ABSTRACT
Lysosomes are compartments for the degradation of both endocytic and autophagic cargoes. The shape of lysosomes changes with cellular degradative demands; however, there is limited knowledge about the mechanisms or significance that underlies distinct lysosomal morphologies. Here, we found an extensive tubular autolysosomal network in Drosophila abdominal muscle remodeling during metamorphosis. The tubular network transiently appeared and exhibited the capacity to degrade autophagic cargoes. The tubular autolysosomal network was uniquely marked by the autophagic SNARE protein Syntaxin17 and its formation depended on both autophagic flux and degradative function, with the exception of the Atg12 and Atg8 ubiquitin-like conjugation systems. Among ATG-deficient mutants, the efficiency of lysosomal tubulation correlated with the phenotypic severity in muscle remodeling. The lumen of the tubular network was continuous and homogeneous across a broad region of the remodeling muscle. Altogether, we revealed that the dynamic expansion of a tubular autolysosomal network synchronizes the abundant degradative activity required for developmentally regulated muscle remodeling.
KEY WORDS: Muscle, Autolysosome, Drosophila, Metamorphosis, Atrophy, Syntaxin17
Summary: Autophagy-dependent formation of an extensive Syntaxin17-marked tubular network synchronizes degradative activity across a broad region of remodeling muscle during Drosophila development.
INTRODUCTION
Lysosomes are membrane-bound compartments for the degradation of both endocytic and autophagic cargoes in the eukaryotic cell. The lumen of lysosomes maintains an acidic pH to allow digestion of materials by a series of acid hydrolases (Lawrence and Zoncu, 2019). In addition to the catabolic function, lysosomes play numerous roles, such as secretion, nutrient sensing and signaling through mechanistic target of rapamycin complex I (mTORC1) and AMP-activated protein kinase (AMPK). Thus, the regulation of lysosomal function is critical for cellular homeostasis. The MiT/TFE family of transcription factors, including TFEB and TFE3 in mammals, are master regulators of the expression of a myriad of lysosomal and autophagic functions needed to meet changing degradative demands (Martina et al., 2014; Sardiello et al., 2009). However, very little is known about the mechanisms that mediate the modulation of lysosomal degradative capacity through coordinated changes in activity, quantity, distribution and morphology (Hipolito et al., 2018).
Although lysosomes are generally thought of as spherical organelles, lysosomal shape undergoes morphological changes in response to certain conditions. The existence of tubulated lysosomes, called tubular lysosomes or nematolysosomes, have been known since the 1970s in various cell types, including macrophages, pancreatic exocrine cells, neurons and muscle cells (Knapp and Swanson, 1990; Swanson et al., 1987; Okada et al., 1986; Robinson et al., 1986; Shi et al., 1992; Araki et al., 1993; Oliver, 1980). Tubulated lysosomes are prominent in lipopolysaccharide (LPS)-activated macrophages and dendritic cells (Hipolito et al., 2018). Recently, the tubular lysosomal network was described in the fly larval body wall muscle (Johnson et al., 2015) and the nematode epidermis during molting (Miao et al., 2020). In general, the extended tubular lysosomes exhibit features of typical functional lysosomes, including the accumulation of acid phosphatases, lysosomal proteases, Lamp and vacuolar H+-ATPase (V-ATPase). Microtubules may template the tubulation. In a model developed in mammals, plus-end-directed kinesin motors and minus-end-directed dynein–dynactin complexes extend lysosomal tubules in opposite directions via functions of the Arl8b–SKIP (also known as PLEKHM2) complex and Rab7–RILP or –FYCO1 complexes, respectively (Mrakovic et al., 2012). However, the stretching model alone cannot explain how the tubular lysosome becomes over 10 µm in length. To date, the mechanisms shaping tubular lysosomes are poorly understood, and the physiological significance of the tubulation remains enigmatic.
Autophagy is an intracellular bulk degradation system in which double-membrane-bound autophagosomes sequester and deliver cytosolic materials to the lysosomes or vacuoles for degradation. Autophagosome formation is mediated by at least 18 core autophagy-related (Atg) proteins acting within six functional units (Mizushima et al., 2011): (1) the ULK (Atg1) protein kinase complex; (2) the autophagy-specific phosphatidylinositol 3-kinase complex; (3) the phosphatidylinositol 3-phosphate-binding protein complex; (4) Atg9; (5) the LC3 (Atg8) conjugation system; and (6) the Atg12 conjugation system. All six units are pivotal for autophagy. However, the Atg8 and Atg12 ubiquitin-like conjugation systems seem to be dispensable for the elongation of the autophagic membrane in mammalian cells (Tsuboyama et al., 2016). The completed autophagosomes then fuse with lysosomes to form autolysosomes, the site for autophagic degradation and subsequent macromolecule efflux. The fusion is mediated by two soluble NSF attachment protein receptor (SNARE) complexes consisting of Qa–Qbc–R SNARES: Syntaxin17–SNAP29–VAMP8 and Syntaxin7–SNAP29–Ykt6 (Itakura et al., 2012; Takáts et al., 2013; Matsui et al., 2018; Takáts et al., 2018). It has been reported that autolysosomes tubulate during lysosome reformation from the autolysosome, a process called autophagic lysosome reformation (ALR) (Yu et al., 2010). The ALR tubule seems to be a kind of tubular lysosome; however, the tubule is neither acidic nor contains acidic hydrolases. Accordingly, it is thought of as a proto-lysosome (Yu et al., 2010).
Differentiated muscle cells, or myofibers, have highly organized and specialized organelles needed for muscle contraction. The contractile system is made up from sarcomeres arrayed into myofibrils. Sarcomere contractions are coordinated by changing levels of cytoplasmic calcium in response to a signal relay along the ‘excitation–contraction coupling’ system: transverse (T)-tubule invaginations of the plasma membrane in junctions with the sarcoplasmic reticulum. Mechanisms must remodel these organelles during ongoing muscle reorganization in response to muscle cell growth, use, damage, atrophy and aging. However, the mechanisms of muscle remodeling remain mostly unknown, in part due to challenges with observing the organellar dynamics within intact muscles. In Drosophila, a set of larval body wall muscles persist throughout metamorphosis as pupal abdominal muscles, called dorsal internal oblique muscles (DIOMs). In the DIOMs, the entire contractile and excitation–contraction coupling systems undergo a developmentally programmed remodeling during metamorphosis (Fujita et al., 2017), providing an excellent experimental model to study mechanisms of synchronous muscle atrophy and hypertrophy (Kuleesha et al., 2016). We recently reported that autophagy plays a critical role in DIOM remodeling (Fujita et al., 2017). Upon disassembly of myofiber organization in DIOMs, the cytoplasmic contents, including mitochondria, are enwrapped by autophagic membranes and delivered into lysosomes for degradation. In the process, not only autophagosome formation but also lysosomal functions must be regulated. However, little is known about the mechanisms that coordinate lysosomal function with cellular remodeling.
Here, we found an extensive, tubular autolysosomal (tAL) network in Drosophila muscles during metamorphosis. The induction of autophagy upon muscle remodeling was necessary for autolysosomal tubulation, which was uniquely marked by the autophagy-related SNARE Syntaxin17 (Stx17). The tubular network was continuous with a homogeneous lumen, and the tAL network extended over a wider range of the remodeling muscle cell than the spherical lysosomes found in the stable myofibers prior to remodeling. We show that the tubular autolysosomal network acts to synchronize activity and meet increased degradative demand during muscle remodeling.
RESULTS
Syntaxin17 marks a tubular network in remodeling muscle cells
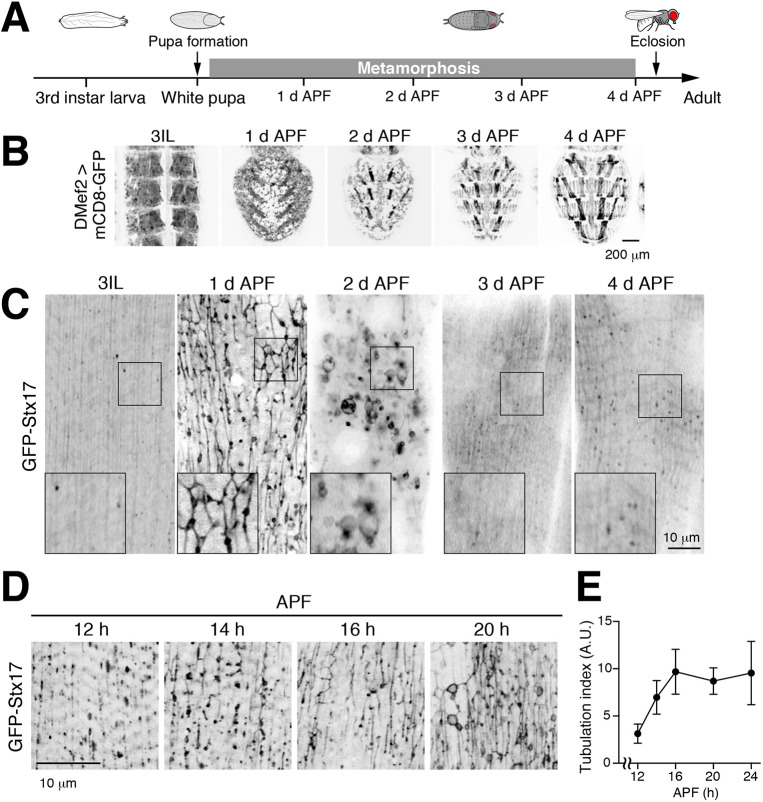
To gain insight into organelle dynamics upon muscle remodeling during metamorphosis (Fig. 1A,B), the localizations of GFP-tagged reporters for different cellular membrane compartments were observed in intact abdominal DIOMs every day after pupal formation (APF). At the third instar larval (3IL) stage, GFP–Stx17, a marker of autophagic membranes in differentiated muscle (Fujita et al., 2017), was detected, as previously described, as vesicular structures. Strikingly, upon metamorphosis we found that GFP–Stx17 appeared as a tubular network present in all DIOMs by 1 d APF (Fig. 1C). Thereafter, the extent of tubulation gradually decreased, and a GFP–Stx17 vesicular pattern was restored by 3 d APF (Fig. 1C). To observe how the network developed, we performed timecourse microscopy. GFP–Stx17 remained in puncta at 12 h APF, then became increasingly tubulated until 16–20 h APF (Fig. 1D,E), indicating a dynamic rearrangement of GFP–Stx17-labeled membranes with the onset of DIOM muscle remodeling (Fujita et al., 2017).
Fig. 1.
Syntaxin17 marks a tubular network in remodeling muscle cells. (A) Timeline of fly development from third instar larva to adult at 25°C (d APF, days after puparium formation). (B) Timecourse microscopy of mCD8–GFP in dorsal abdominal muscles during metamorphosis. (C,D) Timecourse microscopy of GFP–Stx17 in dorsal internal oblique muscle (DIOM) imaged through the cuticle of live wild-type animals from third instar larvae (3IL) to 4 d APF (C) and from 12 h to 20 h APF (D). Boxes in C indicate regions shown magnified as inset images. (E) Quantification of GFP–Stx17-positive tubules in DIOMs from 12 to 24 h APF. The values are the mean±s.d., n=7.
The Syntaxin17 tubular network has characteristics of autolysosomes
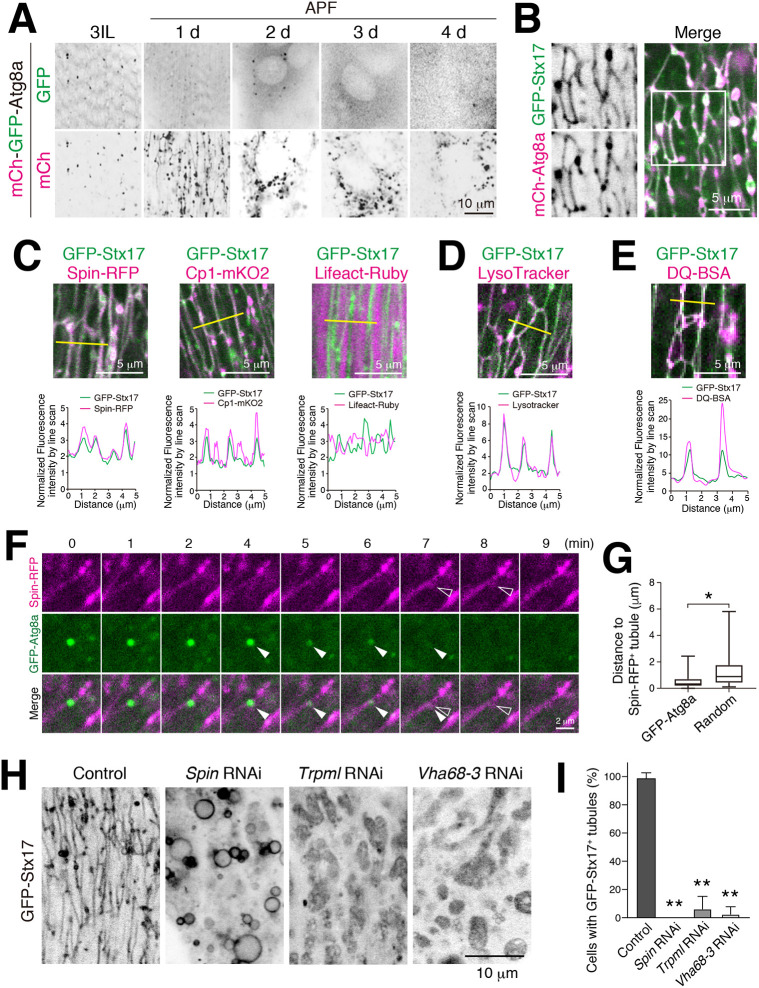
Stx17, a SNARE protein, localizes to the autophagosome and detaches just after fusion with the lysosome (Itakura et al., 2012; Takáts et al., 2013). We have reported that autophagy is robustly induced during DIOM remodeling by 1 d APF (Fujita et al., 2017). Therefore, we postulated that the tubular network was an autophagy-related structure. To test this possibility, we performed timecourse microscopy of autophagic flux over DIOM remodeling. Using an mCherry–GFP–Atg8a reporter, autophagosomes and autolysosomes can be distinguished by GFP-sensitivity to low pH (Kimura et al., 2007). Whereas the mCherry–GFP–Atg8a signal in the GFP channel did not exhibit any tubular structures, the mCherry was distributed to highly tubulated structures (Fig. 2A) that colocalized with GFP–Stx17 (Fig. 2B) in 1 d APF DIOMs. This result indicates that the Stx17-positive tubular network is an autolysosome-related organelle.
Fig. 2.
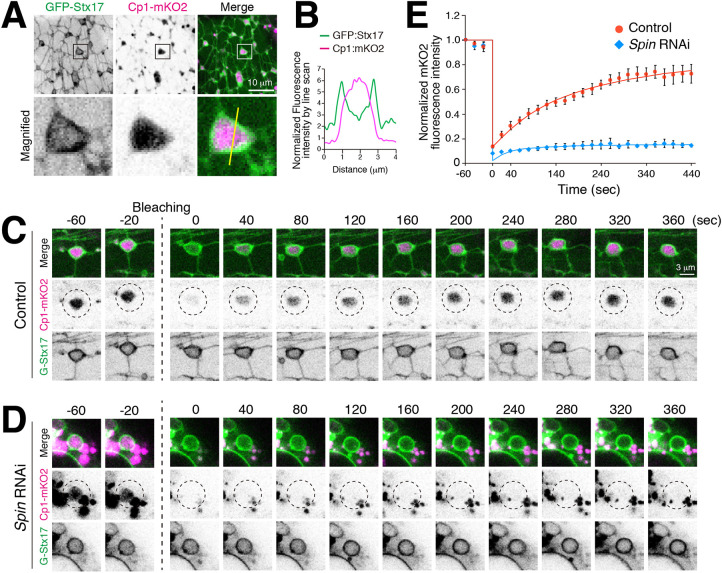
The Syntaxin17 tubular network has characteristics of autolysosomes. (A) Timecourse microscopy of mCherry–GFP–Atg8a in DIOM imaged through the cuticle of live animals from 3IL to 4 d APF. (B) Colocalization between GFP–Stx17 and mCherry–Atg8a in 20 h APF DIOM. Box in the merge image indicates region shown as separate fluorescence channels on the left. (C) Colocalization between GFP–Stx17 and Spin–RFP, Cp1–mKO2 or Lifeact–Ruby in 20 h APF DIOMs. Line plot profiles of the yellow line in each panel are shown. (D,E) Live pupae were injected with LysoTracker Red (D) or DQ Red–BSA (E), and DIOMs were imaged through the cuticle at 20 h APF. Line plot profiles of the yellow line in each panel are shown. (F) Time-lapse imaging of Spin–RFP and GFP–Atg8a in 20 h APF DIOMs. Disappearing GFP punctum, white arrowheads; shape change of tubular lysosome, open arrowheads. (G) Quantification of the distance between a Spin–RFP-positive tubule and the point at which GFP–Atg8a puncta disappeared (GFP:Atg8) or randomly drawn puncta (random). n>30. Boxes show the interquartile range, with the median indicated. Whiskers show the maximum and minimum values. *P<0.05 (Student's t-test). (H,I) GFP–Stx17 localization in DIOMs at 20 h APF from control, spin RNAi, Trpml RNAi or Vha68-3 RNAi-treated animals. (H) Representative images of GFP–Stx17 localization. (I) Mean+s.d. percentage of DIOMs with more than 5 µm GFP–Stx17-positive tubules, n=10. **P<0.001 (Dunnett's test).
We characterized the tubular compartment further. GFP–Stx17-positive tubular structures colocalized with two lysosomal proteins, Spinster (Spin), a lysosomal sugar transporter (visualized using a fusion to RFP, Spin–RFP), and the cathepsin L cysteine protease (visualized using a fusion to mKusabira-Orange2, Cp1–mKO2), but not with F-actin sarcomeres (Lifeact–Ruby) (Fig. 2C; Fig. S1A). Moreover, compartmental acidification and degradative activity – including throughout the tubules – was indicated by GFP–Stx17 colocalization with LysoTracker Red, a dye for acidic organelles, and dye-quenched (DQ) Red–BSA a fluorogenic substrate for proteases, respectively (Fig. 2D,E; Fig. S1B,C). The autolysosomal activity seen within the GFP–Stx17-positive tubules contrasts with the lack of activity described for tubules involved in ALR (Yu et al., 2010; Chen and Yu, 2013). To determine whether the autolysosomal tubules have a capacity to receive and degrade transported materials, we performed live imaging of autophagosomes (using GFP–Atg8a) and the autolysosome network (using Spin–RFP), respectively. We observed initially bright GFP–Atg8a puncta that, over a few minutes, quenched at the site of Spin–RFP-positive tubules (Fig. 2F; Movie 1). Concomitant with quenching of GFP–Atg8a, the shape of the Spin–RFP-positive tubule was transiently distended, indicating autophagosome fusion with the tubule. We noticed that the signal from GFP–Atg8a puncta disappeared near tubules in most cases. Collectively, the distances between GFP–Atg8a puncta to the nearest Spin–RFP tubule was significantly lower than that for randomly simulated puncta (Fig. 2G), suggesting that autophagosomes are delivered to and degraded in tubules of the tAL network.
Altogether, we identified a distinct and highly tubulated autolysosomal compartment that expands with muscle remodeling. This tubulated autolysosome is uniquely marked by Stx17 and exhibits degradative capacity throughout the tubular network. Hereafter, we refer to this structure as the tubular autolysosomal (tAL) network.
Formation of the tubular autolysosomal network requires lysosomal function, independent of mTOR activity
To test whether the autolysosomal degradative function is required for formation of the tubulated network, we examined the effects of knockdown for genes encoding proteins with lysosomal functions, namely spinster, Trpml and Vha68-3 (Dermaut et al., 2005; Wong et al., 2012; Mauvezin et al., 2015). Strikingly, knockdown of each disrupted the GFP–Stx17 tubular network (Fig. 2H,I), suggesting that lysosomal homeostasis and/or cargo degradation is critical for network tubulation. Because uptake of extracellular DQ Red–BSA must occur for eventual colocalization at autolysosomes (Fig. 2E), we postulated that formation of the tAL network may also depend on endocytic delivery to lysosomes (Guha et al., 2003). To test this possibility, we conditionally disrupted shibire, the sole fly ortholog of dynamin, which is involved in endocytic uptake for a significant portion of cell surface cargoes delivered to lysosomes (Kosaka and Ikeda, 1983). Flies with the temperature-sensitive mutation, shits1, were reared at permissive temperature (19°C) until 12 h APF, shifted to restrictive temperature (29°C) and then examined at 20 h APF (Fig. S1D,E). A block in shibire function scarcely affected Spin–RFP organization (Fig. S1E,F), suggesting that dynamin-dependent endocytosis does not substantially contribute to formation of the tAL network.
In the process of ALR, autolysosomal tubulation depends on reactivation of mTORC1 activity in response to efflux of autophagic degradation products (Yu et al., 2010). To test whether mTOR activity is involved in the formation of the tAL network, mTOR activity was forcibly inactivated or activated in DIOMs (Dibble and Cantley, 2015). Inactivation of mTOR by Tor or Rheb RNAi resulted in thinner DIOMs (Fig. S2A,B); however, the formation of the tAL network was largely unaffected at 20 h APF (Fig. S2C,D). In mammalian cells, forced activation of mTOR activity suppresses the loss of ALR tubulation due to spinster RNAi (Rong et al., 2011). Activation of mTOR by Tsc1 RNAi or Rheb overexpression led to thicker DIOMs (Fig. S2E,F); however, it was unable to suppress loss of the tAL network in spin RNAi conditions (Fig. S2G,H). Thus, our data suggest that mTOR activity is not essential for the formation of the tubulated autolysosome network in muscle.
Formation of the tubular autolysosomal network depends on autophagy but not on the Atg12 conjugation system
Next, we asked whether autophagy is required for formation of the tAL network. We tested the requirements for at least one ATG gene from each of the six functional protein units involved in autophagy (Mizushima et al., 2011), as well as genes required for the fusion between autophagosomes and lysosomes (Lőrincz and Juhász, 2019). RNAi of Atg1, FIP200 (also known as Atg17), Atg9, Atg18a, Vps34 (also known as Pi3k59f), Stx17, Snap29 or Vps39 each severely blocked formation of the tubular autolysosomal network (Fig. S3A–D). In contrast, Atg5, Atg7 or Atg12 RNAi showed only a minimal effect on the tubular network. We obtained similar results for the genes tested using either of the tAL compartment markers Spin–RFP (Fig. S3A,B) and mCherry–Stx17 (Fig. S3C,D). These results suggest the importance of autophagy, surprisingly not including the Atg12 conjugation system, for tAL network formation.
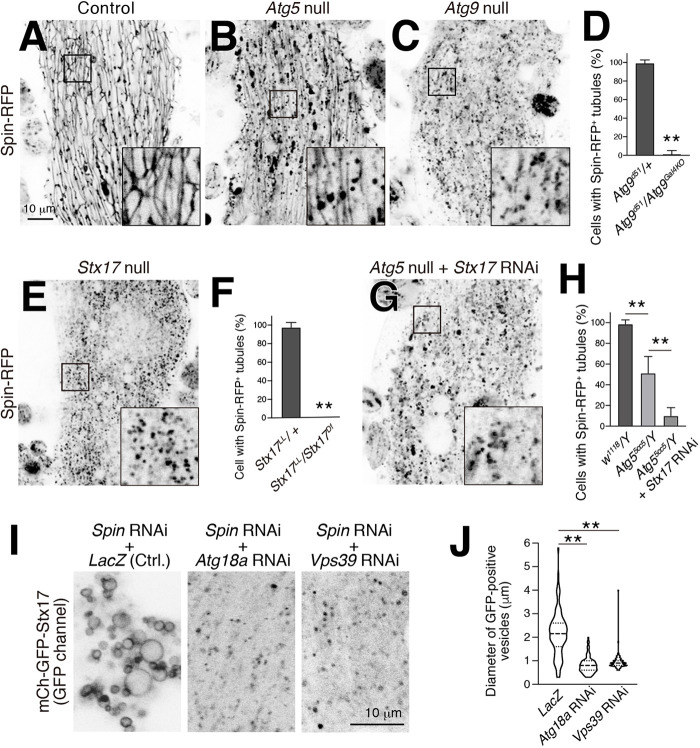
To verify that the categories of ATG gene results were not simply due to variability with hypomorphic RNAi conditions, we examined null mutants for Atg5, Atg9 and Stx17 (Kim et al., 2016; Wen et al., 2017; Takáts et al., 2013). Consistent with the RNAi results, loss of Atg9 or Stx17 functions fully blocked tAL formation, whereas Atg5-null mutants only partially reduced the extent of the tubulated network (Fig. 3A–F,H; Fig. S3E–G). The tubulated autolysosome still present in Atg5-null mutant DIOMs was dependent on Stx17 function (Fig. 3G,H). The size of the Stx17-positive vacuoles seen upon spinster RNAi was significantly reduced by the combined knockdown of autophagy functions encoded by Atg18a or Vps39 (Fig. 3I,J), suggesting that the Stx17 compartment size depends on membrane flux through autophagy. From these results, we conclude that autophagy – independent of the Atg12 conjugation system – is necessary for formation of the tAL network.
Fig. 3.
Formation of the tubular autolysosomal network depends on autophagy but not on the Atg12 conjugation system. (A–C) Spin–RFP-positive tubules in control (A), Atg5-null (B) or Atg9-null (C) DIOMs at 20 h APF. (D) Mean+s.d. percentage of DIOMs with Spin–RFP-positive tubules in control (Atg9d51/+) or Atg9-null mutant (Atg9d51/Atg9Gal4KO) animals, n=10. **P<0.001 (Student's t-test). (E) Spin–RFP-positive tubules in Stx17-null DIOMs at 20 h APF. (F) Mean+s.d. percentage of DIOMs at 20 h APF with Spin–RFP-positive tubules in control (Stx17LL/+) or Stx17-null (Stx17LL/Stx17Df) animals, n=10. **P<0.001 (Student's t-test). (G) Spin–RFP in Stx17-knockdown Atg5-null DIOMs at 20 h APF. (H) Mean+s.d. percentage of DIOMs at 20 h APF with Spin–RFP-positive tubules in control (w1118/Y) , Atg5-null (Atg55cc5/Y), or combination of Atg5-null and Stx17 RNAi-treated (Atg55cc5/Y+Stx17 RNAi) animals, n=10. **P<0.001 (Tukey's test). (I) mCherry–GFP–Stx17 fluorescence following co-RNAi of spin and Atg18a or Vps39 in 20 h APF DIOMs. LacZ, spin RNAi only control. (J) Violin plot of the diameter of mCherry–GFP–Stx17-positive vesicles in each genotype shown in I. n=100. Dashed line indicates the median. Dotted lines indicate the 25th and 75th quartiles. **P<0.001 (Dunnett's test) (J). Boxes in A–C, E and G indicate regions shown at higher magnification in the inset images. Scale bar in A applies to B,C,E and G.
A tubular lysosomal network has been reported in larval body wall muscles (Johnson et al., 2015). The authors reported that autophagy is not a prerequisite for the tubulation, because Atg7 RNAi did not affect it. Atg7 is an E1 enzyme for both the Atg12 and Atg8 ubiquitin-like conjugation systems (Juhász et al., 2007). As in larval muscles, Atg7 also was not required for formation of the tAL network in DIOMs (Fig. S3A,B). Thus, we predicted that the tubular lysosome in larval body wall muscles may also depend on core ATG genes, but not Atg5, Atg7 or Atg12. As reported, Spin–RFP-positive tubular lysosomes were observed close to the muscle cell surface in control and Atg7 RNAi larval body wall muscles (Fig. S3H). In contrast, the tubular network was disrupted by Atg1, Atg18a or Stx17 RNAi (Fig. S3H), indicating that autophagy is also essential for the tubular lysosomal network in larval body wall muscles.
Ultrastructural analysis indicates that autophagosomes are a membrane source for the tAL network
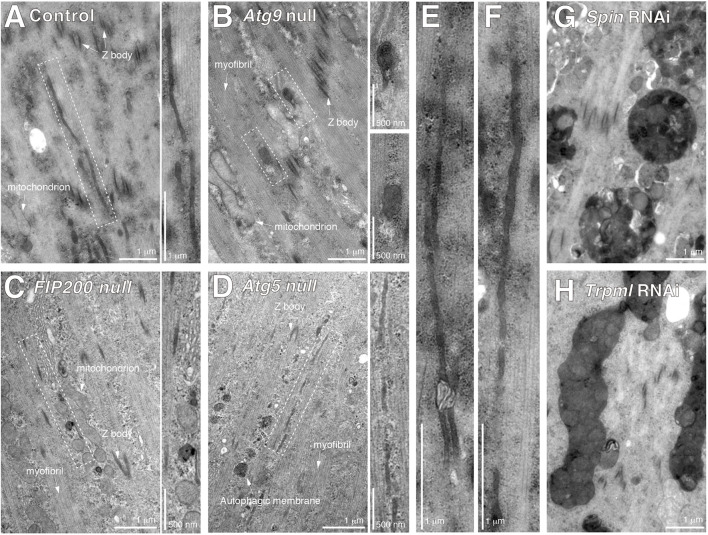
To analyze the ultrastructure of the tAL network, DIOMs were cut longitudinally and examined using transmission electron microscopy (TEM). Lysosomes and autolysosomes appear as electron-dense structures by TEM. Consistent with this, we observed electron-dense tubular structures in both control (Fig. 4A,E,F) and Atg5-null mutant DIOMs (Fig. 4D). The diameter of the tubules seen by TEM ranged between ∼50 and 100 nm. In contrast, mostly short or spherical electron-dense structures were observed in Atg9- or FIP200-null mutants (Kim et al., 2013) (Fig. 4B,C). On the other hand, large vacuolated structures accumulated upon spin or Trpml RNAi (Fig. 4G,H), consistent with the light microscopy results (Fig. 2H). The large vacuolated structures were single membraned and contained cytoplasmic organelles, such as mitochondria; therefore, they shared the features of autolysosomes. Autophagosome-like double-membrane vesicles were observed in Atg5-null DIOMs (Fig. S3I), as similarly reported in ATG3 knockout mammalian cells (Tsuboyama et al., 2016), indicating that Atg5 is not essential for elongation of the autophagic membrane in DIOMs at 1 d APF. Because autophagosome-like double-membrane vesicles and the tAL network were still formed in Atg5-mutant muscles, yet the tAL network depended on Stx17 (Fig. 3I,J), we conclude that autophagic membrane is a primary membrane source for tAL network formation in both wild-type and Atg5-null muscle cells.
Fig. 4.
Ultrastructural analysis supports that autophagosomal membrane is a membrane source for the tAL network. TEM images of 20 h APF DIOMs of control (A,E,F), Atg9-null (B), FIP200-null (C), Atg5-null (D), spin RNAi (G) or Trpml RNAi (H) animals. Typical examples of electron-dense membranous tubular structures in control DIOMs are shown in E and F. Dashed boxes in A–D indicate regions shown at higher magnification to the right.
Dynamics of tAL network formation
To gain insights into the mechanism of how spherical lysosomes are converted into the tubular network, we performed time-lapse imaging of Spin–RFP, a lysosome marker, in 14 h APF DIOMs when the tAL network begins to form (Fig. 1D). In control DIOMs around 14 h APF, Spin–RFP-positive spherical structures were observed to actively tubulate (Fig. 5A,B). Furthermore, we observed that two short tubules fused together to become a network (Fig. 5C). In sharp contrast to the control, the tubulation was not observed in the Stx17 RNAi condition (Fig. 5D). The Spin–RFP-positive spherical lysosomes in the control condition were larger than those in the Stx17 RNAi condition (Fig. 5A,B,D), suggesting that the non-tubulated structures were autolysosomes. Consistent with this notion, almost all of the Spin-positive non-tubulated structures were positive for an indicator of autophagic cargo, mCherry–Atg8a (Fig. S4A). Moreover, live imaging of Spin–RFP and GFP–Atg8a showed that the spherical autolysosomes tubulated without further fusion with GFP–Atg8a-positive autophagosomes (Fig. S4B). These data collectively suggest that autophagosome fusion is a prerequisite for the initial tubulation process in formation of the tAL network.
Fig. 5.
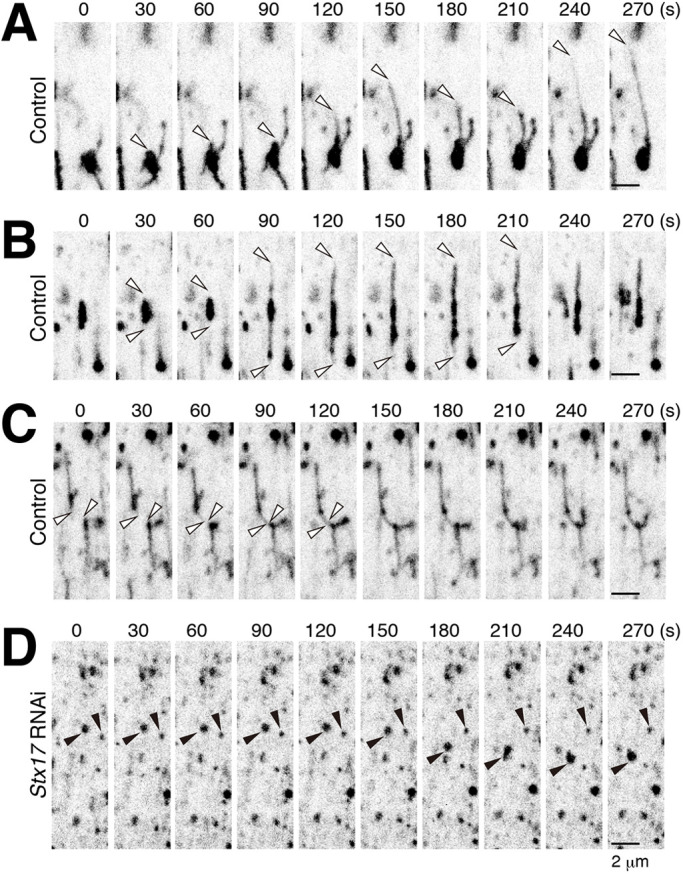
Dynamics of the tAL network formation. (A–C) Time-lapse imaging of Spin–RFP in control DIOMs around 14 h APF. White arrowheads indicate the tips of elongating tubules. (D) Time-lapse imaging of Spin–RFP in Stx17 RNAi DIOM around 14 h APF. Black arrowheads indicate Spin–RFP-positive puncta. Tubulation was not observed in the Stx17 RNAi condition.
The extent of the tubular network correlates with muscle remodeling ability
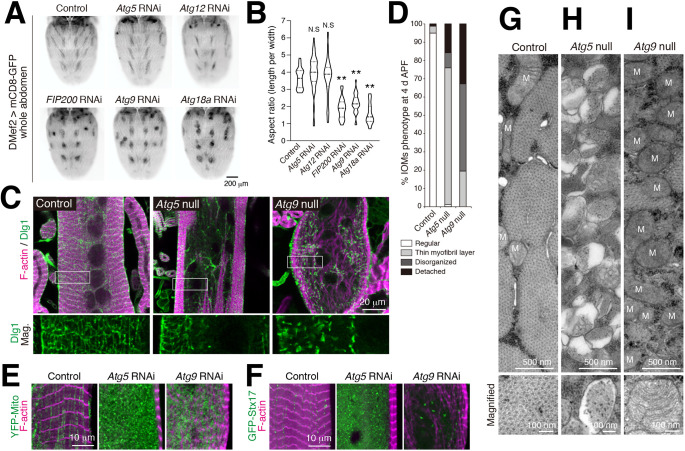
As shown above, there was a significant difference in the effects on efficiency of tAL network formation between disruption of genes core to autophagy versus disruption of genes in the Atg12 conjugation system (Figs 3,4). We next compared whether the same loss-of-function conditions also differentially impacted muscle remodeling. Knockdown of FIP200, Atg9 or Atg18a had a noticeable effect on DIOM shape at 4 d APF, after remodeling is completed. However, knockdown of Atg5 or Atg12 did not affect muscle shape (Fig. 6A,B), demonstrating again that the same two sets of ATG genes associated with distinct phenotypes during DIOM remodeling.
Fig. 6.
The extent of the tubular network correlates with muscle remodeling ability. (A) Effect of RNAi of ATG genes on the shape of DIOMs at 4 d APF. (B) Violin plot of the aspect ratio of DIOMs, as in A. n>50. N.S, not significant; **P<0.001 (Dunnett's test). Dashed line indicates the median. Dotted lines indicate the 25th and 75th quartiles. (C) T-tubule (Dlg1, green) and myofibril (F-actin, magenta) organization in DIOMs of control, Atg5-null or Atg9-null animals. Boxes indicate regions of the higher magnification Dlg1-staining images shown below. (D) Quantification of DIOM phenotypes in control, Atg5-null, or Atg9-null DIOMs, n>10. Regular, straight DIOM with organized myofibrils and T-tubules. Thin myofibril layer, straight DIOM with thin myofibrils. Disorganized, irregular-shaped DIOM with disorganized myofibrils and T-tubules. Detached, detached and rounded DIOM. (E) Mitochondria (YFP:Mito, green) and myofibrils (F-actin, magenta) in 4 d APF DIOMs of control, Atg5 or Atg9 RNAi-treated animals. (F) Autophagic membranes (GFP–Stx17, green) and myofibrils (F-actin, magenta) in 4 d APF DIOMs of control, Atg5 or Atg9 RNAi-treated animals. (G–I) TEM images of 4 d APF DIOM transverse-sections of control (G), Atg5-null (H) or Atg9-null (I) animals. M, mitochondria.
We characterized each of the two phenotypic subgroups further at the organelle level using Atg5 and Atg9 mutants. Following completion of remodeling, control DIOMs had well-organized myofibrils (as visualized by F-actin staining) and T-tubules (as visualized by Dlg1 staining) at 4 d APF. Although the shape was nearly normal for the Atg5-null DIOMs, they contained both an organized peripheral layer of myofibrils and T-tubules and a disorganized central region at 4 d APF (Fig. 6C,D). In contrast, Atg9-null animals had irregularly shaped DIOMs with more extensively disorganized myofibrils and fragmented Dlg1-positive structures throughout the cells (Fig. 6C,D). To assess the contractile function of 4 d APF DIOMs, we performed time-lapse imaging of GCaMP6S, a genetically encoded calcium indicator (Chen et al., 2013). GCaMP6S enables visualization of both the changes in cytosolic Ca2+ levels and the contraction of DIOMs (Fig. S5A). Compared to the control, Atg5 RNAi hardly affected the DIOM contraction upon the elevation of Ca2+ level; however, Atg18a RNAi severely disrupted DIOM contraction (Fig. S5B,C, Movies 2–4). These data show a clear correlation between the phenotypic severity and contractile function in DIOMs.
Mitochondria accumulated in both Atg5 and Atg9 RNAi conditions (Fig. 6E), suggesting a block in mitophagy for both conditions. GFP–Stx17-positive autophagic structures, however, only accumulated in Atg5 RNAi- but not in Atg9 RNAi-treated muscles (Fig. 6F). Furthermore, we performed TEM of transverse sections through remodeled DIOMs (Fig. S6A). Control DIOMs were filled with myofibrils and organized organelles recognizable based on their ultrastructure, such as mitochondria and T-tubules (Fig. 6G; Fig. S6B). The Atg5-null DIOMs instead were filled with thousands of autophagic membranes (Fig. 6H; Fig. S6C,E), likely representative of the numerous GFP–Stx17-positive vesicles also observed to accumulate (Fig. 6F). In contrast, Atg9-null DIOMs were filled with mitochondria but lacked any recognizable autophagic compartments (Fig. 6I; Fig. S6D), similar to phenotypes previously described for Atg1 or Atg18a RNAi (Fujita et al., 2017). Collectively, these data demonstrate that there are two distinct loss of ATG phenotypes in DIOM remodeling, with the efficiency of tAL network formation correlating with the phenotypic severity of muscle remodeling: only a slightly reduced tAL network correlated with partially organized muscle (Atg5 mutant), and a fully disrupted tAL network associated with more completely disorganized muscle (Atg9 mutant).
Furthermore, the muscle remodeling was also affected by spin or Trpml RNAi, which induced loss of the tAL network in 1 d APF DIOMs (Fig. 2H). Both autolysosomes and autophagosomes accumulated in spin or Trpml RNAi DIOMs at 4 d APF (Fig. S6F,G), suggesting the importance of the tAL network in the execution of autophagy during the muscle remodeling.
The tubulated lumen is continuous, allowing synchronization of autolysosomal capacity with muscle remodeling
What is the advantage of having autolysosomes organized into a tubular network versus having numerous isolated spherical vesicles? The interconnected tAL network could enable synchronous degradative activity across broad regions of the relatively large muscle cells. To investigate the continuity of the tAL network, we performed fluorescence recovery after photobleaching (FRAP) analysis of fly cathepsin L, Cp1. At 24 h APF, GFP–Stx17 marked large, rounded intersections between several tubule branches that were filled with Cp1–mKO2 (Fig. 7A,B). In control DIOMs, Cp1–mKO2 signal at the intersecting branchpoints recovered over several minutes after bleaching (Fig. 7C,E). In contrast, the signal of Cp1–mKO2 was not recovered in discontinuous vacuoles in spin RNAi DIOMs (Fig. 7D,E). These results show that the lumen of the tAL network is continuous and allows protein contents between tubules to intermix.
Fig. 7.
The tubulated autolysosome lumen is continuous and intermixed. (A) Colocalization of GFP–Stx17 and Cp1–mKO2 in 24 h APF DIOM. Boxes indicate the region shown in magnified images below. Line indicates the line section shown in B. (B) Line plot profiles of the yellow line in panel A. (C,D) FRAP analysis of Cp1–mKO2 in 24 h APF DIOMs of control (C) or spin RNAi (D) animals. Dashed circle indicates region of photobleaching. (E) Quantification of C and D. The mean±s.d. is shown, n=5.
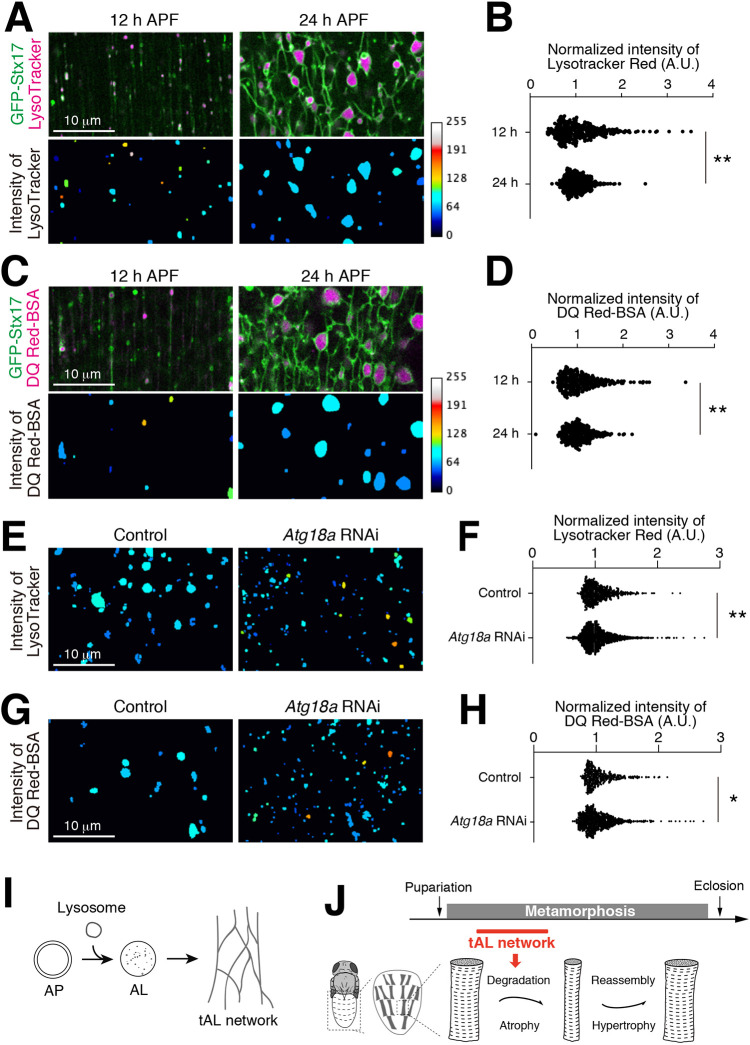
We next tested whether lysosomal activity is more homogeneous across a tAL network than that found amongst multiple individual lysosomes over a similar muscle area. LysoTracker Red or DQ Red–BSA was injected into pupae expressing GFP–Stx17 to stain acidified compartments. Both indicators stained small discontinuous vesicles in 12 h APF DIOMs (Fig. 8A,C) and the intersections of the tAL network in 24 h APF DIOMs (Fig. 8A,C). The acquired confocal images were binarized, objects extracted, and the mean intensities of each LysoTracker Red or DQ Red–BSA-positive object were measured. As predicted, the intensities of LysoTracker Red or DQ Red–BSA were more heterogeneous in discontinuous lysosomes at 12 h APF and more homogeneous in the tAL network at 24 h APF (Fig. 8B,D). In addition, we compared the heterogeneity of degradative compartments in control and Atg18a RNAi DIOMs at 24 h APF. Atg18a RNAi DIOMs lost the tAL network and had discontinuous punctate lysosomes (Fig. S3A–D). Strikingly, the intensities of LysoTracker Red or DQ Red–BSA were more heterogeneous in the Atg18a RNAi condition and more homogeneous in the control condition (Fig. 8E–H). Taken together, these results indicate that the tAL network synchronizes the degradative compartments.
Fig. 8.
The tubular autolysosomal network synchronizes the degradative compartments. (A) Micro-injected LysoTracker Red and GFP–Stx17 in 12 or 24 h APF DIOMs. The intensity maps (bottom) show representative images of the intensities of LysoTracker Red-positive objects. The median intensity in each image was set as 60. (B) The intensities of LysoTracker-positive objects, as described for A, are shown in a scatter plot, n>250. The median intensity for each image was set as one. **P<0.001 (F-test). (C) Micro-injected DQ Red–BSA and GFP–Stx17 in 12 or 24 h APF DIOMs. The intensity maps (bottom) show the intensities of DQ Red–BSA-positive objects. The median intensity in each image was set as 60. (D) The intensities of DQ Red–BSA-positive objects, as described for C, are shown in a scatter plot, n>250. The median intensity for each image was set as one. **P<0.001 (F-test). (E) Micro-injected LysoTracker Red in control or Atg18a RNAi DIOMs at 24 h APF. The intensity maps show representative images of the intensities of LysoTracker Red-positive objects. The median intensity in each image was set as 60. (F) The intensities of LysoTracker-positive objects, as described for E, are shown in a scatter plot, n>250. The median intensity for each image was set as one. **P<0.001 (F-test). (G) Micro-injected DQ Red–BSA and GFP–Stx17 in control or Atg18a RNAi DIOMs at 24 h APF. The intensity maps show the intensities of DQ Red–BSA-positive objects. The median intensity in each image was set as 60. (H) The intensities of DQ Red–BSA-positive objects, as described for G, are shown in a scatter plot, n>250. The median intensity for each image was set as one. *P<0.05 (F-test). (I) Both autophagy and lysosomal degradative activity are required for the tAL network formation. AP, autophagosome; AL, autolysosome. (J) The tAL network functions in the degradation of organelles during the remodeling of Drosophila abdominal muscles.
DISCUSSION
Tubular lysosomes have been known since over 40 years ago, but the mechanisms of how they are formed and their significance are mostly unknown. Here, we found expansion of a tubular autolysosomal network in the Drosophila abdominal muscles during metamorphosis. The tubular network appears transiently and is fully functional as a degradative organelle. The formation of the network depends on not only autophagy but also the degradation capacity of acidic organelles. FRAP analysis of cathepsin L revealed that the lumen of the tubular network is continuous and intermixed. As far as we know, the autophagy-dependent activity of a tubular lysosomal network has not been reported. Accordingly, we designate the structure as the tubular autolysosomal network, or tAL network.
In contrast to the already known tubular lysosomes, the tAL network is an autophagy-related organelle (Fig. 8I). First, we found that the tAL network is uniquely marked with Stx17, previously known as an autophagosomal SNARE (Fig. 1). However, the tAL network has characteristics of autolysosomes and not autophagosomes (Fig. 2). Furthermore, the formation of the tAL network depends on autophagy, and the loss of a series of ATG genes led to the accumulation of only small spherical lysosomes (Figs 3,4; Figs S3 and S7A). To our surprise, components of Atg12 and Atg8 systems were not essential for formation of the tAL network (Figs 3,4; Figs S3 and S7B).
How is the tAL network formed in absence of Atg5, a component of the Atg12 system? The formation of the tAL network in wild-type and Atg5-deficient muscles both depended on Stx17 (Fig. 3E–H). Furthermore, elongated autophagic membranes accumulated in the Atg5-null DIOMs (Figs S3I and S6E). We propose that the fusion between autophagic membranes and lysosomes results in the formation of the tAL network (Fig. S7B). We predict that the autophagic membrane but not the endocytic membrane is the dominant membrane source, because inhibition of the fusion of autophagosomes and lysosomes wholly prevented tAL network formation (Fig. 3; Fig. S3). Consistent with this notion, a block in dynamin-dependent endocytosis hardly affected the tAL network (Fig. S1D–F). The formation of a tAL network in Atg5-null DIOMs could be explained by diminished yet ongoing leaky autophagic flux. Because almost all autophagic flux assays in Drosophila utilize Atg8, lipidation of which depends on Atg5, we could not judge the autophagic flux in the Atg5-null mutant; however, the Atg5-null mutant might degrade autophagic cargoes to some extent. Our TEM analysis revealed the existence of elongated autophagic membrane in the Atg5-null DIOMs (Figs S3I and S6E). Even if the autophagosomal membranes are incompletely closed in the Atg5-null condition, which would block autophagy, membrane fusion with multiple lysosomes could result in autophagosomal membrane closure. It is critical to establish an Atg8-independent autophagic flux assay to test the hypothesis.
It has been reported that Drosophila larval muscles have a tubular lysosomal network (Johnson et al., 2015). Because Atg7 RNAi did not block the tubulation, it was thought that autophagy is dispensable for the tubular network formation. Through the analysis of the tAL network in pupal DIOMs, we found that Atg7 and other components of the two ubiquitin-like conjugation systems were dispensable for formation of the tAL network, the same as the tubular lysosomal network in larval body wall muscle (Figs 3,4). Furthermore, we revealed that the tubular lysosomal network in larval muscles also depended on Atg1, Atg18a and Stx17 (Fig. S3H), in agreement with our result in pupal muscle. Thus, it is reasonable to think that the tAL network and the tubular lysosome in larval muscles are closely related structures. Although we first discovered the tAL network in DIOMs during metamorphosis, we observed a similar structure to varying degrees in other muscles, such as the adult indirect flight muscles that form in the pupal thorax and dorsal longitudinal muscles in the abdomen.
The tAL network has characteristics different to those of ALR tubules, which are proto-lysosomes and do not have degradation capacities (Chen and Yu, 2013). The tAL exhibits acidification and degradative function throughout the tubular network, and both endocytosed DQ Red–BSA and autophagosomes were degraded in the structure (Fig. 2). Although the tAL network and the ALR tubules have these dissimilarities, both are autolysosome-related compartments. More work is needed to address whether the formation of the tAL network also depends on factors involved in ALR, such as clathrin, PI(4,5)P2, microtubules and Kinesin1 (Rong et al., 2012; Du et al., 2016).
To our surprise, Stx17 localized to the autolysosomal compartment in the remodeling muscle (Fig. 2). So far, it is known that Stx17 localizes to the autophagosome and detaches just after fusion with the lysosome in mammalian cells (Tsuboyama et al., 2016). Stx17 is a unique SNARE protein, which has a hairpin-like two α-helixes in its carboxy-terminus. Through the acidic α-helixes, Stx17 is specifically recruited to the autophagic membranes (Itakura et al., 2012; Takáts et al., 2013). Similar to Stx17, the ER morphogens, such as Reticulons and REEPs, have hairpin-like membrane-anchoring domains, which are inserted into and drive deformation of the ER membrane like wedges (Park and Blackstone, 2010). Therefore, it is possible to speculate that Stx17 induces the tubulation of membranes, in a manner similar to the ER morphogens. Further studies are required to reveal the mechanisms shaping positive membrane curvature and the highly branched morphology of the tAL network.
What is the advantage to forming a tubular autolysosomal network? Our data suggests that there are two merits of the tAL network over spherical autolysosomes. First is the expansion of the surface area. TEM data indicated that the diameter of the tubes is ranged between 50 and 100 nm (Fig. 4). If we compare the surface area to volume ratio of a 70-nm-diameter tube and a 500-nm-diameter spherical vesicle, the tube has an approximately five times higher value than that of the vesicle (Fig. S7C). The formation of a tAL network would increase the chance of docking and fusion of autophagosomes with the degradative compartments. Autophagy is massively induced with the onset of DIOM remodeling (Fujita et al., 2017), and autophagosomes fuse with and are degraded in the tAL network (Fig. 2F,G). Thus, the tAL network would allow handling of the extremely high autophagic flux in the relatively large, multinucleated muscle cells. We propose that there is positive feedback on the autophagic degradation in the remodeling DIOMs. The induction of autophagy to higher levels leads to tAL network formation, which is able to more efficiently fuse with and degrade more autophagosomes than spherical lysosomes can. Alternatively, the expanded surface by tubulation may be advantageous for the process of microautophagy, in which the lysosomes directly engulf cytosolic materials by membrane invagination (Oku and Sakai, 2018).
The second benefit to tubulated autolysosomes is the synchronization of the degradative compartments in the cell. FRAP analysis of Cp1–mKO2 revealed that the lumen of the tAL network is continuous and intermixed (Fig. 7C–E). During DIOM remodeling, organelles are disassembled in the early pupal stage and then reassembled in the late pupal stage (Fig. 8J). For the regulated muscle remodeling, all events, such as degradation and signaling from the degradative organelle, must be synchronized. Because muscles are massive cells, it is difficult to synchronize a number of spherical lysosomes over a wide cellular region. The formation of the tubular network would synchronize the degradative compartments in the cell (Fig. S7D). Consistent with this notion, we showed that the tAL network activity is more homogeneous than the activity of discontinuous, spherical lysosomes (Fig. 8A–H; Fig. S7D).
Both Atg5 and Atg9 seem to be pivotal for autophagy in DIOMs; however, the null mutants showed distinct phenotypes of both the tAL network and muscle remodeling (Fig. 6). In contrast to Atg9, Atg5 was required but not essential for the formation of the tAL network (Figs 3,4). Similarly, the Atg5-null mutation induced a milder muscle remodeling phenotype than the Atg9-null mutation (Fig. 6). Furthermore, we observed that the muscle remodeling was also severely affected by spin- or Trpml-RNAi treatments (Fig. S6F,G), which induced loss of the tAL network in 1 d APF DIOMs (Fig. 2H). These correlations suggest that the extent of the tAL network plays an important role in DIOM remodeling (Fig. 8J).
The existence of tubular lysosomes has been known in a variety of tissues; however, the mechanism and biological significance of the tubulation remain obscure and highly speculative. It is likely that mammalian muscles also have tubulated lysosomes (Robinson et al., 1986; Okada et al., 1986). Because the tubular lysosomes, including the tAL network in DIOMs, are fragile and highly sensitive to dissection and fixation processes, live imaging is an essential technique for the analysis of the structure. Because fly abdominal muscles are located close to the transparent overlying cuticle, we succeeded in observing the tubular autolysosome in live animals through the cuticle. Hence, Drosophila is an ideal model system to analyze the tubular lysosomal network and the dynamics of muscle remodeling. Our findings in this study provide new insights into the mechanisms of the morphogenesis of lysosomes as well as regulation of fundamental membrane trafficking pathways, such as autophagy and endocytosis. We predict that the expansion of surface area and synchronization are the keys to understanding the tubular lysosomes. We revealed the close relationship between the extent of tubular lysosomes and the phenotypic severity in muscle remodeling, and we discovered that formation of the tAL compartment is distinct from ALR and occurs via a mechanism that requires autophagy, lysosomal activity and stepwise tubulation and fusion into an extensive network. However, the specific factors that directly tubulate the membrane network remain unknown. Identification of such factors is the next crucial step and would answer the fundamental question of why lysosomes dynamically change shape in certain conditions.
MATERIALS AND METHODS
Reagents and antibodies
The following reagents were used: Alexa Fluor 546–Phalloidin conjugate (1.0 U/ml; Invitrogen), LysoTracker Red DND-99 (Thermo Fisher Scientific, Waltham, MA) and DQ Red–BSA (Thermo Fisher Scientific, Waltham, MA). The following antibodies were used: mouse anti-fly Dlg1 (1:200; clone 4F3; Developmental Studies Hybridoma Bank, Iowa City, IA) and anti-mouse IgG Alexa Fluor 488 conjugate (1:1000; Thermo Fisher Scientific, Waltham, MA).
Drosophila strains
Flies were reared at 25°C, unless stated otherwise. For muscle-targeted gene expression, DMef2-GAL4 driver was used. UAS-LacZ was used as a control in RNAi experiments. Genotypes in figures are described in Table S1. All genetic combinations were generated by standard crosses. Genotypes of flies used in this study include the following: (1) Atg55cc5/FM7 actin-GFP (from Jun H. Lee, University of Michigan, MI; Atg5 null); (2) Atg9Gal4KO/CyO-GFP (from Guang-Chao Chen, Academica Sinica, Taiwan; Atg9 null); (3) Atg9d51/CyO-GFP (from Guang-Chao Chen; Atg9 null); (4) w; FIP2003F5/TM6B-GFP (from Jun H. Lee; FIP200 null); (5) w; FIP2004G7/TM6B-GFP (from Jun H. Lee; FIP200 null); (6) w; UASp-mCherry:GFP:Atg8a (from Harald Stenmark, Oslo University Hospital, Norway); (7) y w; UAS-GFP:Atg8a (from Thomas Neufeld, University of Minnesota, MN); (8) y w; UAS- mCherry:Atg8a (from Thomas Neufeld); (9) w; UAS-Spinster:myc:RFP/CyO (From Graeme Davis, University of California San Francisco, CA); (10) w; UAS-IR-Atg1 (from Guang-Chao Chen); (11) w, shi[1] (Bloomington Drosophila Stock Center, BDSC 7968; shibire temperature-sensitive allele); (12) w; Df(3L)Exel8098/TM6B, Tb[1] (BDSC 7922; Stx17 deficiency); (13) y w; P{w[+mC]=GAL4-Mef2.R}3 (BDSC 27390); (14) w; P{w[+mC]=UAS-lacZ.B}melt[Bg4-1-2] (BDSC 1776); (15) y w; P{y[+t*] w[+mC]=UAS-Lifeact-Ruby}VIE-19A (BDSC 35545); (16) w; P{w[+mC]=UAS-mCD8::GFP.L}LL5, P{UAS-mCD8::GFP.L}2 (BDSC 5137); (17) w; P{w[+mC]=UAS-Rheb.Pa}3 (BDSC 9689); (18) y v; P{y[+t7.7] v[+t1.8]=TRiP.HMS02818}attP40 (TRiP, BDSC 44098; TRPML RNAi); (19) y v; P{y[+t7.7] v[+t1.8]=TRiP.HMS01611}attP2/TM3, Sb[1] (TRiP, BDSC 36918; FIP200 RNAi); (20) y v; P{y[+t7.7] v[+t1.8]=TRiP.HMS01246}attP2 (TRiP, BDSC 34901; Atg9 RNAi); (21) y v; P{y[+t7.7] v[+t1.8]=TRiP.HMS01358}attP2/TM3, Sb[1] (TRiP, BDSC 34369; Atg7 RNAi); (22) y v; P{y[+t7.7] v[+t1.8]=TRiP.HMS01153}attP2 (TRiP, BDSC 34675; Atg12 RNAi); (23) y v; P{y[+t7.7] v[+t1.8]=TRiP.GL00012}attP2 (TRiP, BDSC 35144; Tsc1 RNAi); (24) y v; P{y[+t7.7] v[+t1.8]=TRiP.HMS00923}attP2 (TRiP, BDSC 33966; Rheb RNAi); (25) y v; P{y[+t7.7] v[+t1.8]=TRiP.GL00156}attP2 (TRiP, BDSC 35578; Tor RNAi); (26) y v; P{y[+t7.7] v[+t1.8]=TRiP.JF01937}attP2 (TRiP, BDSC 25896; Stx17 RNAi); (27) y v; P{y[+t7.7] v[+t1.8]=TRiP.JF01883}attP2 (TRiP, BDSC 25862; SNAP29 RNAi); (28) y v; P{y[+t7.7] v[+t1.8]=TRiP.HMS02438} (TRiP, BDSC 42605; Vps39 RNAi); (29) Sco/CyO; P{UAS-Cp1.mKO2}3 (VDRC 309010); (30) P{Mef2-GAL4}3, P{UAS-Cp1.mKO2}3/TM6B, Tb[1] (VDRC 309005), (31) w; UAS-IR-Atg18KK100064 (VDRC 105366; Atg18a RNAi); (32) w; UAS-IR-Spinster (NIG-Fly 8428R-4; Spin RNAi); (33) w; UAS-IR-Vha68-3 (NIG-Fly 5075R-1; Vha68-3 RNAi); (34) w; UAS-IR-Vps34 (NIG-Fly 5373R-2; Vps34 RNAi); (35) w; UAS-IR-Atg5 (NIG-Fly 1643R-2; Atg5 RNAi); (36) w; UAS-mCD8:GFP; DMef2-GAL4, UAS-Dcr2; (37) w; DMef2-GAL4, UAS-Dcr2; (38) w; UAS-Dcr2; DMef2-GAL4, UAS-GFP:Stx174; and (39) w; UASt-GFP:Stx174 (Fujita et al., 2017). New genotypes generated during this study include the following: (40) w; DMef2-GAL4, UAS-GFP:Stx174; (41) w; UAS-Spin:myc:RFP/CyO; DMef2-GAL4, UAS-Dcr2/TM6C Sb Tb; (42) w; Atg9d51, UAS-Spin:myc:RFP/CyO; DMef2-GAL4, UAS-Dcr2/TM6C Sb Tb; (43) w; UAS-Spin:myc:RFP/CyO; DMef2-GAL4, Stx17LL06330/TM6C Sb Tb; (44) w; UAS-Spin:myc:RFP/CyO; DMef2-GAL4, UAS-IR-Stx17TRiP.JF01937/TM6B Hu Tb; (45) w; UAS-IR-Spinster NIG.8428R-4, UAS-mCherry:GFP:Stx17/CyO; DMef2-GAL4, UAS-Dcr2, tub-GAL80ts; (46) w; UAS-Dcr2; DMef2-GAL4, sqh-YFP:MitoBDSC7194/TM6C Sb Tb; (47) w; UAS-Dcr2/CyO; DMef2-GAL4, UAS-Cp1:mKO2, UAS-GFP:Stx17, (48) w; UAS-Dcr2; DMef2-GAL4, UAS-mCherry:Stx17/TM6C Sb Tb; (49) w; Sp/CyO; DMef2-GAL4, UAS-mCherry:Stx17/TM6C Sb Tb; (50) w; Atg9d51/CyO-GFP; DMef2-GAL4, UAS-mCherry:Stx17/TM6C Sb Tb; (51) w; Atg9d51/CyO-GFP; DMef2-GAL4, UAS-mCherry:Stx17/TM6C Sb Tb; and (51) w; UAS-Dcr2; DMef2-GAL4, UAS-GCaMP6S/TM6C Sb Tb. The control line w1118 was from Erina Kuranaga, Tohoku University, Sendai, Japan.
DNA engineering
Standard molecular biology techniques were used to construct plasmid vectors. For multiple-fragment in vitro assembly, pCM43b vector (gift of Barry Dickson, Janelia Research Campus, VA) was digested by EcoRI and NotI. mCherry and GFP–Stx17 were amplified using following primer sets; 5′-AGGGAATTGGGAATTCACCATGGTTTCAAAAGGTGAAG-3′ and 5′-GCTTCCTCCTCCTCCCTTGTACAGCTCGTCCATGCCGCC-3′ for mCherry, 5′-GGAGGAGGAGGAAGCATGGTGAGCAAGGGCGAG-3′ and 5′-TCCTCTAGTGCGGCCTCATTCTGGCTTCTCTTTTAGC-3′ for GFP–Stx17. The three DNA fragments were assembled using an In-fusion HD cloning kit (Takara, Kusatsu, Japan). The resulting DNA construct (pCM43b-mCherry–GFP–Stx17) was validated by sequencing and then injected into embryos for phiC31 insertion.
Muscle preparations and immunofluorescence in Drosophila
Muscle preparations in pupal abdomens were performed as previously described (Ribeiro et al., 2011). Staged pupae were removed from the pupal case and pinned on a sylgard-covered (Dow Corning, Midland, MI) Petri dish in dissection buffer (5 mM HEPES, pH 7.2, 128 mM NaCl, 2 mM KCl, 4 mM MgCl2 and 36 mM sucrose). Abdomens were opened with microscissors, pinned flat and fixed at room temperature for 20 min [4% formaldehyde, 50 mM EGTA, phosphate-buffered saline (PBS)]. Then, the samples were unpinned and blocked at room temperature for 30 min (0.3% bovine serum albumin, 2% goat serum, 0.6% Triton X100 in PBS), incubated with primary antibody overnight at 4°C, washed (0.1% Triton in PBS), then incubated for 2 h at room temperature with Alexa Fluor 488-conjugated secondary antibodies (Thermo Fisher Scientific, Waltham, MA) and counterstained with phalloidin for F-actin as needed. The stained samples were washed and mounted in FluorSave reagent (Merck Millipore, Burlington, MA).
Confocal fluorescence microscopy
For imaging of live pupal DIOMs, staged pupae were removed from the pupal case, mounted between slide-glass and cover-glass following a protocol described by Zitserman and Roegiers (2011), and imaged through the cuticle from the dorsal side. Live DIOMs were observed on a confocal microscope FV1000D (Olympus, Tokyo, Japan) with a 60× oil/1.35 NA UPlanSApo objective (Olympus, Tokyo, Japan) or FV3000 (Olympus, Tokyo, Japan) with a 60× silicone/1.30 NA UPlanSApo objective (Olympus, Tokyo, Japan). The image acquisition software used was Fluoview (Olympus, Tokyo, Japan). The exported images were adjusted and analyzed using ImageJ (NIH, Bethesda, MD).
Electron microscopy
Staged pupae (20 h or 4 d APF) were removed from pupal cases, pinned on a sylgard-covered Petri dish, dissected directly in fixative (2% paraformaldehyde, 2.5% glutaraldehyde, 150 mM sodium cacodylate, pH 7.4) and fixed for 2 h at room temperature and then overnight at 4°C. The dissected fillets were washed with 0.1 M phosphate buffer pH 7.4, post-fixed in 1% OsO4 buffered with 0.1 M phosphate buffer for 2 h, dehydrated in a graded series of ethanol and embedded flat in Epon 812 (TAAB, Aldermaston, UK). Ultrathin sections, the thickness of 70 nm, were collected on copper grids covered with Formvar (Nisshin EM, Tokyo, Japan), double-stained with uranyl acetate and lead citrate and then observed by a transmission electron microscope, H-7100 (Hitachi, Tokyo, Japan).
Time-lapse imaging using a laser scanning confocal microscope
Live imaging of Spin–RFP was performed using an FV3000 with a 60× silicone/1.30 NA UPlanSApo objective (Olympus, Tokyo, Japan). DIOMs at 14 h APF expressing Spin–RFP were imaged through the cuticle from the dorsal side. Live imaging of GCaMP6S was performed using an FV3000 with a 4× air/0.16 NA UPLXAPO objective (Olympus, Tokyo, Japan). Animals at 4 d APF expressing GCaMP6S in muscles were removed from the pupal case, mounted between slide-glasses and imaged from the dorsal side. The image acquisition software used was Fluoview (Olympus, Tokyo, Japan). The exported images were adjusted and analyzed using ImageJ.
Time-lapse imaging using a spinning-disk confocal microscope
DIOMs at 20 h APF expressing GFP–Atg8a and Spin–RFP were imaged every 60 s for 30 min using a 60× silicone/1.30 NA UPlanSApo objective (Olympus, Tokyo, Japan) on an inverted microscope (IX83; Olympus, Tokyo, Japan) with a spinning-disk confocal scanner unit (Dragonfly; Andor Technology, Belfast, UK) and a CMOS camera (Zyla 4.2; Andor Technology, Belfast, UK). Z-series images of each time point were exported using Fusion (Andor Technology, Belfast, UK). Then, the exported files were cropped, thresholded, and analyzed using ImageJ.
FRAP analysis
DIOMs at 24 h APF expressing both Cp1–mKO2 and GFP–Stx17 were imaged using a 60× silicone/1.30 NA UPlanSApo objective on a confocal microscope, FV3000 (Olympus, Tokyo, Japan). Three frames were acquired before photobleaching. Bleaching was performed on equally sized rectangular regions of interest (ROI) with a 568 nm laser at 30.7% power for 2 ms per pixel. Images of each channel were immediately acquired after the bleaching at a 20 s interval for 440 s. ROI mean intensities were measured using ImageJ. FRAP analysis to determine normalized fluorescence intensity was performed as described previously (Goodwin and Kenworthy, 2005).
Fly injection
Micro-injection was carried out using a stereomicroscope (SZX16; Olympus, Tokyo, Japan) with a manipulator (M-152; Narishige, Tokyo, Japan), an injector (IM-400; Narishige, Tokyo, Japan) and a compressor (0.2LE-8SB; Hitachi, Tokyo, Japan). LysoTracker Red was diluted in DMSO to 1 mM, and DQ Red–BSA was diluted in water to 10 mg/ml. The micro-injection needle was loaded with the solutions and then introduced into the abdomen of staged pupae. Although we could not control the exact amount of the solution for each injection, because of limitations imposed by our injection system, we roughly estimate the amount was 5–10 nl. Injected pupae were cultured at 25°C for 15 min for LysoTracker or 3 h for DQ Red–BSA, and observed on an FV3000 microscope.
Image analyses
Quantification of tubules was performed in two ways. The percentage of DIOMs containing at least one tubule >5 μm in length was manually counted. At least eight DIOMs were checked in each animal. More than ten animals were analyzed in each genotype (Figs 2I and 3A–J; Figs S2D and S3A–E). Alternatively, the ‘tubulation index’ was determined as the total tubule area (i.e. the sum of the area of all tubules) per unit area using ImageJ. Intensities were binarized and then skeletonized using LpxLineExtract, which is invoked by Lpx_Filter2d plugin in the LPixel ImageJ plugins package (Kuki et al., 2017). The >5 μm skeletonized lines in the images were extracted using the analyze particles function in Fiji ImageJ. More than ten cropped images were used for the quantification. The total tubule area per unit area was shown as an arbitrary value (Fig. 1E; Fig. S1D).
The distance between points at which GFP–Atg8a puncta disappeared or randomly simulated puncta and the nearest Spin–RFP-positive tubule was measured using the measure function in ImageJ (Fig. 2G). The Spin–RFP-positive tubules in the images were extracted, as mentioned above. As a simulation, we randomly drew puncta in images in which GFP–Atg8a-positive puncta disappeared, using ImageJ. More than 30 cases were used for the quantification.
Quantification of the diameter of vacuoles in Fig. 3L was performed as follows: the mCherry–GFP–Stx17-positive vacuole size was determined by a morphometric analysis using ImageJ. More than 100 vacuoles from at least ten DIOM images were analyzed for each genotype. Images of the GFP channel were used for the quantification.
To categorize DIOM remodeling phenotypes, 4 d APF animals were dissected and stained for Dlg1 and F-actin. More than ten animals were analyzed for each genotype. At least ten DIOMs per animal were visually observed and categorized into these four groups: (1) regular – a straight DIOM with organized myofibrils and T-tubules; (2) thin myofibril layer – a straight DIOM with thin myofibrils; (3) disorganized – irregular-shaped DIOM with disorganized myofibrils and T-tubules; and (4) detached – detached and rounded DIOM (Fig. 5D).
Quantification of the aspect ratio of DIOMs was performed as follows; 4 d APF animals expressing mCD8–GFP in muscle were observed using a fluorescence stereo microscope, SZX16 (Olympus, Tokyo, Japan) with a CMOS camera, ORCA-ER (Hamamatsu Photonics, Hamamatsu City, Japan). More than 50 DIOMs from at least five animals were analysed for each genotype. The aspect ratio, length per width, was determined by a morphometric analysis using ImageJ (Fig. 5B; Fig. S2B,F).
For the comparative analysis of LysoTracker Red and DQ Red–BSA intensities, the acquired raw images were binarized, extracted and measured for the average fluorescence intensity of objects using Fiji ImageJ. The median intensity was set as one in each image. More than 250 objects from at least five images were analyzed for each time point.
DIOM contraction was quantified as follows: time-lapse data of GCaMP6S in 4 d APF DIOMs were obtained using an FV3000. The contraction index was defined as the length of contracted DIOM divided by that of relaxed DIOMs. Four contractions of single DIOM were quantified using ImageJ and averaged. Five DIOMs from five animals were analyzed for each genotype.
Statistics
Each experiment was performed at least three times as biological and technical replicates (at least three different cohorts of unique flies were analyzed in repeat procedures performed on at least three different days). One exception was for TEM analyses, which were performed on two parallel replicates with multiple animals each. All replicate experiments were performed in parallel with wild-type controls. The s.d. was used as error bars for bar charts from the mean value of the data. When more than two genotypes or treatments were used in an experiment, the statistical analysis was performed using Tukey's test or Dunnett's test on Prism8 software. An unpaired two-tailed Student's t-test was used to compare two means. F-test was used to compare the dispersion between the two groups. P<0.05 was regarded as statistically significant. P<0.05 is indicated with single asterisks, and P<0.001 is indicated with double asterisks.
Supplementary Material
Acknowledgements
We are grateful to Guang-Chao Chen (Academia Sinica), Erina Kuranaga (Tohoku University), Jun H. Lee (University of Michigan), Graeme Davis (UCSF), Thomas Neufeld (University of Minnesota), Harald Stenmark (Oslo University Hospital), Bloomington Drosophila Stock Center, DGRC, VDRC, FlyTrap, Kyoto DGGR, and NIG fly for reagents. We are grateful to members of the Fujita lab and Fukuda lab for helpful comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: N.F.; Methodology: T.M., N.F.; Software: T.M., N.F.; Validation: T.M., N.F.; Formal analysis: T.M., N.F.; Investigation: T.M., Y.S., N.F.; Resources: T.M., A.A.K., M.F., N.F.; Data curation: N.F.; Writing - original draft: N.F.; Writing - review & editing: T.M., A.A.K., M.F., N.F.; Supervision: N.F.; Funding acquisition: A.A.K., M.F., N.F.
Funding
This work was supported in part by a Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT; grant number 18K06202 to N.F.), a Grant-in-Aid for Scientific Research (B) from MEXT (grant number 19H03220 to M.F.), a Grant-in-Aid for Scientific Research on Innovative Areas from MEXT (grant number 20H05315 to N.F.), Japan Science and Technology Agency (JST) PRESTO (grant number JPMJPR18H8 to N.F.), JST CREST (grant number JPMJCR17H4 to M.F.), a research grant from the Astellas Foundation for Research on Metabolic Disorders (N.F.), and the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award R01AR073840 (A.A.K.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.248336.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.248336.reviewer-comments.pdf
References
- Araki N., Tenkova T. I., Fujiwara T. and Takashima Y. (1993). The shape and distribution of lysosomes and endocytosis in the ciliary epithelial cells of rats. Cell Tissue Res. 274, 65-70. 10.1007/BF00327986 [DOI] [PubMed] [Google Scholar]
- Chen Y. and Yu L. (2013). Autophagic lysosome reformation. Exp. Cell Res. 319, 142-146. 10.1016/j.yexcr.2012.09.004 [DOI] [PubMed] [Google Scholar]
- Chen T. W., Wardill T. J., Sun Y., Pulver S. R., Renninger S. L., Baohan A., Schreiter E. R., Kerr R. A., Orger M. B., Jayaraman V. et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295-300. 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermaut B., Norga K. K., Kania A., Verstreken P., Pan H., Zhou Y., Callaerts P. and Bellen H. J. (2005). Aberrant lysosomal carbohydrate storage accompanies endocytic defects and neurodegeneration in Drosophila benchwarmer. J. Cell Biol. 170, 127-139. 10.1083/jcb.200412001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble C. C. and Cantley L. C. (2015). Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 25, 545-555. 10.1016/j.tcb.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Su Q. P., Chen Y., Zhu Y., Jiang D., Rong Y., Zhang S., Zhang Y., Ren H., Zhang C. et al. (2016). Kinesin 1 drives autolysosome tubulation. Dev. Cell 37, 326-336. 10.1016/j.devcel.2016.04.014 [DOI] [PubMed] [Google Scholar]
- Fujita N., Huang W., Lin T. H., Groulx J. F., Jean S., Nguyen J., Kuchitsu Y., Koyama-Honda I., Mizushima N., Fukuda M. et al. (2017). Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. Elife 6, 1-30. 10.7554/eLife.23367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin J. S. and Kenworthy A. K. (2005). Photobleaching approaches to investigate diffusional mobility and trafficking of Ras in living cells. Methods 37, 154-164. 10.1016/j.ymeth.2005.05.013 [DOI] [PubMed] [Google Scholar]
- Guha A., Sriram V., Krishnan K. S. and Mayor S. (2003). Shibire mutations reveal distinct dynamin-independent and -dependent endocytic pathways in primary cultures of Drosophila hemocytes. J. Cell Sci. 116, 3373-3386. 10.1242/jcs.00637 [DOI] [PubMed] [Google Scholar]
- Hipolito V. E. B., Ospina-Escobar E. and Botelho R. J. (2018). Lysosome remodelling and adaptation during phagocyte activation. Cell. Microbiol. 20, e12824 10.1111/cmi.12824 [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C. and Mizushima N. (2012). The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256-1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Johnson A. E., Shu H., Hauswirth A. G., Tong A. and Davis G. W. (2015). VCP-dependent muscle degeneration is linked to defects in a dynamic tubular lysosomal network in vivo. Elife 4, 1-20. 10.7554/eLife.07366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G., Érdi B., Sass M. and Neufeld T. P. (2007). Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 21, 3061-3066. 10.1101/gad.1600707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Park H. L., Park H. W., Ro S. H., Nam S. G., Reed J. M., Guan J. L. and Lee J. H. (2013). Drosophila Fip200 is an essential regulator of autophagy that attenuates both growth and aging. Autophagy 9, 1201-1213. 10.4161/auto.24811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Sandford E., Gatica D., Qiu Y., Liu X., Zheng Y., Schulman B. A., Xu J., Semple I., Ro S.-H. et al. (2016). Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife 5, 1-18. 10.7554/eLife.12245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T. and Yoshimori T. (2007). Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452-460. 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- Knapp P. E. and Swanson J. A. (1990). Plasticity of the tubular lysosomal compartment in macrophages. J. Cell Sci. 95, 433-439. [DOI] [PubMed] [Google Scholar]
- Kosaka T. and Ikeda K. (1983). Reversible blockage of membrane retrieval and endocytosis in the garland cell of the temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J. Cell Biol. 97, 499-507. 10.1083/jcb.97.2.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuki H., Higaki T., Yokoyama R., Kuroha T., Shinohara N., Hasezawa S. and Nishitani K. (2017). Quantitative confocal imaging method for analyzing cellulose dynamics during cell wall regeneration in Arabidopsis mesophyll protoplasts. Plant Direct 1, e00021 10.1002/pld3.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleesha Y., Puah W. C. and Wasser M. (2016). A model of muscle atrophy based on live microscopy of muscle remodelling in Drosophila metamorphosis. R. Soc. Open Sci. 3, 150517 10.1098/rsos.150517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence R. E. and Zoncu R. (2019). The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 21, 133-142. 10.1038/s41556-018-0244-7 [DOI] [PubMed] [Google Scholar]
- Lőrincz P. and Juhász G. (2019). Autophagosome-lysosome fusion. J. Mol. Biol. 432, 2462-2482. 10.1016/j.jmb.2019.10.028 [DOI] [PubMed] [Google Scholar]
- Martina J. A., Diab H. I., Lishu L., Jeong-A L., Patange S., Raben N. and Puertollano R. (2014). The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal 7, ra9 10.1126/scisignal.2004754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Jiang P., Nakano S., Sakamaki Y., Yamamoto H. and Mizushima N. (2018). Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J. Cell Biol. 217, 2633-2645. 10.1083/jcb.201712058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvezin C., Nagy P., Juhász G. and Neufeld T. P. (2015). Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 6, 7007 10.1038/ncomms8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao R., Li M., Zhang Q., Yang C. and Wang X. (2020). An ECM-to-nucleus signaling pathway activates lysosomes for C. elegans larval development. Dev. Cell 52, 21-37.e5. 10.1016/j.devcel.2019.10.020 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. and Ohsumi Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107-132. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Mrakovic A., Kay J. G., Furuya W., Brumell J. H. and Botelho R. J. (2012). Rab7 and Arl8 GTPases are necessary for lysosome tubulation in macrophages. Traffic 13, 1667-1679. 10.1111/tra.12003 [DOI] [PubMed] [Google Scholar]
- Okada T., Robinson J. M. and Karnovsky M. J. (1986). Cytochemical localization of acid phosphatase in striated muscle. Histochemistry 85, 177-183. 10.1007/BF00494801 [DOI] [PubMed] [Google Scholar]
- Oku M. and Sakai Y. (2018). Three distinct types of microautophagy based on membrane dynamics and molecular machineries. BioEssays 40, 1-6. 10.1002/bies.201800008 [DOI] [PubMed] [Google Scholar]
- Oliver C. (1980). Cytochemical localization fo acid phosphatase and trimetaphosphatase activities in exocrine acinar cells. J. Histochem. Cytochem 28, 78-81. 10.1177/28.1.6243325 [DOI] [PubMed] [Google Scholar]
- Park S. H. and Blackstone C. (2010). Further assembly required: construction and dynamics of the endoplasmic reticulum network. EMBO Rep. 11, 515-521. 10.1038/embor.2010.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro I., Yuan L., Tanentzapf G., Dowling J. J. and Kiger A. (2011). Phosphoinositide regulation of integrin trafficking required for muscle attachment and maintenance. PLoS Genet. 7, e1001295 10.1371/journal.pgen.1001295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. M., Okada R., Castellot J. J. and Karnovsky M. J. (1986). Unusual lysosomes in aortic smooth muscle cells: presence in living and rapidly frozen cells. J. Cell Biol. 102, 1615-1622. 10.1083/jcb.102.5.1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y., McPhee C., Denga S., Huanga L., Chen L., Liu M., Tracy K., Baehreck E. H., Yu L. and Lenardo M. J. (2011). Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc. Natl. Acad. Sci. USA 108, 7826-7831. 10.1073/pnas.1013800108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y., Liu M., Ma L., Du W., Zhang H., Tian Y., Cao Z., Li Y., Ren H., Zhang C. et al. (2012). Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat. Cell Biol. 14, 924-934. 10.1038/ncb2557 [DOI] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S. et al. (2009). A gene network regulating lysosomal biogenesis and function. Science 325, 473-477. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- Shi Y., Sakai M. and Ogawa K. (1992). Ultracytochemical study on nematolysosomes in neurons of the central nervous system. Act. Histochem. Cytochem 25, 437-445. 10.1267/ahc.25.437 [DOI] [Google Scholar]
- Swanson J., Burke E. and Silverstein S. C. (1987). Tubular lysosomes accompany stimulated pinocytosis in macrophages. J. Cell Biol. 104, 1217-1222. 10.1083/jcb.104.5.1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Nagy P., Varga Á., Pircs K., Kárpáti M., Varga K., Kovács A. L., Hegedus K. and Juhász G. (2013). Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201, 531-539. 10.1083/jcb.201211160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Glatz G., Szenci G., Boda A., Horváth G. V., Hegedűs K., Kovács A. L. and Juhász G. (2018). Non-canonical role of the SNARE protein Ykt6 in autophagosome-lysosome fusion. PLoS Genet. 14, e1007359 10.1371/journal.pgen.1007359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboyama K., Koyama-Honda I., Sakamaki Y., Koike M., Morishita H. and Mizushima N. (2016). The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354, 1036-1041. 10.1126/science.aaf6136 [DOI] [PubMed] [Google Scholar]
- Wen J. K., Wang Y. T., Chan C. C., Hsieh C. W., Liao H. M., Hung C. C. and Chen G. C. (2017). Atg9 antagonizes TOR signaling to regulate intestinal cell growth and epithelial homeostasis in Drosophila. Elife 6, e29338 10.7554/eLife.29338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. O., Li R., Montell C. and Venkatachalam K. (2012). Drosophila TRPML is required for TORC1 activation. Curr. Biol. 22, 1616-1621. 10.1016/j.cub.2012.06.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., McPhee C. K., Zheng L., Mardones G. A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F. et al. (2010). Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942-946. 10.1038/nature09076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitserman D. and Roegiers F. (2011). Live-cell imaging of sensory organ precursor cells in intact Drosophila pupae. J. Vis. Exp. 51, 2706 10.3791/2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.