement of SARS-CoV-2 is nearly 96% similar to that of the SARS-CoV. It was revealed from the SARS-CoV-2 Mpro crystal structure that the non-polar amino acids in the interface of dimer replaced the polar ones. This mutation led to the enhanced catalytic activity of the dimer, whereas the dimer dissociation constants were reported to have remained the same (Zhang, 2020). For the replication and production of virus proteome, protease is an essential requirement, and hence, Mpro is suggested as one of the potential targets for curing COVID-19.
2.4. RNA dependent RNA polymerase of SARS-CoV-2
Very complex protein machinery is involved in the viral genome replication, as well as the transcription of SARS-CoV-2. The proteins associated with this complex are part of a large polypeptide chain and are translated from ORF1a and ORF1b. On the whole, the SARS-CoV-2′s RNA dependent RNA polymerase (RdRp) complex structure corresponds to that of the SARS-CoV. The RNA dependent RNA polymerase, i.e., Nsp 12, is the key component of the enzyme complex. The complex formation is assisted by other co-factors like Nsp7, Nsp8, along with Nsp12 and the development of viral genomic RNA is catalyzed. The template-directed RNA synthesis is catalyzed at the central grove of the complex. The positively charged nascent RNA strands exit path, which is also accessible to the solvents, facilitates the electrostatic interaction between the enzyme complex and nascent RNA strand (Lai, 2020, Subissi, 2014).
The Nsp12 (RNA polymerase unit) can be considered as a reassuring potential target for the development of therapeutic drugs like remedesivir (CID: 121304016) and viral inhibitors. Drugs like remedesivir inhibit the RdRp activities and have now entered into clinical trials for developing a promising cure for COVID-19.
3. Basic molecular mechanism of coronavirus pathogenesis
The transmission of SARS-CoV-2 usually occurs via close contact among individuals and the droplet infection, as well as via stool, sweat, exposure to respiratory secretions, and urine (Guo, 2008, Gu and Korteweg, 2007, Ding, 2004). Upon the entry inside the body, the virus binds to pneumocytes and enterocytes, which are the primary target cells, as a result of which, the virus instigates an infection and replication cycle. CoVs like SARS-CoV-2 may also target other cells, including the kidney’s tubular epithelial cells, neuronal cells, renal epithelial tubules, and also cerebral cells and immune cells (Guo, 2008, Gu and Korteweg, 2007).
Spike proteins play a significant role in the attachment of CoVs like SARS-CoV-2 to the target or the host cells. The attachment is brought about by the interaction of spike protein-host cell protein, i.e., ACE2 receptors (Li, 2003). Along with its nucleocapsid protein, the viral genome gets released into the host cell’s cytoplasm, after the recognition of the receptor by the virus. The viral genomes consist of two genes, namely, ORF1a and ORF1b, that produce two polyproteins (PPs), i.e., pp1a and pp1ab (Te Velthuis et al., 2012). Both pp1a and pp1ab are involved in the development and arrangement of the replication transcription complex by obtaining command over the host ribosomes to facilitate their own process of translation (Te Velthuis et al., 2012, Stobart, 2013). Following the processing of polyproteins by the protease, the production of 16 different NSPs takes place, and each NSP has its specified function for the virus.
The synthesized structural proteins like a spike (S), membrane (M), and envelope (E) enter inside the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) complex and are then employed in developing the viral envelope structure (Narayanan, 2000). On the contrary, the formation of the ribonucleoprotein (RNP) complex takes place by the association of the replicated genome and the nucleocapsid (N) protein. The S, E, and M proteins are involved in the formation of the outer cover (Narayanan, 2000). Ultimately, by forming a structure that resembles a bud, the viral particle buds off and is shot out of the ERGIC complex (de Wit, 2016). At last, vesicle formation is carried out by the mature virions. These virus particles are thus released outside into the extracellular region by fusion of the vesicles, formed by virions, with the plasma membrane (de Wit, 2016, Nieto-Torres, 2011). The mechanism of pathogenesis, as well as the regeneration and life cycle of SARS-CoV-2, is described in Fig. 1 .
Fig. 1.
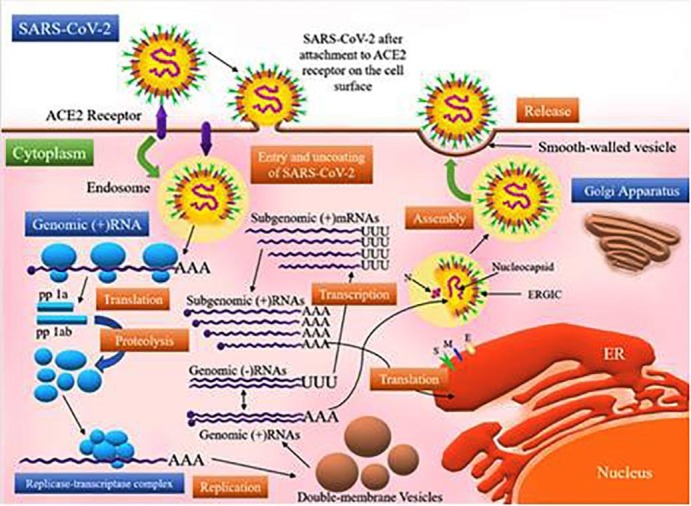
Mechanism of infection and life cycle of SARS-CoV-2 in human cells. The spike (S) glycoproteins of SARS-CoV-2 bind to the angiotensin-converting enzyme 2 (ACE2) receptors expressed on the surface of the host cell. This binding facilitates the entry of the virus inside the host cell via an endosomal pathway. Upon its entry, the virus releases its RNA in the cytoplasm. The polyproteins pp1a and pp1ab are then produced by the translation of ORF 1a and ORF1ab. These polyproteins are further cleaved by the proteases, which results in the production of 16 NSPs. The RTC (replication transcription complex) uses the genomic (+) RNA as the template. The genomic (+) RNA produced via the process of replication becomes the new virus particle’s genome. The subgenomic mRNAs produced by transcription are translated into structural proteins, i.e., spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins, which are responsible for the formation of the viral particle. The S, M, and E proteins enter the ER (endoplasmic reticulum) and the N protein combines with the genomic (+) RNA, forming a nucleoprotein complex. At last, all the viral components are integrated into the ERGIC (ER-Golgi intermediate compartment) and are ultimately released in the extracellular environment through exocytosis.
The coronavirus infection usually induces a surge of pro-inflammatory chemokines and cytokines, thus resulting in functional deterioration and damages to the tissues of infected organs, especially lungs, and even leads to failure of lung and deaths in several cases (Du, 2009).
4. SARS-CoV-2: potential therapeutic drug targets
Based on certain pathways, the therapeutic approaches against the CoVs have been classified into various categories. These therapeutic approaches may include targeting the structural proteins of the virus, which may hinder the self-assembling process of the virus or may block the attachment of the virus to the receptors expressed on the human cells. Some approaches may inhibit the replication and synthesis of viral RNA by targeting the functional proteins and enzymes essentially required by the virus. The therapeutic approaches that aim at blocking the viral entry inside the host cell are based on the idea of directly acting on the enzymes or receptors which are specific for the host. Another therapeutic approach may work on the principle of restoring the innate immunity of the host by introducing certain virulence factors. Several target proteins which can be considered as potential therapeutic targets for curing COVID-19 are Nsp1, Nsp7-Nsp8 complex, Nsp14-Nsp16 complex, Nsp3 (which further includes Nsp3b, Nsp3c, Nsp3e, and PLpro), E (envelope) protein/E-channel, 3CLpro, S (spike) protein, M (membrane) protein, C-terminal RNA binding domain (CRBD), helicase, RNA-dependent RNA polymerase, TMPRSS2, ACE2 receptors, and N-terminal RNA binding domain (NRBD). We analysed the amino acid sequence of S and M protein using bioinformatics tools such as Clustal W with default parameters. Our comprehensive multiple sequence analysis revealed a few conserved putative motifs at the heterogeneous N-terminal polypeptide (Fig. 2 A) region of S-protein from different strains of CoV. Intriguingly, the C-terminus region of S-protein showed more conserved amino acids compared to N-terminus (Fig. 2A). On the other hand, analysis of M-protein from different strains (reported from different regions/country) fetched conserved motifs throughout the polypeptide chain (Fig. 2B) with comparatively less variation at the N-terminus. We are speculating that these motifs probably have a direct role in the viral maturation process, and therefore, targeting these highly conserved regions may open some promising avenues in drug discovery against this novel respiratory pathogen.
Fig. 2.
(A) Organization of functional motifs on the polypeptide chain of S-protein of SARS-CoV-2. Schematics are drawn approximately to the scale and represent the approximate consensus of representative homologs. Two long conserved stretches are represented in green and black at C-terminal domain of polypeptide. Smaller motifs (orange and purple) are also found at N-terminal region. (B) Organization of the functional motif on the polypeptide chain of M-protein of SARS-CoV-2. The highly conserved region (black box) at N-terminal domain of polypeptide could be a potential drug target. Sequence Alignment analysis was performed by CLUSTALW (https://www.genome.jp/tools-bin/clustalw) using the default parameters.
4.1. Viral structural proteins as potential therapeutic targets
The spike (S) protein of SARS-CoV-2 is expressed in a trimeric form on the surface of the virus and imparts the crown-like appearance (hence the name coronavirus), which is the prime structural protein. The host-virus interaction, as well as the determination of tissue tropism or host tropism, is mediated by the spike protein as the attachment of spike protein with the receptors present on the host cell facilitate the invasion of the virus (Millet and Whittaker, 2015). TMPRSS2, which is a host cell protease, cleaves the spike protein into two subunits, i.e., S1 and S2. S1 primarily binds with the surface receptor expressed on the host cell, whereas the virus to cell and cell to cell membrane fusion is carried out by the S2 subunit of the spike protein. Activation by cleavage and structural integrity of the spike protein plays a significant role in the virulence of CoVs and their invasion (Xia, 2014). Spike protein or the host cell receptors can be considered as potential therapeutic targets for developing anti-viral drugs, which can block the viral invasion inside the host cell or the host-virus interaction (Rane, et al., 2020). Some small molecular compounds were used against the spike protein in the virtual ligand screening studies, and the results suggested that some drugs like itraconazole (CID: 55283) and posaconazole (CID: 468595), which are antifungal drugs, anti-coagulant drugs like dabigatran (CID: 216210) etexilate (CID: 135565674), anti-hypertensive drugs such as iloprost (CID: 5311181), prazosin (CID: 4893), and rescinnamine (CID: 5280954) and some anti-bacterial drugs like cefsulodin (CID: 656644), sulfasalazine (CID: 5339), azlocillin (CID: 6479523), and penicillin (CID: 2349) manifested low binding energy and high binding affinity with the spike protein (Wu, 2020).
Among the major structural proteins of SARS-CoV-2, envelope (E)/E-channel protein plays a significant role in the maintenance of the host virulence and structural integrity of the virus. The nucleocapsid (N) protein’s C-terminal RNA binding domain (CRBD) and N-terminal RNA binding domain (NRBD) are essentially required by the N proteins when present inside the host cell, in order to efficiently bind with the RNA of the CoV. Various anti-inflammatory, anti-asthmatic, anti-bacterial, anti-tumor, and antibacterial drugs, were used for study against targets like E proteins and N proteins, especially NRBD and CRBD, and exhibited a decent binding affinity. Hence, the CRBD and NRBD of N protein along with the E protein can be used as possible therapeutic drug targets for designing anti-coronavirus drugs (Wu, 2020).
4.2. Therapeutic targets inhibiting replication and synthesis of viral RNA
In the case of CoVs, NSPs are engaged as a prominent functional protein in transcribing RNA, translating, and synthesizing proteins. Processes like the synthesis of proteins, RNA translation, transcription, modification, processing, host infection, and viral replication are all carried out by NSPs. The functions of the major NSPs are listed in Table 1 . The biological functions of NSPs like RNA dependent RNA polymerase, PLpro, helicase, and 3CLpro are well known, and due to the clarity about their essential active site for the enzyme, they are considered as major targets for developing small-molecular inhibitors. Nsp12-RdRp has been given significant importance in several studies conducted to find out inhibitors for CoVs. Nsp12-RdRp can be considered as a potential drug target since its targeted inhibition could not induce any significant side effects and notable toxicity on host cells (Chu, 2006).
Table 1.
Function of the major non-structural proteins associated with SARS-CoV-2, which can be treated as potential therapeutic targets (Forni, 2016).
| Non-Structural Proteins (NSPs) | Associated Functions |
|---|---|
| Nsp1 |
|
| Nsp2 |
|
| Nsp3 |
|
| Nsp4 |
|
| Nsp5 |
|
| Nsp6 |
|
| Nsp7-Nsp8 Complex |
|
| Nsp9 |
|
| Nsp10, Nsp14, and Nsp16 |
|
| Nsp11 |
|
| Nsp12 |
|
| Nsp13 |
|
| Nsp14 |
|
| Nsp15 |
|
| Nsp16 |
|
4.2.1. RdRp (RNA-dependent RNA polymerase)
Nsp12 is a conserved coronavirus protein which acts as an RNA-dependent RNA polymerase and is a crucial component of the viral transcription and replication complex. The polymerase’s RdRp domain consists of a conserved Ser-Asp-Asp sequence motif and is situated at the C-terminus (Subissi, 2014). A primer of length up to six nucleotides can be de novo synthesized by the aid of Nsp8, which can be employed in RNA synthesis using RdRp. Additionally, the enhancement in Nsp12’s RdRp activity along with an increase in the binding of Nsp12 to the RNA is facilitated by the Nsp7-Nsp8 complex (Imbert, 2006). The results of the virtual ligand screening study suggested that some anti-bacterial drugs like novobiocin (CID: 54675769), chenodeoxycholic acid (CID: 10133), anti-fungal drugs like itraconazole (CID: 55283), anti-tumor drugs including idarubicin (CID: 42890), anti-allergic drugs like cortisone (CID: 222786), liver protective drugs like silybin (CID: 31553), an anticoagulant drug like dabigatran etexilate (CID: 135565674) and muscle relaxants like pancuronium bromide (CID: 27350), anti-viral drugs like Remdesivir (CID: 121304016), Favipiravir, (CID: 492405) Ribavirin (CID: 37542) and Sofosbuvir (CID: 45375806), etc. repurposing as potential RdRp inhibitors (Wu, 2020).
4.2.2. PLpro (Papain-like protease)
The release of Nsp1, Nsp2, and Nsp3 upon the cleavage of replicase polyprotein’s N-terminus is facilitated by PLpro. The released NSPs, i.e., Nsp1, Nsp2, and Nsp3, are crucially required for regulation and correction of replication of the virus (Harcourt, 2004). PLpro has also been reported to remarkably aggravating the innate immunity of the host (Chen, 2014, Yuan, 2015, Li, 2016). PLpro has been a pre-eminent enzyme in carrying out the viral replication as well as host infection, and therefore is considered as one of the crucial targets for inhibition of CoVs. In the results of a virtual ligand screening study, it was revealed that a series of anti-bacterial drugs like cefamandole (CID: 456255), tigecycline (CID: 54686904), chloramphenicol (CID:), anti-viral drugs like thymidine (CID: 5789), ribavirin (CID: 37542), valganciclovir (CID: 135413535), Phaitanthrin D (CID: 44561298), Chrysin (CID: 5281607), anti-tussive drugs like levodropropizine (CID: 65859) and muscle relaxant drugs including carbamate (CID: 276) and chlorphenesin (CID: 7697), contrast agent like Iopromide (CID: 3736) showed a high binding affinity with PLpro. The proficient hydrogen bonding and strong hydrophobic interactions between the anti-viral drug ribavirin (CID: 37542) with the PLpro enzyme suggested that it can be considered as a formidable inhibitor for PLpro (Wu, 2020).
4.2.3. Nsp13/helicase
Nsp13, also known as helicase, which includes a Hel (helicase) domain along with an N-terminal MBD metal-binding domain (MBD), is a protein with multiple functionalities. A total of 26 cysteine residues constitute the N-terminal structures and form a Zn2+ binding domain, whereas the conserved sequence motif at the C-terminus constitutes the Hel domain. Unpacking or unwinding of the double-stranded DNA/RNA is carried out by Nsp13 in an NTP-dependent manner along 5′ to 3′ direction (Ivanov and Ziebuhr, 2004). Notably, the SARS-Nsp13 sequence has been reported to be a vital component for CoV replication and has also been reported to be conserved, which makes it a potential target for developing antiviral drugs. Very few studies and reports have discussed the inhibitors for Nsp13 (Shum and Tanner, 2008, Jang, 2008). As suggested by the results of studies based on the structural modeling of helicase protein and virtual ligand screening, it was revealed that antiviral drugs like simeprevir (CID: 24873435), paritaprevir (CID: 45110509) anti-HIV-1 (human immune deficiency virus – 1) drug like saquinavir (CID: 441243), canrenoic (CID: 656615), which is a diuretic drug, an anti-fungal drug like itraconazole (CID: 55283), dabigatran (CID: 216210), which is an anti-coagulant drug, Ergotamine (CID: 8223) which is a drug used in to prevent vascular headaches and some antibacterial drugs including cefsulodine (CID: 656575), rolitetracycline (CID: 54682938), cefoperazone (CID: 44187), cefpiramide (CID: 636405) and lymecycline (CID:54707177) were anticipated to be potent inhibitors for helicase protein (Wu, 2020).
4.2.4. Mpro/3-Chymotrypsin-like protease (3C-like main protease/3CLpro)
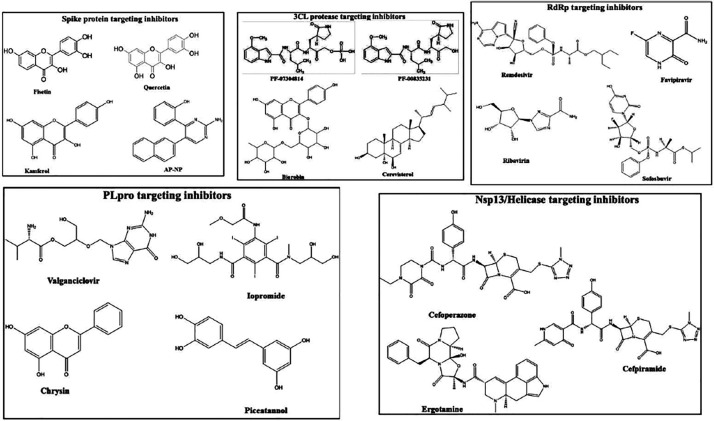
Nsp5 is also recognized as the 3CLpro and is known for its function to produce mature enzymes upon their automatic cleavage from polyproteins. In addition to their cleavage from the polyproteins, they carry out the cleavage of downstream NSPs at 11 different sites for the Nsp4-Nsp16 release (Yang, 2005). The maturation of the NSPs, which are critical for the viral life cycle, is directly effectuated and imparted by 3CLpro. The comprehensive and precise investigation of the mechanism of catalysis and structural organization of 3CLpro makes it an appealing target for the development of anti-coronavirus drugs. Small-molecular or peptide inhibitors are mainly involved in targeting 3CLpro of CoVs (Pillaiyar, 2016). The monomer of 3CLpro primarily has 3 domains, i.e., domain I, domain II and domain III, comprising residues 8–101, 102–184 and 201–303, respectively. Domains II and III are connected via a long loop that is constituted of residues 185 to 200. 3CLpro consists of a CysHis catalytic dyad (i.e., Cys145 and His41), and is situated in the gap between domain I and domain II. Various anti-hypertensive drugs including nicardipine, telmisartan along with anti-bacterial drugs like demeclocycline, lymecycline, oxytetracycline, cerevisterol and doxycycline, antioxidant like Hesperidin, etc. show a very high 3CLpro binding affinity, thus making 3CLpro as an attractive therapeutic target for SARS-CoV-2 treatment (Wu, 2020). Montelukast, which is an anti-asthmatic drug, displayed low binding energy to 3CLpro and fitted well inside the active site of 3CLpro. The stability of the compound at the active site was maintained by the presence of hydrophobic amino acids and H-bonding between the carbonyl group of Montelukast and Asn142 (Wu, 2020). The structure of the compounds which were reported to target the possible drug target sites present in SARS-CoV-2 is shown in Fig. 3 .
Fig. 3.
Chemical structures of the potential drug-like compounds that target the different druggable sites in SARS-CoV-2.
4.2.5. Other NSPs as potential targets
The viral RNA synthesis and replication are supported by several other crucial non-structural proteins (NSPs) like Nsp7-Nsp8 complex, Nsp3b, Nsp3e, Nsp9, Nsp10, Nsp14, Nsp15, and Nsp16. These NSPs can be used as a potential target in developing anti-viral drugs against SARS-CoV-2. The results from the studies based on virtual ligand screening and structural modeling of these proteins have demonstrated that several anti-viral, anti-inflammatory and anti-bacterial drugs documented at the ZINC drug database as well as in-house naturally derived product database have shown assuring results in binding with these target NSPs with admirable binding affinities (Wu, 2020).
4.3. Potential therapeutic targets obstructing host-virus interactions
At present, the host-specific receptor for SARS-CoV-2, i.e., the ACE2 (angiotensin-converting enzyme – 2) is contemplated as a potential host-specific target for treating COVID-19 and hindering the entry of SARS-CoV-2 inside the host cells (Ge, 2013). The sequence of SARS-CoV-2′s RBD is somewhat identical to that of the previously encountered SARS-CoV (Wan, 2020) and recent studies including the virtual screenings have made it clear that several drugs like losartan, which is an anti-hypertensive medication, ergotamine (analgesic drug), anti-diabetes drugs like troglitazone, liver-protective druglike silybin and anti-bacterial drug-like cefmenoxime, etc., showed low binding energy and high binding affinity with the ACE2 receptors.
Additionally, some studies have demonstrated that targeting and inhibiting the enzyme activity of TMPRSS2, which induces cleavage of S protein and facilitates the CoV infection, can bar the entry of coronaviruses inside the host cell (Glowacka, 2011). The results obtained from the virtual ligand screening studies depict that several anti-viral natural compounds like neoandrographolide, phyllaemblicin, kouitchenside I, and phyllaemblicin G7 along with some anti-bacterial drugs including cefoperazone, pivampicillin, clindamycin, and hetacillin displayed properties of a potent TMPRSS2 inhibitor, and thus it can be considered as a possible therapeutic target (Wu, 2020). Integrated multi-omics studies allow high-throughput screening of the potential therapeutic targets and host-pathogen protein–protein interactions. Such unbiased systems-level approaches are found to be extremely promising in the identification of the potential drug targets for this novel coronavirus (Ray and Srivastava, 2020). A recent study by Gordon et al. identified 332 high confidence SARS-CoV-2-human protein–protein interactions using affinity-purification mass spectrometry (Gordon, 2020). Importantly, 66 of these SARS-CoV-2 interacting host factors could be targeted by several existing FDA-approved drugs or drugs in clinical trials. The authors demonstrated those pharmacological inhibitors of mRNA translation and predicted regulators of the Sigma1 and Sigma2 receptors show antiviral activity against this pathogen. It is also important to note that majority of these host factors that interact with SARS-CoV-2 show robust 24-hour oscillations in mammals (Ray and Reddy, 2020). Therefore, the rhythmic patterns in the expression and activity profiles of these potential drug targets need to be considered while designing and dosing the candidate drugs.
4.4. Therapeutic targets restraining the virulence of SARS-CoV-2
ORF7a, Nsp1, and Nsp3c are the three main factors that assist the CoVs in evading the immune system and are responsible for meddling with the innate immunity of the host, thus enhancing the virulence of the virus. The release of freshly-assembled CoVs from the host cell is inhibited by bone marrow matrix antigen 2 (BST-2). The activity of BST-2 is inhibited by ORF7a of the CoV as it directly binds to BST-2 and blocks its glycosylation (Taylor, 2015). Inhibition of the production of type – I interferon and specific degradation of host mRNA is executed by the interaction of Nsp1 and the 40S ribosomal subunit of the host (Kamitani, 2006, Narayanan, 2008). CoVs are assisted by the Nsp3c in evading the innate immune system of the host as Nsp3c attaches with ADP-ribose of the host (Forni, 2016). Since ORF7a, Nsp1, and Nsp3c play a crucial role in imparting virulence; therefore, they can be considered as potential targets for developing anti-coronavirus drugs.
The results obtained from the study of virtual ligand screening involving ORF7a, Nsp1, and Nsp3c suggest that various natural products possessing anti-inflammatory or anti-bacterial properties as well as several clinical drugs showed high binding affinity to these three protein targets. Several anti-biotic compounds like streptomycin (CID: 19649), cefpiramide (CID: 636405), tetracycline (CID: 54675776), lymecycline (CID: 54707177), and piperacillin (CID: 43672) along with some naturally derived compounds showing anti-inflammatory effects like S. baicalensis derived wogonoside (CID: 12004622), Platycodon grandifloras derived platycodin D (CID: 162859), Vitex negundo derived vitexin (CID: 5280441), xanthones (CID: 7020) obtained from Swertia genus, and derivatives of andrographolide (CID: 5318517) showed considerable binding affinity to the protein targets (Wu, 2020). The possible drugs/therapeutic molecules that may be effective in targeting the essential functional molecules associated with SARS-CoV-2 are enlisted in Table 2 .
Table 2.
Potential therapeutic molecules/drugs that may be effective in targeting the various essential functional molecules associated with SARS-CoV-2 (Wu, 2020).
| Functional molecules associated with SARS-CoV-2 that may act as potential targets | Potential therapeutic drugs/molecules acting on the targets |
|---|---|
| Nsp1 |
|
| Nsp3 |
|
| Nsp7-Nsp8 Complex | Arbidol |
| Nsp12 | Remedesivir |
| Nsp13 | Lopinavir |
| Nsp14 | Arbidol |
| Nsp15 | Arbidol |
| Spike (S) protein |
|
| Envelope (E) protein |
|
| NRBD | Lopinavir |
| TMPRSS2 | Remidesivir |
| ACE2 complex | Hesperidin – targets the binding interface of S protein and ACE2 |
5. Conclusion
The structural elucidation of SARS-CoV-2 reveals several significant functional molecules associated with the virus, essentially required for their survival and pathogenesis in humans. These protein-based molecules have shown promising results in several in silico studies and are efficient targets that can assist in developing anti-SARS-CoV-2 drugs. Several drugs and naturally derived compounds possessing anti-biotic activities have shown promising results in studies based on virtual ligand screening and exhibited good binding affinity with the molecules of interest. The number of COVID-19 cases is increasing steadily, and it is of utmost importance that a therapeutic strategy is developed, which can tackle the crisis more effectively. To date, no drug or vaccine has been reported to cure or prevent SARS-CoV-2 infection. Consequently, developing a strategy that directly targets the crucial components of this virus may provide substantial therapeutic benefits.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
KR, TR and SR thank IIEST, Shibpur, Howrah, West Bengal, Bihar Agriculture University, Sabour (Bhagalpur) and Mahatma Gandhi Central University Motihari, Bihar, respectively.
References
- Cheng V.C., et al. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20(4):660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- Zaki A.M., et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- de Groot R.J., et al. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 2013;87(14):7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piyush, R., A. Chatterjee, and S. Ray, Plasma therapy a possible hope to fight against COVID-19. 2020.
- Zhu N., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarshi K., Chatterjee A., Ray S. Combating COVID-19 with mesenchymal stem cell therapy. Biotechnol. Rep, 2020 doi: 10.1016/j.btre.2020.e00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fani, M., Teimoori, A., Ghafari, S.J.F.V., 2020. Comparison of the COVID-2019 (SARS-CoV-2) pathogenesis with SARS-CoV and MERS-CoV infections.
- Yang, J., et al., Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. 2020. 94: p. 91–95. [DOI] [PMC free article] [PubMed]
- Fang, L., Karakiulakis, G., Roth, M.J.T.L.R.M., Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? 2020. 8(4): p. e21. [DOI] [PMC free article] [PubMed]
- World Health Organization. WHO Director-General's opening remarks at the media briefing on COVID-19 - 3 March 2020. 2020 [cited 2020 June 15]; Available from: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---3-march-2020.
- Munster V.J., et al. A novel coronavirus emerging in China – key questions for impact assessment. N. Engl. J. Med. 2020;382(8):692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- Bai Y., et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323(14):1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S.A., et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessler J., et al. Incubation periods of acute respiratory viral infections: a systematic review. Lancet. Infect. Dis. 2009;9(5):291–300. doi: 10.1016/S1473-3099(09)70069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer J.A., Klinkenberg D., Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Eurosurveillance. 2020;25(5):2000062. doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. Does hand hygiene reduce SARS-CoV-2 transmission? Graefe's Arch. Clin. Experim. Ophthalmol. 2020:1–2. doi: 10.1007/s00417-020-04652-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., et al. Aerodynamic characteristics and RNA concentration of SARS-CoV-2 aerosol in Wuhan hospitals during COVID-19 outbreak. BioRxiv. 2020 [Google Scholar]
- Zhao Y., et al. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. BioRxiv. 2020 [Google Scholar]
- Prabakaran P., Xiao X., Dimitrov D.S. A model of the ACE2 structure and function as a SARS-CoV receptor. Biochem. Biophys. Res. Commun. 2004;314(1):235–241. doi: 10.1016/j.bbrc.2003.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane, J.S., et al., Targeting SARS-CoV-2 Spike Protein of COVID-19 with Naturally Occurring Phytochemicals: An in Silco Study for Drug Development, 2020, ChemRxiv. [DOI] [PMC free article] [PubMed]
- Llanes, A., et al., Betacoronavirus genomes: How genomic information has been used to deal with past outbreaks and the COVID-19 pandemic. 2020. [DOI] [PMC free article] [PubMed]
- Bárcena M., et al. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. 2009;106(2):582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y.-T., et al. Identifying SARS-CoV membrane protein amino acid residues linked to virus-like particle assembly. PLoS ONE. 2013;8(5) doi: 10.1371/journal.pone.0064013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virology J. 2019;16(1):69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp, R.A. and S.M. Tompkins, Roles of Host Gene and Non-coding RNA Expression in Virus Infection. Vol. 419. 2018: Springer.
- Hussain S., et al. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79(9):5288–5295. doi: 10.1128/JVI.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier A., et al. The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J. Biol. Chem. 2019;294(39):14406–14421. doi: 10.1074/jbc.RA119.008964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes, L., Coronavirus replication and reverse genetics. Vol. 287. 2004: Springer Science & Business Media.
- Xia S., et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020:1–3. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., et al., Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes and infection, 2020. [DOI] [PMC free article] [PubMed]
- Zhang N., Jiang S., Du L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev. Vaccines. 2014;13(6):761–774. doi: 10.1586/14760584.2014.912134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarshi, K., Chatterjee, A., Ray, S., BCG vaccination strategy implemented to reduce the impact of COVID-19: Hype or Hope? 2020: p. 100049. [DOI] [PMC free article] [PubMed]
- Chan J.F.-W., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., et al. Continuous and discontinuous RNA synthesis in coronaviruses. Annual Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoyen N., et al. High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog. 2016;12(2) doi: 10.1371/journal.ppat.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan R., Zheludev I.N., Das R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses. BioRxiv. 2020 doi: 10.1261/rna.076141.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane, J.S., et al., Targeting virus-host interaction: An in silico approach to develop promising inhibitors against COVID-19. 2020.
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annual Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp, D., et al., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. 2020. 367(6483): p. 1260–1263. [DOI] [PMC free article] [PubMed]
- Cheng J., et al. The S2 subunit of QX-type infectious bronchitis coronavirus spike protein is an essential determinant of neurotropism. Viruses. 2019;11(10):972. doi: 10.3390/v11100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B., Kang X. Activation and maturation of SARS-CoV main protease. Protein Cell. 2011;2(4):282–290. doi: 10.1007/s13238-011-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L., et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., et al. Pathogenetic mechanisms of severe acute respiratory syndrome. Virus Res. 2008;133(1):4–12. doi: 10.1016/j.virusres.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 2007;170(4):1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., et al. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J. Pathol.: A J. Pathol. Soc. Great Britain and Ireland. 2004;203(2):622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Velthuis A.J., van den Worm S.H., Snijder E.J. The SARS-coronavirus nsp7+ nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40(4):1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobart C.C., et al. Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J. Virol. 2013;87(23):12611–12618. doi: 10.1128/JVI.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., et al. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74(17):8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., et al. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., et al. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., et al. Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. Virus Res. 2014;194:200–210. doi: 10.1016/j.virusres.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sinica B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.K., et al. Antiviral activity of nucleoside analogues against SARS-coronavirus (SARS-CoV) Antiviral Chem. Chemother. 2006;17(5):285–289. doi: 10.1177/095632020601700506. [DOI] [PubMed] [Google Scholar]
- Subissi L., et al. SARS-CoV ORF1b-encoded nonstructural proteins 12–16: replicative enzymes as antiviral targets. Antiviral Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., et al. A second, non-canonical RNA-dependent RNA polymerase in SARS Coronavirus. EMBO J. 2006;25(20):4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., et al. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., et al. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5(5):369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., et al. p53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling. J. Biol. Chem. 2015;290(5):3172–3182. doi: 10.1074/jbc.M114.619890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-W., et al. SARS coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6. Int. J. Mol. Sci. 2016;17(5):678. doi: 10.3390/ijms17050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 2004;78(14):7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum K.T., Tanner J.A. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. ChemBioChem. 2008;9(18):3037–3045. doi: 10.1002/cbic.200800491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K.J., et al. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun. 2008;366(3):738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10) doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T., et al. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59(14):6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.-Y., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., et al. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, S. Srivastava, S., COVID-19 Pandemic: Hopes from Proteomics and Multiomics Research. 2020. [DOI] [PubMed]
- Ray S., Reddy A.B. COVID-19 management in light of the circadian clock. Nat. Rev. Mol. Cell Biol. 2020;21(9):494–495. doi: 10.1038/s41580-020-0275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.K., et al. Severe acute respiratory syndrome coronavirus ORF7a inhibits bone marrow stromal antigen 2 virion tethering through a novel mechanism of glycosylation interference. J. Virol. 2015;89(23):11820–11833. doi: 10.1128/JVI.02274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., et al. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. 2006;103(34):12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82(9):4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., et al. Extensive positive selection drives the evolution of nonstructural proteins in lineage C betacoronaviruses. J. Virol. 2016;90(7):3627–3639. doi: 10.1128/JVI.02988-15. [DOI] [PMC free article] [PubMed] [Google Scholar]