Abstract
The H19 long non-coding RNA is involved in the development of tamoxifen resistance in breast cancer. However, the relationship between H19 and the metastatic potential and treatment options for tamoxifen-resistant (TAMR) breast cancer is not completely understood. Curcumin inhibits cellular proliferation, migration and invasiveness in several cancer types, including pancreatic cancer, breast cancer and chronic myeloid leukemia. The present study aimed to investigate the role of H19 in MCF-7/TAMR cell epithelial-mesenchymal transition (EMT), migration and invasiveness, and to assess the ability of curcumin to inhibit H19-mediated effects. Reverse transcription-quantitative PCR and western blot analysis were conducted to detect the gene or protein expression. Cell Counting Kit-8, wound healing and Transwell invasion assays were performed to estimate the capabilities of cell viability, invasion and migration. H19 overexpression enhanced MCF-7/TAMR cell EMT, invasion and migration by upregulating Snail. Furthermore, curcumin notably decreased the expression levels of epithelial marker E-cadherin and markedly increased the expression levels of mesenchymal marker N-cadherin in MCF-7/TAMR cells compared with the control group. In addition, following treatment with curcumin for 48 h, H19 expression was decreased in a dose-dependent manner. Moreover, curcumin treatment for 48 h significantly attenuated H19-induced alterations in N-cadherin and E-cadherin expression levels. Curcumin also prevented H19-induced invasion and migration. The present study indicated that H19 may serve as a promoting factor of EMT, invasion and migration in MCF-7/TAMR cells, suggesting that curcumin may prevent H19-associated metastasis. Therefore, curcumin may serve as a promising therapeutic drug for patients with TAMR breast cancer.
Keywords: tamoxifen-resistance, epithelial-mesenchymal transition, breast cancer, H19 imprinted maternally expressed transcript, curcumin
Introduction
Breast cancer is the most common malignancy and the leading cause of cancer-associated death in women worldwide (1). Among patients with breast cancer, ~60–70% are hormone-responsive, thus endocrine therapy has become the standard treatment strategy for disease management in these patients (2). Tamoxifen, a selective estrogen receptor (ER) modulator that blocks the binding between estrogen and its cognate receptor, is most commonly used to treat ER-positive breast cancer, especially in premenopausal patients (3). A 5-year course of adjuvant therapy with tamoxifen significantly reduces the 15-year risk of recurrence and death in patients with breast cancer (4). Although the initial response to tamoxifen is satisfactory, the majority of patients develop resistance and tumor progression (5). Moreover, tamoxifen-resistant (TAMR) breast cancer is more prone to invasion and metastasis (6), thus the 5-year survival rate of patients with breast cancer drops from 80 to 27% with the occurrence of distant metastasis (7).
Several mechanisms influencing resistance to endocrine therapy have been identified. The identified mechanisms are complex and primarily focused on the epigenetic regulation, mutation, truncation and fusion events of the ER1 gene (8,9). Long non-coding RNAs (lncRNAs) have been shown to function as master regulators for gene expression and play a critical role in various biological functions and disease processes including cancer (10). As such, accumulating evidence has demonstrated that lncRNA H19 serves an important role in the proliferation, metastasis, chemoresistance and endocrine resistance of breast cancer (11,12). Recent research has reported that H19 is substantially upregulated in TAMR breast cancer cell lines and tumor tissues, and is associated with tamoxifen resistance (13). Several other studies have demonstrated that H19 dysregulation influences tumor metastasis by regulating the epithelial-mesenchymal transition (EMT) of tumor cells (14,15). Zhou et al (16) demonstrated that H19 could promote EMT in breast cancer cells. However, to the best of our knowledge, the role of H19 in TAMR breast cancer has not been previously reported.
Curcumin, a natural compound derived from turmeric, has been reported to possess antitumor effects, including the prevention of metastasis and progression in multiple cancer types, including pancreatic cancer, breast cancer and chronic myeloid leukemia (17). Curcumin exerts its anticancer effects by targeting multiple intracellular signaling pathways. In vitro investigation has demonstrated that curcumin can inhibit EMT in pancreatic cancer cells by blocking the hedgehog signaling pathway, and the same inhibitory effect has been identified in breast cancer cells (18,19). In addition, curcumin was reported to enhance the radio-sensitivity of renal cancer cells and increase the response of pancreatic cancer cells to gemcitabine (20). However, the effect of curcumin on EMT in TAMR breast cancer cells is not completely understood. Therefore, the present study aimed to determine whether H19 induced EMT in TAMR breast cancer, and to investigate whether curcumin blocked H19-mediated effects in TAMR breast cancer cells.
Materials and methods
Cell culture, TAMR-MCF-7 cell establishment and curcumin treatment
The MCF-7 human breast cancer cell line was obtained from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. MCF-7/TAMR cells were established by treating MCF-7 cells at 37°C with 1 µM 4-hydroxytamoxifen (Sigma-Aldrich; Merck KGaA) for 3 weeks and then 100 nM 4-hydroxytamoxifen for 6 months, as previously described (21). Cells were cultured in RPMI-1640 (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C with 5% CO2. Curcumin (Sigma-Aldrich; Merck KGaA) was dissolved in DMSO (Sigma-Aldrich; Merck KGaA) to prepare a 10 mM stock solution and aliquots were stored at −20°C. Curcumin stock solution was diluted in culture medium so that the final DMSO content was <0.1%. Cells were treated with curcumin at a dose of 5, 10, 20, 30 or 40 µM for 48 h at 37°C with 5% CO2. Untreated cells were used as a control.
Cell viability
MCF-7/TAMR cells were seeded (5×103 cells/well) into 96-well plates and incubated at 37°C with 5% CO2 for 24 h. Cells were treated with different concentrations of curcumin (0, 5, 10, 20, 30 and 40 µM) for 48 h at 37°C with 5% CO2. Subsequently, 10 µl Cell Counting Kit-8 solution (Sigma-Aldrich; Merck KGaA) was added to each well and incubated at 37°C for 1 h. DMSO (0.1%) was added to the control wells. The optical density was determined at a wavelength of 450 nm using the iMark™ Microplate Absorbance Reader (Bio-Rad Laboratories, Inc.).
H19 knockdown and overexpression in MCF-7/TAMR cells
H19 pcDNA3.1 expression vector (H19-epv), empty vector negative control (H19-epv-NC), scrambled small interfering (si)RNA negative control (siNC cat. no. siN0000001-1-5) and H19 siRNA (5′-CCTGTAACCAAAAGTGACCG-3′) were obtained from Guangzhou RiboBio Co., Ltd. Briely, 2×104 MCF-7/TAMR cells were plated in phenol red-free medium containing 10% FBS in 6-well plates, then transfected with 100 nM siRNA (siRNAH19 or siNC) or 1 µg of H19 expression plasmid (H19-epv or empty vector) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) as previously described (20) when cells reached 70% confluence. Following 24-h incubation at 37°C with 5% CO2, cells were treated with curcumin for subsequent experiments as aforementioned.
Wound healing assay
MCF7/TAMR cells were seeded (5×105 cells/well) into 6-well plates. Following transfection, cells were treated with curcumin for 48 h at 37°C. At 90–100% confluence, a sterile pipette tip was used to form a single scratch across the cell monolayer. After washing with PBS, cells were incubated in RPMI-1640 supplemented with 1% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C with 5% CO2. The width of the wound in each group was examined at 0, 24 and 48 h using a light microscope (magnification, ×40; Olympus Corporation), then analyzed using Image J software (version 1.8.0; National Institutes of Health). The migration rate was calculated as: Migration rate = (Width of the wound at 0 h - Width of the wound at 24 or 48 h) / Width of the wound at 0 h.
Transwell invasion assay
Following transfection and curcumin treatment, cell invasion was assessed using modified Boyden chambers (pore size, 8.0 µm; Costar; Corning, Inc.). The Transwell inserts were pre-coated with Matrigel overnight at 37°C with 5% CO2. A total of 2×104 transfected cells were resuspended in 200 µl serum-free medium and plated into the upper chambers of the Transwell inserts in a 24-well plate. Subsequently, 600 µl RPMI-1640 supplemented with 20% FBS was added to the lower chambers. Following incubation at 37°C for 24 h, non-invading cells were removed with cotton swabs and the filters were rinsed with PBS. Invading cells were fixed with methanol for 20 min at room temperature, stained with 1% crystal violet for 30 min at room temperature. Stained cells were counted in eight random microscopic fields using a light microscope (magnification, ×200; Olympus Corporation).
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Total RNA (1 µg) was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. Subsequently, qPCR was performed using iQ™ SYBR® Green Supermix (Bio-Rad Laboratories, Inc.) and the iQ™5 real-time detection system (Bio-Rad Laboratories, Inc.). The following primers were used for qPCR: H19 forward, 5′-GTCCGGCCTTCCTGAACACCTT-3′ and reverse, 5′-GCTTCACCTTCCAGAGCCGAT-3′; E-cadherin forward, 5′-CAACAAAGACAAAGAAGGCAAGG-3′ and reverse, 5′-TGAGAGAAGAGAGTGTATGTGGC-3′; vimentin forward, 5′-GGAGGAGATGCTTCAGAGAGAG-3′ and reverse, 5′-GGATTTCCTCTTCGTGGAGTTTC-3′; Snail forward, 5′-AGGACCACAGTGGCTCAGAAAGGAPDH-3′ and reverse, 5′-TGATGACCCTTTTGGCTCCC-3′; and GAPDH forward, 5′-GGAAGCTTGTCATCAATGGAAATC-3′ and reverse, 5′-TGATGACCCTTTTGGCTCCC-3′. The following thermocycling conditions were used for qPCR: 95°C for 5 min; followed by 40 cycles at 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec. mRNA expression levels were quantified using the 2−ΔΔCq method (22) and normalized to the internal reference gene GAPDH.
Western blotting
Total protein was extracted from cells using RIPA buffer (Sigma-Aldrich; Merck KGaA) containing 1% PMSF, 0.3% protease inhibitor and 0.1% phosphorylated proteinase inhibitor. Total protein was quantified using the BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). Proteins (20 µg) were separated via 12% SDS-PAGE and transferred to nitrocellulose membranes, which were blocked in blocking buffer [5% non-fat dry milk in TBS with 0.5% Tween, (TBS-T)] at room temperature for 2 h. After washing with TBS-T, the membranes were incubated overnight at 4°C with primary antibodies targeted against: E-cadherin (1:300; Abcam; cat. no. ab40772), N-cadherin (1:300; Abcam; cat. no. ab76011), vimentin (1:300; Abcam; cat. no. ab16700), Snail (1:300; Abcam; cat. no. ab216347) and GAPDH (1:1,000; cat. no. G9545; Sigma-Aldrich; Merck KGaA). Following primary incubation, the membranes were incubated for 2 h at room temperature with a HRP-conjugated Affinipure goat anti-rabbit secondary antibody (1:500; Abcam; cat. no. ab6721). Protein bands were visualized using Immobilon Western Chemilum HRP Substrate (cat. no. WBKLS0100; EMD Millipore) and the expression levels of each protein were analyzed using Image Lab software version 4.1 (Bio-Rad Laboratories, Inc.).
Immunofluorescence assay
Cells were plated onto confocal laser small dishes at a density of 5×104, and treated with curcumin as aforementioned. The cells on chamber slides were washed with PBS for 15 min, fixed in 4% paraformaldehyde for 30 min at room temperature and permeabilized with 0.1% Triton X-100 for 5 min at room temperature. After three washes with PBS (5 min each), non-specific binding was blocked with 3% BSA (Thermo Fisher Scientific, Inc.) for 1 h at room temperature. Subsequently, cells were incubated with primary antibodies targeted against E-cadherin (1:100 in PBS with 1% BSA) and N-cadherin (1:100 in PBS with 1% BSA) for 2 h at room temperature. Following washing with PBS, cells were incubated with Alexa Fluor 488 goat anti-rabbbit IgG (Proteintech, cat. no. SA00006-2) for E-cadherin and Alex Fluor 594 goat anti-rabbbit IgG (Proteintech; cat. no. SA00006-4) secondary antibody for 1 h at room temperature. After three 5-min washes with PBS, cell nuclei were stained with 10 µg/ml Hoechst 33258 for 10 min at room temperature. Following washing with PBS, cells were visualized using a fluorescence microscope (magnification, ×200).
Statistical analysis
Data are presented as the mean ± SEM. Comparisons among multiple groups were analyzed using one-way ANOVA following by the SNK or Tukey's post hoc test. Comparisons between two groups were analyzed using an unpaired Student's t-test. Statistical analyses were performed using SPSS software (version 21.0; IBM Corp.). Each experiment was performed at least three times. P<0.05 was considered to indicate a statistically significant difference.
Results
H19 induces EMT in MCF-7/TAMR cells
To determine the effects of H19 on MCF-7/TAMR cell EMT, H19 was overexpressed and knocked down using HPV-epv and siRNAH19, respectively. The results indicated that following transfection with siRNAH19 for 24 h, H19 expression was significantly decreased by 40-fold, vimentin expression was notably downregulated and E-cadherin expression was markedly upregulated compared with the siRNAH19-NC group (Fig. 1A, C and E). To further clarify the role of H19 in EMT, H19 was overexpressed in MCF-7/TAMR cells. At 24 h post-transfection, a 60-fold significant increase in H19 expression levels was observed in H19-epv-transfected MCF-7/TAMR cells compared with H19-epv-NC-transfected cells (Fig. 1B). Moreover, H19 overexpression notably increased vimentin expression levels and markedly decreased E-cadherin expression levels compared with the H19-epv-NC group (Fig. 1D and E). Collectively, the aforementioned results suggested that H19 promoted EMT in TAMR breast cancer cells.
Figure 1.
H19 induces MCF-7/tamoxifen-resistant cell epithelial-mesenchymal transition. Transfection efficiency of (A) siRNAH19 and (B) H19-epv. The effects of (C) siRNAH19 and (D) H19-epv on E-cadherin and vimentin mRNA expression levels were determined via reverse transcription-quantitative PCR. (E) The effects of siRNAH19 and H19-epv on E-cadherin and vimentin protein expression levels were determined via western blotting. **P<0.05 vs. siRNAH19-NC or H19-epv-NC. E-cad, E-cadherin; siRNA, small interfering RNA; epv, expression vector; NC, negative control.
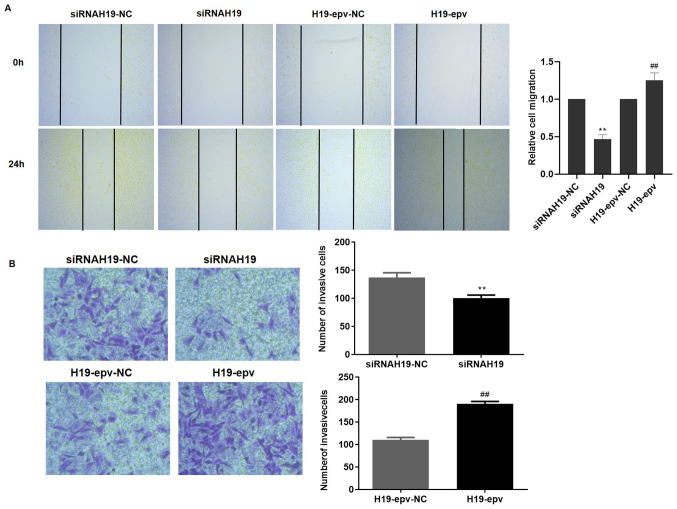
H19 promotes MCF-7/TAMR cell migration and invasion
Wound healing and Transwell assays were performed to determine the influence of H19 expression on MCF-7/TAMR cell migration and invasion. Following transfection with H19-epv or siRNAH19 for 24 h, the results demonstrated that H19 overexpression promoted wound closure, whereas H19 knockdown inhibited wound closure compared with the H19-epv-NC and siRNAH19-NC groups, respectively (Fig. 2A). Furthermore, the Transwell invasion assay results suggested that H19 overexpression significantly promoted MCF-7/TAMR cell invasion compared with the H19-epv-NC group, whereas H19 knockdown significantly decreased cell invasion compared with the siRNAH19-NC group (Fig. 2B). The results indicated that H19 induced TAMR breast cancer cell migration and invasion.
Figure 2.
H19 induces MCF-7/tamoxifen-resistant cell migration and invasion. Cells were transfected with H19-epv or siRNAH19 for 24 h. Cell (A) migration (magnification, ×40) and (B) invasion (Magnification, ×200) were assessed by performing wound healing and Transwell invasion assays, respectively. **P<0.05 vs. siRNAH19-NC, ##P<0.01 vs. H19-epv-NC. NC, negative control; epv, expression vector; siRNA, small interfering RNA.
H19 upregulates Snail, promoting MCF-7/TAMR cell EMT
Snail is a key regulator of the EMT process (23). Therefore, to investigate the mechanisms underlying H19-induced EMT, the mRNA and protein expression levels of Snail in MCF-7/TAMR cells following transfection with H19-epv or siRNAH19 were determined (Fig. 3). H19 overexpression notably increased Snail mRNA and protein expression levels compared with the H19-epv-NC group. However, H19 knockdown markedly decreased the mRNA and protein expression levels of Snail compared with siRNAH19-NC. The results suggested that H19 promoted EMT in TAMR breast cancer cells via Snail upregulation.
Figure 3.
H19 promotes MCF-7/tamoxifen-resistant cell epithelial-mesenchymal transition via regulating Snail. Cells were transfected with H19-epv or siRNAH19 for 24 h. Snail (A) protein and (B) mRNA expression levels were measured via western blotting and reverse transcription-quantitative PCR, respectively. **P<0.05 vs. siRNAH19-NC, ##P<0.05 vs. H19-epv-NC. NC, negative control; epv, expression vector; siRNA, small interfering RNA.
Curcumin inhibits H19-induced EMT in MCF-7/TAMR cells
The impact of curcumin on MCF-7/TAMR cells proliferation was investigated. As demonstrated in Fig. 4A, administration of MCF-7/TAMR cells with curcumin for 48 h inhibited proliferation in a dose-dependent manner, with an IC50 value of 31.7 µM. No significant differences in MCF-7/TAMR cell viability were observed at curcumin concentrations up to 20 µM. The expression levels of EMT biomarkers (E-cadherin and N-cadherin) were subsequently assessed via RT-qPCR and western blotting. Compared with the control group, following treatment with curcumin for 48 h, the expression levels of the mesenchymal marker N-cadherin were notably reduced, whereas the expression levels of the epithelial marker E-cadherin were markedly increased (Fig. 4B and C). The results indicated that curcumin inhibited MCF-7/TAMR cell EMT.
Figure 4.
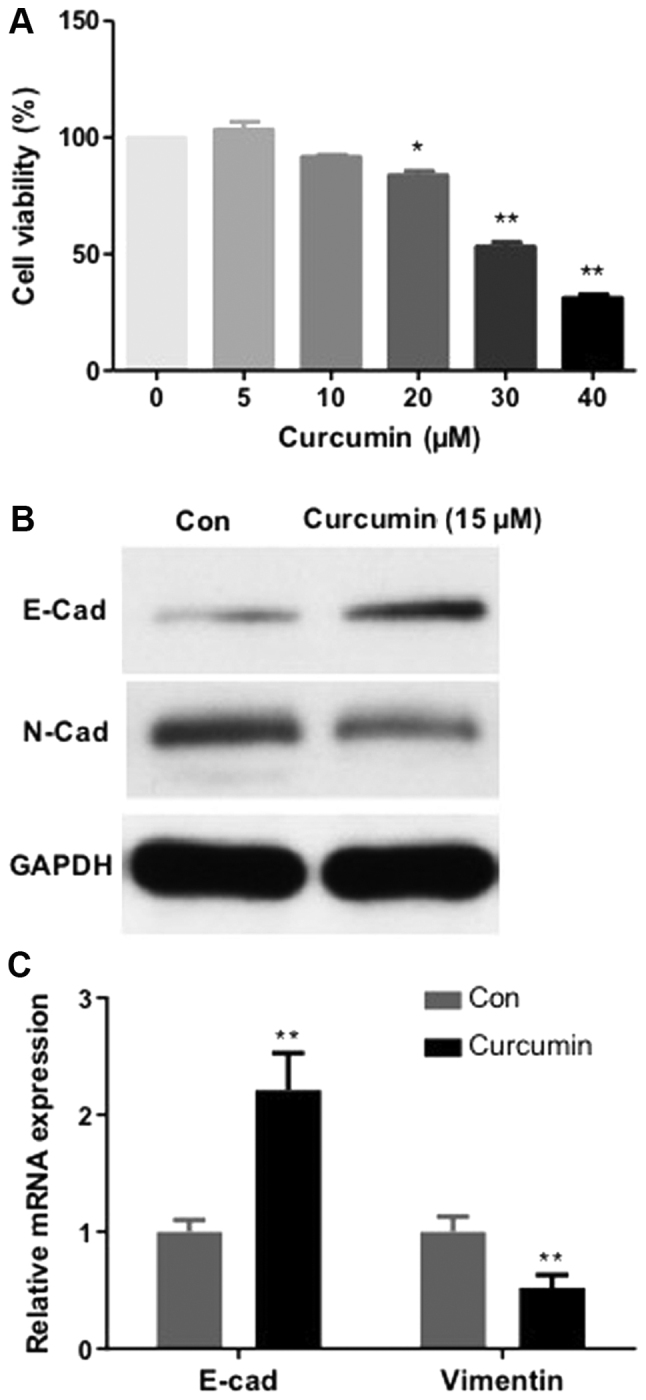
Curcumin influences MCF-7/tamoxifen-resistant cell viability and epithelial-mesenchymal transition. (A) The effect of curcumin on cell viability was determined by performing a Cell Counting Kit-8 assay. *P<0.05 and **P<0.01 vs. 0 µM curcumin groups. E-cadherin and N-cadherin (B) protein and (C) mRNA expression levels were determined via western blotting and reverse transcription-quantitative PCR, respectively. **P<0.01 vs. Con. Con, control; E-cad, E-cadherin; N-cad, N-cadherin.
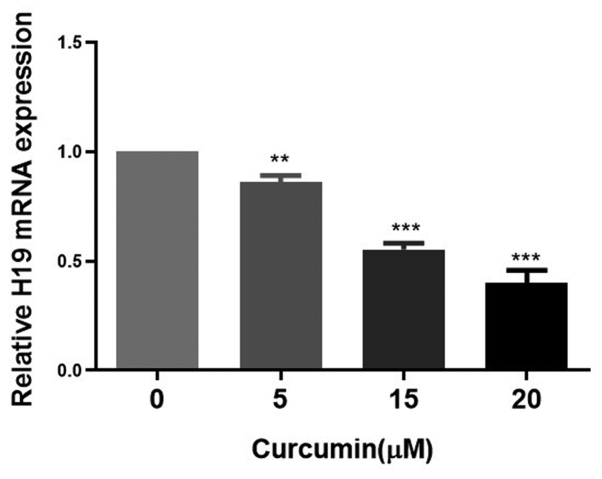
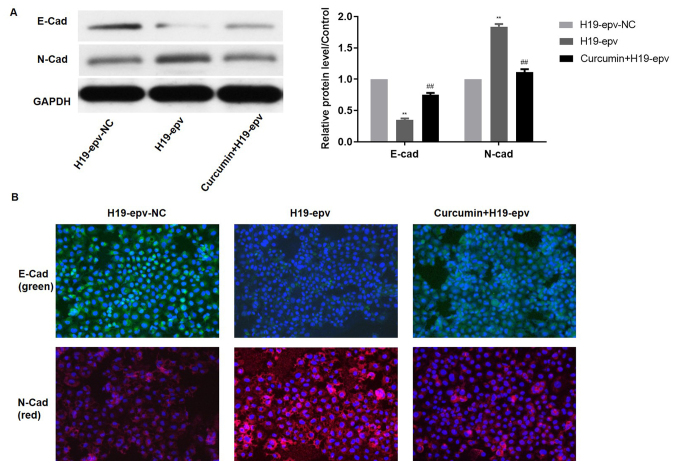
Subsequently, the effects of different concentrations of curcumin on the expression of H19 in MCF-7/TAMR cells were assessed. H19 expression was significantly decreased in a dose-dependent manner by concentrations of curcumin between 5 and 20 µM for 48 h (Fig. 5). Therefore, 15 µM curcumin was selected for use in subsequent experiments. To verify the influence of curcumin on H19-induced EMT, the expression levels of E-cadherin and N-cadherin were evaluated in MCF-7/TAMR cells following H19 overexpression in the presence or absence of curcumin. Compared with the H19-epv group, the protein expression levels of N-cadherin were notably decreased and E-cadherin expression levels were significantly increased in H19-epv-transfected cells treated with curcumin for 48 h (Fig. 6A). Immunofluorescence analysis indicated that H19-overexpression cells displayed notably lower levels of E-cadherin expression and considerably higher levels of N-cadherin expression compared with the H19-epv-NC group. However, following treatment with curcumin for 48 h, the expression levels of E-cadherin and N-cadherin in H19-overexpression cells were recovered (Fig. 6B). The results suggested that curcumin reversed H19-induced effects on EMT.
Figure 5.
Curcumin inhibits H19 mRNA expression in MCF-7/tamoxifen-resistant cells. Cells were treated with various concentrations of curcumin for 48 h. Subsequently, H19 mRNA expression levels were determined via reverse transcription-quantitative PCR. **P<0.05, ***P<0.01 vs. 0 µM curcumin.
Figure 6.
Curcumin attenuates H19-induced epithelial-mesenchymal transition in MCF-7/tamoxifen-resistant cells. Following transfection with H19-epv, cells were treated with or without curcumin for 48 h. (A) E-cadherin and N-cadherin protein expression levels were determined via western blotting. **P<0.01 vs. H19-epv-NC; ##P<0.01 vs. H19-epv. (B) Cytolocalization of E-cadherin and N-cadherin in MCF-7/TAMR cells was detected by performing immunofluorescence staining. Magnification, ×200. E-cad, E-cadherin; N-cad, N-cadherin; epv, expression vector; NC, negative control.
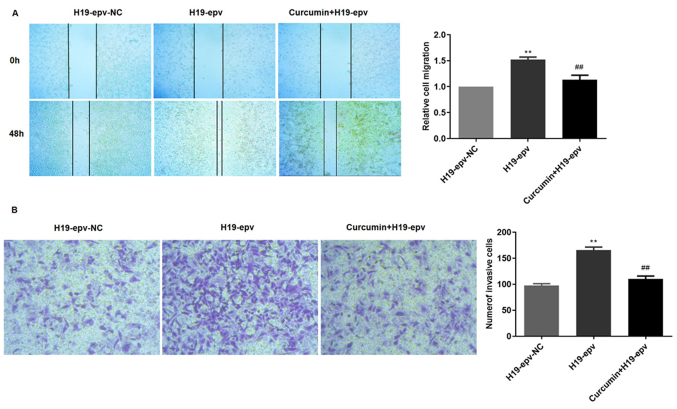
Curcumin inhibits H19-induced MCF-7/TAMR cell migration and invasion
To verify the influence of curcumin on H19-induced migration and invasion, MCF-7/TAMR cells were transfected with H19-epv for 24 h, and then incubated in the presence or absence of curcumin for 48 h. The results demonstrated that cell migration in the H19-epv group was significantly increased compared with H19-epv-NC. However, treatment with curcumin for 48 h significantly decreased wound closure in H19-overexpression cells (Fig. 7A). The Transwell invasion assay results also indicated that the number of invading cells was higher in the H19-overexpression group compared with the NC group, but curcumin treatment significantly decreased H19 overexpression-induced cell invasion (Fig. 7B). Collectively, the results indicated that curcumin inhibited MCF-7/TAMR cell migration and invasion resulting from H19-induced EMT.
Figure 7.
Curcumin inhibits H19-induced MCF-7/tamoxifen-resistant cell migration and invasion. Following transfection with H19-epv for 24 h, cells were treated with or without curcumin for 48 h. Cell (A) migration (magnification, ×40) and (B) invasion (magnification, ×200) were determined by performing wound healing and Transwell invasion assays, respectively. **P<0.05 vs. H19-epv-NC; ##P<0.01 vs. 19-epv. NC, negative control; epv, expression vector.
Discussion
Despite its wide use for the treatment of breast cancer, a third of patients with breast cancer develop resistance to tamoxifen, which results in tumor progression (24). Additionally, TAMR breast cancer is more prone to invasion and metastasis (6), thus identifying the mechanisms underlying drug-resistant tumor metastasis, as well as novel therapeutic strategies is critical to improve patient survival. H19 was previously reported to contribute to tamoxifen resistance in breast cancer cells via various molecular mechanisms, including the regulation of autophagy, and Notch and hepatocyte growth factor signaling (13).
Previous studies have indicated that H19 is upregulated in a variety of cancer cell types, including breast cancer, gastric cancer and glioma cells, and that H19 knockdown inhibits tumor growth, migration and invasion (25,26). EMT is important for tumor cell migration and invasion (27), and E-cadherin and vimentin are important markers of EMT. During EMT, E-cadherin and vimentin expression levels are downregulated and upregulated, respectively (28). H19 also mediates migration and EMT-related activity in lung cancer and cisplatin-resistance ovarian cancer cells (24,26). Collectively, the aforementioned studies suggested that H19 upregulation is associated with invasion, metastasis and EMT in different types of cancer cells. In the present study, the influence of H19 on migration, invasion and EMT was investigated by performing H19 overexpression and knockdown in TAMR breast cancer cells. Compared with the H19-epv-NC group, H19 overexpression promoted migration and invasion, increased vimentin expression and decreased E-cadherin expression. H19 knockdown displayed the opposite effect, inhibiting migration and invasion, increasing E-cadherin expression levels and decreasing vimentin expression levels compared with the siRNAH19-NC group. The results suggested that H19 promoted EMT in MCF-7/TAMR cells via regulating the expression of vimentin and E-cadherin. As a zinc finger transcriptional repressor, Snail is a key regulator of EMT that downregulates and upregulates the expression of epithelial and mesenchymal genes, respectively (29). The results of the present study suggested that H19, as an upstream factor, upregulated the expression levels of Snail, which decreased E-cadherin expression, thus triggering EMT in MCF-7/TAMR cells.
Accumulating evidence has demonstrated that curcumin exerts anticancer effects via multiple signaling pathways. Hu et al (30) demonstrated that curcumin significantly inhibited breast cancer cell proliferation by inhibiting the phosphorylation of AKT/mTOR signaling pathway-related proteins. Curcumin was also reported to induce autophagy and activate lysosomal functioning by inhibiting the AKT/mTOR signaling pathway and activating transcription factor EB (30). In hepatoma cells, curcumin inhibits TGF-β1-induced EMT by suppressing Smad2 signaling pathway activation (31). A recent study also indicated that curcumin inhibits superoxide dismutase-induced migration, invasion and EMT-related gene expression in pancreatic cancer (32). In the present study, curcumin exerted an inhibitory effect on viability and EMT in MCF-7/TAMR cells and significantly attenuated H19-induced migration, invasion and EMT. The present study provided several insights into the mechanism underlying curcumin-mediated regulation of H19-induced EMT and migration. Firstly, compared with the siRNAH19-NC group, H19 overexpression notably upregulated vimentin expression and markedly downregulated E-cadherin expression, which indicated that H19 promoted EMT in TAMR breast cancer cells. Furthermore, the wound healing and Transwell invasion assays demonstrated that H19 overexpression promoted migration and invasion in MCF-7/TAMR cells compared with the H19-epv-NC group. The present study had several limitations. The role of curcumin on H19-induced EMT requires further investigation. For instance, an animal model should be applied to validate the molecular mechanism related to the H19/Snail/E-cadherin axis in regulation of MCF-7/TAMR EMT and demonstrate the therapeutic potential of curcumin. Therefore, the mechanism of the present study requires further investigation using in vivo models of TAMR breast cancer.
In conclusion, the present study suggested that H19 promoted EMT, migration and invasion in MCF-7/TAMR cells, whereas curcumin inhibited H19-induced effects. Therefore, the use of curcumin to inhibit the H19/Snail/E-cadherin axis may serve as a promising therapeutic option for patients with TAMR breast cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Fujian Science and Technology Innovation Joint Fund Project (grant no. 2017Y9067), the Medical Science Research Project (grant no. BJBQEKYJJ-B19001CS), the Young and Middle-Aged Backbone Talents Project (grant no. 2019-ZQN-35), the Science and Technology Department of Fujian Province (grant no. 2017Y9098), the High-Level Hospital Foster Grants from Fujian Provincial Hospital (grant no. 2019HSJJ06), the Fujian Natural Science Foundation Project (grant no. 2019J01177) and the Startup Fund for Scientific Research, Fujian Medical University (grant nos. 2016QH106 and 2017XQ1024).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JC and JZ conceptualized the study. JC, BZ and HS designed the study. JC, HS, MX and GZ carried out the experiments and curated the data. JC, CX and XH analyzed the data. JC and HS wrote the manuscript. JZ reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Poggio F, Ceppi M, Lambertini M, Bruzzi P, Ugolini D, Bighin C, Levaggi A, Giraudi S, D'Alonzo A, Vaglica M, et al. Concurrent versus sequential adjuvant chemo-endocrine therapy in hormone-receptor positive early stage breast cancer patients: A systematic review and meta-analysis. Breast. 2017;33:104–108. doi: 10.1016/j.breast.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Liu Y. Endocrine therapy for breast cancer: Past and present. Zhonghua Yi Shi Za Zhi. 2015;45:28–32. (In Chinese) [PubMed] [Google Scholar]
- 4.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S, McGale P, Pan HC, Taylor C, et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level metaanalysis of randomised trials. Lancet. 2011;378:771–784. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burstein HJ, Temin S, Anderson H, Buchholz TA, Davidson NE, Gelmon KE, Giordano SH, Hudis CA, Rowden D, Solky AJ, et al. Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: American society of clinical oncology clinical practice guideline focused update. J Clin Oncol. 2014;32:2255–2269. doi: 10.1200/JCO.2013.54.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang M. Tamoxifen resistance in breast cancer. Biomol Ther (Seoul) 2012;20:256–267. doi: 10.4062/biomolther.2012.20.3.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park M, Kim J, Phuong NTT, Park JG, Park JH, Kim YC, Baek MC, Lim SC, Kang KW. Involvement of the P2X7 receptor in the migration and metastasis of tamoxifen-resistant breast cancer: Effects on small extracellular vesicles production. Sci Rep. 2019;9:11587. doi: 10.1038/s41598-019-47734-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Marchi T, Liu NQ, Stingl C, Timmermans MA, Smid M, Look MP, Tjoa M, Braakman RB, Opdam M, Linn SC, et al. 4-protein signature predicting tamoxifen treatment outcome in recurrent breast cancer. Mol Oncol. 2016;10:24–39. doi: 10.1016/j.molonc.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Zhou C, Jiang H, Liang L, Shi W, Zhang Q, Sun P, Xiang R, Wang Y, Yang S. ZEB1 induces ER-α promoter hypermethylation and confers antiestrogen resistance in breast cancer. Cell Death Dis. 2017;8:e2732. doi: 10.1038/cddis.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD, Kang ZJ, Fan WJ, Zhu LL, Li M, et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017;8:e2569. doi: 10.1038/cddis.2016.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu QN, Wang G, Guo Y, Peng Y, Zhang R, Deng JL, Li ZX, Zhu YS. LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. Oncotarget. 2017;8:91990–92003. doi: 10.18632/oncotarget.21121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basak P, Chatterjee S, Bhat V, Su A, Jin H, Lee-Wing V, Liu Q, Hu P, Murphy LC, Raouf A. Long non-coding RNA H19 acts as an estrogen receptor modulator that is required for endocrine therapy resistance in ER+ breast cancer cells. Cell Physiol Biochem. 2018;51:1518–1532. doi: 10.1159/000495643. [DOI] [PubMed] [Google Scholar]
- 14.Wu Y, Zhou Y, He J, Sun H, Jin Z. Long non-coding RNA H19 mediates ovarian cancer cell cisplatin-resistance and migration during EMT. Int J Clin Exp Pathol. 2019;12:2506–2515. [PMC free article] [PubMed] [Google Scholar]
- 15.Ye Y, Guo J, Xiao P, Ning J, Zhang R, Liu P, Yu W, Xu L, Zhao Y, Yu J. Macrophages-induced long noncoding RNA H19 up-regulation triggers and activates the miR-193b/MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Lett. 2020;469:310–322. doi: 10.1016/j.canlet.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li LY, Guan GH, Liu Q, Qian YH, Xie D. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10:eaak9557. doi: 10.1126/scisignal.aak9557. [DOI] [PubMed] [Google Scholar]
- 17.Shanmugam MK, Rane G, Kanchi MM, Arfuso F, Chinnathambi A, Zayed ME, Alharbi SA, Tan BK, Kumar AP, Sethi G. The multifaceted role of curcumin in cancer prevention and treatment. Molecules. 2015;20:2728–2769. doi: 10.3390/molecules20022728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao L, Xiao X, Lei J, Duan W, Ma Q, Li W. Curcumin inhibits hypoxia-induced epithelial mesenchymal transition in pancreatic cancer cells via suppression of the hedgehog signaling pathway. Oncol Rep. 2016;35:3728–3734. doi: 10.3892/or.2016.4709. [DOI] [PubMed] [Google Scholar]
- 19.Gallardo M, Calaf GM. Curcumin inhibits invasive capabilities through epithelial mesenchymal transition in breast cancer cell lines. Int J Oncol. 2016;49:1019–1027. doi: 10.3892/ijo.2016.3598. [DOI] [PubMed] [Google Scholar]
- 20.Li G, Wang Z, Chong T, Yang J, Li H, Chen H. Curcumin enhances the radiosensitivity of renal cancer cells by suppressing NF-κB signaling pathway. Biomed Pharmacother. 2017;94:974–981. doi: 10.1016/j.biopha.2017.07.148. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Xie S, Yang J, Xiong H, Jia Y, Zhou Y, Chen Y, Ying X, Chen C, Ye C, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol. 2019;12:81. doi: 10.1186/s13045-019-0747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Shi J, Chai K, Ying X, Zhou BP. The Role of Snail in EMT and Tumorigenesis. Curr Cancer Drug Targets. 2013;13:963–972. doi: 10.2174/15680096113136660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mills JN, Rutkovsky AC, Giordano A. Mechanisms of resistance in estrogen receptor positive breast cancer: Overcoming resistance to tamoxifen/aromatase inhibitors. Curr Opin Pharmacol. 2018;41:59–65. doi: 10.1016/j.coph.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Si H, Chen P, Li H, Wang X. Long non-coding RNA H19 regulates cell growth and metastasis via miR-138 in breast cancer. Am J Transl Res. 2019;11:3213–3225. [PMC free article] [PubMed] [Google Scholar]
- 26.Liao S, Yu C, Liu H, Zhang C, Li Y, Zhong X. Long non-coding RNA H19 promotes the proliferation and invasion of lung cancer cells and regulates the expression of E-cadherin, N-cadherin, and vimentin. OncoTargets Ther. 2019;12:4099–4107. doi: 10.2147/OTT.S185156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang GM, Wang HS, Zhang F, Zhang KS, Liu ZC, Fang R, Wang H, Cai SH, Du J. Histone deacetylase inhibitor induction of epithelial-mesenchymal transitions via up-regulation of Snail facilitates cancer progression. Biochim Biophys Acta. 2013;1833:663–671. doi: 10.1016/j.bbamcr.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Kokkinos MI, Wafai R, Wong MK, Newgreen DF, Thompson EW, Waltham M. Vimentin and epithelial-mesenchymal transition in human breast cancer - observations in vitro and in vivo. Cells Tissues Organs. 2007;185:191–203. doi: 10.1159/000101320. [DOI] [PubMed] [Google Scholar]
- 29.Al Khatib AM, Stepan AE, Margaritescu C, Simionescu C, Ciurea RN. E-cadherin and snail immunoexpression in colorectal adenocarcinomas. Curr Health Sci J. 2019;45:204–209. doi: 10.12865/CHSJ.45.02.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu S, Xu Y, Meng L, Huang L, Sun H. Curcumin inhibits proliferation and promotes apoptosis of breast cancer cells. Exp Ther Med. 2018;16:1266–1272. doi: 10.3892/etm.2018.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao MT, Liu HF, Liu ZG, Xiao P, Chen JJ, Tan Y, Jiang XX, Jiang ZC, Qiu Y, Huang HJ, et al. Curcumin downregulates the expression of Snail via suppressing Smad2 pathway to inhibit TGF-β1-induced epithelial-mesenchymal transitions in hepatoma cells. Oncotarget. 2017;8:108498–108508. doi: 10.18632/oncotarget.22590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Jiang Z, Xiao X, Wang Z, Wu Z, Ma Q, Cao L. Curcumin inhibits superoxide dismutase-induced epithelial-to-mesenchymal transition via the PI3K/Akt/NF-κB pathway in pancreatic cancer cells. Int J Oncol. 2018;52:1593–1602. doi: 10.3892/ijo.2018.4295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.