Abstract
Long non-coding RNAs (lncRNAs) have been discovered to serve important roles in a variety of types of cancer, including cervical cancer. The low expression of lncRNA long intergenic non-protein coding RNA 861 (LINC00861) is related to poor prognosis in ovarian cancer. However, the effects and underlying mechanisms of LINC00861 in cervical cancer remain largely unknown. The present study aimed to examine the role of LINC00861 in the development and progression of ovarian cancer and its underlying mechanisms. The expression levels of LINC00861 and microRNA (miR)-513b-5p were analyzed using reverse transcription-quantitative PCR analysis. Cell proliferation, migration and invasion were measured by using Cell Counting Kit-8, colony formation, wound healing and Transwell assays, respectively. A luciferase assay was used to determine whether miR-513b-5p targeted LINC00861 and PTEN. The expression of protein was measured by using western blot assay. The results of the present study discovered that LINC00861 expression levels were significantly downregulated in cervical cancer tissues and CaSki and ME-180 cell lines. Downregulated LINC00861 expression levels were identified to be associated with an advanced-stage, lymph node metastasis and the poor survival of patients with cervical cancer. Gene Set Enrichment Analysis revealed that the PI3K/AKT/mTOR signaling pathway was significantly enriched in cervical tumors expressing low expression levels of LINC00861 compared with tumors expressing high levels of LINC00861. The overexpression of LINC00861 reduced cervical cancer cell proliferation, migration, invasion and epithelial-mesenchymal transition (EMT) processes, upregulated PTEN protein expression levels and downregulated phosphorylated (p)-AKT and p-mTOR protein expression levels. The regulatory relationship between LINC00861, microRNA (miR)-513b-5p and PTEN was validated using a dual luciferase reporter gene assay. PTEN expression levels were significantly downregulated in the miR-513b-5p mimic group and significantly upregulated in the miR-513b-5p inhibitor group compared with the mimic NC and inhibitor NC in both cell lines. Furthermore, LINC00861 was suggested to serve as a competing endogenous RNA by sponging miR-513b-5p and consequently upregulating the expression levels of PTEN in cervical cancer cells. The expression of PTEN, the phosphorylation of Akt and mTOR and and the EMT phenotype were rescued following co-transfection with LINC00861 and miR-513b-5p mimics. In conclusion, the findings of the present study indicated that the LINC00861/miR-513b-5p axis may inhibit the progression of cervical cancer cells through the PTEN/AKT/mTOR signaling pathway to suppress the EMT process.
Keywords: cervical cancer, long intergenic non-protein coding RNA 861, microRNA-513b-5p, PTEN/AKT/mTOR, epithelial-mesenchymal transition
Introduction
Cervical cancer is the fourth most common type of malignancy and a prominent cause of cancer-related deaths in women worldwide (1). Indeed, cervical cancer was responsible for 570,000 cases and 311,000 deaths in 2018 worldwide, ranking after breast cancer (2.1 million cases), colorectal cancer (0.8 million) and lung cancer (0.7 million) (1). Clinical characteristics, such as advanced tumor stage and lymph node metastasis, have been associated with poor prognosis and poor therapeutic outcomes (2). The 5-year overall survival for women with locally advanced cervical cancers is ~70% following completion of concurrent chemoradiotherapy (2). Despite a steady decline in cervical cancer incidence and mortality rates in recent years due to human papillomavirus (HPV)-based screening programs and HPV vaccination programs, cervical cancer continues to pose a considerable threat to middle-aged women (3). Therefore, there remains an urgent requirement to discover novel prognostic biomarkers and to determine the important signaling pathways that are activated or inactivated in the event of cervical cancer. In addition, more effective therapeutic strategies for anticancer therapy in cervical cancer need to be developed.
Long non-coding RNAs (lncRNAs) are transcripts of >200 nucleotides in length that do not encode proteins (4). Previous studies have reported that lncRNAs have functions in numerous biological activities, including cell cycle regulation (5), stem cell differentiation (6), the immune response (7), cancer progression (8) and chemotherapy resistance (9). A few studies have reported that the lncRNA, long intergenic non-protein coding RNA 861 (LINC00861), was involved in the progression and prognosis of a wide range of types of cancer. For example, the downregulated expression levels of LINC00861 were reportedly associated with a poor prognosis in ovarian cancer (10). LINC00861 expression levels were also demonstrated to be downregulated in an early recurrence group of patients with hepatocellular carcinoma compared with a non-early recurrence group of patients (11). In addition, in another previous study, LINC00861 was able to bind to hsa-microRNA (miRNA/miR)-510, and it was associated with clinical biomarkers, such as the receptor tyrosine protein kinase ERBB-2, estrogen receptor and progesterone receptor, in breast cancer (12). Emerging evidence has revealed that lncRNAs serve as competing endogenous RNAs (ceRNAs) to regulate the epithelial-mesenchymal transition (EMT) process, such as the lncRNAs plasmacytoma variant translocation 1 and colon cancer associated transcript-1, in cervical cancer (13,14). However, the expression and underlying mechanism of LINC00861 in cervical cancer progression remain unclear.
In the current study, the expression levels of LINC00861 were discovered to be significantly downregulated in cervical cancer tissues, as well as CaSki and ME-180 cell lines. The downregulated LINC00861 expression levels were associated with an advanced-stage, lymph node metastasis and the poor survival of patients with cervical cancer. LINC00861 overexpression was also demonstrated to inhibit cervical cancer cell proliferation, migration, invasion and EMT via functioning as a ceRNA for miR-513b-5p, which subsequently regulated the PTEN/AKT/mTOR signaling pathway.
Materials and methods
Clinical specimens
A total of 56 cervical cancer tissues from patients (age, 35–59) with cervical cancer and their matched adjacent normal tissues were obtained from the Department of Gynecology at Yantai Hospital of Traditional Chinese Medicine from April 2017 to December 2018. All samples were collected, snap-frozen in liquid nitrogen and kept at −80°C until use. Patients were included if they were pathologically diagnosed with cervical cancer and had not received anticancer therapy prior to tumor excision. Patients were excluded if they had other malignant tumors. Written informed consent was obtained from each patient. The study protocol was approved by the Research Ethics Committee of Yantai Hospital of Traditional Chinese Medicine (approval no. YTSZYYY2017-112).
All diagnoses were histologically confirmed independently by at least two experienced pathologists following the examination of the sections following hematoxylin and eosin (H&E) staining. Tissues were fixed in 4% paraformaldehyde for 24 h at 25°C, dehydrated, embedded in paraffin, and cut into 4-µm slices for histological staining. These sections were stained with hematoxylin (Beyotime Institute of Biotechnology) for 15 min and eosin (Beyotime Institute of Biotechnology) for 5 min at 25°C. Samples were examined under a light microscope (magnification, ×400; Nikon Corporation).
Cell culture and transfection
Human cervical cancer cell lines, CaSki and ME-180, and the human cervical epithelial cell line, Ect1/E6E7, were obtained from the American Type Culture Collection. All cells were maintained in RPMI-1640 medium (HyClone; Cytiva) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), and maintained at 37°C in a humidified atmosphere with 5% CO2.
CaSki and ME-180 cells (1.5×106 cells/ml) were seeded into 6-well plates and transfected with 50 nM pcDNA3.1-LINC00861 or empty vector pcDNA3.1 as a negative control (NC), (both Shanghai GenePharma Co., Ltd.) using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) 37°C for 6 h, according to the manufacturer's protocol. miR-513b-5p expression levels were overexpressed or knocked down by transfecting CaSki and ME-180 cells (1.5×106 cells/ml) with 50 nM miR-513b-5p mimic or miR-513b-5p inhibitor, respectively, or their respective NCs, mimic NC and inhibitor NC (all Shanghai GenePharma Co., Ltd.). The sequences are listed in Table I. The transfections were also performed using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions at 37°C for 6 h. CaSki and ME-180 cells were harvested at 48 h after transfection.
Table I.
Oligonucleotide sequences.
| Nucleic acids name | Sequence (5′→3′) |
|---|---|
| MicroRNA-513b-5p | UUCACAAGGAGGUGUCAUUUAU |
| mimic | AAAUGACACCUCCUUGUGAAUU |
| Mimic NC | UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT |
| MicroRNA-513b-5p inhibitor | AUAAAUGACACCUCCUUGUGAA |
| Inhibitor NC | CAGUACUUUUGUGUAGUACAA |
NC, negative control.
Cell Counting Kit-8 (CCK-8) assay
CaSki and ME-180 cells were seeded into 96-well plates at a density of 2×103 cells/100 µl and incubated at 37°C for 24, 48 and 72 h. Subsequently, 10 µl CCK-8 solution (Dojindo Molecular Technologies, Inc.) was added to each well and incubated for 2 h. The absorbance at a wavelength of 450 nm was measured in each well using a microplate reader (Bio-Rad Laboratories, Inc.).
Colony formation assay
CaSki and ME-180 cells were plated into six-well plates at a density of 1×103 cells/100 µl. Following incubation for 12 days at 37°C, the cells were fixed with 4% paraformaldehyde at 25°C for 15 min and stained with 0.1% crystal violet solution at 25°C for 20 min. Colonies were counted under a light microscope (magnification, ×100; Nikon Corporation), with >50 cells considered a colony.
Wound healing assay
CaSki and ME-180 cells were seeded in six-well plates at a density of 4×105 cells/well. The cell monolayers were scratched using a 200-µl pipette tip once the cultured cells reached 100% confluence and then cultured in serum-free RPMI-1640 medium. At 0 and 24 h, the width of the wound was visualized under a light microscope (magnification, ×100; Nikon Corporation). The wound healing rate was calculated by using the formula: The wound healing rate (%) = (width of the wound at 0 h-width of the wound at 24 h)/width of the wound at 0 h ×100%.
Transwell invasion assay
CaSki and ME-180 cells at a density of 4×103 cells/100 µl in serum-free RPMI-1640 medium were plated into the upper chambers of 24-well Transwell plates (Corning, Inc.) precoated with Matrigel (EMD Millipore) at 37°C for 60 min. The lower chambers were filled with 500 µl RPMI-1640 medium supplemented with 10% FBS. Following 24 h of incubation at 37°C, the invasive cells were fixed with 75% methanol at 25°C for 15 min and then stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) for at 25°C 15 min. The invasive cells were visualized under a light microscope (magnification, ×100; Nikon Corporation).
Dual luciferase reporter assay
The Diana Tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php) and TargetScan 7.2 (http://www.targetscan.org/vert_72/) bioinformatics databases were used to identify potential binding sites between LINC00861, miR-513b-5p and PTEN. The mutations in the miR-513-5p binding sites were introduced using the GeneArt™ site-directed mutagenesis system (Thermo Fisher Scientifc, Inc.). The LINC00861 wild-type (WT) or LINC00861 mutant (MUT) and PTEN-WT or PTEN-MUT binding miR-513-5p were synthesized and cloned into the Renilla psiCHECK2 vector (Promega Corporation). Subsequently, CaSki and ME-180 cells at 5×104 cells/well in 24-well plates were co-transfected with 500 ng the LINC00861-WT, LINC00861-MUT, PTEN-WT or PTEN-MUT vectors and the miR-513-5p mimic or mimic NC, using Lipofectamine 2000 reagent. Following 48 h of transfection at 37°C, the relative luciferase activity was measured using a Dual Luciferase Reporter assay system (Promega Corporation). Renilla luciferase activity was used as the normalization.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from frozen tissues and cultured cells (CaSki, ME-180, and Ect1/E6E7) using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). 1 µg RNA was reverse transcribed into cDNA using PrimeScript™ RT reagent kit with gDNA Eraser (cat. no. RR047A; Takara Bio, Inc.). qPCR was subsequently performed on an Applied Biosystems 7500 Real-Time PCR system (Thermo Fisher Scientific, Inc.) using the Power SYBR™ Green PCR Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR was amplified using the following thermocycling conditions: i) Initial denaturation at 95°C for 30 sec; followed by 35 cycles of denaturation at 95°C for 5 sec, annealing at 55°C for 20 sec and extension at 72°C for 20 sec. The primer sequences used for qPCR are listed in Table II. U6, which is a small nuclear RNA, and GAPDH were used as the internal controls for miRNA and mRNA, respectively. Data were analysed using the 2−ΔΔCq method (15).
Table II.
Primers used for reverse transcription-quantitative PCR.
| Gene | Primer sequence (5′→3′) |
|---|---|
| Long intergenic non- | F: ATGGTTAGGCACATGGGGTG |
| protein coding RNA 861 | R: CCAGGCTCTGGGCAACATTA |
| MicroRNA-513b-5p | F: TGGTACTGATGTGATGGACT |
| R: TCATATCACACAGCACCGAT | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT | |
| GAPDH | F: ATGGGGAAGGTGAAGGT |
| R: AAGCTTCCCGTTCTCAG |
F, forward; R, reverse.
Western blotting
Total protein was extracted from the CaSki and ME-180 cell lines using RIPA lysis buffer (Thermo Fisher Scientific, Inc.). Total protein was quantified by using an Enhanced BCA Protein Assay kit (Beyotime Institute of Biotechnology) and 50 µg protein/lane was separated via 10% SDS-PAGE. The separated proteins were subsequently transferred onto PVDF membranes (EMD Millipore) and blocked by using 5% non-fat milk for 1 h at room temperature. The membranes were then incubated overnight at 4°C with the following primary antibodies (all Cell Signaling Technology, Inc.): Anti-PTEN (1:2,000; cat. no. 9188), anti-phosphorylated (p)-AKT (1:2,000; cat. no. 4060), anti-AKT (1:2,000; cat. no. 9272), anti-p-mTOR (1:2,000; cat. no. 5536), anti-mTOR (1:2,000; cat. no. 2972), anti-E-cadherin (1:2,000; cat. no. 3195), anti-Snail (1:2,000; cat. no. 3879), anti-vimentin (1:2,000; cat. no. 5741) and anti-GAPDH (1:2,000; cat. no. 5174). Following washing with TBST, the membranes were incubated with the HRP-conjugated anti-rabbit secondary antibodies (1:3,000, A0208; Beyotime Institute of Biotechnology). Protein bands were visualized using enhanced chemiluminescence kit (Sigma-Aldrich; Merck KGaA) and imaged using a gel imaging system. The optical densities of the protein bands were measured using Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.).
Bioinformatics analysis
Overall survival of cervical cancer patients for LINC00861 was done using gene expression profiling interactive analysis (http://gepia2.cancer-pku.cn). The cervical cancer patients were divided into high LINC00861 expression group (LINC00861 expression > median; n=145) and low LINC00861 expression group (LINC00861 expression ≤ median; n=146). Gene Set Enrichment Analysis (GSEA) (16) was performed on The Cancer Genome Atlas (TCGA) database dataset (CESC; http://cancergenome.nih.gov/) of LINC00861 expression by using R package clusterProfiler (17). Gene sets (h.all.v7.1.symbols.gmt) were downloaded from the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb/genesets.jsp; Broad Institute, Inc.).
Statistical analysis
All experiments were performed ≥3 times. Statistical data are presented as the mean ± SD and statistical analysis was performed using GraphPad Prism 7 software (GraphPad Software, Inc.). A two-tailed paired Student's t-test was used to determine the statistical differences in the LINC00861 expression levels between cervical cancer and matched adjacent normal tissues. The survival analysis was performed using a Kaplan-Meier test and a log-rank test was used to determine the statistical significance between the two groups. A χ2 test was performed to analyze the association between LINC00861 expression levels and the clinicopathological features of the patients with cervical cancer. The remaining two group comparisons were performed using an unpaired Student's t-test. The comparisons among ≥3 groups were conducted using a one-way ANOVA followed by a Tukey's multiple comparisons test. P<0.05 was considered to indicate a statistically significant difference.
Results
LINC00861 expression levels are significantly downregulated in cervical cancer tissues and are associated with prognosis
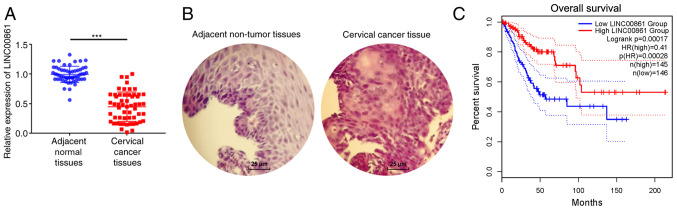
To investigate the role of LINC00861 in cervical cancer progression, RT-qPCR analysis was performed to analyze LINC00861 expression levels in 56 cervical cancer and adjacent normal tissues. LINC00861 expression levels were significantly downregulated in the cervical cancer tissues compared with the adjacent normal tissues (Fig. 1A). In addition, the expression levels of LINC00861 were associated with lymph-node metastasis and advanced tumor stage. No associations were identified between LINC00861 levels and age, tumor size, histology, and differentiation (Table III). Representative H&E staining images are shown in Fig. 1B. Cervical cancer tissues displayed enlarged and hyperchromatic nuclei, little cytoplasm, and atypical nuclei. TCGA data indicated that patients with low LINC00861 expression levels had a shorter survival time compared with patients with high LINC00861 expression levels (Fig. 1C).
Figure 1.
Expression levels of LINC00861 in cervical cancer tissues. (A) Reverse transcription-quantitative PCR was used to determine that LINC00861 expression levels were downregulated in cervical cancer tissues compared with normal adjacent tissues. ***P<0.001. (B) Representative hematoxylin and eosin staining images of cervical cancer tissues and their matched adjacent normal tissues. Scale bar, 25-µm. (C) The cervical cancer patients were divided into high LINC00861 expression group (LINC00861 expression > median; n=145) and low LINC00861 expression group (LINC00861 expression ≤ median; n=146). Patients with cervical cancer with low LINC00861 expression had a poorer overall survival compared with patients with higher expression. LINC00861, long intergenic non-protein coding RNA 861.
Table III.
Association between LINC00861 expression levels and the clinicopathological features of patients with cervical cancer.
| LINC00861 expression | ||||
|---|---|---|---|---|
| Variable | n | High (n=28) | Low (n=28) | P-value |
| Age, years | 0.179 | |||
| <45 | 25 | 15 | 10 | |
| ≥45 | 31 | 13 | 18 | |
| Tumor size, cm | 0.284 | |||
| <4 | 26 | 11 | 15 | |
| ≥4 | 30 | 17 | 13 | |
| Histology | 0.313 | |||
| Squamous cell carcinoma | 45 | 24 | 21 | |
| Adenocarcinoma | 11 | 4 | 7 | |
| Differentiation | 0.237 | |||
| Well + moderate | 40 | 22 | 18 | |
| Poor | 16 | 6 | 10 | |
| Lymph node metastasis | 0.035a | |||
| No | 41 | 24 | 17 | |
| Yes | 15 | 4 | 11 | |
| Tumor stage | 0.016a | |||
| Ib-IIa | 29 | 19 | 10 | |
| IIb-IIIa | 27 | 9 | 18 | |
P<0.05. LINC00861, long intergenic non-protein coding RNA 861.
Overexpression of LINC00861 attenuates cervical cancer cell proliferation
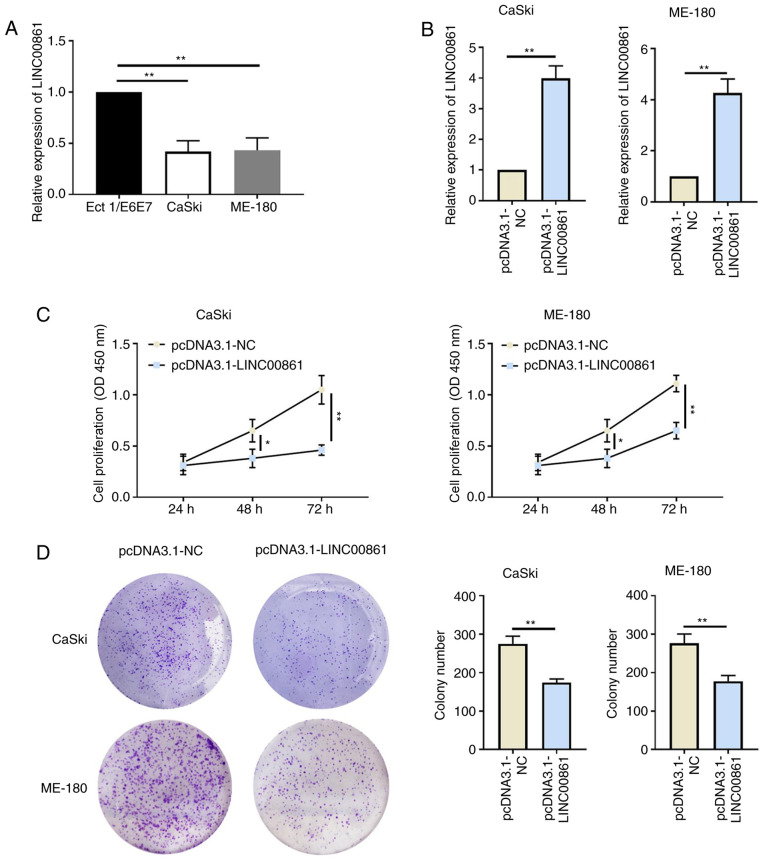
The expression levels of LINC00861 were significantly downregulated in cervical cancer cells (CaSki and ME-180) compared with the human cervical epithelial cell line, Ect1/E6E7 (Fig. 2A). Cervical cancer cells were subsequently transfected with either pcDNA3.1-NC or pcDNA3.1-LINC00861 and the transfection efficiency was verified using RT-qPCR (Fig. 2B). The proliferative ability of the cervical cancer cells was significantly attenuated following the overexpression of LINC00861 compared with the cells transfected with pcDNA3.1-NC, as determined using CCK-8 and colony formation assays (Fig. 2C and D).
Figure 2.
Overexpression of LINC00861 attenuates cervical cancer cell proliferation. (A) RT-qPCR analysis of LINC00861 expression levels in cervical cancer cells (CaSki and ME-180) and the human cervical epithelial cell line Ect1/E6E7. (B) Transfection efficiency of pcDNA3.1-LINC00861 in CaSki and ME-180 cell lines was confirmed using RT-qPCR. (C) Cell proliferative rate of cervical cancer cells transfected with pcDNA3.1-LINC00861 or pcDNA3.1-NC was analyzed using a Cell Counting Kit-8 assay. (D) Proliferative ability of cervical cancer cells following LINC00861 overexpression was analyzed using a colony formation assay in CaSki and ME-180 cells. *P<0.05, **P<0.01. LINC00861, long intergenic non-protein coding RNA 861; NC, negative control; OD, optical density; RT-qPCR, reverse transcription-quantitative PCR.
Overexpression of LINC00861 attenuates cervical cancer cell migration and invasion
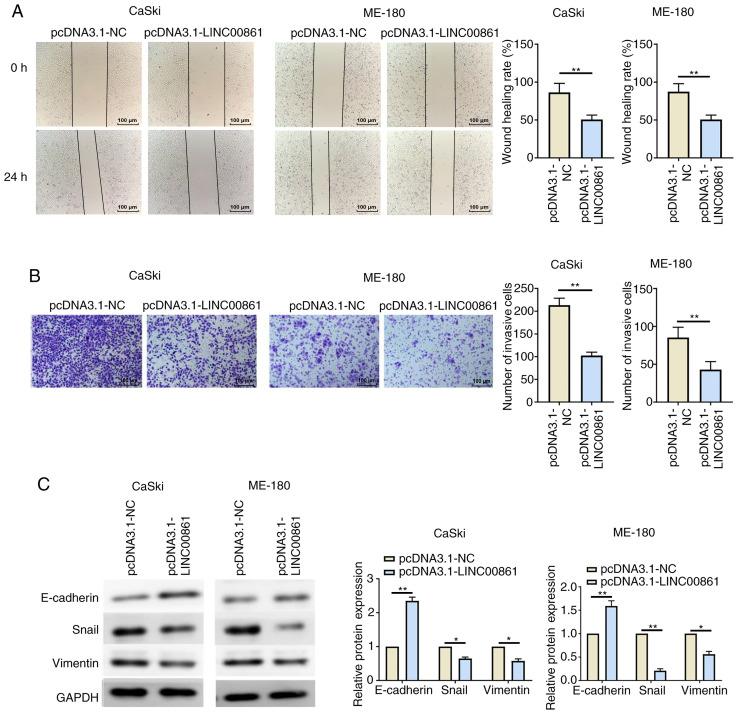
In addition to proliferation, the effects of LINC00861 on migration and invasion were also investigated. Wound healing and Transwell invasion assays revealed that the transfection with pcDNA3.1-LINC00861 resulted in significantly reduced cell migratory and invasive abilities compared with cells transfected with pcDNA3.1-NC (Fig. 3A and B). The expression levels of several EMT-related proteins were further analyzed using western blotting. The results revealed that the overexpression of LINC00861 led to the significant upregulation of E-cadherin expression levels and the significant downregulation of Snail and vimentin expression levels in both cervical cancer cell lines compared with the pcDNA3.1-NC-transfected cells (Fig. 3C).
Figure 3.
Overexpression of LINC00861 attenuates cervical cancer cell migration and invasion. (A) Wound healing assay was used to determine the migratory ability of cervical cancer cells transfected with pcDNA3.1-NC or pcDNA3.1-LINC00861. Scale bar, 100-µm. (B) Invasive ability of cervical cancer cells following LINC00861 overexpression was analyzed using a Transwell assay. Scale bar, 100-µm. (C) LINC00861 overexpression regulated the expression levels of epithelial-mesenchymal transition-related proteins, E-cadherin, N-cadherin and Snail, in cervical cancer cells. *P<0.05, **P<0.01. LINC00861, long intergenic non-protein coding RNA 861; NC, negative control.
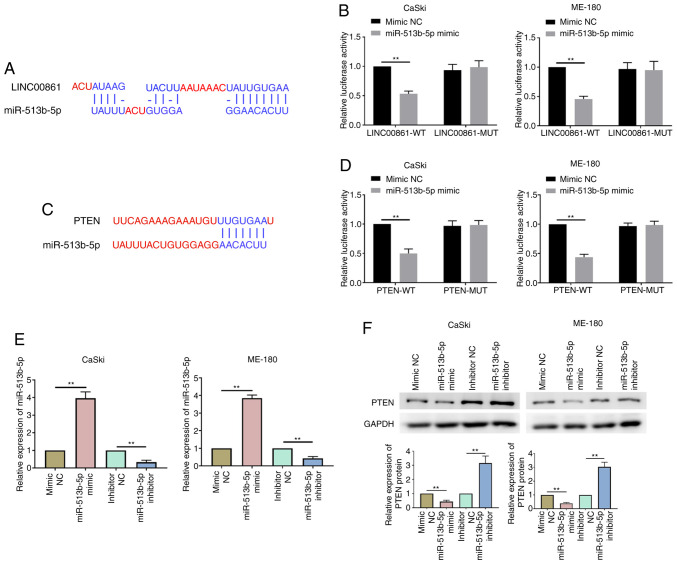
LINC00861 competitively binds to miR-51b-5p and regulates PTEN expression levels
The online bioinformatics software, Diana Tools and TargetScan, predicted that miR-513b-5p contained putative binding sites within the 3′-untranslated region (UTR) of LINC00861 and PTEN (Fig. 4A and C). The dual luciferase reporter assay identified that the co-transfection with the miR-513b-5p mimic could significantly decrease the relative luciferase activity of the LINC0086-WT and PTEN-WT groups compared with the co-transfection with the mimic NC (Fig. 4B and D). However, miR-513b-5p mimic transfection had no effect on the luciferase activity of reporter vector containing MUT 3′UTR of LINC0086 and PTEN. RT-qPCR analysis demonstrated that the miR-513b-5p mimic significantly upregulated miR-513b-5p expression levels, whereas the miR-513b-5p inhibitor significantly downregulated the expression levels of miR-513b-5p in cervical cancer cells compared with their respective NCs (Fig. 4E). Western blotting further revealed that PTEN expression levels were significantly downregulated in the miR-513b-5p mimic group compared with the mimic NC group in both cell lines (Fig. 4F). Conversely, the transfection with the miR-513b-5p inhibitor significantly upregulated the expression levels of PTEN compared with the inhibitor NC in both cell lines. These data indicated that LINC00861 may serve as a ceRNA of miR-513b-5p in cervical cancer progression.
Figure 4.
LINC00861 competitively binds to miR-513b-5p and regulates PTEN expression levels. (A) LINC00861 3′-UTR sequence containing the putative miR-513b-5p binding sites. (B) Relative luciferase activity of cervical cancer cells was analyzed following the co-transfection with LINC00861-WT or LINC00861-MUT 3′-UTR and the miR-513b-5p mimic or mimic NC. (C) Putative miR-513b-5p binding site in the 3′-UTR sequence of PTEN. (D) Relative luciferase activity was analyzed following the co-transfection with PTEN-WT or PTEN-MUT 3′-UTR and the miR-513-5p mimic or mimic NC into cervical cancer cells. (E) Expression levels of miR-513b-5p were analyzed using reverse transcription-quantitative PCR in cervical cancer cells transfected with the miR-513b-5p mimic, inhibitor or the respective NCs. (F) Expression levels of PTEN were analyzed using western blotting in cervical cancer cells transfected with the miR-513b-5p mimic, inhibitor or respective NCs. **P<0.01. LINC00861, long intergenic non-protein coding RNA 861; NC, negative control; UTR, untranslated region; miR, microRNA; WT, wild-type; MUT, mutant.
Both LINC00861 and miR-513b-5p overexpression modulate the PTEN/AKT/mTOR signaling pathway
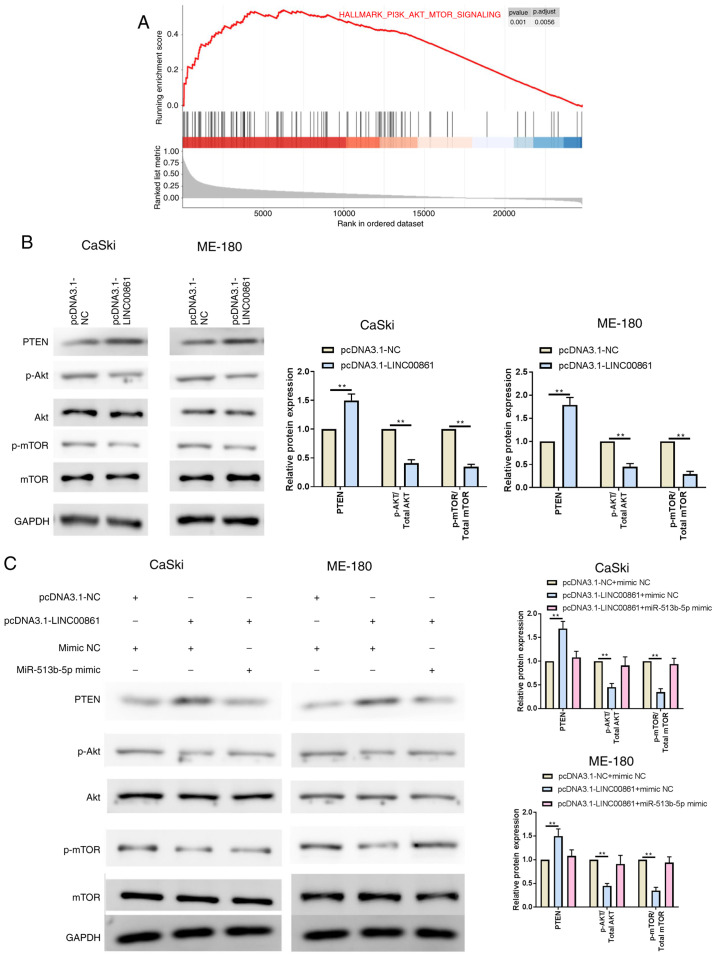
To understand the underlying mechanism of LINC00861 in cervical cancer progression, GSEA was performed on TCGA cervical cancer datasets. The results revealed that downregulation of LINC00861 was associated with the PI3K/AKT/mTOR signaling (Fig. 5A), thereby indicating that the PI3K/AKT/mTOR signaling pathway may be involved in the inhibition effect of LINC00861 on proliferation, migration and invasion of cervical cancer cells. The overexpression of LINC00861 significantly upregulated PTEN expression levels, while downregulating p-AKT and p-mTOR expression levels, compared with the pcDNA3.1-NC group in both cell lines (Fig. 5B). Cervical cancer cells were subsequently co-transfected with LINC00861 and miR-513b-5p mimic; as expected, the expression levels of PTEN, p-AKT and p-mTOR were recovered following the co-transfection with pcDNA3.1-LINC00861 and miR-513b-5p compared with the pcDNA3.1-LINC00861 + mimic NC group in both cell lines (Fig. 5C). Thus, it was suggested that the miR-513b-5p mimic may block the effect of the overexpression of LINC00861.
Figure 5.
Both LINC00861 and miR-513b-5p overexpression modulates the PTEN/AKT/mTOR signaling pathway. (A) Gene Set Enrichment Analysis revealed that the PI3K/AKT/mTOR signaling pathway was enriched in patients from The Cancer Genome Atlas dataset with high LINC00861 expression vs. low LINC00861 expression. (B) Expression levels of PTEN, p-AKT/total AKT, p-mTOR/total mTOR in cervical cancer cell lines transfected with pcDNA3.1-LINC00861 or pcDNA3.1-NC cells were analyzed using western blotting. (C) Western blotting analysis of the relative protein expression levels of PTEN, p-AKT, AKT, p-mTOR and mTOR in cervical cancer cells following the co-transfection with pcDNA3.1-LINC00861 and the miR-513b-5p mimic. **P<0.01. LINC00861, long intergenic non-protein coding RNA 861; NC, negative control; miR, microRNA; p-, phosphorylated.
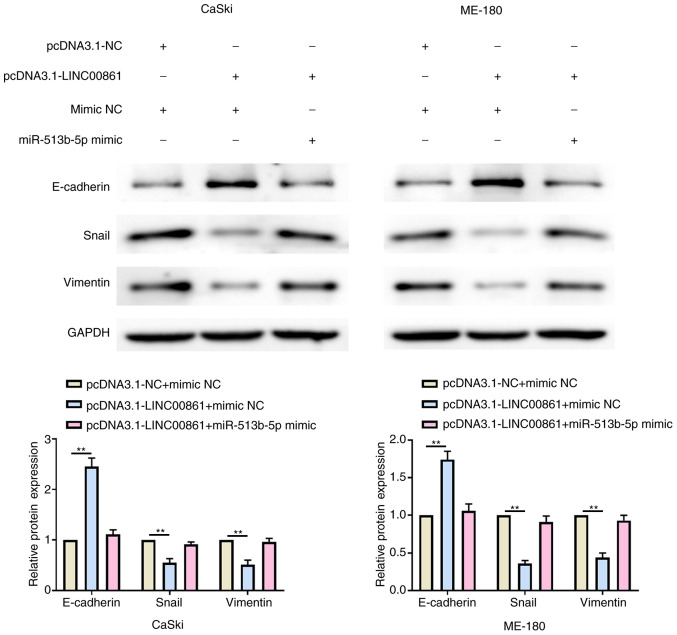
Both LINC00861 and miR-513b-5p overexpression modulate the EMT phenotype
Western blotting revealed that the overexpression of LINC00861 significantly upregulated E-cadherin expression levels and downregulated Snail and vimentin expression levels compared with the pcDNA3.1-NC + mimic NC group in both cell lines (Fig. 6). Conversely, the expression levels of E-cadherin, Snail and vimentin were recovered following the co-transfection with the pcDNA3.1-LINC00861 and miR-513b-5p mimic in both cell lines.
Figure 6.
Both LINC00861 and miR-513b-5p overexpression modulates the epithelial-mesenchymal transition phenotype. Western blotting analysis of the relative protein expression levels of E-cadherin, Snail and vimentin in cervical cancer cells following the co-transfection with pcDNA3.1-LINC00861 and the miR-513b-5p mimic. **P<0.01. LINC00861, long intergenic non-protein coding RNA 861; NC, negative control; miR, microRNA.
Discussion
The abnormal expression of lncRNAs has been strongly implicated in a wide range of cancer types, such as prostate (18), colorectal (19) and cervical cancer (20). Several studies have reported that lncRNAs could serve as either oncogenes or tumor suppressors (21). For example, lncRNA nuclear paraspeckle assembly transcript 1 functions as an oncogene to drive aggressive endometrial cancer progression (22). LINC00589 and lncRNA maternally expressed gene 3 are tumor suppressor (23,24). Previous evidence has suggested that lncRNAs may serve as ceRNAs to regulate miRNAs, which consequently regulates the expression of target genes (25). For example, the expression levels of LINC01234 were discovered to be significantly upregulated in gastric cancer tissues and associated with poor clinical outcomes (26). In addition, the knockdown of LINC01234 inhibited cell proliferation in vitro by serving as a ceRNA for miR-204-5p and subsequently regulating core-binding factor subunit β expression in gastric cancer (26). Another previous study reported that the overexpression of lncRNA LINC00673 promoted non-small cell lung cancer (NSCLC) proliferation, migration, invasion and EMT by sponging miR-150-5p (27). Furthermore, the lncRNA proteasome 20S subunit α 3 antisense RNA 1 promoted esophageal cancer progression by sponging miR-101 to upregulate Histone-lysine N-methyltransferase EZH2 expression (28). In the present study, the expression levels of LINC00861 were revealed to be significantly downregulated in cervical cancer tissues, and were associated with an advanced-stage, lymph node metastasis and poor survival in patients with cervical cancer. In addition, the overexpression of LINC00861 was demonstrated to inhibit cervical cancer cell proliferation, migration, invasion and EMT by functioning as a ceRNA for miR-513b-5p and upregulating PTEN expression levels. Consistent with these results, a recent report illustrated that circ_La-related protein 4 (LARP4) inhibited cell proliferation and migration in ovarian cancer by sponging miR-513b-5p to regulate the expression of LARP4 (29). Wang et al (30) reported that miR-513b-5p promoted p53 protein expression by targeting interferon regulatory factor 2, thereby suppressing testicular embryonal carcinoma cell proliferation and inducing apoptosis in vitro. It seems contradictory that miR-513b-5p may serve two completely opposite roles in testicular embryonal carcinoma and cervical cancer. In the present study, miR-513b-5p reversed the inhibitory effects of LINC00861 on proliferation of cervical cancer cells. However, several studies have revealed that the functions of miRNAs may be specific to individual tissues; for example, miR-182 served an oncogenic role in endometrial carcinoma cells and promoted cell proliferation, migration and invasion by targeting forkhead box protein F2 (31). Meanwhile, Lv et al (32) demonstrated that miR-182 inhibited the proliferation of laryngeal squamous cell carcinoma cells by targeting cisplatin resistance-related protein 9. These two aforementioned studies supported the findings of the present study, that is, that miR-513b-5p may serve as an oncogenic miRNA in cervical cancer.
PTEN, an important tumor suppressor gene, is one of the most commonly deleted or mutated tumor suppressors in a variety of types of human cancer, including breast (33), prostate (34), and cervical cancer (35). PTEN inactivates the PI3K/AKT/mTOR signaling pathway, which is involved in an abundance of essential cellular processes, such as cell growth, differentiation, proliferation and migration; most of these processes have also been associated with cancer progression (36). For example, syntaxin binding protein 5-AS1 reduced cervical cancer cell proliferation and invasion by regulating the miR-96-5p/PTEN axis (37). Wang et al (38) reported that long intergenic non-protein coding RNA, p53 induced transcript alleviated the progression of NSCLC by sponging miR-543 and subsequently inducing PTEN expression. Shi et al (39) have suggested that the knockdown of miR-17-5p may suppresses the malignant behavior of thyroid cancer by upregulating PTEN expression levels and by inactivating the AKT/mTOR signaling pathway. The findings of the present study supported the results of previous studies.
Tumor metastasis is the cause of cancer mortality in >90% of cases (40). The EMT process can be hijacked by epithelial cancer cells to promote abnormal migration and invasion (41). EMT is characterized by epithelial cell markers such as E-cadherin are down-regulated, which leads to loss of cell-cell adhesion, while mesenchymal markers such as vimentin, Snail and N-cadherin are up-regulated, thereby allowing the cells to migrate to different organs (42). Currently, numerous lncRNAs, including lncRNAs pro-transition associated RNA and GATA binding protein 6-AS1, have been reported to be involved in the EMT process in ovarian and gastric cancer, respectively (43,44). In the current study, the lncRNA LINC00861 was indicated to inhibit the migration and invasion of cervical cancer cells by inhibiting the EMT process of cervical cancer cells.
In conclusion, the results of the present study indicated that LINC00861 overexpression may inhibit the proliferation, migration and invasion of cervical cancer cells by sponging miR-513b-5p, which subsequently regulated the PTEN/AKT/mTOR signaling pathway. Therefore, the results suggested that LINC00861 may serve as a tumor suppressor in the development and progression of cervical cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XS conceived and designed the study; HL, LZ and XD performed the experiments, analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The experimental protocol involving patients was approved by the Research Ethics Committee of Yantai Hospital of Traditional Chinese Medicine (approval no. YTSZYYY2017-112). All patients provided written informed consent for participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393:169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- 3.Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J, Bray F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob Health. 2020;8:e191–e203. doi: 10.1016/S2214-109X(19)30482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Hu YW, Kang CM, Zhao JJ, Nie Y, Zheng L, Li HX, Li X, Wang Q, Qiu YR. LncRNA PLAC2 down-regulates RPL36 expression and blocks cell cycle progression in glioma through a mechanism involving STAT1. J Cell Mol Med. 2018;22:497–510. doi: 10.1111/jcmm.13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, Li Y, Gong R, Gao M, Feng C, Liu T, Sun Y, Jin M, Wang D, Yuan Y, et al. The long non-coding RNA-ORLNC1 regulates bone mass by directing mesenchymal stem cell fate. Mol Ther. 2019;27:394–410. doi: 10.1016/j.ymthe.2018.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu X, Goswami S, Qiu J, Chen Q, Laverdure S, Sherman BT, Imamichi T. Profiles of long non-coding RNAs and mRNA expression in human macrophages regulated by interleukin-27. Int J Mol Sci. 2019;20:6207. doi: 10.3390/ijms20246207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang M, Zhou L, Yu F, Zhang Y, Li P, Wang K. The functional roles of exosomal long non-coding RNAs in cancer. Cell Mol Life Sci. 2019;76:2059–2076. doi: 10.1007/s00018-019-03018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang F, Zhang L, Zhang C. Long noncoding RNAs and tumorigenesis: Genetic associations, molecular mechanisms, and therapeutic strategies. Tumour Biol. 2016;37:163–175. doi: 10.1007/s13277-015-4445-4. [DOI] [PubMed] [Google Scholar]
- 10.Zheng M, Hu Y, Gou R, Nie X, Li X, Liu J, Lin B. Identification three LncRNA prognostic signature of ovarian cancer based on genome-wide copy number variation. Biomed Pharmacother. 2020;124:109810. doi: 10.1016/j.biopha.2019.109810. [DOI] [PubMed] [Google Scholar]
- 11.Lv Y, Wei W, Huang Z, Chen Z, Fang Y, Pan L, Han X, Xu Z. Long non-coding RNA expression profile can predict early recurrence in hepatocellular carcinoma after curative resection. Hepatol Res. 2018;48:1140–1148. doi: 10.1111/hepr.13220. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Li Y, Wang Q, Zhang X, Wang D, Tang HC, Meng X, Ding X. Identification of an lncRNAmiRNAmRNA interaction mechanism in breast cancer based on bioinformatic analysis. Mol Med Rep. 2017;16:5113–5120. doi: 10.3892/mmr.2017.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen CJ, Cheng YM, Wang CL. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J Drug Target. 2017;25:637–644. doi: 10.1080/1061186X.2017.1307379. [DOI] [PubMed] [Google Scholar]
- 14.Li R, Liu J, Qi J. Knockdown of long non-coding RNA CCAT1 suppresses proliferation and EMT of human cervical cancer cell lines by down-regulating Runx2. Exp Mol Pathol. 2020;113:104380. doi: 10.1016/j.yexmp.2020.104380. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu R, Lu Z. Long non-coding RNA HCP5 promotes prostate cancer cell proliferation by acting as the sponge of miR-4656 to modulate CEMIP expression. Oncol Rep. 2020;43:328–336. doi: 10.3892/or.2019.7404. [DOI] [PubMed] [Google Scholar]
- 19.Bai J, Xu J, Zhao J, Zhang R. LncRNA NBR2 suppresses migration and invasion of colorectal cancer cells by downregulating miRNA-21. Hum Cell. 2020;33:98–103. doi: 10.1007/s13577-019-00265-1. [DOI] [PubMed] [Google Scholar]
- 20.Liang H, Zhang C, Guan H, Liu J, Cui Y. LncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5p. J Cell Physiol. 2019;234:7266–7278. doi: 10.1002/jcp.27484. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–2100. doi: 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong P, Xiong Y, Yue J, Xu D, Ihira K, Konno Y, Kobayashi N, Todo Y, Watari H. Long noncoding RNA NEAT1 drives aggressive endometrial cancer progression via miR-361-regulated networks involving STAT3 and tumor microenvironment-related genes. J Exp Clin Cancer Res. 2019;38:295. doi: 10.1186/s13046-019-1306-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Li Z, Liu L, Wang Q, Li S, Chen D, Hu Z, Yu T, Ding J, Li J, et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology. 2018;67:171–187. doi: 10.1002/hep.29405. [DOI] [PubMed] [Google Scholar]
- 24.Ghafouri-Fard S, Taheri M. Maternally expressed gene 3 (MEG3): A tumor suppressor long non coding RNA. Biomed Pharmacother. 2019;118:109129. doi: 10.1016/j.biopha.2019.109129. [DOI] [PubMed] [Google Scholar]
- 25.Yao Y, Zhang T, Qi L, Zhou C, Wei J, Feng F, Liu R, Sun C. Integrated analysis of co-expression and ceRNA network identifies five lncRNAs as prognostic markers for breast cancer. J Cell Mol Med. 2019;23:8410–8419. doi: 10.1111/jcmm.14721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P, Chen Q, Wei C, Fu H, Xu T, et al. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer. Clin Cancer Res. 2018;24:2002–2014. doi: 10.1158/1078-0432.CCR-17-2376. [DOI] [PubMed] [Google Scholar]
- 27.Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H, Kong J, Ding K, Shen HM, Wu H, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16:118. doi: 10.1186/s12943-017-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu BQ, Lin XH, Ye XD, Huang W, Pei X, Xiong D, Long X, Zhu SQ, Lu F, Lin K, et al. Long non-coding RNA PSMA3-AS1 promotes malignant phenotypes of esophageal cancer by modulating the miR-101/EZH2 axis as a ceRNA. Aging. 2020;12:1843–1856. doi: 10.18632/aging.102716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin W, Ye H, You K, Chen L. Up-regulation of circ_LARP4 suppresses cell proliferation and migration in ovarian cancer by regulating miR-513b-5p/LARP4 axis. Cancer Cell Int. 2020;20:5. doi: 10.1186/s12935-019-1071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Zhang X, Wang G, Wang L, Lin Y, Sun F. Hsa-miR-513b-5p suppresses cell proliferation and promotes P53 expression by targeting IRF2 in testicular embryonal carcinoma cells. Gene. 2017;626:344–353. doi: 10.1016/j.gene.2017.05.033. [DOI] [PubMed] [Google Scholar]
- 31.Yao H, Kong F, Zhou Y. MiR-182 promotes cell proliferation, migration and invasion by targeting FoxF2 in endometrial carcinoma cells. Int J Clin Exp Pathol. 2019;12:1248–1259. [PMC free article] [PubMed] [Google Scholar]
- 32.Lv Y, Ye D, Qiu S, Zhang J, Shen Z, Shen Y, Deng H. MiR-182 regulates cell proliferation and apoptosis in laryngeal squamous cell carcinoma by targeting the CRR9. Biosci Rep. 2019;39:BSR20191348. doi: 10.1042/BSR20191348. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Ngeow J, Sesock K, Eng C. Breast cancer risk and clinical implications for germline PTEN mutation carriers. Breast Cancer Res Treat. 2017;165:1–8. doi: 10.1007/s10549-015-3665-z. [DOI] [PubMed] [Google Scholar]
- 34.Patel R, Brzezinska EA, Repiscak P, Ahmad I, Mui E, Gao M, Blomme A, Harle V, Tan EH, Malviya G, et al. Activation of β-catenin cooperates with loss of Pten to drive AR-independent castration-resistant prostate cancer. Cancer Res. 2020;80:576–590. doi: 10.1158/0008-5472.CAN-19-1684. [DOI] [PubMed] [Google Scholar]
- 35.Kaur G, Balasubramaniam SD, Lee YJ. IGFBP-2 in cervical cancer development. Exp Mol Pathol. 2020;113:21. doi: 10.1016/j.yexmp.2019.104362. [DOI] [PubMed] [Google Scholar]
- 36.Li A, Qiu M, Zhou H, Wang T, Guo W. PTEN, insulin resistance and cancer. Curr Pharm Des. 2017;23:3667–3676. doi: 10.2174/1381612823666170704124611. [DOI] [PubMed] [Google Scholar]
- 37.Shao S, Wang C, Wang S, Zhang H, Zhang Y. LncRNA STXBP5-AS1 suppressed cervical cancer progression via targeting miR-96-5p/PTEN axis. Biomed Pharmacother. 2019;117:15. doi: 10.1016/j.biopha.2019.109082. [DOI] [PubMed] [Google Scholar]
- 38.Wang S, Jiang W, Zhang X, Lu Z, Geng Q, Wang W, Li N, Cai X. LINC-PINT alleviates lung cancer progression via sponging miR-543 and inducing PTEN. Cancer Med. 2020;9:1999–2009. doi: 10.1002/cam4.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi YP, Liu GL, Li S, Liu XL. miR-17-5p knockdown inhibits proliferation, autophagy and promotes apoptosis in thyroid cancer via targeting PTEN. Neoplasma. 2020;67:249–258. doi: 10.4149/neo_2019_190110N29. [DOI] [PubMed] [Google Scholar]
- 40.Fu BM. Tumor metastasis in the microcirculation. Adv Exp Med Biol. 2018;1097:201–218. doi: 10.1007/978-3-319-96445-4_11. [DOI] [PubMed] [Google Scholar]
- 41.Ombrato L, Malanchi I. The EMT universe: Space between cancer cell dissemination and metastasis initiation. Crit Rev Oncog. 2014;19:349–361. doi: 10.1615/CritRevOncog.2014011802. [DOI] [PubMed] [Google Scholar]
- 42.Odero-Marah V, Hawsawi O, Henderson V, Sweeney J. Epithelial-mesenchymal transition (EMT) and prostate cancer. Adv Exp Med Biol. 2018;1095:101–110. doi: 10.1007/978-3-319-95693-0_6. [DOI] [PubMed] [Google Scholar]
- 43.Liang H, Yu T, Han Y, Jiang H, Wang C, You T, Zhao X, Shan H, Yang R, Yang L, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer. 2018;17:119. doi: 10.1186/s12943-018-0870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li ZT, Zhang X, Wang DW, Xu J, Kou KJ, Wang ZW, Yong G, Liang DS, Sun XY. Overexpressed lncRNA GATA6-AS1 inhibits LNM and EMT via FZD4 through the Wnt/β-catenin signaling pathway in GC. Mol Ther Nucleic Acids. 2019;19:827–840. doi: 10.1016/j.omtn.2019.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.