Abstract
Cryptosporidium spp. and Giardia duodenalis are common gastrointestinal parasites with a broad range of hosts, including humans, livestock, and wildlife. To examine the infection status and assess the zoonotic potential of Cryptosporidium spp. and G. duodenalis in dairy cattle in Gansu, China, a total of 1414 fecal samples were collected from the rectum, with one sample collected from each individual animal. All the samples were tested using nested PCR based on the small subunit ribosomal RNA (SSU rRNA) gene of Cryptosporidium spp. and G. duodenalis. The overall infection rates of Cryptosporidium spp. and Giardia duodenalis were 4.2% (n = 59) and 1.0% (n = 14), respectively. Four Cryptosporidium species were identified: C. andersoni (n = 42), C. parvum (n = 12), C. bovis (n = 5), and C. ryanae (n = 1). In further analyses of subtypes of C. parvum isolates based on the 60 kDa glycoprotein (gp60) gene, five were successfully subtyped as IIdA19G1 (n = 4) and IIdA15G1 (n = 1). All 14 G. duodenalis isolates were identified as assemblage E using the triosephosphate isomerase (tpi) gene. The relatively low positive rates of Cryptosporidium spp. and G. duodenalis detected here and the predominance of non-human pathogenic species/assemblages of these parasites indicated their unique transmission dynamics in this area and the low level of threat posed to public health. However, continuous monitoring and further studies of these parasites should be conducted for the prevention and control of these pathogens.
Keywords: Cryptosporidium spp., Giardia duodenalis, Dairy cattle, Species, Assemblage, Prevalence
Abstract
Cryptosporidium spp. et Giardia duodenalis sont des parasites gastro-intestinaux communs avec une large gamme d’hôtes, y compris les humains, le bétail et la faune. Pour examiner le statut infectieux et évaluer le potentiel zoonotique de Cryptosporidium spp. et G. duodenalis chez les bovins laitiers à Gansu, Chine, un total de 1414 échantillons fécaux ont été prélevés dans le rectum, avec un échantillon prélevé individuellement sur chaque animal. Tous les échantillons ont été testés en utilisant une PCR nichée basée sur le gène de l’ARN ribosomal de petite sous-unité (ARNr SSU) de Cryptosporidium spp. et G. duodenalis. Les taux globaux d’infection par Cryptosporidium spp. et Giardia duodenalis étaient de 4,2 % (n = 59) et 1,0 % (n = 14), respectivement. Quatre espèces de Cryptosporidium ont été identifiées : C. andersoni (n = 42), C. parvum (n = 12), C. bovis (n = 5) et C. ryanae (n = 1). Dans d’autres analyses des sous-types d’isolats de C. parvum basés sur le gène de la glycoprotéine de 60 kDa (gp60), cinq ont été sous-typés avec succès comme IIdA19G1 (n = 4) et IIdA15G1 (n = 1). Les 14 isolats de G. duodenalis ont été identifiés comme étant l’assemblage E à l’aide du gène de la triosephosphate isomérase (tpi). Les taux positifs relativement faibles de Cryptosporidium spp. et de G. duodenalis détectés ici et la prédominance des espèces/assemblages non pathogènes pour les humains de ces parasites ont indiqué leur dynamique de transmission unique dans cette zone et le faible niveau de menace pour la santé publique. Cependant, une surveillance continue et des études plus approfondies de ces parasites devraient être menées pour la prévention et le contrôle de ces agents pathogènes.
Introduction
Cryptosporidium spp. and Giardia duodenalis are common gastrointestinal parasites with a broad range of hosts, including humans, livestock, and wildlife [6]. These pathogens are spread by the fecal-oral route, and they can also be transmitted through contaminated food or water [10]. When infected with these parasites, most immunocompetent individuals are asymptomatic; however, immunocompromized individuals such as HIV-positive patients may suffer self-limiting diarrhea or a severe wasting and sometimes life-threatening disease [32].
At least 44 Cryptosporidium species and more than 70 genotypes have been described using molecular diagnostic tools based on the small subunit ribosomal RNA (SSU rRNA) gene [15, 38]. Previous studies indicate that more than 10 genotypes/species of Cryptosporidium spp. have been identified in dairy cattle, with C. parvum, C. bovis, C. andersoni and C. ryanae being the most common species [14]. Additionally, C. scrofarum, C. felis, C. suis, and C. hominis also have been detected in dairy cattle [27]. Cryptosporidium parvum has been classified into at least 19 subtype families: IIa to IIi and IIk to IIt, by means of sequencing analysis of the gp60 gene [12, 17]. In China, only the IId subtype family is found in dairy cattle, including IIdA15G1 and IIdA19G1 subtypes [31]. In other countries, IIaA15G2R1 is the most common subtype in dairy cattle, and this subtype has also been commonly detected in humans. Other subtype families such as IId and IIl are uncommon and have only been reported in small numbers in cattle [34].
According to molecular characterization based on sequence analyses of SSU rRNA, G. duodenalis was classified into eight distinct assemblages (A–H), of which assemblages A and B can infect humans and various animals [10]. Assemblages C–H have strong host specificity: assemblages C and D mostly infect dogs; assemblages E, F, G, and H are specific to hoofed livestock, cats, rats, and seals, respectively [10]. However, human cases with Assemblages C, D, E and F have also been reported [5]. In dairy cattle, assemblages A, B, and E have been detected worldwide, and assemblage E is the primary assemblage in most countries [10, 35].
In China, dairy cattle have been regarded as a significant reservoir for human acquisition of cryptosporidiosis and giardiasis [11]. The distribution of Cryptosporidium spp. and G. duodenalis in pre-weaned dairy calves varies among different areas in China. The predominant species of Cryptosporidium spp. in pre-weaned calves in Ningxia is C. parvum, whereas in Henan and Shandong, it is C. bovis, and C. andersoni is the primary species in Shaanxi Province [17, 22, 24]. Only G. duodenalis assemblage E has been detected in Beijing [18], while G. duodenalis assemblage E (95.9%, 47/49) and assemblage E mixed with A (4.1%, 2/49) have been found in Hebei and Tianjin [16]; assemblages E (86.2%, 25/29) and B (13.8%, 4/29) have been found in the Ningxia Hui Autonomous Region (NXHAR) [17]. Cryptosporidium spp. and G. duodenalis infections in dairy cattle could not only be potential sources for humans acquiring cryptosporidiosis and giardiasis but also could cause great economic losses [10, 31].
Gansu Province, located on the northwestern plateau of China, is one of the key zones of both the ancient Silk Road and the New Silk Road Economic Belt. The main climatic feature of Gansu is arid conditions with scarce precipitation, making it one of the driest places in the world. According to official statistics, there are approximately 25.6 million people and 4.4 million dairy cattle in Gansu Province. To date, however, there have been few epidemiological investigations, and little is known about the prevalence and molecular characterization of parasites in dairy cattle in Gansu province. The purpose of the present study was to assess the prevalence and determine the species and genotypes of Cryptosporidium spp. and G. duodenalis in dairy cattle in Gansu province.
Materials and methods
Ethics statement
This study was conducted in accordance with the Chinese Laboratory Animal Administration Act of 1988. The research protocol was reviewed and approved by the Research Ethics Committee of Henan Agricultural University. The farmers gave permission to collect fecal samples, and no animals were injured during the collection.
Sample collection
Fresh fecal samples were collected directly from the rectum using disposable gloves, and the fecal samples were placed in containers with ice packs and quickly transported to the laboratory. All fecal samples were placed in the refrigerator at 4 °C until DNA extraction. A total of 1414 samples were collected between April 2015 and July 2015 from nine dairy cattle farms in Gansu (Fig. 1), and one sample was collected from each individual animal.
Figure 1.
Geographic map of the sampling locations in Gansu, China. The figure was originally designed by the authors under ArcGIS 10.2 software. The original vector diagram imported in ArcGIS was adapted from Natural Earth (http://www.naturalearthdata.com).
DNA extraction
We used an E.Z.N.A.™ Stool DNA Kit (Omega Bio-Tek Inc., Norcross, GA, USA) to extract the genomic DNA, of which the first step is adding 200 mg X glass beads and 540 SLX-Mlus buffer to stool samples, then vortex at maximum speed for 10 min. The extracted DNA was stored at −20 °C before PCR amplification.
PCR amplification
Cryptosporidium spp. were identified based on the small subunit (SSU) rRNA gene by nested PCR [37], and C. parvum was subtyped with nested PCR targeting the gp60 gene [1]. Giardia duodenalis was identified based on the SSU rRNA [2] and triosephosphate isomerase (tpi) genes by nested PCR [4]. The primers are shown in Table 1. The products of the secondary PCR were detected by 1% agarose gel electrophoresis containing DNAGREEN (Tiandz, Inc., Beijing, China). With the Applied Biosystems 2720 Thermal Cycler (Applied Biosystems, Foster City, CA, USA), the amplifications for the Cryptosporidium spp. SSU rRNA gene and C. parvum gp60 gene were performed in 25 μL volume, including 1 μL template DNA or primary PCR product, 2.5 μL 10× KOD-Plus PCR buffer, 2.5 μL dNTPs (2 mM), 1.5 μL MgSO4 (25 mM), 0.5 μL of each primer (25 μM), 16 μL double distilled water, and 0.5 μL KOD-Plus amplification enzyme (1 unit/μL) (ToYoBo Co., Ltd., Osaka, Japan). A total of 35 cycles were carried out; each of these consisted of 94 °C for 45 s, 55 °C for 45 s, and 72 °C for 1 min. There was also an initial hot start at 94 °C for 3 min and a final extension at 72 °C for 7 min. The secondary cycling conditions were identical to those used in the primary PCR. The PCR reaction mixtures for the SSU rRNA and tpi loci of G. duodenalis were conducted: 2.5 μL 10× PCR buffer, 2 μL dNTPs (1.25 mM), 0.3 μL each primer (25 μM), 0.2 μL rTaq DNA polymerase (1 unit/μL) (Takara Shuzo Co., Ltd), 2 μL DNA sample, 16.7 μL double distilled water, and 1 μL of bovine serum albumin (10 mg/mL). Thirty-five PCR cycles (96 °C for 45 s, 55 °C for 30 s, 72 °C for 45 s) with an initial hot start (96 °C for 4 min) and final extension (72 °C for 4 min) were carried out. The secondary PCR cycle conditions were identical to the primary round except that the annealing temperature was increased to 59 °C.
Table 1.
Primer sequences and reaction conditions used in nested PCR amplifications.
| Locus | Primer sequences (5′ – 3′) | Nucleotide fragment (bp) | Annealing temperature (°C) | Reference |
|---|---|---|---|---|
| Cryptosporidium spp. SSU rRNA | SSU-F2: TTCTAGAGCTAATACATGCG | ~1325 | 55 | Xiao et al. [37] |
| SSU-R2: CCCATTTCCTTCGAAACAGGA | ||||
| SSU-F3: GGAAGGGTTGTATTTATTAGATAAAG | 826–864 | 55 | ||
| SSU-R4: CTCATAAGGTGCTGAAGGAGTA | ||||
| C. parvum gp60 | AL3531: ATAGTCTCCGCTGTATTC | 1280 | 52 | Alves et al. [1] |
| AL3535: GGAAGGAACGATGTATCT | ||||
| AL3532: TCCGCTGTATTCTCAGCC | 800–850 | 50 | ||
| AL3534: GCAGAGGAACCAGCATC | ||||
| G. duodenalis SSU rRNA | Gia2029: AAGTGTGGTGCAGACGGACTC | 497 | 55 | Appelbee et al. [2] |
| Gia2150c: CTGCTGCCGTCCTTGGATGT | ||||
| RH11: CATCCGGTCGATCCTGCC | 292 | 59 | ||
| RH4: AGTCGAACCCTGATTCTCCGCCCAGG | ||||
| G. duodenalis tpi | AL3543: AAATIATGCCTGCTCGTCG | 605 | 50 | Cacciò and Ryan [4] |
| AL3546: CAAACCTTITCCGCAAACC | ||||
| AL3544: CCCTTCATCGGIGGTAACTT | 530 | 50 | ||
| AL3545: GTGGCCACCACICCCGTGCC |
Sequencing and phylogenetic analysis
The positive secondary PCR products were sequenced bidirectionally by SinoGenoMax Biotechnology Co., Ltd (Beijing, China). To confirm different species or genotypes, sequences obtained in this study were aligned with reference sequences downloaded from GenBank (http://blast.ncbi.nlm.nih.gov) using Clustal X 2.10 (http://www.clustal.org/).
To infer the phylogenetic relationships of the detected samples, neighbor-joining (NJ) trees were constructed with the MEGA X program (http://www.megasoftware.net) based on evolutionary distances calculated with the Kimura 2-parameter model. The reliability of these trees was assessed with a bootstrap analysis of 1000 replicates.
Statistical analysis
The prevalence of parasitic infections, with the 95% confidence interval (CI), was calculated. The chi-square test was used to compare differences in infection rates between different age groups and clinical symptoms. A two-tailed p-value < 0.05 was considered statistically significant.
Accession numbers
The representative nucleotide sequences obtained from this study were submitted to GenBank under the following accession numbers: ;MT820515–;MT820519, ;MT821528, and ;MT821529.
Results
Prevalence of Cryptosporidium spp. and G. duodenalis
Of the 1414 fecal samples taken from dairy cattle of nine farms, 60 specimens were PCR-positive for Cryptosporidium spp., and the overall infection rate was 4.2%. The highest infection rate (13.5%, n = 27) was detected on a farm in Hezheng county, followed by Minle county (5.3%, n = 3) and Ganzhou city (3.6%, n = 2). Equal infection rates (3.2%) were detected in Shandan (n = 4) and Jinta counties (n = 3), and the infection rates in Gaotai, Linze, and Sunan counties, and Yuzhong district were 2.9% (n = 4), 2.4% (n = 3), 2.3% (n = 9), and 2.2% (n = 5), respectively. Fourteen out of 1414 fecal samples were positive for G. duodenalis, with an overall infection rate of 1.0%. The highest infection rate (2.9%, n = 4) of G. duodenalis was observed on a farm in Gaotai, followed by Linze (2.4%, n = 3) and Jinta (2.1%, n = 2). Equal infection rates (1.8%, n = 1) were detected in Ganzhou and Minle, and the infection rates in Shandan and Sunan were 0.8% (n = 1) and 0.5% (n = 2), respectively. There was no G. duodenalis detected in the Yuzhong or Hezheng (Table 2).
Table 2.
Species, assemblage, and genotype distribution of three enteric pathogens in cattle in Gansu.
| Sampling sites | Overall infection rate (%) (no. positive/no. of samples) | Infection rate (%) (n) |
Species/subtype/assemblage |
|||
|---|---|---|---|---|---|---|
| Cryptosporidium spp. | G. duodenalis | Cryptosporidium spp. | C. parvum | G. duodenalis | ||
| Shandan | 4.8 (5/125) | 3.2 (4) | 0.8 (1) | C. parvum (1), C. andersoni (1), C. ryanae (1), C. bovis (1) | IIdA15G2R1 (1) | Assemblage E (1) |
| Ganzhou | 5.4 (3/55) | 3.6 (2) | 1.8 (1) | C. bovis (2) | Assemblage E (1) | |
| Jinta | 5.3 (5/94) | 3.2 (3) | 2.1 (2) | C. bovis (1), C. parvum (2) | IIdA19G1 (2) | Assemblage E (2) |
| Linze | 4.8 (6/125) | 2.4 (3) | 2.4 (3) | C. parvum (1), C. andersoni (2) | Assemblage E (3) | |
| Gaotai | 5.8 (8/137) | 2.9 (4) | 2.9 (4) | C. parvum (4) | Assemblage E (4) | |
| Minle | 7.0 (4/57) | 5.3 (3) | 1.8 (1) | C. andersoni (2), C. bovis (1) | Assemblage E (1) | |
| Yuzhong | 2.2 (5/226) | 2.2 (5) | 0 (0) | C. parvum (3), C. andersoni (2), | IIdA19G1 (2) | |
| Hezheng | 13.5 (27/200) | 13.5 (27) | 0 (0) | C. andersoni (26), C. parvum (1) | ||
| Sunan | 2.8 (11/395) | 2.3 (9) | 0.5 (2) | C. andersoni (9) | Assemblage E (2) | |
| Total | 5.2 (74/1414) | 4.2 (60) | 1.0 (14) | C. andersoni (42), C. parvum (12), C. bovis (5), C. ryanae (1) | IIdA15G2R1 (1), IIdA19G1 (4) | Assemblage E (14) |
Distribution of Cryptosporidium species/subtypes and G. duodenalis assemblages
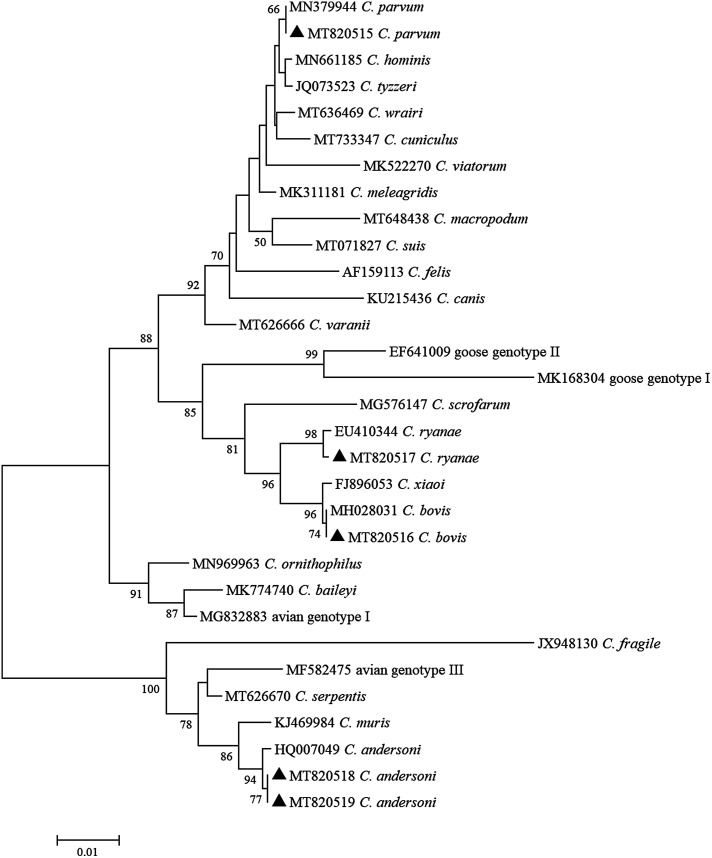
By DNA sequence analysis of the SSU rRNA gene, the Cryptosporidium spp. isolates were identified as four species: C. andersoni, C. parvum, C. bovis, and C. ryanae. Cryptosporidium andersoni (n = 42), the predominant species, accounted for 71.2% of all Cryptosporidium spp. infections and was detected on six farms, while C. parvum (n = 12), C. bovis (n = 5), and C. ryanae (n = 1) were detected on six, three, and one farms, respectively. In the further analyses of subtypes of the 12 C. parvum isolates, five isolates were successfully subtyped, including subtype IIdA15G1 (n = 1) and subtype IIdA19G1 (n = 4) (Table 2).
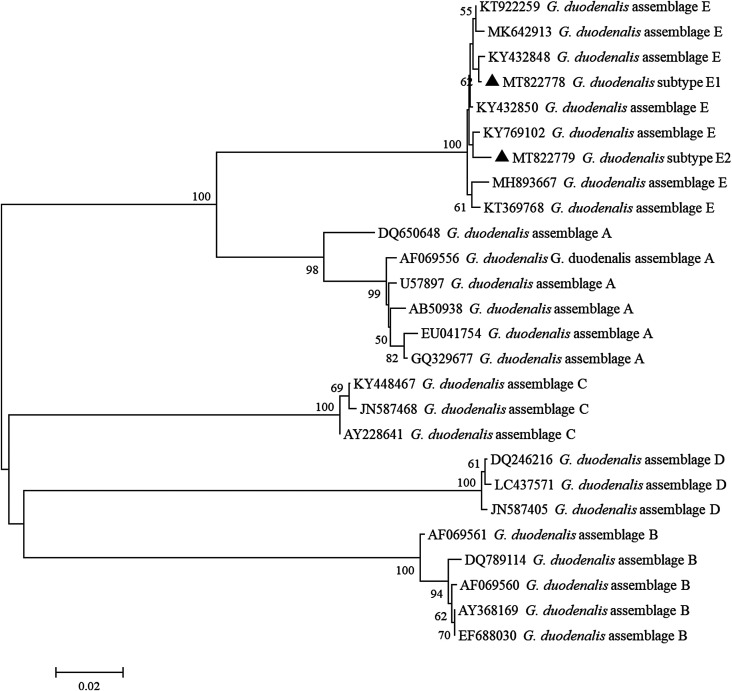
Sequencing analyses of the G. duodenalis SSU rRNA gene showed that all 14 positive samples belonged to assemblage E. Two distinct genotypes from 12 assemblage E isolates were observed including E1 (n = 10) and E2 (n = 2) by amplification and sequencing of the tpi gene.
Concerning the ages of cattle, C. andersoni was found in all age groups except the <3-month group, while C. parvum was only found in <3-month-old dairy cattle. Cryptosporidium bovis and C. ryanae were only detected in 3–11-month-old dairy cattle. With the exception of >24-month-old dairy cattle, G. duodenalis assemblage E was detected in all age groups, including nine in <3-month-old dairy cattle, three in 3–11-month-old dairy cattle, and two in 12–24-month-old dairy cattle (Table 3).
Table 3.
Infection rates and distribution of Cryptosporidium spp. and Giardia duodenalis among cattle of different ages and with or without diarrhea.
| Variable | Overall infection rate (%) (no. positive/no. of samples) | Infection rate (%) (n) and correlation |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
Cryptosporidium spp. |
G. duodenalis |
||||||||
| Infection rate (%) (n) | Cryptosporidium spp. | p-value | OR (95% CI) | Infection rate (%) (n) | G. duodenalis assemblages | p-value | OR (95% CI) | ||
| Age (month) | |||||||||
| <3 | 17.5 (21/120) | 10.0 (12) | C. parvum (12) | 0.011 | 1.00 | 7.5 (9) | E (9) | <0.001 | 1.00 |
| 3–11 | 6.8 (11/162) | 4.9 (8) | C. andersoni (3), C. bovis (4), C. ryanae (1) | 0.158 | 0.47 (0.18–1.18) | 1.8 (3) | E (3) | 0.033 | 0.23 (0.06–0.88) |
| 12–24 | 4.4 (24/546) | 4.0 (22) | C. andersoni (22) | 0.019 | 0.38 (0.18–0.79) | 0.4 (2) | E (2) | <0.001 | 0.05 (0.01–0.22) |
| >24 | 2.9 (17/586) | 2.9 (17) | C. andersoni (17) | 0.001 | 0.27 (0.12–0.58) | 0 | |||
| Symptoma | |||||||||
| With diarrhea | 27.6 (16/58) | 17.2 (10) | C. parvum (10) | 1.00 | 10.3 (6) | E (6) | 1.00 | ||
| Without diarrhea | 8.1 (5/62) | 3.2 (2) | C. parvum (2) | 0.014 | 0.16 (0.03–0.76) | 4.8 (3) | E (3) | 0.312 | 0.44 (0.10–1.85) |
Only samples from <3 month-old group are included here.
Phylogenetic analysis
From the Cryptosporidium spp. SSU rRNA locus, two nucleotide sequences of C. andersoni were generated and exhibited 0 and one single nucleotide polymorphism (SNP) compared to the reference sequence ;MK796098. The sequences belonging to C. parvum, C. bovis, and C. ryanae had 100% homology to the reference sequences ;MT002720, ;MH028031 and ;MK982468, respectively. Of the G. duodenalis tpi gene, E1 and E2 exhibited 100% homology to the reference sequences ;KY769101 and ;KY432851, respectively. To estimate phylogenetic inferences among the identified positive samples, NJ trees were inferred in MEGA X (Figs. 2 and 3).
Figure 2.
Phylogenetic tree depicting evolutionary relationships among Cryptosporidium spp. sequences at the SSU rRNA locus. The phylogenetic tree was inferred by a neighbor-joining analysis of genetic distances calculated by the Kimura 2-parameter mode. Percent bootstrap values greater than 50% from 1000 replicates are shown to the left of nodes. Species identified in this study are indicated by filled triangles.
Figure 3.
Phylogenetic tree depicting evolutionary relationships among Giardia duodenalis sequences at the tpi locus. The phylogenetic tree was inferred by a neighbor-joining analysis of genetic distances calculated by the Kimura 2-parameter mode. Percent bootstrap values greater than 50% from 1000 replicates are shown to the left of nodes. Species identified in this study are indicated by filled triangles.
Correlation analysis
The highest Cryptosporidium spp. infection rate (10.0%, n = 12) was observed in the <3-month-old group, followed by the 3–11-month-old group (4.9%, n = 8), the 12–24-month-old group (4.0%, n = 22), and the >24-month-old group (2.9%, n = 17). Compared with the <3-month-old group, an OR of 0.47 (95% CI: 0.18–1.18; p = 0.158), an OR of 0.38 (95% CI: 0.18–0.79; p = 0.019), and an OR of 0.27 (95% CI: 0.12–0.58; p = 0.001) were associated with the 3–11-month-old group (4.9%, n = 8), 12–24-month-old group, and >24-month-old group, respectively. The infection rates of G. duodenalis were 7.5% (n = 9), 1.8% (n = 3), and 0.4% (n = 2) in <3-month-old, 3–11-month-old, and 12–24-month-old animals, respectively. There was a significant negative correlation between the G. duodenalis infection rate and age in dairy cattle, as an OR of 0.23 (95% CI: 0.06–0.88; p = 0.033) was associated with the 3–11-month-old group, and 0.05 (95% CI: 0.01–0.22, p < 0.001) was associated with the 12–24-month-old group (Table 3).
In <3-month-old dairy cattle group, a lower prevalence of Cryptosporidium spp. was observed in calves without diarrhea (3.2%, 2/62) compared to calves with diarrhea (17.2%, 10/58), with an OR of 0.16 (95% CI: 0.03–0.76, p = 0.014) (Table 3). However, the infection rates of G. duodenalis in <3-month-old dairy cattle group with diarrhea (10.3%, 6/58) and without diarrhea (4.8%, 3/62) were not statistically significant (p = 0.312) (Table 3).
Discussion
In the present study, the overall infection rate of Cryptosporidium spp. was 4.2%. Similar or slightly lower infection rates have been reported in previous studies of Cryptosporidium spp. in dairy cattle in other provinces such as NXHAR (1.7%, 23/1366) [17], Qinghai (2.5%, 26/1027) [25], Shaanxi (2.6%, 32/1224) [42], and Gansu province (4.6%, 58/1257) [41]. The infection rate was lower than those in Shanghai (12.5%, 55/440) [7], Henan (13.0%, 276/2116) [28, 29], Anhui (14.9%, 52/350) [7], Heilongjiang (15.0%, 99/658) [20, 40], and Jiangsu (20.7%, 251/1215) [7]. In our research, the infection rate of G. duodenalis was 1.0%, which was consistent with the infection rates reported in NXHAR (2.1%, 29/1366) [17], and was lower than those reported in dairy cattle in Hebei and Tianjin (4.7%, 49/1040) [16], Heilongjiang (5.2%, 42/814) [21], and Jiangsu (20.6%, 281/1366) [33]. These data further confirmed that the infection rates of Cryptosporidium spp. and G. duodenalis in dairy cattle may vary according to geographical location. Moreover, water-borne transmission is an important route of Cryptosporidium spp. and G. duodenalis cross-species transmission [36], and Gansu has a unique geographical location and climatic conditions that may be the reasons for the lower infection rate in this study. Additionally, sampling time, number of samples, breeding methods, animal health status, and experimental methods also have a certain impact on the infection rate.
The dominant Cryptosporidium species in <3-month-old, 3–11-month-old, 12–24-month-old and > 24-month-old groups were C. parvum, C. bovis, C. andersoni and C. andersoni, respectively. Remarkably, the distribution of the four most common Cryptosporidium species is related to the age of dairy cattle. Cryptosporidium parvum is often detected in pre-weaned calves, which currently result in pre-weaned calf diarrhea. Cryptosporidium bovis and C. ryanae are commonly seen in post-weaned calves and yearlings, and C. bovis is more common than C. ryanae [26]. However, C. andersoni is more likely to infect adult dairy cattle, which normally leads to lower milk production and weight loss [8].
Of the 12 C. parvum isolates, 5 were identified as subtype IIdA19G1 (n = 4) or subtype IIdA15G1 (n = 1) based on sequencing analysis of the gp60 gene, and these 2 subtypes have also frequently been found in bovines [31]. In addition, subtypes IIdA19G1 and IIdA15G1 have also been detected in ruminants (sheep, goats, deer), alpacas, horses, rodents (mice, hamsters, squirrels), pigs, carnivores (dogs, gray wolves, raccoon dogs), and in wastewater around the world [3, 19]. Meanwhile, previous reports have shown that subtypes IIdA15G1 and IIdA19G1 can also cause human infections [13, 30]. Thus, despite the low positive rates of these two subtypes, the threat posed to public health cannot be neglected.
In this study, G. duodenalis assemblage E was the dominant assemblage, consistent with the results reported worldwide [23]. These results were in accordance with the previous definition of assemblage E as a bovine-specific assemblage. However, assemblage E has also been found in several human cases in Brazil and Australia [9, 39]. Therefore, more advanced epidemiological studies are required to assess the risk of assemblage E infecting humans. It is necessary to prevent and control G. duodenalis transmission for the protection of animals and breeders in the area, although the zoonotic risk of G. duodenalis was limited in this study.
In conclusion, the infection rates of Cryptosporidium spp. and G. duodenalis were relatively low in this area. Moreover, the study revealed the presence of C. parvum subtypes IIdA19G1 and IIdA15G1, and only G. duodenalis assemblage E was identified in dairy cattle in Gansu province, indicating that zoonotic transmission of Cryptosporidium spp. and G. duodenalis is possible but unlikely. Therefore, it is necessary to adopt effective strategies to prevent and control the transmission of Cryptosporidium spp. and G. duodenalis among dairy cattle and humans in Gansu province.
Conflict of interest
The authors declare that they have no conflicts of interest.
Acknowledgments
This study was supported in part by the National Natural Science Foundation of China (U1904203, 31672548) and the Leading talents of the Thousand Talents Program of Central China (19CZ0122). The sponsors played no role in study design, in the collection, analysis, or interpretation of the data, in writing the report, or in the decision to submit the article for publication. We thank LetPub (https://www.letpub.com) for linguistic assistance during the preparation of this manuscript.
Cite this article as: Wang Y, Cao J, Chang Y, Yu F, Zhang S, Wang R & Zhang L. 2020. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Gansu, northwest China. Parasite 27, 62.
References
- 1.Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F. 2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. Journal of Clinical Microbiology, 41(6), 2744–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appelbee AJ, Frederick LM, Heitman TL, Olson ME. 2003. Prevalence and genotyping of Giardia duodenalis from beef calves in Alberta, Canada. Veterinary Parasitology, 112(4), 289–294. [DOI] [PubMed] [Google Scholar]
- 3.Baroudi D, Zhang H, Amer S, Khelef D, Roellig DM, Wang Y, Feng Y, Xiao L. 2018. Divergent Cryptosporidium parvum subtype and Enterocytozoon bieneusi genotypes in dromedary camels in Algeria. Parasitology Research, 117(3), 905–910. [DOI] [PubMed] [Google Scholar]
- 4.Cacciò SM, Ryan U. 2008. Molecular epidemiology of giardiasis. Molecular & Biochemical Parasitology, 160(2), 75–80. [DOI] [PubMed] [Google Scholar]
- 5.Cacciò SM, Lalle M, Svärd SG. 2018. Host specificity in the Giardia duodenalis species complex. Infection Genetics and Evolution, 66, 335–345. [DOI] [PubMed] [Google Scholar]
- 6.Cai Y, Zhang N, Gong Q, Zhao Q, Zhang X. 2019. Prevalence of Cryptosporidium in dairy cattle in China during 2008–2018: A systematic review and meta-analysis. Microbial Pathogenesis, 132, 193–200. [DOI] [PubMed] [Google Scholar]
- 7.Chen F, Huang K. 2012. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle from farms in China. Journal of Veterinary Science, 13(1), 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteban E, Anderson BC. 1995. Cryptosporidium muris: prevalence, persistency, and detrimental effect on milk production in a drylot dairy. Journal of Dairy Sciences, 78(5), 1068–1072. [DOI] [PubMed] [Google Scholar]
- 9.Fantinatti M, Bello AR, Fernandes O, Da-Cruz AM. 2016. Identification of Giardia lamblia assemblage E in humans points to a new anthropozoonotic cycle. Journal of Infectious Diseases, 214(8), 1256–1259. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Xiao L. 2011. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clinical Microbiology Reviews, 24(1), 110–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Xiao L. 2017. Molecular epidemiology of cryptosporidiosis in China. Frontiers in Microbiology, 8, 1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-R JC, French N, Pita A, Velathanthiri N, Shrestha R, Hayman D. 2017. Local and global genetic diversity of protozoan parasites: spatial distribution of Cryptosporidium and Giardia genotypes. PLoS Neglected Tropical Diseases, 11(7), e0005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-R JC, Pita AB, Velathanthiri N, French NP, Hayman DTS. 2020. Species and genotypes causing human cryptosporidiosis in New Zealand. Parasitology Research, 119(7), 2317–2326. [DOI] [PubMed] [Google Scholar]
- 14.Gong C, Cao X, Deng L, Li W, Huang X, Lan J, Xiao Q, Zhong Z, Feng F, Zhang Y, Wang W, Guo P, Wu K, Peng G. 2017. Epidemiology of Cryptosporidium infection in cattle in China: a review. Parasite, 24, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holubová N, Tůmová L, Sak B, Hejzlarová A, Konečný R, McEvoy J, Kváč M. 2020. Description of Cryptosporidium ornithophilus n. sp. (Apicomplexa: Cryptosporidiidae) in farmed ostriches. Parasite & Vectors, 13(1), 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu S, Liu Z, Yan F, Zhang Z, Zhang G, Zhang L, Jian F, Zhang S, Ning C, Wang R. 2017. Zoonotic and host-adapted genotypes of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in dairy cattle in Hebei and Tianjin, China. Veterinary Parasitology, 248, 68–73. [DOI] [PubMed] [Google Scholar]
- 17.Huang J, Yue D, Qi M, Wang R, Zhao J, Li J, Shi K, Wang M, Zhang L. 2014. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Veterinary Research, 10, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F, Wang H, Zhang Z, Li J, Wang C, Zhao J, Hu S, Wang R, Zhang L, Wang M. 2016. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Beijing, China. Veterinary Parasitology, 219, 61–65. [DOI] [PubMed] [Google Scholar]
- 19.Li N, Wang R, Cai M, Jiang W, Feng Y, Xiao L. 2019. Outbreak of cryptosporidiosis due to Cryptosporidium parvum subtype IIdA19G1 in neonatal calves on a dairy farm in China. International Journal for Parasitology, 49(7), 569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu A, Wang R, Li Y, Zhang L, Shu J, Zhang W, Feng Y, Xiao L, Ling H. 2009. Prevalence and distribution of Cryptosporidium spp. in dairy cattle in Heilongjiang Province, China. Parasitology Research, 105(3), 797–802. [DOI] [PubMed] [Google Scholar]
- 21.Liu A, Zhang X, Zhang L, Wang R, Li X, Shu J, Zhang X, Shen Y, Zhang W, Ling H. 2012. Occurrence of bovine giardiasis and endemic genetic characterization of Giardia duodenalis isolates in Heilongjiang Province, in the Northeast of China. Parasitology Research, 111(2), 655–661. [DOI] [PubMed] [Google Scholar]
- 22.Ma J, Li P, Zhao X, Xu H, Wu W, Wang Y, Guo Y, Wang L, Feng Y, Xiao L. 2015. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Veterinary Parasitology, 207(3–4), 220–227. [DOI] [PubMed] [Google Scholar]
- 23.Onder Z, Simsek E, Duzlu O, Yetismis G, Ciloglu A, Okur M, Kokcu ND, Inci A, Yildirim A. 2020. Molecular prevalence and genotyping of Giardia duodenalis in cattle in Central Anatolia Region of Turkey. Parasitology Research, 119, 2927–2934. [DOI] [PubMed] [Google Scholar]
- 24.Qi MZ, Fang YQ, Wang XT, Zhang LX, Wang RJ, Du SZ, Guo YX, Jia YQ, Yao L, Liu QD, Zhao GH. 2015. Molecular characterization of Cryptosporidium spp. in pre-weaned calves in Shaanxi Province, north-western China. Journal of Medical Microbiology, 64(Pt 1), 111–116. [DOI] [PubMed] [Google Scholar]
- 25.Ren M, Wu F, Wang D, Li L, Chang J, Lin Q. 2019. Molecular typing of Cryptosporidium species identified in fecal samples of yaks (Bos grunniens) of Qinghai province, China. Journal of Parasitology, 105(2), 195–198. [PubMed] [Google Scholar]
- 26.Santín M, Trout JM, Fayer R. 2008. A longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Veterinary Parasitology, 155(1–2), 15–23. [DOI] [PubMed] [Google Scholar]
- 27.Trout J, Santín M. 2008. Molecular epidemiology, in Cryptosporidium and cryptosporidiosis, Fayer R, Xiao L, Editors. CRC Press and IWA Publishing: Boca Raton: p. 451–483. [Google Scholar]
- 28.Wang R, Ma G, Zhao J, Lu Q, Wang H, Zhang L, Jian F, Ning C, Xiao L. 2011. Cryptosporidium andersoni is the predominant species in post-weaned and adult dairy cattle in China. Parasitology International, 60(1), 1–4. [DOI] [PubMed] [Google Scholar]
- 29.Wang R, Wang H, Sun Y, Zhang L, Jian F, Qi M, Ning C, Xiao L. 2011. Characteristics of Cryptosporidium transmission in preweaned dairy cattle in Henan, China. Journal of Clinical Microbiology, 49(3), 1077–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Zhang H, Zhao X, Zhang L, Zhang G, Guo M, Liu L, Feng Y, Xiao L. 2013. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. Journal of Clinical Microbiology, 51(2), 557–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang R, Zhao G, Gong Y, Zhang L. 2017. Advances and perspectives on the epidemiology of bovine Cryptosporidium in China in the past 30 years. Frontiers in Microbiology, 8, 1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R, Li J, Chen Y, Zhang L, Xiao L. 2018. Widespread occurrence of Cryptosporidium infections in patients with HIV/AIDS: epidemiology, clinical feature, diagnosis, and therapy. Acta Tropica, 187, 257–263. [DOI] [PubMed] [Google Scholar]
- 33.Wang R, Li N, Jiang W, Guo Y, Wang X, Jin Y, Feng Y, Xiao L. 2019. Infection patterns, clinical significance, and genetic characteristics of Enterocytozoon bieneusi and Giardia duodenalis in dairy cattle in Jiangsu, China. Parasitology Research, 118(10), 3053–3060. [DOI] [PubMed] [Google Scholar]
- 34.Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Experimental Parasitology, 124(1), 80–89. [DOI] [PubMed] [Google Scholar]
- 35.Xiao L, Fayer R. 2008. Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. International Journal for Parasitology, 38(11), 1239–1255. [DOI] [PubMed] [Google Scholar]
- 36.Xiao L, Feng Y. 2017. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food and Waterborne Parasitology, 8–9, 14–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao L, Singh A, Limor J, Graczyk TK, Gradus S, Lal A. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Applied and Environmental Microbiology, 67(3), 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu F, Li D, Chang Y, Wu Y, Guo Z, Jia L, Xu J, Li J, Qi M, Wang R, Zhang L. 2019. Molecular characterization of three intestinal protozoans in hospitalized children with different disease backgrounds in Zhengzhou, central China. Parasites & Vectors, 12(1), 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zahedi A, Field D, Ryan U. 2017. Molecular typing of Giardia duodenalis in humans in Queensland – first report of Assemblage E. Parasitology, 144(9), 1154–1161. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W, Wang R, Yang F, Zhang L, Cao J, Zhang X, Ling H, Liu A, Shen Y. 2013. Distribution and genetic characterizations of Cryptosporidium spp. in pre-weaned dairy calves in northeastern China’s Heilongjiang province. PLoS One, 8(1), e54857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Tan Q, Zhou D, Ni X, Liu G, Yang Y, Zhu X. 2015. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle, northwest China. Parasitology Research, 114(7), 2781–2787. [DOI] [PubMed] [Google Scholar]
- 42.Zhao GH, Ren WX, Gao M, Bian QQ, Hu B, Cong MM, Lin Q, Wang RJ, Qi M, Qi MZ, Zhu XQ, Zhang LX. 2013. Genotyping Cryptosporidium andersoni in cattle in Shaanxi Province, Northwestern China. PLoS One, 8(4), e60112. [DOI] [PMC free article] [PubMed] [Google Scholar]