Abstract
Lung cancer is the most prevalent and observed type of cancer in Xuanwei County, Yunnan, South China. Lung cancer in this area is called Xuanwei lung cancer. However, its pathogenesis remains largely unknown. To date, a number of studies have shown that microRNA (miR)-218 functions as a tumor suppressor in multiple types of cancer. However, the role of miR-218 and its regulatory gene network in Xuanwei lung cancer have yet to be investigated. The current study identified that the expression levels of miR-218 in XWLC-05 cells were markedly lower compared with those in immortalized lung epithelial BEAS-2B cells. The present study also demonstrated that overexpression of miR-218 could decrease cell proliferation, invasion, viability and migration in Xuanwei lung cancer cell line XWLC-05 and NSCLC cell line NCI-H157. Additionally, the results revealed that overexpression of miR-218 could induce XWLC-05 and NCI-H157 cell apoptosis by arresting the cell cycle at G2/M phase. Finally, the present study demonstrated that overexpression of miR-218 could lead to a significant increase in phosphatase and tensin homolog (PTEN) and YY1 transcription factor (YY1), and a decrease in B-cell lymphoma 2 (BCL-2) and BMI1 proto-oncogene, polycomb ring finger (BMI-1) at the mRNA and protein level in XWLC-05 and NCI-H157 cell lines. However, we did not observe any remarkable difference in the roles of miR-218 and miR-218-mediated regulation of BCL-2, BMI-1, PTEN and YY1 expression in the progression of Xuanwei lung cancer. In conclusion, miR-218 could simultaneously suppress cell proliferation and tumor invasiveness and induce cell apoptosis by increasing PTEN and YY1 expression, while decreasing BCL-2 and BMI-1 in Xuanwei lung cancer. The results demonstrated that miR-218 might serve a vital role in tumorigenesis and progression of Xuanwei lung cancer and overexpression of miR-218 may be a novel approach for the treatment of Xuanwei lung cancer.
Keywords: microRNA-218, Xuanwei lung cancer, phosphatase and tensin homolog, YY1 transcription factor, B-cell lymphoma 2, BMI1 proto-oncogene, polycomb ring finger
Introduction
Lung cancer is the leading cause of death, accounting for one third of all cancer-related deaths and seriously threatens the lives and health of people worldwide (1). Xuanwei, a county-level city in Yunnan province, China, is one of the areas with the highest lung cancer occurrence and mortality rates in the world (2,3). Previous etiological and epidemiological studies have confirmed that lung cancer in Xuanwei County has its own unique epidemiological characteristics mainly due to polycyclic aromatic hydrocarbons (PAHs) and indoor coal-fired particles. First, lung cancer rates in Xuanwei tended to be higher in rural areas compared with that in urban areas according to the data of Chinese national retrospective surveys (4). Second, the incidence of lung cancer in Xuanwei women (120 per 100,000) was much higher compared with the national average (22.9 per 100,000 women) (4). Third, the age of onset of lung cancer in Xuanwei has been reported to be 15–25 years younger than that of places with high incidences of lung cancer, such as Shanghai (4). Lung cancer epidemiology demonstrated family aggregation and indicated that genetic variation might serve an important role in Xuanwei lung cancer tumorigenesis and progression (5–7). However, there were few studies on genes unique in Xuanwei lung cancer tumorigenesis and progression. The Xuanwei lung cancer cell line XWLC-05 was established by F.C. Yan et al in 2007 (8). It was derived from a female patient who was a 68-year-old Xuanwei permanent resident and diagnosed with moderately differentiated lung adenocarcinoma. XWLC-05 has since been used in various research studies including antitumor drug screening and cancer molecular-targeted therapy (9,10). However, the molecular mechanisms of lung cancer progression in Xuanwei County remain to be elucidated. Despite the efforts made in treatment in recent years, the 5-year overall survival rate of patients with non-small cell lung cancer (NSCLC) is still ~18% although ~60–70% of them have been diagnosed in the early stages of lung cancer (11,12). Therefore, elucidating the molecular mechanisms of occurrence and metastasis of lung cancers is of great significance for the clinical treatment of lung cancer in Xuanwei populations.
MicroRNAs (miRNAs) are highly conserved endogenous non-coding single-stranded RNAs with a length of 18–25 bp that regulate gene expression by binding to the 3′ untranslated region (3′-UTR) of target transcripts leading to mRNA degradation or suppressing translation into protein (13–15). Previous studies suggest that miRNAs serve critical regulatory roles in essential biological and pathological processes via complicated but precise regulation networks (16–18). Therefore, the deletion, mutation or abnormal expression of miRNAs are closely linked to tumor progression. Numerous studies have suggested the roles of miRNAs in carcinogenesis and progression of lung cancer (19–22).
Through high-throughput sequencing, the present study identified that miR-218 expression levels in NSCLC patients were significantly lower than those in paired distal control tissues. Previous studies confirm that miR-218 acts as a tumor-suppressor miRNA in a number of types of cancer, including lung cancer, breast cancer, glioma, hepatocellular carcinoma, gastric cancer, colorectal cancer and prostate cancer (23–29). As a tumor-suppressor miRNA, miRNA (miR)-218 is downregulated in tumor progression and is associated with prognosis in NSCLC (30–32). Furthermore, miR-218 can participate in tumor progression by regulating target genes including PXN, IL6R, JAK3, SLUG, ZEB2, EGFR, MEF2D, CDCP1, RUNX2, HMGB1, ETK, HOXA1, CDK6 and ROBO1 (31–42). The present study revealed for the first time, to the best of the authors' knowledge, the expression levels and therapeutic potential of miR-218 in XWLC-05. Furthermore, it was demonstrated that overexpression of miR-218 could suppress cell proliferation, cell migration and invasion and induce cell apoptosis by regulating B-cell lymphoma 2 (BCL-2), BMI1 proto-oncogene, polycomb ring finger (BMI-1), phosphatase and tensin homolog (PTEN) and YY1 transcription factor (YY1) in NSCLC. BCL-2 is an oncogene and is also central in the apoptosis pathway. Overexpression of BCL-2 has been reported in a number of types of human cancer, including breast, gastric and lung cancer (43–45). Oncogene BMI-1 is overexpressed in various types of human cancer and serves a critical role in malignant transformation, proliferation, cell cycle, apoptosis and distant metastasis (46,47). A previous study revealed that BMI-1 is involved in the self-renewal, differentiation and tumor initiation of cancer stem cells (48). As a powerful tumor suppressor, PTEN is frequently mutated in lung cancer and is the negative regulator of the PI3K/mTOR/AKT oncogenic signaling pathway. A number of studies have confirmed that PTEN is crucial for cell proliferation, invasion and survival, and loss of function of PTEN is frequently observed in a number of types of cancer (49–51). Yin Yang 1 (YY1) (also known as NF-E1, UCRBP and CF1) is a ubiquitous and multifunctional zinc finger transcription factor and can regulate multiple genes associated with multiple cellular processes including cellular differentiation, DNA repair, autophagy and cell survival (52–54). However, the role of YY1 in NSCLC progression remains controversial. Notably, a previous study confirmed that YY1 can act as both oncogene and tumor-suppressor gene in breast cancer and lung cancer (52–54).
The current study evaluated the underlying roles and mechanisms of miR-218 in NSCLC progression. It was identified that the expression of miR-218 in XWLC-05 was significantly lower compared with that in BEAS-2B. Overexpression of miR-218 could suppress cell proliferation, invasion and migration, and induce cell apoptosis in XWLC-05 and NCI-H157 cells. The regulatory mechanisms between miR-218 and its downstream BCL-2, BMI, PTEN and YY1 gene axis were further explored in XWLC-05 and NCI-H157 cells to investigate the specificity and similarity of Xuanwei NSCLC as compared with other NSCLC. The findings revealed that miR-218 could be a potential therapeutic target for NSCLC. Furthermore, the present study will provide a theoretical basis for lung cancer treatment in high-risk areas worldwide.
Materials and methods
Cell strains and cell lines
Normal human lung epithelial cell line BEAS-2B, Xuanwei lung adenocarcinoma cell line XWLC-05 and human lung squamous cell carcinoma cell line NCI-H157 were all provided by Yunnan Cancer Hospital (The Third Affiliated Hospital of Kunming Medical University) and confirmed via short tandem repeat profiling. Lung adenocarcinoma cell line NCI-H1975 was provided by the Stem Cell Bank, Chinese Academy of Sciences. Large cell lung cancer cell line NCI-H460SM was kindly provided by Dr Ming-Sound Tsao (University of Toronto, Canada) (55).
Major reagents
RPMI-1640 medium, phosphate buffer and 0.25% trypsin-EDTA were purchased from HyClone (Cytiva). Fetal bovine serum (FBS), DMEM and Opti-MEM were purchased from Gibco (Thermo Fisher Scientific, Inc.). miRcute miRNA Isolation kit, miRcute miRNA First-strand cDNA Synthesis kit and miRcute miRNA qPCR kit were purchased from Tiangen Biotech Co., Ltd. iTaq™ Universal SYBR® Green Supermix and iScript™ cDNA Synthesis kit were purchased from Bio-Rad Laboratories, Inc. miR-218 (cat. no. HmiRQP0327) and U6 (cat. no. HmiRQP9001) primers were bought from GeneCopoeia, Inc. YY1 and BMI-1 monoclonal antibodies were purchased from Abcam. BCL-2 and PTEN monoclonal antibodies were purchased from ProteinTech Group, Inc. Lipofectamine® 2000 transfection reagent was purchased from Invitrogen (Thermo Fisher Scientific, Inc.). Matrigel was purchased from BD biosciences. pGpU6/EGFP/Neo-miR-218 and pGpU6/EGFP/Neo-miR-NC were purchased from Shanghai GenePharma Co., Ltd. The insert sequence of miR-218 was 5′-TGCTGTTGTGCTTGATCTAACCATGTGTTTTGGCCACTGACTGACACATGGTTACAAGCACAA-3′. The insert sequence of miR-NC was 5′-AATTCGTTCTCCGAACGTGTCACGTGTTTTGGCCACTGACTGACACGTGACATTCGGAGAAA-3′.
Cell culture
Cell culture and maintenance were conducted by the following established procedures: The cell lines were expanded at low passages and stored in liquid nitrogen until use. Cell lines were cultured in RPMI-1640 or DMEM accordingly and supplemented with 10% FBS at 37°C in a 5% CO2 incubator till they reached 70–80% confluence. Cells were washed once with 1X PBS and digested with 0.25% trypsin-EDTA, then harvested for the subsequent analysis.
Bioinformatics analysis
To predict possible targets of miR-218, a bioinformatical analysis was performed using TargetScan 7.2 (http://www.targetscan.org/vert_72/). In the meantime, MiRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and StarBase V3.0 databases (http://starbase.sysu.edu.cn/) were used to predict the relationship between mir-218 and its possible target genes.
Cell transfection
Transfection was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). XWLC-05 and NCI-H157 cells were cultured and grown to 70–80% confluence at 37°C in a 5% CO2 incubator. Then 1 ml Trypsin-EDTA (0.25% trypsin, 1 mM EDTA) was added for digestion at 37°C for 3 min following washing with 1X PBS. Then 10 ml 10% FBS complete medium was added to terminate digestion. The cells were counted using the hemocytometer and then diluted to the desired density. The density of 2×105 cells/ml was used for reverse transcription-quantitative (RT-q) PCR and 2×106 cells/ml for western blot analysis. Then appropriate cells were added into a 6-well cell culture plate. When the cells were cultured at 37°C in a 5% CO2 incubator overnight and reached 70–80% confluence, the RPMI-1640 culture medium containing 10% FBS was removed and cells were washed with 1X PBS twice. The transfection of pGpU6/EGFP/Neo-miR-218 or pGpU6/EGFP/Neo-miR-NC involved 0.5 ml of Opti-MEM at a final concentration of 8 µg/ml and 16 µl of Lipofectamine® 2000 per well. Then, 6 h following transfection, the culture medium was replaced with RPMI-1640 containing 10% FBS. At 48 h following transfection, the cell status and enhanced green fluorescent protein (EGFP) expression were observed under a light and fluorescence microscope at ×40 and ×100 magnification, and the cells of each group were collected for subsequent RT-qPCR detection. Each cell line was divided into three groups: (i) A blank control group (containing only culture medium); (ii) a negative control (NC) group (transfected with pGpU6/EGFP/Neo-miR-NC); and (iii) a transfection group (transfected with pGpU6/EGFP/Neo-miR-218).
RT-qPCR detection for miR-218, BCL-2, BMI-1, PTEN and YY1
The total RNA of 1×106 cells was extracted by using miRcute miRNA Isolation kit. RNA concentration (>300 ng/µl) and purity (OD 260/OD 280 ~2.0) were measured using a NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.), and then the integrity of the RNA was detected by 1.0% agarose gel electrophoresis with ethidium bromide staining. cDNA was synthesized according to the protocols of the cDNA synthesis kit, with a reaction volume at 20 µl, including a 4 µl 5X iScript reaction mix, 1 µl iScript reverse transcriptase, 1 µg RNA template, 0.5 µl 10 µmol/l miR-218 or U6 primer and RNase-free water. The reverse transcription conditions were 25°C for 5 min, 42°C for 30 min and 85°C for 5 min. Then, qPCR was performed according to the manufacturer's protocols. The reaction system was 20 µl, including 10 µl 2X Supermix, 1 µl 10 µmol/l each upstream and downstream primers, 2 µl cDNA template and 6 µl RNase-free water. Reaction conditions were: 95°C for 30 sec, 95°C for 15 sec and 60°C for 1 min with 40 cycles. The levels of miR-218 (with U6 as reference gene) and the mRNA levels of PTEN, BCL-2, BMI-1 and YY1 (with RPS13 as the reference) were then detected in the cells 48 h following transfection. The primers used for the amplification of each gene are given in Table I. In the above experiment, two wells were set for each condition and all reactions were repeated three times. The experimental results were calculated with the formula 2−ΔΔCq to obtain the amount of the relative gene expression (56).
Table I.
Primer sequences for qPCR.
| Gene | Primer sequence (5′→3′) |
|---|---|
| BCL-2 | F: TCCAGGATAACGGAGGCT |
| R: GCCAAACTGAGCAGAGTCTTC | |
| BMI-1 | F: GTCCTATTTGTGATGTCCAAGTTC |
| R: GCAACCTCTCCTCTATCTTCATTA | |
| PTEN | F: CCGAAAGGTTTTGCTACCATTCT |
| R: AAAATTATTTCCTTTCTGAGCATTCC | |
| YY-1 | F: GCACAAAGATGTTCAGGGATAA |
| R: AAGGGCTTCTCTCCAGTATGA | |
| RPS13 | F: GTTGCTGTTCGAAAGCATCTTG |
| R: AATATCGAGCCAAACGGTGAA |
qPCR, quantitative; PCR F, Forward primer; R, Reverse primer.
Cell viability detected by MTT assay
At 48 h following transfection, the cells of each group were digested with 0.25% trypsin-EDTA for ~2–3 min until the adherent cells were detached. The trypsinization process was terminated with medium containing 10% FBS. Then the cells were transferred into a centrifuge tube and centrifuged at 300 × g for 5 min at room temperature following cell counting using a hemocytometer. Each well of the 96-well cell culture plate was seeded with 200 μl cell suspension (1×104 cells/ml) and each group was repeated with 6 replicates. The medium was removed and 20 μl MTT label reagent was added to each well and the liquid in the well was carefully removed after 4 h incubation at 37°C. DMSO (150 μl) was added to each well and the 96-well cell culture plate was placed on an oscillator for 10 min to completely dissolve. The absorbance was determined by the microplate reader at 490 nm wavelength and the plate was assayed each day for 7 days.
Cell migration detected by scratch assay
The cells of each group were digested 48 h after transfection and seeded into the 6-well cell culture plate for 24 h at 37°C. After inoculation, RPMI-1640 culture medium containing 10% FBS was removed and monolayer cells were scratched with 10 µl pipette tips using the same amount of force, making sure each scratch was as wide as possible. Loose cells were then gently washed away with 1X PBS buffer. RPMI-1640 culture medium supplemented with 3% FBS was added and images were captured under a light microscope according to previous studies (22,57,58). The cells were returned to the cell incubator for 72 h at 37°C and images were captured every 24 h at the same position as the previous image. Finally, the migration ability was then calculated by the following formula according to the change in the scratch distance that was measured by Image J software. Migration rate (%) = (the scratch distance at time 0-the scratch distance at indicated time point)/the scratch distance at time 0)x100%.
Cell invasion ability detected by Transwell assay
The cells were collected by trypsinization 48 h after transfection and the cell density was adjusted to 5×105/ml using serum-free medium. A total of 60 µl Matrigel (diluted 7 times with serum-free medium) was added into the upper chambers and incubated at 37°C in 5% CO2 atmosphere for 1 h. Then, 200 µl of cell suspension (5×105 cells/ml) was inoculated into the Matrigel-coated upper chamber (8 µm pore size; Costar; Corning, Inc.). Subsequently, 600 µl culture medium containing 20% FBS was added to the lower chamber. After incubation at 37°C in 5% CO2 atmosphere for 24 h, cells were fixed and stained with 0.2% crystal violet for 1 h at room temperature. The excess stain was then washed away slowly with water and the images were obtained using a light microscope at ×40 and ×100 magnification. The cell number was counted, and the result was compared among all groups.
Cell apoptosis detected by flow cytometry and transmission electron microscopy
DUTP-FITC/propidium iodide (PI) staining was used for detection of apoptotic cells and the Coulter DNA Prep reagents kit for DNA cell cycle analysis. Cells were collected 48 h after transfection and adjusted at 5×106/ml before being centrifuged at 300 × g for 5 min at room temperature and the supernatant discarded. The precipitated cells were washed once with 3 ml ice-cold PBS and centrifuged at 300 × g for 5 min at 4°C. Precooled 70% ethanol was added and the cells maintained at 4°C for 2 h. Then cells were pelleted by centrifugation at 300 × g for 5 min at 4°C and 3 ml ice-cold PBS was added to re-suspend cells for 5 min. The cell suspension was filtered through a 400-mesh screen and then centrifuged at 300 × g for 5 min at 4°C. The PBS was then discarded. In a dark room, 1 ml PI stain was used at 4°C for 30 min. Cell cycle and apoptosis were measured using a Beckman-Coulter flow cytometer (Beckman Coulter, Inc.) and WINCYCLE software version 3.0 (Beckman Coulter, Inc.).
Another batch of cells was centrifuged at 300 × g for 5 min at room temperature and collected at ~5×106/ml. Following washing with PBS twice, the supernatant was discarded, and 1 ml 2.5% glutaraldehyde was added to suspend cells and then transferred into a 1.5 ml Eppendorf tube. The cells were centrifuged at room temperature at 1,200 × g for 15 min and fixed for 1 h with 2.5% glutaraldehyde, then washed for 2 h in phosphate buffer to remove the glutaraldehyde as much as possible and fixed for 1 h at room temperature with 1% osmium solution. Following fixation, 50% ethanol was added to dehydrate for 10 min, 70% ethanol for 10 min, 90% ethanol for 10 min, 90% acetone for 10 min and 100% acetone for three times (10 min each). Acetone and epoxy resin at a 1:1 mixture was used to embed the cells for 2 h and then pure embedding agent (fully impregnated with epoxy resin) added for several hours or overnight. Epoxy resin was used for embedding at 62°C for 2 days. Following sectioning, ultrathin sections (70 nm) were double stained with 2% lead acetate uranium for 20 min at room temperature. Changes in cell morphology were observed and images were captured by transmission electron microscopy (TEM) at ×8,000 and ×10,000 magnification.
Western blotting
Cells were digested 48 h after transfection and washed with ice-cold 1X PBS twice and then lysed with 200 µl RIPA buffer (Beyotime Institute of Biotechnology) containing proteinase inhibitors (Roche Diagnostics) and the protein concentration was determined using the BCA protein assay kit (Beyotime Institute of Biotechnology). Samples with equal amounts of protein (80 µg) per lane were taken and mixed with 4X loading buffer and then denatured. Proteins were separated using 10% SDS-polyacrylamide gels and transferred onto a PVDF membrane (EMD Millipore). The membranes were blocked for 1 h at room temperature with 5% BSA (Beijing Solarbio Science & Technology Co., Ltd.) and then incubated at 4°C overnight with the primary antibodies: Anti-phosphatase and tensin homolog (PTEN) (ProteinTech Group, Inc., cat. no. 60300-1-Ig, dilution 1:1,00), anti-BCL-2 (ProteinTech Group, Inc., cat. no. 60178-1-Ig, dilution 1:1,000), anti-polycomb complex protein BMI-1 (BMI-1) (Abcam, cat. no. ab126783, dilution 1:1,000), anti-transcriptional repressor protein YY1 (YY1) (Abcam, cat. no. ab109237, dilution 1:2,000). After washing the membranes with TBST, the membranes were incubated for 2 h at room temperature with 1:4,000 dilution of the horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology, Inc., cat. no. 7076) or with 1:2,000 dilution of the horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology, Inc., cat. no. 7074). GAPDH (Abmart Pharmaceutical Technology Co., Ltd. cat. no. M20028; 1:5,000) was used as a loading control. Protein expression was detected using the enhanced chemiluminescence (ECL-Plus) reagents (EMD Millipore). The relative protein levels were analyzed using Image J software (version 1.52a; National Institutes of Health).
Statistical methods
All experimental data were imaged with GraphPad Prism 7 (GraphPad Software, Inc.) and Image J (National Institutes of Health). All data were analyzed with GraphPad Prism 7. All results represent the average of triplicate experiments expressed as the mean ± standard deviation. Statistical analysis was performed using one-way ANOVA with Dunnett's post-hoc test where all comparisons were against a single control, with Tukey's post-hoc test where all groups were compared with one another. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-218 expression levels are markedly decreased in NSCLC cell lines
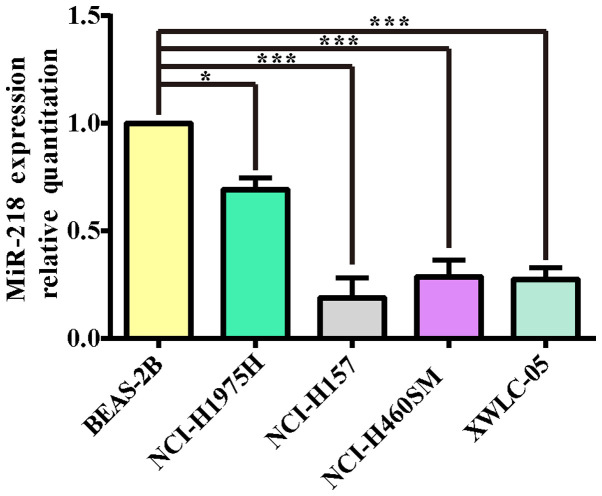
The total RNA of lung cancer cell lines (NCI-H1975, NCI-H157, NCI-H460SM, XWLC-05) and the normal lung cell line BEAS-2B were extracted and their OD260/OD280 values were all >1.9. The results of electrophoresis demonstrated that the integrity of RNA was reliable and could be used as a template for cDNA synthesis. In order to determine the expression of miR-218 in various types of lung cancer cells, BEAS-2B, a normal lung cell line, was selected as the calibrated strain in the present study. qPCR was performed to detect miR-218 expression in the 4 lung cancer cell lines. As shown in Fig. 1, the expression levels of miR-218 in non-small lung cancer cell lines were lower compared with normal human lung epithelial cell line BEAS-2B (P<0.05 and P<0.001 respectively). These results also indicated that XWLC-05 and NCI-H157 cells could be used for the subsequent miR-218 overexpression experiments.
Figure 1.
miR-218 expression levels in four types of lung cancer cell lines (NCI-H1975, NCI-H157, NCI-H460SM and XWLC-05) and a normal lung cell line BEAS-2B detected by qPCR. miR-218 abundance was normalized to U6 as a reference gene and the expression values were normalized to BEAS-2B cells (n=3, *P<0.05, ***P<0.001 vs. BEAS-2B). The experiment was repeated three times. miR, microRNA; qPCR, quantitative PCR.
Transfection optimization
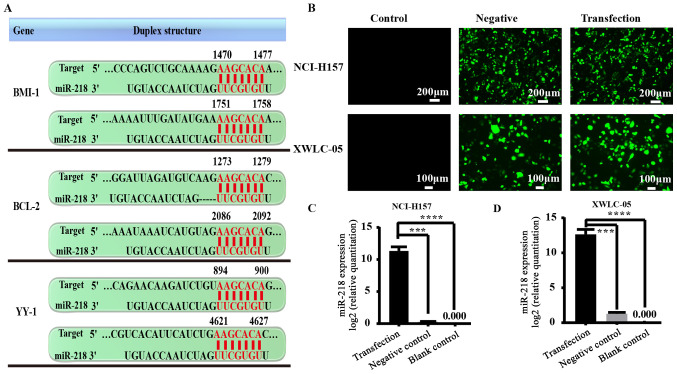
TargetScan algorithm revealed that BMI-1, BCL-2 and YY-1 mRNA were potential targets for miR-218 (Fig. 2A). It was therefore essential to optimize transfection conditions in vitro and evaluate the efficacy of miR-218 overexpression. XWLC-05 and NCI-H157 cells were transfected for 24, 48 and 72 h respectively, then EGFP levels were detected by fluorescence microscope and miR-218 expression levels were measured by qPCR. Fluorescence microscope observation revealed that pGpU6/EGFP/Neo-miR-218 or pGpU6/EGFP/Neo-miR-NC plasmid group showed higher EGFP-positive cells in both cell lines at 48 h after transfection, and demonstrated high gene transfection efficiency (Fig. 2B). qPCR results demonstrated that miR-218 expression level was significantly increased by the pGpU6/EGFP/Neo-miR-218 plasmid in both cell lines at 48 h after transfection (P<0.001; Fig. 2C).
Figure 2.
miR-218 target genes and cell transfection efficiency. (A) The 3′-UTR of BMI-1, BCL-2 and YY1 mRNA are potential targets for miR-218 and the seed matching sequences are marked in red, as predicted by TargetScan. (B) The expression of green fluorescent protein transfected with pGpU6/EGFP/Neo-miR-218 or pGpU6/EGFP/Neo-miR-NC for 48 h was observed using a fluorescence inverted microscope (magnification ×40; scale bar, 200 µm for NCI-H157; magnification, ×100; scale bar, 50 µm for XWLC-05). (C) The expression levels of miR-218 in NCI-H157 cells were analyzed by qPCR at 48 h post-transfection shown in the bar graphs. (D) The expression levels of miR-218 in XWLC-05 cells were analyzed by qPCR at 48 h post-transfection shown in the bar graphs. miR-218 abundance was normalized to U6 as a reference gene (n=3, ***P<0.001, ****P<0.0001 vs. control). Negative, negative control (transfected with pGpU6/EGFP/Neo-miR-NC plasmid). The experiment was repeated three times. miR, microRNA; UTR, untranslated region; qPCR, quantitative PCR.
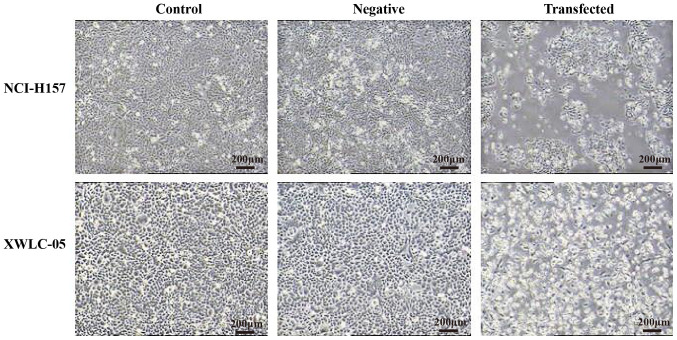
Cell growth status in each group
It was observed under the optical microscope that XWLC-05 and NCI-H157 cells of the negative control group and the blank control group grew well with normal attachment and normal morphology. However, the cells of the transfected group were mostly semi-adherent or of a floating, rounded shape; the number of adherent cells was significantly reduced and the cell morphology was also changed. These findings indicated that miR-218 overexpression could affect cell growth (Fig. 3).
Figure 3.
Upregulation of miR-218 in XWLC-05 and NCI-H157 cells causes changes in cell morphology. Morphologic changes of XWLC-05 and NCI-H157 cells were observed in response to miR-218 overexpression (magnification, ×40; scale bar, 200 µm). Negative, negative control (transfected with pGpU6/EGFP/Neo-miR-NC plasmid). miR, microRNA.
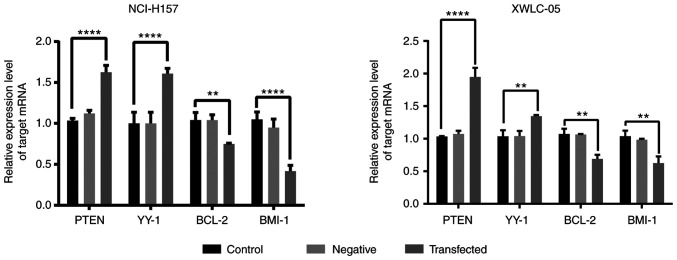
Overexpression of miR-218 inhibits PTEN, BCL-2, BMI-1 and YY1 mRNA expression in NSCLC cells
PTEN, BCL-2, BMI-1 and YY1 mRNA levels were detected using qPCR after the overexpression of miR-218 for 48 h in XWLC-05 and NCI-H157 cells. The results demonstrated that the mRNA levels of PTEN and YY1 were significantly unregulated, while BCL-2 and BMI-1 were significantly downregulated in miR-218 overexpressing cells (Fig. 4).
Figure 4.
mRNA expression levels of PTEN, BCL-2, BMI-1 and YY1 in XWLC-05 and NCI-H157 cells. The mRNA expression levels of PTEN, BCL-2, BMI-1 and YY1 were detected by qPCR in 48 h transfected cells. mRNA abundance was normalized to RPS13 (n=3, **P<0.01, ****P<0.0001 vs. the control). The experiment was repeated three times. miR, microRNA; qPCR, quantitative PCR. PTEN, phosphatase and tensin homolog; YY1, anti-transcriptional repressor protein YY1; BCL-2, B-cell lymphoma 2; BMI-1, BMI1 proto-oncogene, polycomb ring finger.
Effect of protein expression on NSCLC cells transfected with miR-218
As shown in Fig. 5, the expression of PTEN and YY1 protein levels in XWLC-05 and NCI-H157 cells were elevated in the transfected group compared with the negative control group and the blank control group, while the Bcl-2 and BMI-1 protein expression levels were decreased in the transfected group compared with the negative control group and the blank control group. The above results confirmed that miR-218 overexpression could inhibit the expression of oncogenes BCL-2 and BMI-1 but increase the expression of tumor-suppression genes PTEN and YY1 in both XWLC-05 and NCI-H157 cell lines.
Figure 5.
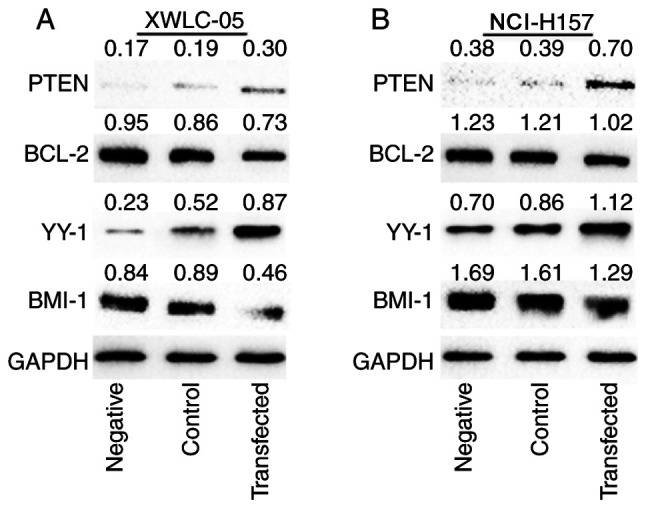
Protein expression of PTEN, BCL-2, BMI-1 and YY1 following elevated expression of miR-218 in (A) XWLC-05 and (B) NCI-H157 cells. PTEN, BCL-2, BMI-1 and YY1 protein levels were determined by western blot analysis in 48 h transfected cells. GAPDH level was used as a reference protein expression. Negative indicated negative control (transfected with pGpU6/EGFP/Neo-miR-NC plasmid). PTEN, anti-phosphatase and tensin homolog; BMI-1, anti-polycomb complex protein BMI-1; YY1, anti-transcriptional repressor protein YY1; miR, microRNA.
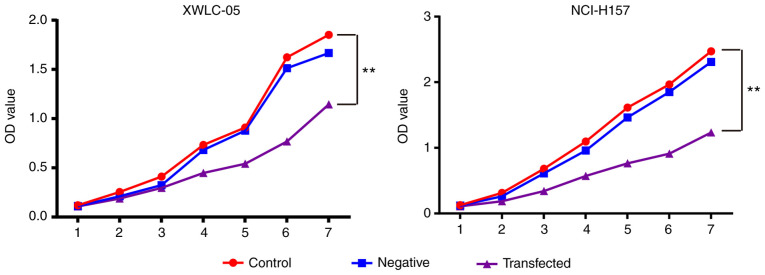
Effect of miR-218 on NSCLC cell proliferation
In order to confirm the role of miR-218 on NSCLC cell proliferation and viability, NCI-H157 and XWLC-05 cells were transfected with miR-218 overexpression plasmid or empty plasmid. The cells were incubated for different time periods and the cell proliferation was assessed by MTT assay. As shown in Fig. 6, the proliferation of both NCI-H157 and XWLC-05 cells transfected with pGpU6/EGFP/Neo-miR-218 was significantly inhibited relative to the remaining groups (P<0.01), but the proliferation in the negative control group demonstrated no significant change compared with the blank control group (P>0.05).
Figure 6.
Overexpression of miR-218 suppresses NSCLC cell proliferation. Cell proliferation rate was measured by MTT assay. XWLC-05 and NCI-H157 cells transfected with pGpU6/EGFP/Neo-miR-218 plasmid demonstrated a significant decreased cell growth rate (n=3, **P<0.01 vs. the control). The experiment was repeated three times. miR, microRNA; NSCLC, non-small cell lung cancer.
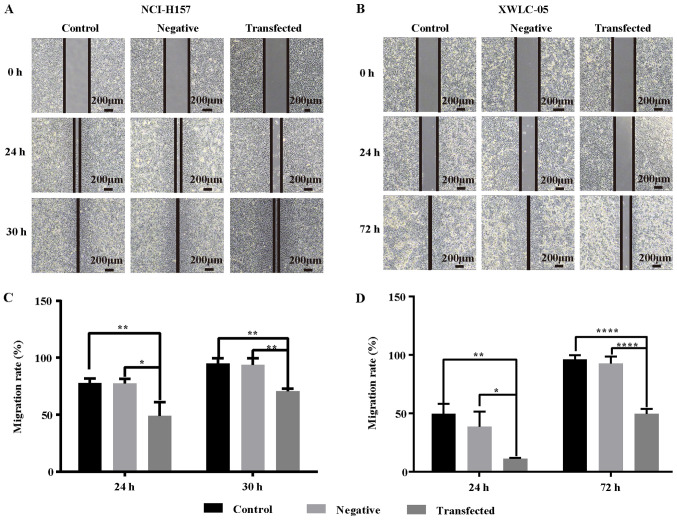
Role of miR-218 in NSCLC cell migration ability
The results of the scratch assay demonstrated that the wound healing ability of XWLC-05 and NCI-H157 cells in the transfection group was significantly reduced. The wounds of NCI-H157 cells were fully healed at 30 h in the blank control group and the negative control group, while the wounds in the transfected group (141.15±8.12 µm) were not yet healed. Similarly, the wounds of XWLC-05 cells in both the blank control group and the negative control group were completely healed at 72 h, while the wounds in the transfected group were still a certain width (287.41±24.81 µm; Fig. 7A and B). Furthermore, there was a significant difference in migration rate at 24 h between the transfection group and the negative control group (P<0.05; Fig. 7C and D). These findings indicated that overexpression of miR-218 weakened the migration ability of NSCLC cells.
Figure 7.
Overexpression of miR-218 inhibits the migration of NCI-H157 and XWLC-05 cells. Wound healing assay was used to assess the migration ability of NCI-H157 and XWLC-05 cells transfected with plasmids. Uniform scratches were made and the serial images were captured at 0, 24, 30 and 72 h respectively following transfection. Representative images of wound healing area of (A) NCI-157 and (B) of XWLC-05 cells in each group are shown (magnification, ×40; scale bar, 200 µm). Histograms indicated (C) NCI-H157 and (D) XWLC-05 cell migration rate in each group. Negative, negative control (transfected with pGpU6/EGFP/Neo-miR-NC plasmid). The experiment was repeated three times (n=3, *P<0.05, **P<0.01, ****P<0.0001 vs. the control). miR, microRNA.
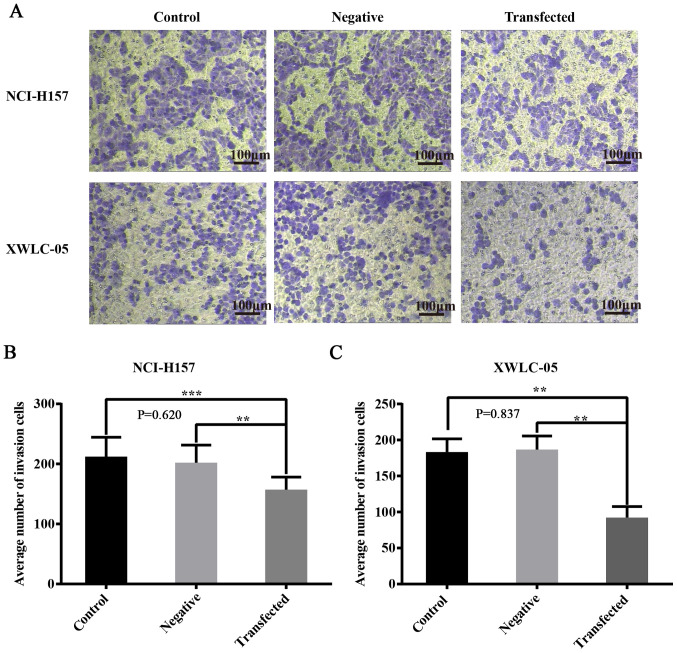
Function of miR-218 in NSCLC cell invasion
As shown in Fig. 8A and B, the Transwell invasion assay demonstrated that a large number of XWLC-05 cells in both the negative control group and the blank control group passed through the membrane (the average number of cells in 5 fields was 183.6±4.24 and 187.7±2.08). However, the number of membrane-passed XWLC-05 cells in the transfection group (the average number of cells in 5 fields was 91.8±3.42) was significantly decreased (P<0.01). Furthermore, the transfection group (the average number of cells in 5 fields was 165.2±9.03) demonstrated significantly fewer membrane-passed NCI-H157 cells compared with both the negative control group and the blank control group (the average number of cells in 5 fields was 207.75±5.24 and 226.75±2.56; P<0.01, P<0.001). The above data confirmed that miR-218 overexpression reduced NSCLC cell invasion.
Figure 8.
Overexpression of miR-218 inhibits the invasion of NCI-H157 and XWLC-05 cells. The number of invasive cells was determined by Transwell invasion assays and counted under an inverted microscope. Invasive cells were stained with 0.2% crystal violet and appear purple. The average number of invasive cells was calculated from 5 random fields. (A) Representative images of each group (magnification, ×100; scale bar, 20 µm). (B and C) Statistical calculations. Negative, negative control (transfected with pGpU6/EGFP/Neo-miR-NC plasmid) and data represent mean ± SD of three independent repeats. The experiment was repeated three times (n=3, **P<0.01, ***P<0.001 vs. control or negative control). miR, microRNA.
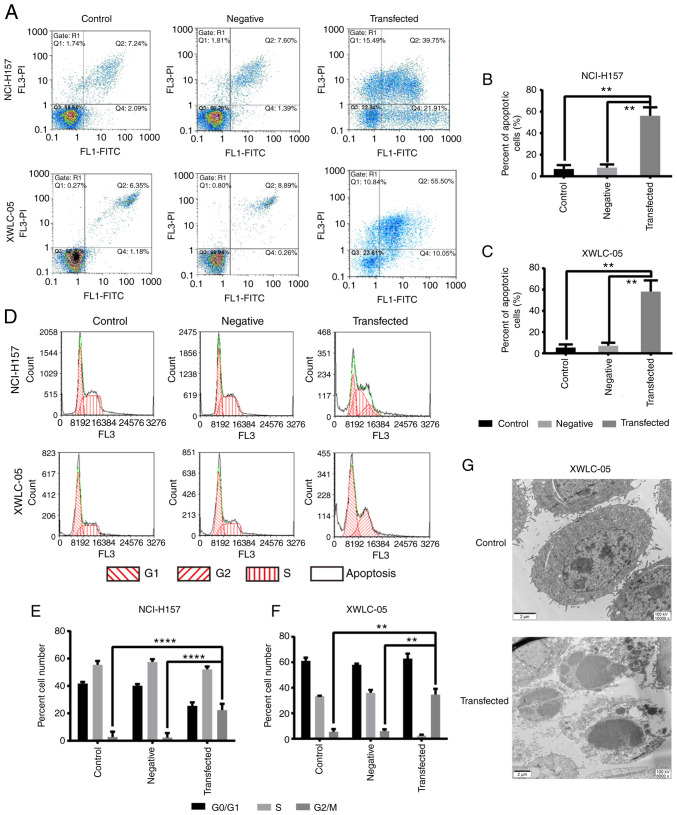
Overexpression of miR-218 induces NSCLC cell apoptosis
The apoptosis rate was measure by flow cytometry. As shown in Fig. 9A-C, the apoptosis rate of both XWLC-05 and NCI-H157 cells in the transfection group was significantly higher compared with the negative control group and the blank control group (P<0.01). Furthermore, overexpression of miR-218 in XWLC-05 and NCI-H157 cells resulted in significant cell cycle arrest in the G2/M phases (Fig. 9D-F). The results of transmission electron microscopy demonstrated that lung cancer XWLC-05 cells in the transfection group exhibited apoptotic alterations in morphology (Fig. 9G). For example, cell plasma membranes were concentrated; ribosome, mitochondria and other organelles were aggregated; cell sizes were reduced and cell structure became denser. There were several apoptotic bodies at different sizes in the apoptotic cells. The findings indicated that overexpression of miR-218 induced the apoptosis of XWLC-05 and NCI-H157 cells.
Figure 9.
Overexpression of miR-218 induces G2/M phase cell cycle arrest and apoptosis in NCI-H157 and XWLC-05 cells. (A) Flow cytometry was used to analyze the apoptosis rate of cells in each group at a qualitative level. (B and C) The percentage of apoptotic cells in each group. Data represented mean ± SD of three independent repeats. (n=3, **P<0.01 vs. control or negative control). Negative, negative control (transfected with pGpU6/EGFP/Neo-miR-NC plasmid). (D) Representative images of cell cycle distribution detected by flow cytometry. (E and F) The percentage of cells at various phases of the cell cycle as measured by flow cytometry. Negative, the negative control group (transfected with pGpU6/EGFP/Neo-miR-NC plasmid) and data represented mean ± SD of three independent repeats. (n=3, **P<0.01, ****P<0.0001 vs. the control or negative control). (G) Morphological changes of XWLC-05 cells induced by miR-218 overexpression by transmission electron microscopy. Magnification, ×8,000 (bottom) and ×10,000 (top); scale bar, 2 µm. XWLC-05 cells maintaining lung cancer cell morphology from the blank control group (top). XWLC-05 cells exhibiting apoptotic cell morphology from the pGpU6/EGFP/Neo-miR-NC plasmid transfected group at 48 h post-transfection (bottom). The experiment was repeated three times. miR, microRNA.
Discussion
Lung cancer is the leading cause of cancer-related death worldwide and a high incidence and mortality rate occurs in Xuanwei County, a county-level city in Yunnan Province, China (1,2). Therefore, the present study focused on Xuanwei lung cancer for its regional specificity. This disease has attracted attention worldwide and a number of studies have been performed on reducing the incidence and mortality of Xuanwei lung cancer (5–7,10,59). However, the survival rate remains low and the precise mechanisms of lung cancer progression in Xuanwei County remain to be elucidated (2,60). Therefore, novel strategies for this regional-specific lung cancer treatment are urgently required.
It is known that the abnormal expression of microRNAs is associated with carcinogenesis. A number of studies report that the dysregulations of miR-155 (61), miR-21 (62), miR-32 (63) and miR-34 (64), in addition to miR-218, serve an important role in the progression of lung cancer (39). The regulation of multiple target genes by miR-218 has been experimentally validated (65). The present study, for the first time to the best of the authors' knowledge, investigated the regulatory mechanisms between miR-218 and BCL-2, BMI, PTEN and YY1 in XWLC-05 cells.
The present study demonstrated that miR-218 overexpression significantly suppressed cell apoptosis in XWLC-05 cells and a similar result was observed in NCI-H157 cells; this was consistent with previous studies (66–68). In order to further investigate the mechanism behind miR-218-induced apoptosis, the protein and mRNA expression levels of BCL-2, BMI, and PTEN were examined in XWLC-05 and NCI-H157 cells following miR-218 transfection. Bioinformatics analysis demonstrated that BCL-2 was a potential target of miR-218 and previous findings confirm that BCL-2 can serve key roles in cell apoptosis (43,44). The results of the present study demonstrated that overexpression of miR-218 could induce cell apoptosis in XWLC-05 and NCI-H157 cells partly by reducing the mRNA and protein expression levels of BCL-2, further emphasizing that BCL-2 expression was regulated by multiple miRNAs in lung cancer (22). In addition, a previous study notes that overexpression of miR-218 induces cell apoptosis in colon cancer by direct regulation of BMI-1 (66). The results of the present study also revealed that overexpression of miR-218 induced cell apoptosis partly by decreasing the mRNA and protein expression levels of BMI-1, while increasing the mRNA and protein expression levels of PTEN in XWLC-05 and NCI-H157 cells. BMI-1 acts as an oncogene by repressing the tumor-suppressor PTEN that can exert a critical negative effect on the activity of the PI3K/AKT pathway (69–71). The findings of the present study indicated that miR-218 could inhibit BMI-1, while increasing PTEN, and may inactivate the PI3K/AKT pathway, consequently inducing apoptosis of lung cancer cells. The present study used the MTT assay to evaluate the effect of miR-218 on XWLC-05 and NCI-H157 cell proliferation. MTT assay demonstrated that cell proliferation was significantly inhibited by increasing the expression of exogenous miR-218 in both XWLC-05 and NCI-H157 cells, which was consistent with other studies (32,34,41). The effect of miR-218 on cell cycle progression was further investigated using flow cytometry. The results demonstrated that miR-218 overexpression led to a significant increase in the number of cells accumulating in the G2 phase, suggesting that miR-218 could reduce cell proliferative capacity with G2 cell cycle arrest in lung cancer. Given the critical function of BMI-1, YY1 and PTEN in cell proliferation, the protein and mRNA expression levels of BMI-1, YY1 and PTEN were examined in XWLC-05 and NCI-H157 cells following miR-218 transfection to identify the potential mechanism responsible for the observed effects of miR-218 on cell growth in lung cancer cells (69–72). The results indicated that miR-218 could inhibit BMI-1 expression, while elevating PTEN expression, and may inactivate the PI3K/AKT pathway, thus inhibiting the proliferation of lung cancer cells. In addition, miR-218 is a direct target of YY1 and it suppresses the proliferation of glioma cells by downregulating YY1 expression (72). However, in the present study overexpression of miR-218-5p significantly increased the expression of endogenous YY1 resulting in the inhibition of the proliferation of lung cancer cells. The role and mechanisms of YY1 in tumorigenesis and development remains controversial (52). Research has identified that YY1 and AP1 synergistically induce the expression of tumor-suppressor gene HLJ1 in lung cancer, thereby inhibiting lung cancer invasion (73). However, research shows that YY1 serves an oncogene role in the occurrence and development of lung cancer (54). The findings of the present study indicated that miR-218 could suppress lung cancer progression partly via miR-218-directed regulation of YY1 expression; YY1 may act as a tumor suppressor under certain conditions (74). It has been confirmed that c-MYC works synergistically with BMI-1, which is a direct target gene of miR-218 (75). Furthermore, c-MYC can suppress the expression levels of YY1 and c-MYC function can also be inhibited by YY1 (76,77). The findings of the present study also indicated that miR-218 might not directly activate YY1 expression, but indirectly suppressed c-MYC, resulting in elevated YY1 expression levels, thereby inhibiting the biological characteristics of malignant tumors.
Next, whether miR-218 contributed to the migration and invasion ability of XWLC-05 and NCI-H157 cells was examined. The wound-healing assay and Transwell invasion assay revealed that ectopic expression of miR-218 markedly repressed the migration and invasion of XWLC-05 and NCI-H157 cells. However, since cells should be grown in low-serum or serum-free media during the wound-healing assay to avoid cell proliferation, further work is needed to optimize the wound-healing assay with a low concentration of serum. Furthermore, it was demonstrated that miR-218 repressed the expression of BMI-1, but increased the expression of PTEN and YY1 in XWLC-05 and NCI-H157 cells, leading to decreased cancer migration and invasion. Repression of BMI-1 was found to suppress cancer cell migration and invasion, including in lung cancer (47,78). Various miRNAs have been identified as suppressing BMI-1 to inhibit tumor invasion (79–81) and studies reveal that miR-218 functioned as a tumor suppressor via negatively regulating BMI-1 in glioma (82) and colorectal cancers (29). Consistent with these findings, the results of the present study demonstrated that miR-218-inhibited BMI-1 was the key to migration and invasion both in XWLC-05 and NCI-H157 cells. BMI-1 can inhibit PTEN resulting in activation of the PI3K/AKT pathway, leading to overexpression of MMP-2, MMP-9 and VEGF and promotion of colon and liver cancer invasion and metastasis (29,83). The findings of the present study indicated that miR-218 could inhibit BMI-1 expression, while indirectly increasing PTEN expression, thereby inhibiting PI3K-AKT pathway activity and lung cancer invasion. The exact mechanism of this requires further study. The present study did not observe any significant difference in the role of miR-218 and its regulation of BCL-2, BMI-1, PTEN and YY1 expression in XWLC-05 and NCI-H157 cells. These results were consistent with our finding (data not shown) and other clinical sample findings (84,85). The findings of the present study indicated that miR-218 might serve a common function in the progression of Xuanwei NSCLC and other NSCLC, which raises doubts about the current ideas about these regional-specific diseases.
Thus far, the precise regulatory mechanisms of lung cancer progression in Xuanwei County have not been fully understood. The specific epidemiological characteristics in Xuanwei County led some researchers to believe that there may be unique molecular mechanisms in the progression of Xuanwei lung cancer (86,87). For example, some studies confirm that mutations in the MUC16 gene are observed in 50% of lung cancer patients residing in Xuanwei and MUC16 participates in the progression of Xuanwei lung cancer (88,89). Moreover, miR-144 was remarkably decreased in Xuanwei NSCLC (90). These findings suggested that air pollution-related genes such as MUC16 and miR-144 might be critical in Xuanwei lung cancer progression (6,87–91). However, the results from the present study emphasized that the dysregulation of miR-218 was also essential for the progression of both Xuanwei NSCLC and other NSCLC. More importantly, the present and previous studies indicated that dysregulations of key miRNAs, including miR-218, miR-21 and miR-34, were the common events both in the progression of the regional-specific NSCLC and other NSCLC (22,92–94).
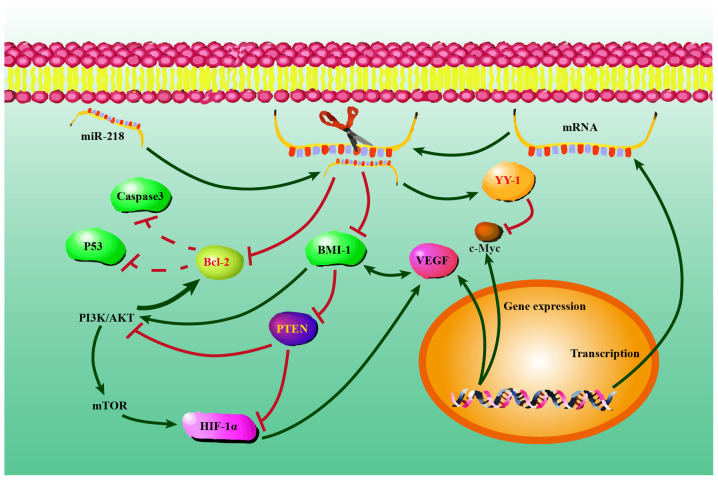
In summary, the results of the present study confirmed that miR-218 could suppress BCL-2 and BMI-1 expression, while increasing PTEN and YY1 expression, leading to the suppression of NSCLC progression. Furthermore, the findings further underline the pivotal roles served by miR-218, providing new insight into the progression of NSCLC. The possible regulatory mechanisms of miR-218 are demonstrated in Fig. 10. Further studies will be required to elucidate the precise mechanisms involved in the downstream molecules of BCL-2, BMI-1, PTEN and YY1 that could contribute to miR-218-mediated suppression of lung cancer progression. Notably, the roles of miR-218 and miR-218-mediated regulation of BCL-2, BMI-1, PTEN and YY1 expression in the progression of Xuanwei NSCLC have no significant difference from other NSCLC. These findings were consistent with our previous studies and suggest that the roles and regulatory mechanisms of certain key miRNAs, including miR-218, miR-21 and miR-34a, on gene expression were similar in the progression of Xuanwei NSCLC and other NSCLC (22,95). It is difficult to say whether the high incidence of Xuanwei NSCLC in South China could only be attributed to scale-specific effects of environmental variables in the area or specific molecular genetic variation. Further studies are required on the precise regulatory network involved in the downstream molecules of BCL-2, BMI-1, PTEN and YY1 that could contribute to miR-218-mediated tumor suppression for the clinical treatment of Xuanwei NSCLC.
Figure 10.
Possible regulatory mechanisms between miR-218 and PTEN, YY1, BCL-2 and BMI-1 in NSCLC cells transfected with pGpU6/EGFP/Neo-miR-218. At the post-transcriptional level, PTEN, YY1, BCL-2 and BMI-1 expression levels were regulated by miR-218. Overexpression of miR-218 activated PTEN and YY1, and repressed BCL-2 and BMI-1 at the mRNA and protein levels. PTEN, YY1, BCL-2 and BMI-1 may have single or combined effects on NSCLC cell proliferation, migration, invasion and apoptosis at the mRNA and protein levels. miR, microRNA; NSCLC, non-small cell lung cancer; PTEN, anti-phosphatase and tensin homolog; BMI-1, anti-polycomb complex protein BMI-1; YY1, anti-transcriptional repressor protein YY1; VEGF, vascular endothelial growth factor.
Acknowledgements
The authors thank Dr Xicai Wang (Yunnan Cancer Hospital and The Third Affiliated Hospital of Kunming Medical University and Yunnan Cancer Center, Yunnan, China) for their technical assistance.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81560380), Yunnan Medical Discipline Leader Project (grant no. D-201601) and the Project of Medical and Health Technology Development Program of Yunnan Province (grant no. 2017NS203).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QZ, YC, JY and CC designed the experiments. ZX, HL, LZ, YZ, TW, DC, GL, SL, QY and CY performed the experiments. ZX and QC analyzed the data. YC, CH, CC and LC interpreted the data. QZ, YC, CG and LC contributed reagents/materials/analysis tools. YC, QZ and QC wrote the manuscript. CG made substantial contributions to the aquisition of data. All authors participated in revising the manuscript critically for important intellectual content and final approval of the version to be published. All authors agreed to be accountable for the work in ensuring that questions related to the integrity of any part of the work are appropriately investigated and resolved. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Guo W, Ran J, Tang R, Lin H, Chen X, Ning B, Li J, Zhou Y, Chen LC, et al. Five-year lung cancer mortality risk analysis and topography in Xuan Wei: A spatiotemporal correlation analysis. BMC Public Health. 2019;19:173. doi: 10.1186/s12889-019-6490-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao Y, Gao H. Prevalence and causes of air pollution and lung cancer in Xuanwei City and Fuyuan County, Yunnan Province, China. Front Med. 2012;6:217–220. doi: 10.1007/s11684-012-0192-8. [DOI] [PubMed] [Google Scholar]
- 4.Xiao Y, Shao Y, Yu X, Zhou G. The epidemic status and risk factors of lung cancer in Xuanwei City, Yunnan Province, China. Front Med. 2012;6:388–394. doi: 10.1007/s11684-012-0233-3. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Duan Y, Meng QH, Gong R, Guo C, Zhao Y, Zhang Y. Integrated analysis of DNA methylation profiling and gene expression profiling identifies novel markers in lung cancer in Xuanwei, China. PLoS One. 2018;13:e0203155. doi: 10.1371/journal.pone.0203155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Ran J, Chen LC, Costa M, Huang Y, Chen X, Tian L. Bituminous coal combustion and Xuan Wei Lung cancer: A review of the epidemiology, intervention, carcinogens, and carcinogenesis. Arch Toxicol. 2019;93:573–583. doi: 10.1007/s00204-019-02392-y. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y, Chen K, Zhou Y, Hu Z, Chen S, Huang Y. Application of serum microRNA-9-5p, 21-5p, and 223-3p combined with tumor markers in the diagnosis of non-small-cell lung cancer in Yunnan in southwestern China. OncoTargets Ther. 2018;11:587–597. doi: 10.2147/OTT.S152957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan FC, Wang QQ, Ruan YH, Ma LJ, Jia JT, Jin KW, Chin J. Establishment and biological characteristics of lung cancer cell line XWLC-05. Ai Zheng. 2007;26:21–25. [PubMed] [Google Scholar]
- 9.Lei J, Li QH, Yang JL, Liu F, Wang L, Xu WM, Zhao WX. The antitumor effects of oncolytic adenovirus H101 against lung cancer. Int J Oncol. 2015;47:555–562. doi: 10.3892/ijo.2015.3045. [DOI] [PubMed] [Google Scholar]
- 10.Xiong G, Chen X, Zhang Q, Fang Y, Chen W, Li C, Zhang J. RNA interference influenced the proliferation and invasion of XWLC-05 lung cancer cells through inhibiting aquaporin 3. Biochem Biophys Res Commun. 2017;485:627–634. doi: 10.1016/j.bbrc.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, He S, Mei R, Kang Y, Duan J, Wei R, Xiang C, Wu Y, Lu X, Cai Z, et al. miR 29a suppresses IL 13 induced cell invasion by inhibiting YY1 in the AKT pathway in lung adenocarcinoma A549 cells. Oncol Rep. 2018;39:2613–2623. doi: 10.3892/or.2018.6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai L, Lin S, Girard L, Zhou Y, Yang L, Ci B, Zhou Q, Luo D, Yao B, Tang H, et al. LCE: An open web portal to explore gene expression and clinical associations in lung cancer. Oncogene. 2019;38:2551–2564. doi: 10.1038/s41388-018-0588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S, Guo X, Wang B, Gang Y, Zhang Y, et al. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 2010;6:e1000879. doi: 10.1371/journal.pgen.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 15.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 16.Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y, Ren J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019;76:441–451. doi: 10.1007/s00018-018-2940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mestdagh P, Boström AK, Impens F, Fredlund E, Van Peer G, De Antonellis P, von Stedingk K, Ghesquière B, Schulte S, Dews M, et al. The miR-17-92 microRNA cluster regulates multiple components of the TGF-β pathway in neuroblastoma. Mol Cell. 2010;40:762–773. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Naeli P, Yousefi F, Ghasemi Y, Savardashtaki A, Mirzaei H. The role of microRNAs in lung cancer: Implications for diagnosis and therapy. Curr Mol Med. 2020;20:90–101. doi: 10.2174/1566524019666191001113511. [DOI] [PubMed] [Google Scholar]
- 20.Uddin A, Chakraborty S. Role of miRNAs in lung cancer. J Cell Physiol. 2018 Apr 20; doi: 10.1002/jcp.26607. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 21.Lin PY, Yu SL, Yang PC. MicroRNA in lung cancer. Br J Cancer. 2010;103:1144–1148. doi: 10.1038/sj.bjc.6605901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu LF, Wu ZP, Chen Y, Zhu QS, Hamidi S, Navab R. MicroRNA-21 (miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK and Bcl-2 in lung squamous carcinoma, Gejiu City, China. PLoS One. 2014;9:e103698. doi: 10.1371/journal.pone.0103698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng M, Zeng C, Lu X, He X, Zhang R, Qiu Q, Zheng G, Jia X, Liu H, He Z. miR-218 suppresses gastric cancer cell cycle progression through the CDK6/Cyclin D1/E2F1 axis in a feedback loop. Cancer Lett. 2017;403:175–185. doi: 10.1016/j.canlet.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Guan B, Wu K, Zeng J, Xu S, Mu L, Gao Y, Wang K, Ma Z, Tian J, Shi Q, et al. Tumor-suppressive microRNA-218 inhibits tumor angiogenesis via targeting the mTOR component RICTOR in prostate cancer. Oncotarget. 2017;8:8162–8172. doi: 10.18632/oncotarget.14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jun GJ, Zhong GG, Ming ZS. miR-218 inhibits the proliferation of glioma U87 cells through the inactivation of the CDK6/cyclin D1/p21Cip1/Waf1 pathway. Oncol Lett. 2015;9:2743–2749. doi: 10.3892/ol.2015.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu K, Li C, Zheng X, Yang W, Yao Y, Liu Q. Prognostic significance of miR-218 in human hepatocellular carcinoma and its role in cell growth. Oncol Rep. 2014;32:1571–1577. doi: 10.3892/or.2014.3386. [DOI] [PubMed] [Google Scholar]
- 27.Liu B, Tian Y, Li F, Zhao Z, Jiang X, Zhai C, Han X, Zhang L. Tumor-suppressing roles of miR-214 and miR-218 in breast cancer. Oncol Rep. 2016;35:3178–3184. doi: 10.3892/or.2016.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T, Xu L, Jia R, Wei J. MiR-218 suppresses the metastasis and EMT of HCC cells via targeting SERBP1. Acta Biochim Biophys Sin (Shanghai) 2017;49:383–391. doi: 10.1093/abbs/gmx017. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Shi H, Tang H, Fang Z, Wang J, Cui S. miR-218 inhibits the invasion and migration of colon cancer cells by targeting the PI3K/Akt/mTOR signaling pathway. Int J Mol Med. 2015;35:1301–1308. doi: 10.3892/ijmm.2015.2126. [DOI] [PubMed] [Google Scholar]
- 30.Davidson MR, Larsen JE, Yang IA, Hayward NK, Clarke BE, Duhig EE, Passmore LH, Bowman RV, Fong KM. MicroRNA-218 is deleted and downregulated in lung squamous cell carcinoma. PLoS One. 2010;5:e12560. doi: 10.1371/journal.pone.0012560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu DW, Cheng YW, Wang J, Chen CY, Lee H. Paxillin predicts survival and relapse in non-small cell lung cancer by microRNA-218 targeting. Cancer Res. 2010;70:10392–10401. doi: 10.1158/0008-5472.CAN-10-2341. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Ding L, Hu Q, Xia J, Sun J, Wang X, Xiong H, Gurbani D, Li L, Liu Y, et al. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol Cancer. 2017;16:141. doi: 10.1186/s12943-017-0710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi ZM, Wang L, Shen H, Jiang CF, Ge X, Li DM, Wen YY, Sun HR, Pan MH, Li W, et al. Downregulation of miR-218 contributes to epithelial-mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signaling. Oncogene. 2017;36:2577–2588. doi: 10.1038/onc.2016.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu K, Ding H, Wang W, Liao Z, Fu Z, Hong Y, Zhou Y, Zhang CY, Chen X. Tumor-suppressive miR-218-5p inhibits cancer cell proliferation and migration via EGFR in non-small cell lung cancer. Oncotarget. 2016;7:28075–28085. doi: 10.18632/oncotarget.8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song L, Li D, Zhao Y, Gu Y, Zhao D, Li X, Bai X, Sun Y, Zhang X, Sun H, et al. miR-218 suppressed the growth of lung carcinoma by reducing MEF2D expression. Tumour Biol. 2016;37:2891–2900. doi: 10.1007/s13277-015-4038-2. [DOI] [PubMed] [Google Scholar]
- 36.Chiu KL, Kuo TT, Kuok QY, Lin YS, Hua CH, Lin CY, Su PY, Lai LC, Sher YP. ADAM9 enhances CDCP1 protein expression by suppressing miR-218 for lung tumor metastasis. Sci Rep. 2015;5:16426. doi: 10.1038/srep16426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie J, Yu F, Li D, Zhu X, Zhang X, Lv Z. MicroRNA-218 regulates cisplatin (DPP) chemosensitivity in non-small cell lung cancer by targeting RUNX2. Tumour Biol. 2016;37:1197–1204. doi: 10.1007/s13277-015-3831-2. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, Ge S, Hu C, Yang N, Zhang J. MiRNA-218, a new regulator of HMGB1, suppresses cell migration and invasion in non-small cell lung cancer. Acta Biochim Biophys Sin (Shanghai) 2013;45:1055–1061. doi: 10.1093/abbs/gmt109. [DOI] [PubMed] [Google Scholar]
- 39.Zeng F, Wang Q, Wang S, Liang S, Huang W, Guo Y, Peng J, Li M, Zhu W, Guo L. Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate Etk expression. Oncogene. 2020;39:293–307. doi: 10.1038/s41388-019-0984-2. [DOI] [PubMed] [Google Scholar]
- 40.Jin X, Liu X, Zhang Z, Guan Y. lncRNA CCAT1 acts as a microRNA-218 sponge to increase gefitinib resistance in NSCLC by targeting HOXA1. Mol Ther Nucleic Acids. 2020;19:1266–1275. doi: 10.1016/j.omtn.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Lu C, Zhao G, Han X, Dong K, Wang C, Guan J-Z, Wang Z. Downregulation of miR-218 by nicotine promotes cell proliferation through targeting CDK6 in non-small cell lung cancer. J Cell Biochem. 2019;120:18370–18377. doi: 10.1002/jcb.29148. [DOI] [PubMed] [Google Scholar]
- 42.Li YJ, Zhang W, Xia H, Zhang BS, Chen P, Zhao YL, Li J. miR-218 suppresses epithelial-to-mesenchymal transition by targeting Robo1 and Ecop in lung adenocarcinoma cells. Future Oncol. 2017;13:2571–2582. doi: 10.2217/fon-2017-0398. [DOI] [PubMed] [Google Scholar]
- 43.Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: The balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol. 2019;20:175–193. doi: 10.1038/s41580-018-0089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radha G, Raghavan SC. BCL2: A promising cancer therapeutic target. Biochim Biophys Acta Rev Cancer. 2017;1868:309–314. doi: 10.1016/j.bbcan.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Hata AN, Engelman JA, Faber AC. The BCL2 family: Key mediators of the apoptotic response to targeted anticancer therapeutics. Cancer Discov. 2015;5:475–487. doi: 10.1158/2159-8290.CD-15-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M-C, Li C-L, Cui J, Jiao M, Wu T, Jing LI, Nan K-J. BMI-1, a promising therapeutic target for human cancer. Oncol Lett. 2015;10:583–588. doi: 10.3892/ol.2015.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng X, Wang Y, Zheng X, Liu C, Su B, Nie H, Zhao B, Zhao X, Yang H. shRNA-mediated knockdown of Bmi-1 inhibit lung adenocarcinoma cell migration and metastasis. Lung Cancer. 2012;77:24–30. doi: 10.1016/j.lungcan.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 48.Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: Preclinical and clinical evidences. Stem Cells. 2012;30:372–378. doi: 10.1002/stem.1035. [DOI] [PubMed] [Google Scholar]
- 49.Gkountakos A, Sartori G, Falcone I, Piro G, Ciuffreda L, Carbone C, Tortora G, Scarpa A, Bria E, Milella M, et al. PTEN in lung cancer: Dealing with the problem, building on new knowledge and turning the game around. Cancers (Basel) 2019;11:1141. doi: 10.3390/cancers11081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Álvarez-Garcia V, Tawil Y, Wise HM, Leslie NR. Mechanisms of PTEN loss in cancer: It's all about diversity. Semin Cancer Biol. 2019;59:66–79. doi: 10.1016/j.semcancer.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 51.Papa A, Pandolfi PP. The PTEN-PI3K axis in cancer. Biomolecules. 2019;9:153. doi: 10.3390/biom9040153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarvagalla S, Kolapalli SP, Vallabhapurapu S. The two sides of YY1 in cancer: A friend and a foe. Front Oncol. 2019;9:1230. doi: 10.3389/fonc.2019.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang CC, Tsai MF, Hong TM, Chang GC, Chen CY, Yang WM, Chen JJW, Yang PC. The transcriptional factor YY1 upregulates the novel invasion suppressor HLJ1 expression and inhibits cancer cell invasion. Oncogene. 2005;24:4081–4093. doi: 10.1038/sj.onc.1208573. [DOI] [PubMed] [Google Scholar]
- 54.Huang T, Wang G, Yang L, Peng B, Wen Y, Ding G, Wang Z. Transcription factor YY1 modulates lung cancer progression by activating lncRNA-PVT1. DNA Cell Biol. 2017;36:947–958. doi: 10.1089/dna.2017.3857. [DOI] [PubMed] [Google Scholar]
- 55.Liu J, Blackhall F, Seiden-Long I, Jurisica I, Navab R, Liu N, Radulovich N, Wigle D, Sultan M, Hu J, et al. Modeling of lung cancer by an orthotopically growing H460SM variant cell line reveals novel candidate genes for systemic metastasis. Oncogene. 2004;23:6316–6324. doi: 10.1038/sj.onc.1207795. [DOI] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 57.Li W, Wang W, Ding M, Zheng X, Ma S, Wang X. MiR-1244 sensitizes the resistance of non-small cell lung cancer A549 cell to cisplatin. Cancer Cell Int. 2016;16:30. doi: 10.1186/s12935-016-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang CC, Park AY, Guan JL. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 59.Vermeulen R, Downward GS, Zhang J, Hu W, Portengen L, Bassig BA, Hammond SK, Wong JYY, Li J, Reiss B, et al. Constituents of household air pollution and risk of lung cancer among never-smoking women in Xuanwei and Fuyuan, China. Environ Health Perspect. 2019;127:97001. doi: 10.1289/EHP4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen G, Sun X, Ren H, Wan X, Huang H, Ma X, Ning B, Zou X, Hu W, Yang G. The mortality patterns of lung cancer between 1990 and 2013 in Xuanwei, China. Lung Cancer. 2015;90:155–160. doi: 10.1016/j.lungcan.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 61.Shao C, Yang F, Qin Z, Jing X, Shu Y, Shen H. The value of miR-155 as a biomarker for the diagnosis and prognosis of lung cancer: A systematic review with meta-analysis. BMC Cancer. 2019;19:1103. doi: 10.1186/s12885-019-6297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bica-Pop C, Cojocneanu-Petric R, Magdo L, Raduly L, Gulei D, Berindan-Neagoe I. Overview upon miR-21 in lung cancer: Focus on NSCLC. Cell Mol Life Sci. 2018;75:3539–3551. doi: 10.1007/s00018-018-2877-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou B, Yuan W, Li X. Long intergenic noncoding RNA 319 (linc00319) promotes cell proliferation and invasion in lung cancer cells by directly downregulating the tumor suppressor miR-32. Oncol Rese. 2017 Aug 11; doi: 10.3727/096504017X15016337254650. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 64.Zhang L, Liao Y, Tang L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res. 2019;38:53. doi: 10.1186/s13046-019-1059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu YF, Zhang L, Waye MMY, Fu WM, Zhang JF. MiR-218 mediates tumorigenesis and metastasis: Perspectives and implications. Exp Cell Res. 2015;334:173–182. doi: 10.1016/j.yexcr.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 66.He X, Dong Y, Wu CW, Zhao Z, Ng SSM, Chan FKL, Sung JJY, Yu J. MicroRNA-218 inhibits cell cycle progression and promotes apoptosis in colon cancer by downregulating BMI1 polycomb ring finger oncogene. Mol Med. 2013;18:1491–1498. doi: 10.2119/molmed.2012.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu Y, Xu K, Yagüe E. miR-218 targets survivin and regulates resistance to chemotherapeutics in breast cancer. Breast Cancer Res Treat. 2015;151:269–280. doi: 10.1007/s10549-015-3372-9. [DOI] [PubMed] [Google Scholar]
- 68.Zarogoulidis P, Petanidis S, Kioseoglou E, Domvri K, Anestakis D, Zarogoulidis K. miR-205 and miR-218 expression is associated with carboplatin chemoresistance and regulation of apoptosis via Mcl-1 and Survivin in lung cancer cells. Cell Signal. 2015;27:1576–1588. doi: 10.1016/j.cellsig.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 69.Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu LJ, Kong QL, Xu LH, Zhang X, Liu WL, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest. 2009;119:3626–3636. doi: 10.1172/JCI39374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choi Y, Zhang J, Murga C, Yu H, Koller E, Monia BP, Gutkind JS, Li W. PTEN, but not SHIP and SHIP2, suppresses the PI3K/Akt pathway and induces growth inhibition and apoptosis of myeloma cells. Oncogene. 2002;21:5289–5300. doi: 10.1038/sj.onc.1205650. [DOI] [PubMed] [Google Scholar]
- 71.Lu XX, Cao LY, Chen X, Xiao J, Zou Y, Chen Q. PTEN inhibits cell proliferation, promotes cell apoptosis, and induces cell cycle arrest via downregulating the PI3K/AKT/ hTERT pathway in lung adenocarcinoma A549 cells. BioMed Res Int. 2016;2016:2476842. doi: 10.1155/2016/2476842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao Y, Sun L, Wu Z, Xuan C, Zhang J, You Y, Chen X. miR-218 inhibits the proliferation of human glioma cells through downregulation of Yin Yang 1. Mol Med Rep. 2018;17:1926–1932. doi: 10.3892/mmr.2017.8063. [DOI] [PubMed] [Google Scholar]
- 73.Wang CC, Tsai MF, Dai TH, Hong TM, Chan WK, Chen JJW, Yang PC. Synergistic activation of the tumor suppressor, HLJ1, by the transcription factors YY1 and activator protein 1. Cancer Res. 2007;67:4816–4826. doi: 10.1158/0008-5472.CAN-07-0504. [DOI] [PubMed] [Google Scholar]
- 74.Tan H, Huang S, Zhang Z, Qian X, Sun P, Zhou X. Pan-cancer analysis on microRNA-associated gene activation. EBioMedicine. 2019;43:82–97. doi: 10.1016/j.ebiom.2019.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shrivastava A, Saleque S, Kalpana GV, Artandi S, Goff SP, Calame K. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science. 1993;262:1889–1892. doi: 10.1126/science.8266081. [DOI] [PubMed] [Google Scholar]
- 77.Austen M, Cerni C, Lüscher-Firzlaff JM, Lüscher B. YY1 can inhibit c-Myc function through a mechanism requiring DNA binding of YY1 but neither its transactivation domain nor direct interaction with c-Myc. Oncogene. 1998;17:511–520. doi: 10.1038/sj.onc.1201968. [DOI] [PubMed] [Google Scholar]
- 78.Guo BH, Feng Y, Zhang R, Xu LH, Li MZ, Kung HF, Song LB, Zeng MS. Bmi-1 promotes invasion and metastasis, and its elevated expression is correlated with an advanced stage of breast cancer. Mol Cancer. 2011;10:10. doi: 10.1186/1476-4598-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu L, Li Y, Yan D, He J, Liu D. MicroRNA-183 inhibits gastric cancer proliferation and invasion via directly targeting Bmi-1. Oncol Lett. 2014;8:2345–2351. doi: 10.3892/ol.2014.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He Z, Xia Y, Pan C, Ma T, Liu B, Wang J, Chen L, Chen Y. Up-regulation of miR-452 inhibits metastasis of non-small cell lung cancer by regulating BMI1. Cell Physiol Biochem. 2015;37:387–398. doi: 10.1159/000430362. [DOI] [PubMed] [Google Scholar]
- 81.Guo S, Xu X, Tang Y, Zhang C, Li J, Ouyang Y, Ju J, Bie P, Wang H. miR-15a inhibits cell proliferation and epithelial to mesenchymal transition in pancreatic ductal adenocarcinoma by down-regulating Bmi-1 expression. Cancer Lett. 2014;344:40–46. doi: 10.1016/j.canlet.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 82.Tu Y, Gao X, Li G, Fu H, Cui D, Liu H, Jin W, Zhang Y. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res. 2013;73:6046–6055. doi: 10.1158/0008-5472.CAN-13-0358. [DOI] [PubMed] [Google Scholar]
- 83.Li X, Yang Z, Song W, Zhou L, Li Q, Tao K, Zhou J, Wang X, Zheng Z, You N, et al. Overexpression of Bmi-1 contributes to the invasion and metastasis of hepatocellular carcinoma by increasing the expression of matrix metalloproteinase (MMP) 2, MMP-9 and vascular endothelial growth factor via the PTEN/PI3K/Akt pathway. Int J Oncol. 2013;43:793–802. doi: 10.3892/ijo.2013.1992. [DOI] [PubMed] [Google Scholar]
- 84.Zhou Y, Wang X, Huang Y, Chen Y, Zhao G, Yao Q, Jin C, Huang Y, Liu X, Li G. Down-regulated SOX4 expression suppresses cell proliferation, metastasis and induces apoptosis in Xuanwei female lung cancer patients. J Cell Biochem. 2015;116:1007–1018. doi: 10.1002/jcb.25055. [DOI] [PubMed] [Google Scholar]
- 85.Wang D, Hao T, Pan Y, Qian X, Zhou D. Increased expression of SOX4 is a biomarker for malignant status and poor prognosis in patients with non-small cell lung cancer. Mol Cell Biochem. 2015;402:75–82. doi: 10.1007/s11010-014-2315-9. [DOI] [PubMed] [Google Scholar]
- 86.Li R, Liu Y, Wang T, Tang J, Xie L, Yao Z, Li K, Liao Y, Zhou L, Geng Z, et al. The characteristics of lung cancer in Xuanwei County: A review of differentially expressed genes and noncoding RNAs on cell proliferation and migration. Biomed Pharmacother. 2019;119:109312. doi: 10.1016/j.biopha.2019.109312. [DOI] [PubMed] [Google Scholar]
- 87.Hu Z, Wang X, Yang Y, Zhao Y, Shen Z, Huang Y. MicroRNA expression profiling of lung adenocarcinoma in Xuanwei, China: A preliminary study. Medicine (Baltimore) 2019;98:e15717. doi: 10.1097/MD.0000000000015717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu XJ, Yang MJ, Zhou B, Wang GZ, Huang YC, Wu LC, Cheng X, Wen ZS, Huang JY, Zhang YD, et al. Characterization of somatic mutations in air pollution-related lung cancer. EBioMedicine. 2015;2:583–590. doi: 10.1016/j.ebiom.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kanwal M, Ding XJ, Song X, Zhou GB, Cao Y. MUC16 overexpression induced by gene mutations promotes lung cancer cell growth and invasion. Oncotarget. 2018;9:12226–12239. doi: 10.18632/oncotarget.24203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pan HL, Wen ZS, Huang YC, Cheng X, Wang GZ, Zhou YC, Wang ZY, Guo YQ, Cao Y, Zhou GB. Down-regulation of microRNA-144 in air pollution-related lung cancer. Sci Rep. 2015;5:14331. doi: 10.1038/srep14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou G. Tobacco, air pollution, environmental carcinogenesis, and thoughts on conquering strategies of lung cancer. Cancer Biol Med. 2019;16:700–713. doi: 10.20892/j.issn.2095-3941.2019.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Markou A, Zavridou M, Lianidou ES. miRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer (Auckl) 2016;7:19–27. doi: 10.2147/LCTT.S60341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li YL, Liu XM, Zhang CY, Zhou JB, Shao Y, Liang C, Wang HM, Hua ZY, Lu SD, Ma ZL. MicroRNA-34a/EGFR axis plays pivotal roles in lung tumorigenesis. Oncogenesis. 2017;6:e372. doi: 10.1038/oncsis.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu S, Shen W, Pan Y, Zhu M, Xie K, Geng L, Wang Y, Liang Y, Xu J, Cao S, et al. Genetic variations in key MicroRNAs are associated with the survival of nonsmall cell lung cancer. Medicine (Baltimore) 2015;94:e2084. doi: 10.1097/MD.0000000000002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liuxin Z. Yunnan University; 2018. Role of miR-34a in local lung cancer cell lines XWLC-05 and YTMLC-90 (unpublished PhD thesis) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.