Abstract
Endometrial cancer (EC) is a common malignant gynecological tumor arising from the endometrium, with an annually increasing morbidity and mortality. The present study aimed to investigate the functions of cullin 4A (CUL4A) in EC, as well as the underlying mechanisms. CUL4A expression was assessed in several human EC cells and normal human endometrial epithelial cells (hEECs) via reverse transcription-quantitative polymerase chain reaction and western blotting. Subsequently, short hairpin (sh)RNA-CULA4 was transfected into cells, and cell proliferation, invasion and migration were detected using Cell Counting kit-8, Transwell and wound healing assays, respectively. The STRING database identified that CSN6 interacted with CULA4, and immunoprecipitation was performed to verify the interaction. Subsequently, following CUL4A knockdown, pcDNA3.1-CSN6 was transfected into cells and its effects on cell proliferation, invasion and migration were assessed. The expression levels of matrix metallopeptidase (MMP)2, MMP9 and p53 were evaluated via western blotting. The results indicated that CUL4A was highly expressed in EC cells, compared with hEECs. CULA4-knockdown notably inhibited EC cell proliferation, invasion and migration. The expression levels of MMP2 and MMP9 were significantly decreased, while p53 expression was enhanced following CUL4A-knockdown. The immunoprecipitation assay verified that COP9 signalosome subunit 6 (CSN6) interacted with CULA4. Furthermore, CSN6-overexpression alleviated the inhibitory effects of CUL4A-knockdown on EC cell proliferation, invasion and migration. Similarly, CSN6 overexpression reversed CUL4A-knockdown-mediated effects on the expression of MMP2, MMP9 and p53. In summary, the results demonstrated that CUL4A regulated EC cell proliferation, invasion and migration by interacting with CSN6.
Keywords: endometrial cancer, cullin 4A, proliferation, invasion, migration
Introduction
Endometrial cancer (EC) is a common malignant gynecological tumor arising from the endometrium and is the fourth most common cause of cancer among females (1–3). It currently accounts for ~50% of newly diagnosed gynecological malignancies (4). The morbidity and mortality of EC are increasing year by year, which negatively affects the health of females worldwide (5,6). Although significant advances in therapeutic strategies have been achieved, including hormonal agents, cytotoxic chemotherapy and surgery combined with adjuvant chemotherapy, which contribute towards improvements in the survival rates of patients with EC, the prognosis of patients with EC remains poor (7,8). Therefore, understanding the pathogenesis of EC is important to identify preventative and therapeutic strategies for EC.
Cullin 4A (CUL4A), a member of the cullin family, acts as a scaffold protein to bind DNA damage-binding protein 1 and subsequently form a ubiquitin ligase E3 complex to mediate the degradation of numerous substrates (9,10). Therefore, CUL4A mediates multiple cellular processes, including proliferation, DNA replication, apoptosis and hematopoiesis (11,12). Emerging evidence supports that CUL4A is responsible for the regulation of certain tumor suppressor genes, including p53 and p27 (13,14). Increasing evidence suggests that CUL4A expression was highly upregulated in a variety of different types of human cancer, including non-small cell lung, colorectal, ovarian and breast cancer, which promoted tumor cell proliferation, invasion and migration (15–18). However, the biological functions of CUL4A in EC are not completely understood.
In the present study, the expression of CUL4A in several EC cell lines and human endometrial epithelial cells (hEECs), as well as the potential underlying mechanisms, were examined. The results indicated that CUL4A regulated EC cell proliferation, invasion and migration by interacting with COP9 signalosome subunit 6 (CSN6), which provided a theoretical basis and potential therapeutic strategy for EC.
Materials and methods
The Cancer Genome Atlas (TCGA) database analysis
Human RNA-sequencing data from uterine corpus endometrial carcinoma (UCEC) projects, which included 546 patients with EC and 35 normal tissues were obtained from TCGA analysis in UALCAN database (ualcan.path.uab.edu/). Student's t-test was used to determine statistical significance of CUL4A expression between normal and tumor samples.
Cell culture
Several human endometrial cancer cell lines (Ishikawa, KLE, RL95-2 and AN3CA) and a normal human endometrial epithelial cell line (hEECs) were obtained from Shanghai Institute for Biological Sciences (Shanghai, China. Cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2.
Cell transfection
The short hairpin (sh) RNAs targeted against CUL4A (shRNA-CUL4A-1 and shRNA-CUL4A-2) and a negative control (NC) scrambled shRNA (shRNA-NC) were provided by Shanghai GenePharma Co., Ltd.). In addition, a CSN6-overexpression plasmid (pcDNA-CSN6) and its empty vector (pcDNA-NC) were purchased from of Shanghai Integrated Biotech Solutions Co., Ltd. Cells were plated into 6-well plates (1×106 cells/well) overnight. Subsequently, cells were transfected using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 24 h post-transfection, transfection efficiency was assessed via western blotting and reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Cell Counting kit-8 (CCK-8) assay
Cell proliferation was assessed using the CCK-8 kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), according to the manufacturer's protocol. In brief, transfected RL95-2 cells were cultured on a 96-well plate and incubated for 24, 48 or 72 h at 37°C. Subsequently, 10 µl CCK-8 solution (Dojindo Molecular Technologies, Inc., Rockville, MD, USA) was added to each well. Following incubation for 2 h at 37°C, the optical density at a wavelength of 450 nm was detected using a microplate reader (BioTek Instruments, Inc., Winooski, VT, USA).
Transwell invasion assay
A Transwell invasion assay was conducted using 8-µm Transwell chambers (Corning Inc.) coated with diluted BD Matrigel (BD Biosciences) overnight at 37°C. In brief, 2×105 cells were cultured in the upper chamber with serum-free DMEM, and medium containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) was plated in the lower chamber. Following incubation for 24 h at 37°C, cells on the lower surface of the membrane were fixed with methanol for 20 min at 37°C and stained with crystal violet for 30 min at 37°C. The number of invaded cells was counted and images were captured using an inverted light microscope (magnification, ×200).
Wound healing assay
Cell migration was assessed by conducting a wound healing assay. Cells were plated onto 6-well plates and cultured overnight at 37°C in a humidified atmosphere. At 80% confluence, a linear wound was gently created in the cell monolayer using a pipette tip and the cells were washed three times with phosphate-buffered saline (PBS). Cells were incubated in FBS-free DMEM for 48 h at 37°C. Migratory cells were observed using an IX711 phase contrast microscope (Olympus Corporation) at 0 and 48 h (magnification, ×200).
Co-immunoprecipitation (IP) assay
For co-IP assays, RL95-2 cells were washed in 2 ml PBS (Beyotime Institute of Biotechnology) and centrifuged at 850 × g at room temperature for 5 min to collect the cells. Then, cells were lysed in lysis buffer for IP (Beyotime Institute of Biotechnology). Lysates were incubated with 1 µg antibodies against CUL4A (1:1,000; cat. no. 2699T; Cell Signaling Technology, Inc.), CSN6 (1:1,000; cat. no. sc-393023; Santa Cruz Biotechnology, Inc.) and control lgG (1:1,000; cat. no. 3900S; Cell Signaling Technology, Inc) plus Protein A/G beads (Santa Cruz Biotechnology, Inc.) at 4°C for 2 h. After washing the beads with PBS (Beyotime Institute of Biotechnology) three times, immunoprecipitates were analyzed via western blotting.
RT-qPCR
Total RNA was isolated from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into cDNA using a Reverse Transcription kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's instructions. Subsequently, qPCR was performed using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) and an ABI 7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used: Pre denaturation at 95°C for 10 min, then 40 cycles of denaturation at 95°C for 15 sec and annealing at 60°C for 1 min. The following primer sequences were used: CUL4A, forward, 5′-AAGAGCAGGCAACAAAGAAGCCAC-3′ and reverse 5′-TTGGCCAGTAGCCCATTGTGAGTA-3′; CSN6, forward, 5′-TCATCGAGAGCCCCCTCTTT-3′ and reverse, 5′-CCAATGCGTTCCGCTTCCT-3′; and GAPDH, forward, 5′-GAGTCAACGGATTTGGTCG-3′ and reverse, 5′-TTGATTTTGGAGGGATCTCG-3′. Relative expression was quantified using the 2−ΔΔCq method and normalized to the internal reference gene GAPDH (19).
Western blotting
Total protein was extracted using RIPA buffer (Beyotime Institute of Biotechnology). Total protein was quantified using a BCA kit. Subsequently, equal amounts of protein (40 µg/lane) were separated via 10% SDS-PAGE and transferred onto polyvinylidene membranes (EMD Millipore; Merck KGaA), which were blocked with 5% skimmed fat milk overnight at 4°C. The membranes were incubated with primary antibodies. Following primary incubation, the membranes were incubated with a horseradish peroxidase-conjugated secondary antibody (1:3,000; cat. no. 7074S; Cell Signaling Technology, Inc.) at room temperature for 1.5 h. Proteins were visualized using Image Quant LAS 4000 (Cytiva) and scanned using ImageJ software (National Institutes of Health) with GAPDH as the loading control. Anti-CUL4A (cat. no. 2699T), anti-matrix metallopeptidase (MMP)2 (cat. no. 40994S), anti-MMP9 (cat. no. 13667T), anti-p53 (cat. no. 2527T) and anti-GAPDH (cat. no. 5174T) antibodies (all 1:1,000) were obtained from Cell Signaling Technology, Inc.
Statistical analysis
All experiments were repeated three times independently. Data are presented as the mean ± standard deviation. Statistical analyses were conducted using SPSS (version 19.0; IBM Corp.) and GraphPad Prism (version 6; GraphPad Software, Inc.) software. Comparisons between two groups were analyzed using the unpaired Student's t-test. Comparisons among multiple groups were analyzed using one-way analysis of variance followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
CUL4A is highly expressed in EC tissues and cells
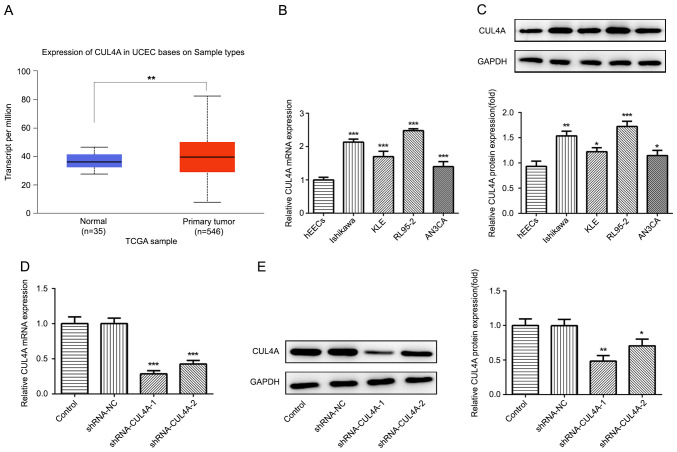
To investigate the functions of CUL4A in EC, TCGA database was used to analyze the level of CUL4A in EC tumor tissues (n=546) and adjacent non-tumor tissues (n=35). As presented in Fig. 1A, the expression level of CUL4A was increased in EC tissues compared with that in adjacent non-tumor tissues. Next, Ishikawa, KLE, RL95-2 and AN3CA EC cell lines, as well as one normal human endometrial epithelial cell line (hEECs) were employed to determine the level of CUL4A via RT-qPCR and western blotting. The mRNA and protein expression of CUL4A was markedly upregulated in EC cell lines, compared with hEECs (Fig. 1B and C). The expression of CUL4A was highest in RL95-2 cells; therefore, RL95-2 cells were selected for subsequent experiments.
Figure 1.
CUL4A is highly expressed in EC tissues and cell lines. (A) TCGA database was used to analyze the mRNA expression level of PITPNA-AS1 in NSCLC tumor tissues and adjacent non-tumor tissues. **P<0.01 vs. normal. CUL4A expression in EC cell lines (Ishikawa, KLE, RL95-2 and AN3CA) and normal human endometrial epithelial cells (hEEC) was examined via (B) RT-qPCR and (C) western blotting. *P<0.05, **P<0.01 and ***P<0.001 vs. hEEC. CUL4A expression following transfection with shRNA-CUL4A-1 or shRNA-CUL4A-2 was detected via (D) RT-qPCR and (E) western blotting. *P<0.05, **P<0.01 and ***P<0.001 vs. shRNA-NC. CUL4A, cullin 4A; EC, endometrial cancer; TCGA, The Cancer Genome Atlas; NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR; shRNA, short hairpin RNA; NC, negative control.
CUL4A-knockdown inhibits EC proliferation, invasion and migration
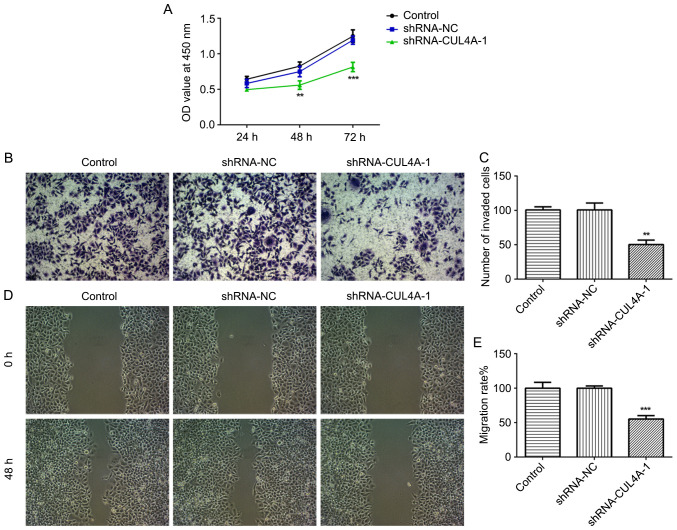
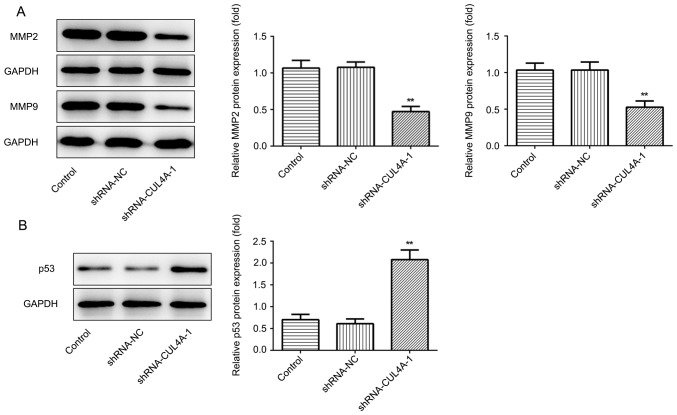
The effects of CUL4A on the functions of EC cells were investigated in the present study. To begin with, RL95-2 cells were transfected with shRNA-CUL4A-1 or shRNA-CUL4A-2. The levels of CUL4A were significantly decreased following transfection with shRNA-CUL4A, compared with shRNA-NC (Fig. 1D and E). shRNA-CUL4A-1 induced lower expression levels of CUL4A, compared with shRNA-CUL4A-2, and was used for subsequent experiments. The CCK-8 assay indicated that CUL4A-knockdown inhibited RL95-2 cell proliferation, compared with shRNA-NC (Fig. 2A). In addition, RL95-2 cell invasion and migration were markedly suppressed by shRNA-CUL4A-1, compared with shRNA-NC (Fig. 2B-E). Furthermore, western blotting was performed to detect the expression of migration-related proteins MMP2 and MMP9. CUL4A-knockdown decreased the expression of MMP2 and MMP9, compared with shRNA-NC (Fig. 3A). The results demonstrated that CUL4A-knockdown inhibited EC cell proliferation, invasion and migration.
Figure 2.
CUL4A-knockdown inhibits RL95-2 cell proliferation, invasion and migration. (A) The Cell Counting kit-8 assay was performed to assess cell proliferation. (B and C) Cell invasion was measured using Transwell assays. (D and E) A wound healing assay was performed to assess cell migration. **P<0.01 and ***P<0.001 vs. shRNA-NC. CUL4A, cullin 4A; shRNA, short hairpin RNA; NC, negative control; OD, optical density.
Figure 3.
CUL4A-knockdown decreases the expression of MMP2 and MMP9, but increases the expression of p53 in RL95-2 cells. (A) The expression of MMP2 and MMP9 was determined using western blot analysis. (B) Western blotting was used to detect the expression of p53. **P<0.01 vs. shRNA-NC. CUL4A, cullin 4A; MMP, matrix metallopeptidase; shRNA, short hairpin RNA; NC, negative control.
CUL4A-knockdown promotes the expression of p53
Western blotting was performed to assess the effect of CUL4A-knockdown on the expression of p53, a tumor suppressor gene regulated by CUL4A. shRNA-CUL4A transfection notably upregulated p53 protein expression, compared with shRNA-NC (Fig. 3B). The results indicated that CUL4A-knockdown increased p53 expression.
CSN6 interacted with CUL4A
To further investigate the molecular mechanisms underlying CUL4A-mediated EC cell proliferation, invasion and migration, the STRING database was applied to detect the potential proteins interacting with CULA4. The results indicated that CSN6 interacted with CULA4 (Fig. 4A). CSN6 is a pivotal subunit of the constitutive photomorphogenesis 9 (COP9) signalosome (CSN). To further verify the interaction between CUL4A and CSN6, a Co-IP assay was performed. The results indicated that there was a strong interaction between CUL4A and CSN6 (Fig. 4B). Taken together, these results indicated that CSN6 interacted with CUL4A in RL95-2 cells.
Figure 4.
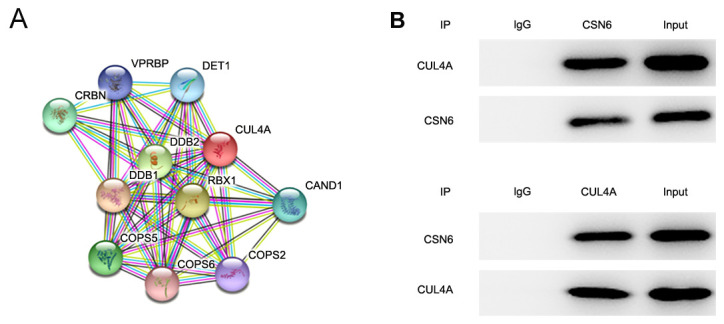
CSN6 interacts with CUL4A. (A) The protein-protein interaction network of CUL4A-related proteins. (B) Immunoprecipitation was performed with indicated antibodies in RL95-2 cells. CSN6, COP9 signalosome subunit 6; CUL4A, cullin 4A.
CSN6-overexpression attenuates the inhibitory effects of CUL4A-knockdown on EC cell proliferation, invasion and migration
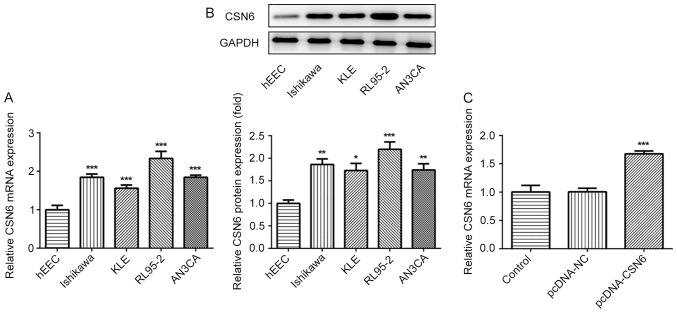
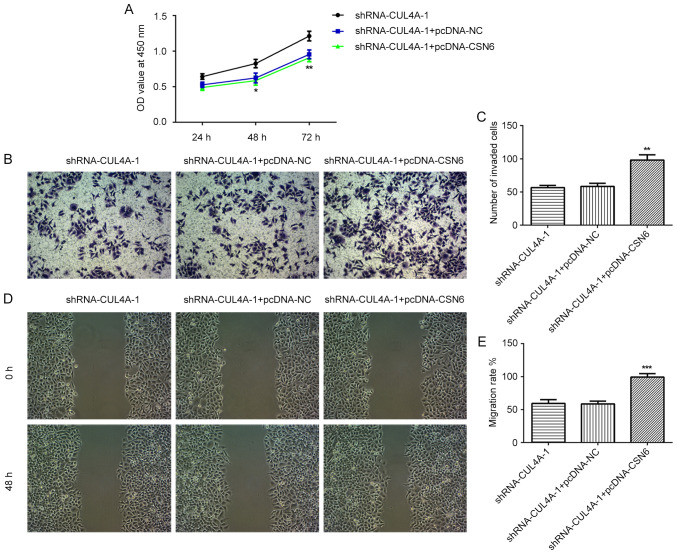
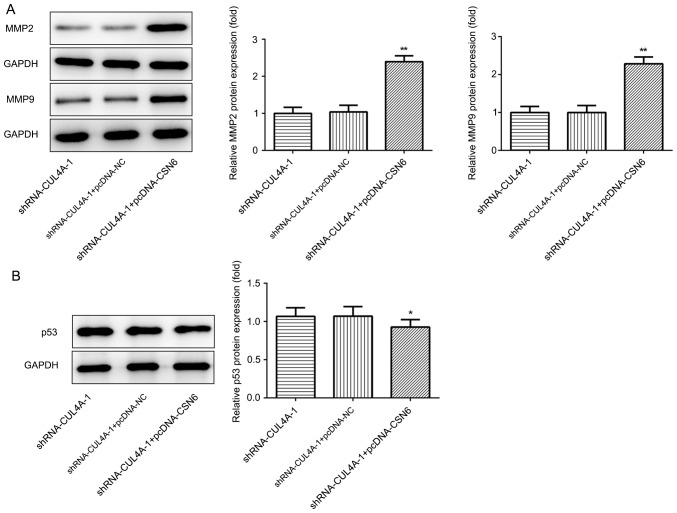
The expression of CSN6 in EC cell lines was evaluated via RT-qPCR and western blotting. The level of CSN6 was markedly increased in EC cell lines, compared with hEECs (Fig. 5A and B). Subsequently, a CSN6-overexpression plasmid was transfected into cells, which notably increased CSN6 expression, compared with pcDNA-NC (Fig. 5C). Subsequently, the effects of CSN6-overexpression on CUL4A-knockdown RL95-2 cell proliferation, invasion and migration were detected. CSN6-overexpression relieved the inhibitory effects of CUL4A-knockdown on RL95-2 cell proliferation (Fig. 6A). Furthermore, CSN6-overexpression increased cell invasion (Fig. 6B and C) and migration (Fig. 6D and E) in CUL4A-knockdown RL95-2 cells. The expression levels of MMP2 and MMP9 presented the same trend as aforementioned (Fig. 7A). Taken together, these results indicated that CSN6-overexpression attenuated the inhibitory effects of CUL4A knockdown on EC cell proliferation, invasion and migration.
Figure 5.
CSN6 expression is upregulated in EC cell lines. The expression of CSN6 was evaluated via (A) RT-qPCR and (B) western blotting. *P<0.05, **P<0.01 and ***P<0.001 vs. hEEC. (C) The expression of CSN6 following transfection with CSN6-overexpression plasmid was measured via RT-qPCR. ***P<0.001 vs. shRNA-NC. CSN6, COP9 signalosome subunit 6; EC, endometrial cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; shRNA, short hairpin RNA; NC, negative control.
Figure 6.
CSN6-overexpression attenuates the inhibitory effects of CUL4A-knockdown on EC cell proliferation, invasion and migration. (A) The Cell Counting Kit-8 assay was used to assess cell proliferation. (B and C) Cell invasion was examined by performing a Transwell assay. (D and E) Wound healing assays were conducted to assess cell migration. *P<0.05, **P<0.01 and ***P<0.001 vs. shRNA-CUL4A-1 + pcDNA-NC. CSN6, COP9 signalosome subunit 6; CUL4A, cullin 4A; EC, endometrial cancer; shRNA, short hairpin RNA; NC, negative control; OD, optical density.
Figure 7.
CSN6-overexpression increases the expression of MMP2 and MMP9, but decreases the expression of p53, compared with CUL4A-knockdown alone. (A) The expression of MMP2 and MMP9 was measured using western blot analysis. (B) Western blotting was used to examine the expression of p53.*P<0.05 and **P<0.01 vs. shRNA-CUL4A-1 + pcDNA-NC. CSN6, COP9 signalosome subunit 6; MMP, matrix metallopeptidase; CUL4A, cullin 4A; shRNA, short hairpin RNA; NC, negative control.
CSN6-overexpression downregulates the expression of p53, compared with CUL4A-knockdown alone
To further clarify the regulatory mechanisms underlying CUL4A in EC, the effects of CSN6-overexpression on p53 expression were examined via RT-qPCR and western blotting. As shown in Fig. 7B, CSN6-overexpression decreased the expression levels of p53 protein in CUL4A-knockdown RL95-2 cells. The results indicated that CUL4A regulated the expression of p53 by interacting with CSN6.
Discussion
As one of the most common gynecological malignancies worldwide, the morbidity and mortality of EC are increasing. Although a number of studies have suggested that diverse aberrantly expressed genes in EC may aggravate malignant behavior (20,21), the mechanism underlying the progression and metastasis of EC requires further investigation. To the best of our knowledge, the present study was the first to investigate the expression status, functions and molecular mechanisms underlying CUL4A in EC. The results indicated that CUL4A regulated EC cell proliferation, invasion and migration by interacting with CSN6, which provided a theoretical basis and potential therapeutic target for the treatment of EC.
Increasing evidence has indicated that the expression of CUL4A is markedly increased in a variety of types of human cancer, and may serve as a potential oncogene (17,22). For example, CUL4A was overexpressed in the tissues of patients with non-small cell lung cancer, and CUL4A-knockdown suppressed lung cancer cell invasion and metastasis (23). CUL4A inhibition also restrained the progression of breast cancer (24). Additionally, CUL4A is highly expressed in ovarian cancer and functions as an oncogene (25). However, the functions of procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 in EC have not been reported and require further investigation. Increasing evidence has suggested that uncontrolled proliferation, invasion and migration are dominant features of cancer cells, which exert crucial effects in the development process of human cancer (26,27). In the present study, notably upregulated CUL4A expression was observed in EC cells, and CUL4A-knockdown refrained EC cell proliferation, invasion and migration, which was consistent with previous studies (28,29). Taken together, these results indicated that CUL4A-knockdown serves protective roles in EC.
CUL4A participates in the proteolysis of p53, and CUL4A depletion results from an accumulation of p53 (13). p53 is a tumor suppressor gene, which is involved in the growth and metastasis of various types of cancer (30–32). CSN6, a pivotal subunit of the constitutive COP9 CSN, is notably upregulated in a number of different types of human cancer (33,34). Previous studies have highlighted the importance of CSN6 as a regulator of the degradation of cancer-related protein p53 (35,36). In the present study, the STRING database identified that CSN6 interacted with CULA4, and the Co-IP assay verified this interaction. The expression of CSN6 was significantly enhanced in EC cells, and CSN6 overexpression attenuated the inhibitory effects of CUL4A-knockdown on EC cell proliferation, invasion and migration. Furthermore, CSN6 overexpression markedly reduced p53 expression in CUL4A-knockdown EC cells. The results suggested that CUL4A interacted with CSN6 and further modulated p53 expression.
In conclusion, to the best of our knowledge, the present study was the first to investigate the pivotal roles of CUL4A in the functions of EC cells. CUL4A regulated EC cell proliferation, invasion and migration by interacting with CSN6 and further modulating p53 expression. The results also suggested that CUL4A may serve as a potential biomarker and therapeutic target for the treatment of EC, which provides an innovative perspective for the clinical therapy of EC. However, the usage of only one EC cell line, lack of pull down experiment using the purified CUL4 and CSN6 proteins and the addition of EtBr to the co-IP are limitations of the present study, a comprehensive analysis resolving these issues is required in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
XW searched the literature, designed the experiments, analyzed the data and wrote the manuscript. XW and TC performed the experiments. TC revised the manuscript. All authors read and approval the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Di Tucci C, Capone C, Galati G, Iacobelli V, Schiavi MC, Di Donato V, Muzii L, Panici PB. Immunotherapy in endometrial cancer: New scenarios on the horizon. J Gynecol Oncol. 2019;30:e46. doi: 10.3802/jgo.2019.30.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shu S, Liu X, Xu M, Gao X, Fan J, Liu H, Li R. MicroRNA-424 regulates epithelial-mesenchymal transition of endometrial carcinoma by directly targeting insulin-like growth factor 1 receptor. J Cell Biochem. 2018 doi: 10.1002/jcb.27528. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 3.Sun P, Mao X, Gao M, Huang MM, Chen LL, Ruan GY, Huang WY, Braicu EI, Sehouli J. Novel endocrine therapeutic strategy in endometrial carcinoma targeting estrogen-related receptor b by XCT790 and siRNA. Cancer Manag Res. 2018;10:2521–2535. doi: 10.2147/CMAR.S168043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Bendifallah S, Ballester M, Darai E. Endometrial cancer: Predictive models and clinical impact. Bull Cancer. 2017;104:1022–1031. doi: 10.1016/j.bulcan.2017.06.017. (in French) [DOI] [PubMed] [Google Scholar]
- 6.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 7.Tran AQ, Gehrig P. Recent advances in endometrial cancer. F1000Res. 2017;6:81. doi: 10.12688/f1000research.10020.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright JD, Burke WM, Wilde ET, Lewin SN, Charles AS, Kim JH, Goldman N, Neugut AI, Herzog TJ, Hershman DL. Comparative effectiveness of robotic versus laparoscopic hysterectomy for endometrial cancer. J Clin Oncol. 2012;30:783–791. doi: 10.1200/JCO.2011.36.7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–889. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- 10.Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 11.Sharma P, Nag A. CUL4A ubiquitin ligase: A promising drug target for cancer and other human diseases. Open Biol. 2014;4:130217. doi: 10.1098/rsob.130217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin X, Ma YC, Zhu WY, Fan L. CUL4A expression is associated with tumor stage and prognosis in nasopharyngeal carcinoma. Medicine (Baltimore) 2019;98:e18036. doi: 10.1097/MD.0000000000018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nag A, Bagchi S, Raychaudhuri P. Cul4A physically associates with MDM2 and participates in the proteolysis of p53. Cancer Res. 2004;64:8152–8155. doi: 10.1158/0008-5472.CAN-04-2598. [DOI] [PubMed] [Google Scholar]
- 14.Li B, Jia N, Kapur R, Chun KT. Cul4A targets p27 for degradation and regulates proliferation, cell cycle exit, and differentiation during erythropoiesis. Blood. 2006;107:4291–4299. doi: 10.1182/blood-2005-08-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bian WG, Zhou XN, Song S, Chen HT, Shen Y, Chen P. Reduced miR-363-3p expression in non-small cell lung cancer is associated with gemcitabine resistance via targeting of CUL4A. Eur Rev Med Pharmacol Sci. 2019;23:649–659. doi: 10.26355/eurrev_201901_16879. [DOI] [PubMed] [Google Scholar]
- 16.Sui XM, Zhou H, Zhu L, Wang DQ, Fan SM, Zhao W. CUL4A promotes proliferation and metastasis of colorectal cancer cells by regulating H3K4 trimethylation in epithelial-mesenchymal transition. OncoTargets Ther. 2017;10:735–743. doi: 10.2147/OTT.S118897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han XN, Fang ZL, Wang H, Jiao RF, Zhou J, Fang N. CUL4A functions as an oncogene in ovarian cancer and is directly regulated by miR-494. Biochem Biophys Res Commun. 2016;480:675–681. doi: 10.1016/j.bbrc.2016.10.114. [DOI] [PubMed] [Google Scholar]
- 18.Wang YS, Liu XY, Zheng H, Wang Q, An L, Wei GW. Suppression of CUL4A attenuates TGF-β 1-induced epithelial-to-mesenchymal transition in breast cancer cells. Int J Mol Med. 2017;40:1114–1124. doi: 10.3892/ijmm.2017.3118. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Cai L, Wang H, Yang Q. CRKL overexpression promotes cell proliferation and inhibits apoptosis in endometrial carcinoma. Oncol Lett. 2017;13:51–56. doi: 10.3892/ol.2016.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin QY, Chen H, Zhang MF, Xiong HZ, Jiang QP. Knocking down FAM83B inhibits endometrial cancer cell proliferation and metastasis by silencing the PI3K/AKT/mTOR pathway. Biomed Pharmacother. 2019;115:108939. doi: 10.1016/j.biopha.2019.108939. [DOI] [PubMed] [Google Scholar]
- 22.Zhang TJ, Xue D, Zhang CD, Zhang ZD, Liu QR, Wang JQ. Cullin 4A is associated with epithelial to mesenchymal transition and poor prognosis in perihilar cholangiocarcinoma. World J Gastroenterol. 2017;23:2318–2329. doi: 10.3748/wjg.v23.i13.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung MS, Chen YC, Lin PY, Li YC, Hsu CC, Lung JH, You L, Xu Z, Mao JH, Jablons DM, Yang CT. Cul4A modulates invasion and metastasis of lung cancer through regulation of ANXA10. Cancers (Basel) 2019;11:618. doi: 10.3390/cancers11050618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang YS, Liu XY, Zheng H, Wang Q, An L, Wei GW. [Corrigendum] Suppression of CUL4A attenuates TGF-β 1-induced epithelial-to-mesenchymal transition in breast cancer cells. Int J Mol Med. 2017;40:1611. doi: 10.3892/ijmm.2017.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu RF, Cai LM, Chi YG, Ding XC, Wu XQ. miR-377 targets CUL4A and regulates metastatic capability in ovarian cancer. Int J Mol Med. 2018;41:3147–3156. doi: 10.3892/ijmm.2018.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, He W, Gao J, Luo J, Huang X, Gao C. Computational prediction and experimental validation of a novel synthesized pan-PIM inhibitor PI003 and its apoptosis-inducing mechanisms in cervical cancer. Oncotarget. 2015;6:8019–8035. doi: 10.18632/oncotarget.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie PM, Wang XL, Kong M, Bai XY, Jiang T. TRAF4 promotes endometrial cancer cell growth and migration by activation of PI3K/AKT/Oct4 signaling. Exp Mol Pathol. 2019;108:9–16. doi: 10.1016/j.yexmp.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Gong Y, Xiang XJ, Feng M, Chen J, Fang ZL, Xiong JP. CUL4A promotes cell invasion in gastric cancer by activating the NF-κB signaling pathway. Biologics. 2017;11:45–53. doi: 10.2147/BTT.S127650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu YY, Wang YS, Ma GX, Wang Q, Wei GW. CUL4A is overexpressed in human pituitary adenomas and regulates pituitary tumor cell proliferation. J Neurooncol. 2014;116:625–632. doi: 10.1007/s11060-013-1349-2. [DOI] [PubMed] [Google Scholar]
- 30.Zhao ZH, Wang WW, Niu LH, Zhang JY, Jiang GZ. p53-induced long non-coding RNA PGM5-AS1 inhibits the progression of esophageal squamous cell carcinoma through regulating miR-466/PTEN axis. IUBMB Life. 2019;71:1492–1502. doi: 10.1002/iub.2069. [DOI] [PubMed] [Google Scholar]
- 31.Zhao J, Chen HQ, Yang HF, Li Y, Chen DJ, Huang YJ, He LX, Zheng CF, Wang LQ, Wang J, et al. Epigenetic silencing of ALX4 regulates microcystin-LR induced hepatocellular carcinoma through the P53 pathway. Sci Total Environ. 2019;683:317–330. doi: 10.1016/j.scitotenv.2019.05.144. [DOI] [PubMed] [Google Scholar]
- 32.De U, Son JY, Sachan R, Park YJ, Kang D, Yoon K, Lee BM, Kim IS, Moon HR, Kim HS. A new synthetic histone deacetylase inhibitor, MHY2256, induces apoptosis and autophagy cell death in endometrial cancer cells via p53 acetylation. Int J Mol Sci. 2018;19:2743. doi: 10.3390/ijms19092743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jhan JH, Lee YC, Li WM, Chang LL, Hsu WC, Lin HH, Liang PI, Hsu YL, Wu WJ, Lee HY, et al. The prognostic value of CSN6 expression in upper tract urothelial carcinomas. Kaohsiung J Med Sci. 2019;35:559–565. doi: 10.1002/kjm2.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du WQ, Liu ZX, Zhu WT, Li TT, Zhu Z, Wei L, Song J, Pei DS. CSN6 promotes tumorigenesis of gastric cancer by ubiquitin-independent proteasomal degradation of p16INK4a. Cancer Biol Med. 2019;16:514–529. doi: 10.20892/j.issn.2095-3941.2018.0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou J, Deng Q, Zhou J, Zou J, Zhang Y, Tan P, Zhang W, Cui H. CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR. Oncogene. 2017;36:1134–1144. doi: 10.1038/onc.2016.280. [DOI] [PubMed] [Google Scholar]
- 36.Wang WQ, Tang M, Zhang L, Xu X, Qi X, Yang Y, Jin F, Chen B. Clinical implications of CSN6 protein expression and correlation with mutant-type P53 protein in breast cancer. Jpn J Clin Oncol. 2013;43:1170–1176. doi: 10.1093/jjco/hyt148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.