Abstract
In this study, compound microbial inoculants, including three Bacillus strains and one Yeast strain, were inoculated into swine manure composting to explore the effects on antibiotic resistance genes (ARGs) and mobile genetic elements (MGEs), microbial community structure, and pathogenic bacteria. The results indicated that the abundances of the detected ARGs ranged from 3.6 × 103 to 1.13 × 108 copies/g. The ARGs with the highest abundance was sul2, and the lowest was blaCTX. Composting removes most of the ARGs and MGEs by 22.8–99.7%. These ARGs were significantly reduced during the thermophilic phase of compost. The removal rate of ARGs at the different layers of compost pile was different as follows: middle layer > upper layer > lower layer. But some ARGs proliferated significantly in the maturation phase of compost, especially the sulfonamide resistance genes. Compound microbial inoculants increased the temperature of compost, accelerated water loss, nitrogen fixation, and increased the removal rate of β-lactamase resistance genes, the transposon gene tn916 and part of tetracycline resistance genes by 3.7–23.8% in compost. Compound microbial inoculants changed the community structure and increased the Bacillus abundance in the thermophilic phase of compost. And it was helpful for removing pathogens during composting. The addition of compound microbial inoculants causes the decrease of Firmicutes and the increase of Bacteroidetes, which may be related to the removal and proliferation of ARGs.
Keywords: swine manure, aerobic composting, compound microbial inoculants, antibiotic resistance genes, 16S rRNA sequencing
Introduction
With the rapid development of intensive culture, antibiotics have been widely used in feed for promoting growth and for disease prevention purposes. However, antibiotics in animal tissues are absorbed incompletely; parts of antibiotics are excreted in urine and feces (Qiu et al., 2016). Antibiotic residues cause serious pollution to the environment, pose a threat to human health, and even have toxic side effects (Sarmah et al., 2006). If the residual antibiotics in the environment do not degrade effectively, these residues may help to maintain or develop antibiotic-resistant microbial populations (Witte, 1998). Additionally, the migration of antibiotics in the environment has led to the spread of resistant genes as a new type of environmental pollutant. Manure carrying antibiotic resistance genes (ARGs) can enter soil, following direct land application (Fang et al., 2015). Many studies have shown that the diversity and abundance of ARGs in soil with long-term application of livestock manure has increased significantly (Ji et al., 2012; Chen et al., 2016), and ARGs in farmland have also increased significantly (Wang et al., 2014; Mu et al., 2015). Therefore, the removal of ARGs from swine manure has emerged as an environmental issue before its application to soil.
Aerobic composting is one of the most effective ways to realize the harmless and resourceful treatment of livestock and poultry manure. Aerobic composting can significantly reduce the concentration of antibiotics in animal manure (Ray et al., 2017), but there is little research on whether ARGs can undergo compost removal. Previous studies show that aerobic composting has various effects on different ARGs (Joy et al., 2014; Su et al., 2015). It has been found that the relative abundances of the tetracycline resistance genes and sulfonamide resistance genes were significantly reduced during the swine manure composting, while quinolone resistance gene still partially remained (Selvam et al., 2012). Thus, composting is most likely an effective way to remove ARGs.
Microbes are the main carriers of ARGs. Several studies have used 16S rRNA sequencing technology to analyze the relationship between ARGs and microbial communities in compost (Jia et al., 2015; Zhang et al., 2016). A large number of studies have shown that exogenous microbial agents can accelerate the composting process and shorten the composting cycle (Nakasaki et al., 2013; Ravindran et al., 2019). However, the current research focuses more on the observation of the application effect of a single functional strain (Zeng et al., 2010). And the impact of inoculation on ARGs is even less studied. Only Duan et al. (2019) studied the effects of Bacillus subtilis on ARGs and mobile genetic elements (MGEs) during cattle manure composting. But in practical application, the addition of compound microbial agents is often used to accelerate the composting process. Previous study also showed that compound microbial inoculants can improve composting efficiency of organic solid wastes (Ravindran et al., 2019). Our previous studies have found that Bacillus is the main dominant bacterium in the high-temperature period of primitive composting (Yi et al., 2012). Exogenous Bacillus spp. may prolong the high-temperature composting time and better kill resistant microorganisms. Thus, in the present study, we delved into the effects of Bacillus compound microbial inoculants on composting microbial communities and ARGs. At present, research on these effects has not been reported.
The main aims of the present study were: (1) to investigate the quantitative variation and spatial distribution of ARGs in different periods and different height layers in compost, (2) to use 16S rRNA gene sequencing technology to investigate the dynamic changes of bacterial community structure and composition in compost with the addition of a compound microbial inoculants, and (3) to analyze the correlation between microbial community changes and ARGs. We also detected the relative abundances of pathogenic bacteria during composting. The results of this study may help with the development of high-efficiency microbial agents for the control of ARGs, which is of great significance to guide the application of microbial agents in composting and to further improve composting efficiency.
Materials and Methods
Description of Raw Materials
Fresh swine manure was taken from the boutique pig farm of Huazhong Agricultural University. Sawdust from the Veterinary Hospital of Huazhong Agricultural University was approximately 0.5–1 cm in size. The total carbon content of the swine manure was 360.8 g/kg, the total nitrogen content was 30.4 g/kg, and the water content was 69.25%. The total carbon content in sawdust was 471.9 g/kg, the total nitrogen content was 2.2 g/kg, and the water content was 9.38%.
Composting Process
This composting experiment was conducted in November 2018 at the National Animal Engineering Technology Research Center of Huazhong Agricultural University. Each composting device was a 60-L rectangular foam container (280 × 540 × 400 cm). And it was perforated with one ventilation hole (2 × 2 cm) on the side of the insulation wall. The composting device is very similar to the research from Duan et al. (2019). Two composting experiments were labeled as CK (swine manure + sawdust) and AB (swine manure + sawdust + 1% compound microbial agents − Bacillus subtilis: Bacillus licheniformis: Bacillus megaterium: Yeasts = 1:1:1:2). The initial volume of the compost material is about 50 L, and the fresh weight is about 17 kg. Accordingly, B. subtilis have been deposited in China Center for Type Culture Collection (CCTCC) and the CCTCC NO: M2019185. These strains were isolated from previous swine manure composting samples. The C/N of the two experimental materials was adjusted to ⁓20, and the water content was ⁓60%. The composting device was connected to the blower, the blast volume was 5 L/kg∙min, and the ventilation was 5 min per hour.
Compost Sample Collection
Composting samples were collected individually from the upper, central, and lower portions of the compost to achieve high representativeness of samples in different height layers. Samples collected on days 0 (initial phase), 1 (mesophilic phase), 2 (thermophilic phase), 5 (cooling phase), 12 (maturation phase), and 32 (maturation phase) were designated as detection ARGs samples. At the same time, samples collected on days 1 (mesophilic phase), 2 (thermophilic phase), and 32 (maturation phase) were used to analyze changes in microbial community structure. Each sample was split into two parts: one part was stored at −80°C for subsequent DNA extraction, and another part was stored at 4°C for later chemical analysis.
Physicochemical Index Analyses
The temperature at the center of each composting container was measured at 9:00 AM daily using a thermometer. At the same time, the room temperature was also recorded daily. Each compost sample and deionized water were mixed at a ratio of 1:10 W(g): V(ml) and shaken at room temperature for 1 h. The pH of the extract was measured. Moisture content was measured by drying in an oven at 105°C for 24 h or until no change in dry weight. Analysis of the total nitrogen (TN), total carbon (TC), and C/N ratios of the samples was performed by using an elemental analyzer (Vario MAX-CN Germany Elementar).
DNA Extraction and qPCR
Total genomic DNA was extracted using the Fecal DNA Kit (Simgen, Hangzhou, China) according to the manufacturer’s instructions. Nine tetracycline resistance genes (tetA, tetC, tetG, tetH, tetL, tetM, tetO, tetQ, and tetW), five macrolide resistance genes (ermA, ermB, ermC, ermF, and ermQ), two sulfonamide resistance genes (sul1 and sul2), two β-lactamase resistance genes (blaCTX and blaTEM), the integrase gene intI1, and the transposon gene tn916 were analyzed by qPCR (Table 1). The qPCR mixtures (total volume, 10 μl) consisted of 5 μl of Hieff qPCR SYBR Green Master Mix (Yeasen, China), 1 μl of template DNA, 0.2 μl of each primer (10 μM), and 3.6 μl of ddH2O. qPCR was performed using a LightCycler®96 system (Roche, Switzerland). Each reaction is repeated three times. The qPCR conditions comprised 5 min at 95°C, followed by 40 cycles for 30 s at 95°C, 30 s at the annealing temperatures shown in Table 1, and then 30 s at 72°C. The calibration curve of qPCR was generated using plasmids that carried the target genes. The copy number of ARGs was calculated by the square of the related coefficient (R2) of the standard curve >0.99 using the external reference method.
Table 1.
PCR primer sequences and annealing temperature.
| Gene | Forward (5'-3') | Reverse (5'-3') | AT(°C) |
|---|---|---|---|
| 16SrRNA | CCTACGGGAGGCAGCAG | ATTACCGCGGCTGCTGG | 55 |
| tetA | GCTACTCCTGCTTGCCTTC | CATAGATCGCCGTGAAGAGG | 57 |
| tetC | GCGGGATATCGTCCATTCCG | GCGTAGAGGATCCACAGGACG | 60 |
| tetG | GCAGAGCAGGTCGCTGG | CCTGCAAGAGAAGCCAGAAG | 57 |
| tetH | CAACCCATTACGGTGTGCTA | AAGTGTGGTTGAGAATGCCA | 57 |
| tetL | TCGTTAGCGTGCTGTCATTC | GTATCCCACCAATGTAGCCG | 55 |
| tetM | ACAGAAAGCTTATTATATAAC | TGGCGTGTCTATGATGTTCAC | 55 |
| tetO | GATGGCATACAGGCACAGACC | GCCCAACCTTTTGCTTCACTA | 60 |
| tetQ | AGAATCTGCTGTTTGCCAGTG | CGGAGTGTCAATGATATTGCA | 60 |
| tetW | GAGAGCCTGCTATATGCCAGC | GGGCGTATCCACAATGTTAAC | 60 |
| ermA | AAGCGGTAAACCCCTCTGA | TTCGCAAATCCCTTCTCAAC | 57 |
| ermB | AAAACTTACCCGCCATACCA | TTTGGCGTGTTTCATTGCTT | 57 |
| ermC | GAAATCGGCTCAGGAAAAGG | TAGCAAACCCGTATTCCACG | 60 |
| ermF | TCGTTTTACGGGTCAGCACTT | CAACCAAAGCTGTGTCGTTT | 57 |
| ermQ | CACCAACTGATATGTGGCTAG | CTAGGCATGGGATGGAAGTC | 60 |
| sul1 | CACCGGAAACATCGCTGCA | AAGTTCCGCCGCAAGGCT | 55 |
| sul2 | GCGCTCAAGGCAGATGGCATT | GCGTTTGATACCGGCACCCGT | 60 |
| blaCTX | CTATGGCACCACCAACGATA | ACGGCTTTCTGCCTTAGGTT | 60 |
| blaTEM | TCGGGGAAATGTGCG | GGAATAAGGGCGACA | 60 |
| qnrA | ATTTCTCACGCCAGGATTTG | GCAGATCGGCATAGCTGAAG | 55 |
| copB | GGTTGGTCAACAGGATGTCGTACT | TTCCTGCTCGACCAGTTGGAATAC | 57 |
| pcoA | GCTGCAGATGGCCAGTATGTAAA | CCCTCGAGCGTAACCGGTCC | 57 |
| pcoD | ATCAGCAGGCAGGACAATAC | CTGATGTGGGTATTAGCTGGATT | 55 |
| czrC | TAGCCACGATCATAGTCATG | ATCCTTGTTTTCCTTAGTGACTT | 60 |
| czcA | TCGACGGTGCCGTGGTGATCGTCGAGAA | GTCATCGCCATCGGACGGAACA | 60 |
| tcrB | CATCACGGTAGCTTTAAGGAGATTTTC | ATAGAGGACTCCGCCACCATTG | 55 |
| intI1 | GGCTTCGTGATGCCTGCTT | CATTCCTGGCCGTGGTTCT | 60 |
| tn916 | TCCTACAGCGACAGCCAGTGA | TGCGTTGCTTTGGTCTGCTGGT | 55 |
16S rRNA Gene High-Throughput Sequencing
Six sets of samples collected from two groups, named CK_D1, AB_D1, CK_D2, AB_D2, CK_D32, and AB_D32, were used to identify the microbial communities by performing 16S rRNA gene-based high-throughput sequencing. D1 represents the heating period, D2 represents the high-temperature period, D32 represents the ripening period, and there were three replicates of each group of samples for a total of 18 samples. The V3–V4 hypervariable regions of the bacterial 16S rRNA gene were amplified with primers 338F (5'-ACTCCTACGGGAGGCAGCAG-3') and 806R (5'-GGACTACHVGGGTWTCTAAT-3') by a PCR thermocycler system (GeneAmp 9700, ABI, United States). The high-throughput sequencing method is consistent with Li et al. (2020). Purified amplicons were pooled in equimolar amounts and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, United States) according to standard protocols by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
Statistical Analysis
GraphPad Prism 7 was used to generate various figures. Averages and SDs of ARGs were determined using Microsoft Excel 2010. The 16S rRNA gene sequencing data were analyzed on the free online I-Sanger Cloud Platform of Majorbio. In addition, we compared the resulting sequences with the constructed Human Pathogenic Bacteria (HPB) database. The HPB database was provided by Shanghai Majorbio Bio-pharm Technology Co., Ltd. The HPB relative abundances were calculated at the genus level.
Results
Changes of Conventional Composting Indicators
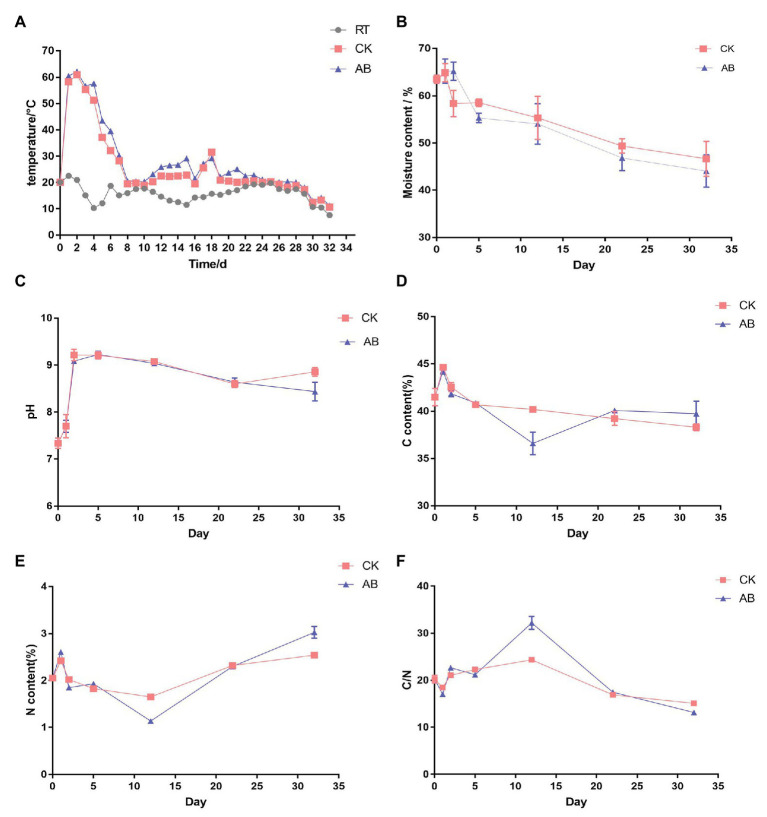
The key indicators of composting are temperature, pH, moisture, total carbon (TC), total nitrogen (TN), and C/N. The results are shown in Figure 1. Swine manure aerobic composting was performed for 32 days. The maximum temperature in AB was 62.1°C, the maximum temperature in CK was 60.9°C, and the temperatures of the two groups both lasted more than 3 days above 55°C (Figure 1A). It reached the standard of harmless treatment of reactor compost. The moisture content continued to decline during composting and that of the AB group dropped faster than the CK group (Figure 1B). This is because the microbial activity in the AB group compost was stronger, the temperature during the thermophilic phase was higher, and the high temperature lasted longer, so the water loss rate was more rapid. The pH increased gradually in the early stage of composting and declined slowly and stabilized (Figure 1C). In the early stage of composting, the ammonification was strong, the pH value rose rapidly, and the nitrification increased in the maturation phase; then, the pH value began to decrease, and the final pH value was stable between 8 and 9. The TC content in composting process showed a downward trend (Figure 1D). The carbon content of the AB group declined rapidly during the thermophilic phase, which may indicate strong microbial activity in the early stage of composting and converting the organic matter into carbon dioxide or minerals. The change in TN content of the composting piles first decreased and then increased (Figure 1E). The content of TN in the AB group was higher than that in the CK group in the maturation phase, which may be attributable to compound microbial inoculants playing a role in nitrogen preservation. The initial carbon and nitrogen ratio of the material was ⁓20. At the end of composting, the carbon to nitrogen ratio of the AB group was 13.15 and that of the CK group was 15.08 (Figure 1F). Both compost groups reached a mature state. The above results show that adding compound microbial inoculants can increase the maximum temperature of compost, prolong the high-temperature period, and accelerate water loss and nitrogen fixation during swine manure composting. The addition of compound microbial inoculants can accelerate and strengthen the composting process and improve the fertilizer efficiency of composting products.
Figure 1.
Physicochemical changes during swine manure composting. (A) Temperature, (B) moisture, (C) pH, (D) TC, (E) TN, and (F) C/N in the two treatment conditions during the composting process. CK: the swine manure composting. AB: add 1% compound microbial agents in the swine manure composting.
Changes of ARGs and MGEs During Composting
Eighteen ARGs and two MGEs were detected during swine manure composting. ARGs showed the same trend in the two groups (Figures 2, 3). The abundances of ARGs ranged from 3.6 × 103 to 1.13 × 108 copies/g. The ARGs with the highest abundance was sul2, and the lowest was blaCTX. For nine tetracycline resistance genes, the abundance of tetW gene is the highest, and the tetG abundance is the lowest (Figure 2). As the compost goes on, the reservoir of 11 ARGs (tetH, tetL, tetM, tetO, tetQ, tetW, ermB, ermQ, blaCTX, blaTEM, and tn916) are reduced at the end of composting. The total removal rates are 22.8–99.7%. tetM, tetO, tetQ, ermB, ermQ, and blaTEM were significantly reduced at the end of the thermophilic phase on the 5th day. At the end of composting, the reduction effect of ARGs (tetH, tetL, tetM, blaCTX, and tn916) in the AB group was significantly higher than that in the CK group. Compound microbial inoculants increased these ARGs removal in swine manure with increasing rates of 3.7–23.8%. At the same time, we also studied the spatial distribution and variation characteristics of ARGs during composting. We found that the contents of ARGs at different layers of the composting piles were significantly different. The abundance of ARGs in the middle and lower layers of compost showed the same trend. Overall, the reduction effect of ARGs was as follows: middle layer > upper layer > lower layer. On the other hand, we found that tetA, tetC, tetG, ermA, ermC, ermF, sul1, sul2, and intI1 proliferated. These ARGs began to proliferate on the 5th day, and the abundance increased significantly on the 12th day. This may be due to the proliferation of bacteria carrying these ARGs during the maturation phase.
Figure 2.
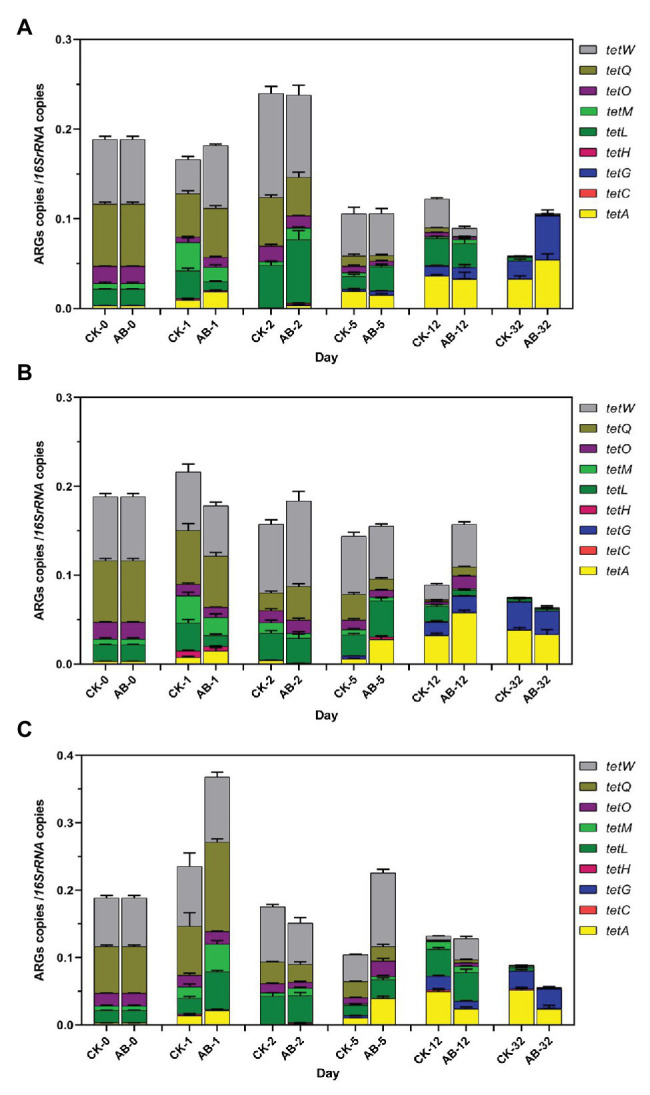
Abundances of tetracycline resistance genes during swine manure composting. (A–C) refer to the upper, middle, and lower layers of compost, respectively. CK: the swine manure composting. AB: add 1% compound microbial agents in the swine manure composting. 0, 1, 2, 5, 12, and 32 are the days for composting.
Figure 3.
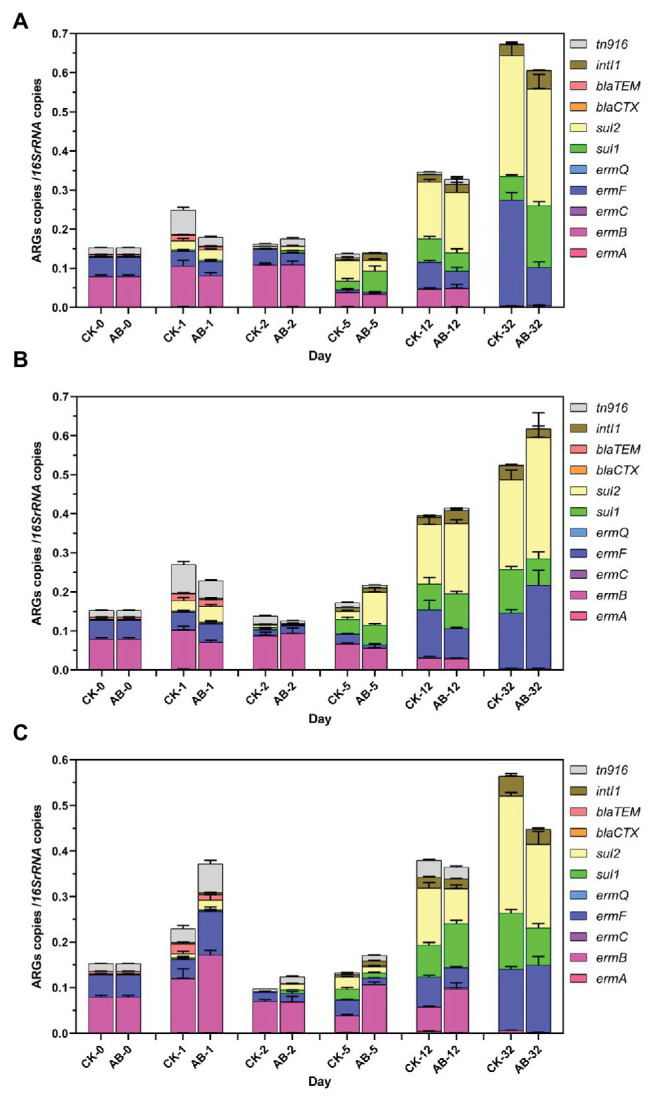
Abundances of ARGs and MGEs during swine manure composting. (A–C) refer to the upper, middle, and lower layers of compost, respectively. CK: the swine manure composting. AB: add 1% compound microbial agents in the swine manure composting. 0, 1, 2, 5, 12, and 32 are the days for composting.
Variation of Microbial Community
Change of microbial community at the phylum level during composting is shown in Figure 4. The sequences data reported in this study have been deposited in NCBI SRA with the accession number PRJNA663659 (https://www.ncbi.nlm.nih.gov/sra/; PRJNA663659). Firmicutes, Actinobacteria, Bacteroidetes, Spirochaetae, and Proteobacteria were the most dominant flora during the two kinds of composting. However, their relative abundances varied significantly at different phases of composting (p < 0.05). There was no significant difference in the structural composition of AB and CK between the mesophilic phase and thermophilic phase. Firmicutes and Proteobacteria were dominant flora during the mesophilic phase. However, Firmicutes accounted for more than 95% during the thermophilic phase. In the maturation phase, there was a significant difference in community structure between AB and CK. Bacteroidetes and Actinobacteria in group AB were 23.8 and 3.1% higher than those in group CK, respectively. But Firmicutes and Proteobacteria in group AB were 17.3 and 9.5% lower than those in group CK, respectively.
Figure 4.
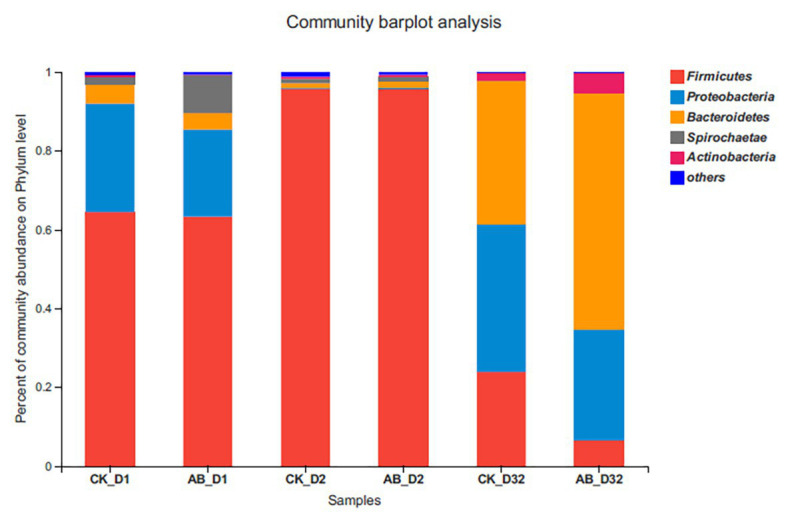
Structure and abundance of flora at the phylum level. CK: the swine manure composting. AB: add 1% compound microbial agents in the swine manure composting. D1 represents the mesophilic phase, D2 represents the thermophilic phase, and D32 represents the maturation phase.
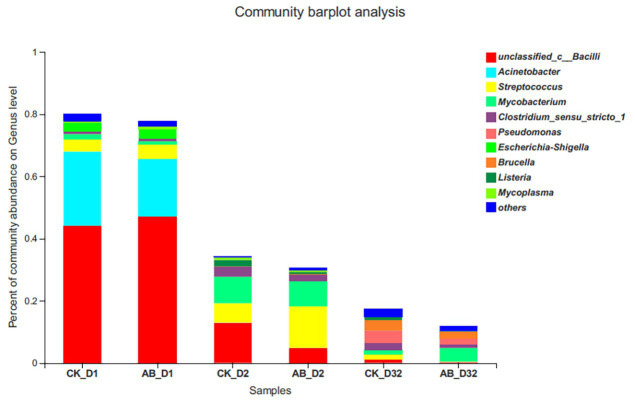
At the same time, we also analyzed the changes of microbial communities during composting at the genus level (Figure 5). In the mesophilic phase, the composition of CK and AB flora was similar. Kurthia showed the highest abundance, followed by Acinetobacter, Escherichia, Shigella, Streptococcus, and Treponema. In the thermophilic phase (day 2), Bacillus showed the highest abundance. The Bacillus abundance of the AB group was higher than that of the CK group, which may be due to the addition of compound microbial inoculants. At the end of composting, the abundance of Bacillus dropped dramatically, replaced by Sphingobacterium, Ochrobactrum, Cellvibrio, etc. From the results, the microbial community structure at different stages of the composting was quite different. The addition of compound microbial inoculants changed the community structure during composting.
Figure 5.
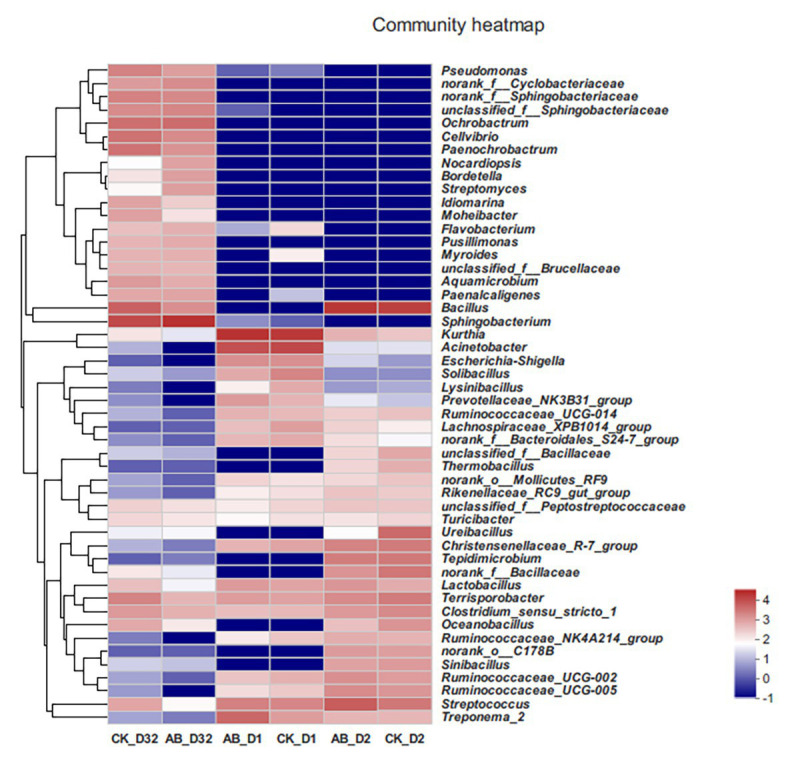
Heat map of species abundance at the genus level in compost. CK: the swine manure composting. AB: add 1% compound microbial agents in the swine manure composting. D1 represents the mesophilic phase, D2 represents the thermophilic phase, and D32 represents the maturation phase.
Variation of Pathogens During Composting
We analyzed the change of HPB relative abundance during the composting process (Figure 6). HPB assigned to 10 genera were detected during composting, among which Unclassified Bacilli, Streptococcus, Acinetobacter, and Escherichia/Shigella were dominant. The abundance of pathogens decreased sharply during the thermophilic phase on the 2nd day of composting. At the end of composting, the relative abundance of HPB in the CK group decreased by 78.16%, and the AB group decreased by 86.77%. At the end of composting, the composition of composting bacteria in the two groups was different. The CK group mainly consisted of Pseudomonas and Brucella, while the AB group was mainly Mycobacterium.
Figure 6.
Relative abundance changes of human pathogenic bacteria (HPB) during composting. CK: the swine manure composting. AB: add 1% compound microbial agents in the swine manure composting. D1 represents the mesophilic phase, D2 represents the thermophilic phase, and D32 represents the maturation phase.
Associations of Bacterial Community With Environmental Factors
CCA analysis is mainly used to reflect the correlation between bacterial community and environmental factors (Figure 7). The results showed that the selected environmental variables explained a total of 79.48% of species changes. Temperature, moisture, TC, TN, and C/N were significantly correlated with microbial communities, and pH had little effect on the community. Actinobacteria and Tenericutes were more affected by environmental factors. The change of TN content had a greater influence on Proteobacteria and Bacteroidetes. Temperature and C/N changes were closely related to Firmicutes. Combined with the stage of composting, TC, and moisture played important roles in mesophilic phase. At the thermophilic phase, temperature and C/N played important roles. During the maturation phase, TN and pH affected microbial community changes.
Figure 7.
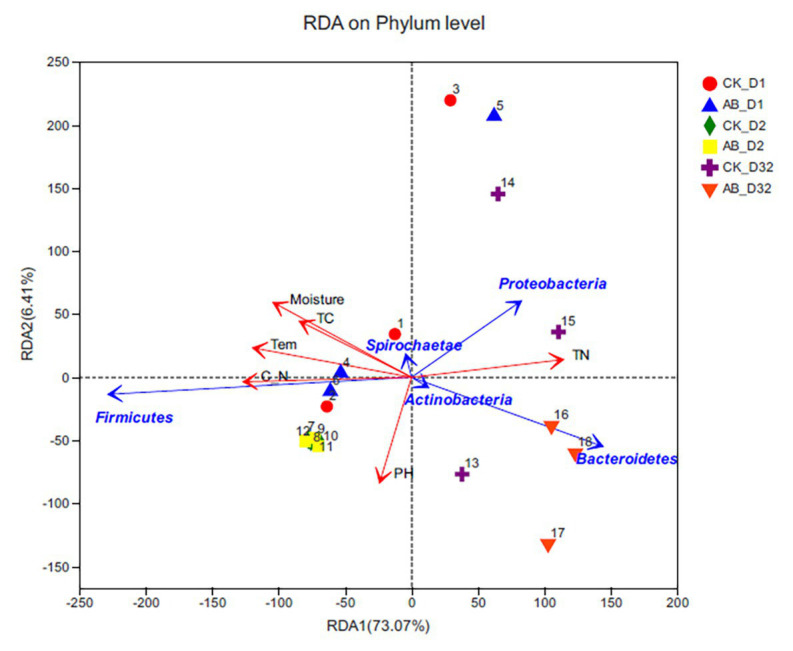
Redundancy analysis between environmental factors and microbial community structure (phylum level). CK: the swine manure composting. AB: add 1% compound microbial agents in the swine manure composting. D1 represents the mesophilic phase, D2 represents the thermophilic phase, and D32 represents the maturation phase.
Correlations Between ARGs, MGEs, and Microbial Community
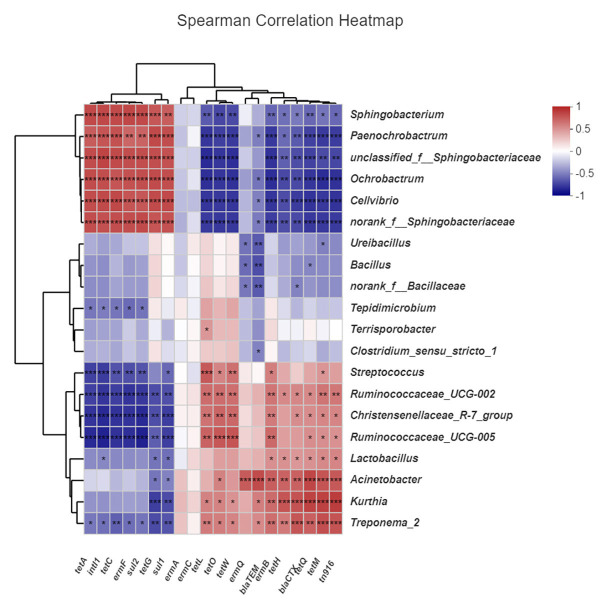
We analyzed the correlations between ARGs, MGEs, and microbial community structure (genus level, top 20 abundances). As shown in Figure 8, tetL, tetO, tetW, and intI1 were significantly positively correlated with Streptococcus, Christensenellaceae, and Ruminococcaceae. ermQ and blaTEM were significantly positively correlated with Acinetobacter. ermB, tetH, blaCTX, tetQ, tetM, and tn916 were significantly positively correlated with Acinetobacter, Kurthia, and Treponema_2. These microflora occupy a dominant position in the early stage of composting and were greatly reduced in the maturation phase, so these ARGs were also reduced. On the other hand, tetA, tetC, tetG, ermF, sul1, and sul2 were significantly positively correlated with Sphingobacterium, Paenochrobactrum, Ochrobactrum, and Cellvibrio. These microflora occupy a dominant position during the maturation phase of composting, so these ARGs proliferate at the end of composting. However, ermA and ermC failed to match the related flora, and possibly because of the low abundance of host bacteria, they could not enter the forefront of species matching.
Figure 8.
Correlation between antibiotic resistance genes (ARGs), mobile genetic elements (MGEs), and microbial community. CK: the swine manure composting. AB: add 1% compound microbial agents in the swine manure composting. D1 represents the mesophilic phase, D2 represents the thermophilic phase, and D32 represents the maturation phase.
Discussion
Composting is an effective way to remove ARGs. Some ARGs were significantly reduced during the thermophilic phase. This demonstrated that thermophilic phase of aerobic composting plays important roles in the removal of ARGs, which is consistent with the previous studies (Cui et al., 2016; Zhang et al., 2016, 2020). But some ARGs proliferated significantly in the late period of composting. Our results show that three tetracycline ARGs, three macrolide ARGs, and two sulfonamide ARGs and intI1 proliferated during the late composting period. It has been found that intI1 is closely related to macrolide ARGs and sulfonamide ARGs (Cheng et al., 2013; Su et al., 2015). So the proliferation of ARGs may be involved in the intI1-mediated HGT (Horizontal gene transfer) process.
This study found that the addition of compound microbial inoculants could remove more ARGs and MGEs than natural composting. It could be that the compound microbial inoculants may kill the host bacteria carrying ARGs or high temperature inhibited HGT and eliminated some ARG hosts. However, composting process had a greater impact on ARGs than compound microbial inoculants. In this study, the composting process removes most of ARGs and MGEs by 22.8–99.7%, but compound microbial inoculants only increased the removal rate of part of ARGs by 3.7–23.8% in compost. This is consistent with the research results of Duan et al. (2019). And composting could not remove the ARGs completely. Some ARGs showed more proliferation in the AB group than in the CK group at the end of composting. This may be related to the change of the community structure of compost with the addition of compound microbial inoculants. Microorganisms carrying ARGs proliferate in large numbers in the maturation phase of composting. Some thermophilic bacteria may be the hosts of ARGs (Qian et al., 2016).
Similar to Awasthi et al. (2019) and Wu et al. (2020), microbial community composition was the most direct factor affecting ARGs during composting. The richness and diversity of microbial communities in the composting process showed a downward trend in the maturation phase of composting, which also explained why most ARGs were reduced. First, the decrease of microbial density constrains the horizontal transfer process of ARGs (Ma et al., 2016). Second, due to the decrease of bacterial diversity, the host bacteria of ARGs were also reduced (Wang et al., 2017). At the same time, our research also indicated that the relative abundance of Firmicutes decreased, but Bacteroidetes, Proteobacteria and Actinobacteria were increased in the maturation phase of compost. Duan et al. (2019) pointed out that Firmicutes are the main cause of ARGs changes during cattle manure composting. It has been reported that a decrease in the relative abundance of Firmicutes might explain the removal of most ARGs in compost (Huerta et al., 2013; Stegmann et al., 2015). Luo et al. (2017) showed that more than 50% of potential hosts of ARGs and MGEs belonged to Bacteroidetes and Proteobacteria. Zhang et al. (2020) indicated that Actinobacteria were the primary potential host of ARGs during commercial livestock manure composting. It follows that the proliferation of some ARGs may be related to the increase of Bacteroidetes, Proteobacteria, and Actinobacteria in maturation phase. The analysis at the genus level verifies the above conclusions. The abundance of Bacillus has decreased significantly, and it has been replaced by Sphingobacterium, Ochrobactrum, Cellvibrio, etc., in maturation phase, which are closely related to the proliferation of part ARGs. These bacteria belong to Bacteroidetes or Proteobacteria.
The addition of compound microbial inoculants altered the microbial community in the compost. The Bacillus abundance was higher in the thermophilic phase than that of the control group. Bacillus is the most important component in the compound microbial inoculants. This also directly proves the effect and experimental reliability of adding compound microbial inoculants. At the same time, our research also indicated that the relative abundance of Firmicutes and Proteobacteria decreased, but Bacteroidetes and Actinobacteria were increased in the maturation phase of compost by adding compound microbial inoculants. Among them, Firmicutes and Bacteroidetes change greatly. The succession of Firmicutes and Bacteroidetes during the composting may be the potential driver for ARGs alteration. The relative abundance of Firmicutes in group AB was significantly lower than that in group CK. This also explains that the removal of ARGs in the AB group was higher than that in the CK group.
Most environmental factors were significantly correlated with microbial community. Zhang et al. (2019) pointed out that environmental factors may not directly affect ARGs, but may indirectly affect the changes of ARGs by influencing microbial community. Duan et al. (2019) found B. subtilis at 0.5% could cause a change in compost’s pH, thereby reducing ARGs during cattle manure composting. However, there was no significant difference in pH between the two groups in this study, which had little impact on the microbial community. We also found that there were large numbers of pathogens in the compost material by analyzed the changes of microbial communities at the genus level. Previous study has also shown that most of the host bacteria of ARGs are pathogens (Li et al., 2015). Our results showed that compost removes most of the pathogens and the addition of compound microbial inoculants helped with the removal of pathogens. Fan et al. (2020) also found the relative abundance of potential HPB decreased by 74.6 and 91.4% at the end of vacuum-type composting and positive-pressure composting, respectively. But a recent study shows that the total relative abundance of pathogens did not obviously decline with the composting progressing during commercial livestock manure composting (Zhang et al., 2020). This result is inconsistent with ours.
Conclusion
Composting removes most of the ARGs and MGEs. ARGs were significantly reduced during the thermophilic phase, and the reduction effect of ARGs at the different layer of compost pile was as follows: middle layer > upper layer > lower layer. But some ARGs proliferated significantly in the maturation phase of composting, especially the sulfonamide resistance genes. Compound microbial inoculants increased the maximum temperature of compost, accelerated water loss and nitrogen fixation, and changed the community structure during swine manure composting. Inocula increased the removal rate of β-lactamase resistance genes, the transposon gene tn916, and part of tetracycline resistance genes in compost. The dominant phyla were Firmicutes, Proteobacteria, and Bacteroidetes in swine manure compost. ARGs were significantly positively correlated with microbial communities. Compound microbial inoculants changed the community structure and were helpful for removing pathogens during composting.
Data Availability Statement
The sequences data reported in this study have been deposited in NCBI SRA with the accession number PRJNA663659 (https://www.ncbi.nlm.nih.gov/sra/PRJNA663659).
Author Contributions
JW and KL contributed to the study conception and design. KL, RC, SM, and RY conducted the experiments and analyzed data. The first draft of the manuscript was written by KL and JW. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported by the National Key R&D Program of China (Program No. 2018YFD0500204), the Fundamental Research Funds for the Central Universities (Program No. 2662019FW012), and the Science and Technology Program of Wuhan, China (Program No. 2016020101010091).
References
- Awasthi M. K., Liu T., Chen H., Verma S., Duan Y., Awasthi S. K., et al. (2019). The behavior of antibiotic resistance genes and their associations with bacterial community during poultry manure composting. Bioresour. Technol. 280, 70–78. 10.1016/j.biortech.2019.02.030, PMID: [DOI] [PubMed] [Google Scholar]
- Chen Q., An X., Li H., Su J., Ma Y., Zhu Y. G. (2016). Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 92–93, 1–10. 10.1016/j.envint.2016.03.026, PMID: [DOI] [PubMed] [Google Scholar]
- Cheng W., Chen H., Su C., Yan S. (2013). Abundance and persistence of antibiotic resistance genes in livestock farms: a comprehensive investigation in eastern China. Environ. Int. 61, 1–7. 10.1016/j.envint.2013.08.023, PMID: [DOI] [PubMed] [Google Scholar]
- Cui E., Wu Y., Zuo Y., Chen H. (2016). Effect of different biochars on antibiotic resistance genes and bacterial community during chicken manure composting. Bioresour. Technol. 203, 11–17. 10.1016/j.biortech.2015.12.030, PMID: [DOI] [PubMed] [Google Scholar]
- Duan M., Zhang Y., Zhou B., Wang Q., Gu J., Liu G., et al. (2019). Changes in antibiotic resistance genes and mobile genetic elements during cattle manure composting after inoculation with Bacillus subtilis. Bioresour. Technol. 292:122011. 10.1016/j.biortech.2019.122011, PMID: [DOI] [PubMed] [Google Scholar]
- Fan H., Wu S., Woodley J., Zhuang G., Bai Z., Xu S., et al. (2020). Effective removal of antibiotic resistance genes and potential links with archaeal communities during vacuum-type composting and positive-pressure composting. J. Environ. Sci. 89, 277–286. 10.1016/j.jes.2019.09.006, PMID: [DOI] [PubMed] [Google Scholar]
- Fang H., Wang H., Cai L., Yu Y. (2015). Prevalence of antibiotic resistance genes and bacterial pathogens in long-term manured greenhouse soils as revealed by metagenomic survey. Environ. Sci. Technol. 49, 1095–1104. 10.1021/es504157v, PMID: [DOI] [PubMed] [Google Scholar]
- Huerta B., Marti E., Gros M., López P., Pompêo M., Armengol J., et al. (2013). Exploring the links between antibiotic occurrence, antibiotic resistance, and bacterial communities in water supply reservoirs. Sci. Total Environ. 456–457, 161–170. 10.1016/j.scitotenv.2013.03.071, PMID: [DOI] [PubMed] [Google Scholar]
- Ji X., Shen Q., Liu F., Ma J., Xu G., Wang Y. (2012). Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China. J. Hazard. Mater. 235–236, 178–185. 10.1016/j.jhazmat.2012.07.040, PMID: [DOI] [PubMed] [Google Scholar]
- Jia S., Shi P., Hu Q., Li B., Zhang T., Zhang X. X. (2015). Bacterial community shift drives antibiotic resistance promotion during drinking water chlorination. Environ. Sci. Technol. 49, 12271–12279. 10.1021/acs.est.5b03521, PMID: [DOI] [PubMed] [Google Scholar]
- Joy S. R., Li X., Snow D. D., Gilley J. E., Woodbury B., Bartelt-Hunt S. L. (2014). Fate of antimicrobials and antimicrobial resistance genes in simulated swine manure storage. Sci. Total Environ. 481, 69–74. 10.1016/j.scitotenv.2014.02.027, PMID: [DOI] [PubMed] [Google Scholar]
- Li Y., Xu H., Hua D., Zhao B., Mu H., Jin F., et al. (2020). Two-phase anaerobic digestion of lignocellulosic hydrolysate: focusing on the acidification with different inoculum to substrate ratios and inoculum sources. Sci. Total Environ. 699:134226. 10.1016/j.scitotenv.2019.134226, PMID: [DOI] [PubMed] [Google Scholar]
- Li B., Yang Y., Ma L., Ju F., Guo F., Tiedje J. M., et al. (2015). Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 9, 2490–2502. 10.1038/ismej.2015.59, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Li B., Li L. G., Zhang T., Angelidaki I. (2017). Antibiotic resistance genes and correlations with microbial community and metal resistance genes in full-scale biogas reactors as revealed by metagenomic analysis. Environ. Sci. Technol. 51, 4069–4080. 10.1021/acs.est.6b05100, PMID: [DOI] [PubMed] [Google Scholar]
- Ma L., Xia Y., Li B., Yang Y., Li L. G., Tiedje J. M., et al. (2016). Metagenomic assembly reveals hosts of antibiotic resistance genes and the shared resistome in pig, chicken, and human feces. Environ. Sci. Technol. 50, 420–427. 10.1021/acs.est.5b03522, PMID: [DOI] [PubMed] [Google Scholar]
- Mu Q., Li J., Sun Y., Mao D., Wang Q., Luo Y. (2015). Occurrence of sulfonamide-, tetracycline-, plasmid-mediated quinolone- and macrolide-resistance genes in livestock feedlots in Northern China. Environ. Sci. Pollut. Res. Int. 22, 6932–6940. 10.1007/s11356-014-3905-5, PMID: [DOI] [PubMed] [Google Scholar]
- Nakasaki K., Araya S., Mimoto H. (2013). Inoculation of Pichia kudriavzevii RB1 degrades the organic acids present in raw compost material and accelerates composting. Bioresour. Technol. 144, 521–528. 10.1016/j.biortech.2013.07.005, PMID: [DOI] [PubMed] [Google Scholar]
- Qian X., Sun W., Gu J., Wang X. J., Zhang Y. J., Duan M. L., et al. (2016). Reducing antibiotic resistance genes, integrons, and pathogens in dairy manure by continuous thermophilic composting. Bioresour. Technol. 220, 425–432. 10.1016/j.biortech.2016.08.101, PMID: [DOI] [PubMed] [Google Scholar]
- Qiu J., Zhao T., Liu Q., He J., He D., Wu G., et al. (2016). Residual veterinary antibiotics in pig excreta after oral administration of sulfonamides. Environ. Geochem. Health 38, 549–556. 10.1007/s10653-015-9740-x, PMID: [DOI] [PubMed] [Google Scholar]
- Ravindran B., Nguyen D. D., Chaudhary D. K., Chang S. W., Kim J., Lee S. R., et al. (2019). Influence of biochar on physico-chemical and microbial community during swine manure composting process. J. Environ. Manag. 232, 592–599. 10.1016/j.jenvman.2018.11.119, PMID: [DOI] [PubMed] [Google Scholar]
- Ray P., Chen C., Knowlton K. F., Pruden A., Xia K. (2017). Fate and effect of antibiotics in beef and dairy manure during static and turned composting. J. Environ. Qual. 46, 45–54. 10.2134/jeq2016.07.0269, PMID: [DOI] [PubMed] [Google Scholar]
- Sarmah A. K., Meyer M. T., Boxall A. B. A. (2006). A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65, 725–759. 10.1016/j.chemosphere.2006.03.026, PMID: [DOI] [PubMed] [Google Scholar]
- Selvam A., Xu D., Zhao Z., Wong J. W. C. (2012). Fate of tetracycline, sulfonamide and fluoroquinolone resistance genes and the changes in bacterial diversity during composting of swine manure. Bioresour. Technol. 126, 383–390. 10.1016/j.biortech.2012.03.045, PMID: [DOI] [PubMed] [Google Scholar]
- Stegmann E., Frasch H. J., Kilian R., Pozzi R. (2015). Self-resistance mechanisms of actinomycetes producing lipid II-targeting antibiotics. Int. J. Med. Microbiol. 305, 190–195. 10.1016/j.ijmm.2014.12.015, PMID: [DOI] [PubMed] [Google Scholar]
- Su J. Q., Wei B., Ou-Yang W. Y., Huang F. Y., Zhao Y., Xu H. J., et al. (2015). Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ. Sci. Technol. 49, 7356–7363. 10.1021/acs.est.5b01012, PMID: [DOI] [PubMed] [Google Scholar]
- Wang H., Sangwan N., Li H. Y., Su J. Q., Oyang W. Y., Zhang Z. J., et al. (2017). The antibiotic resistome of swine manure is significantly altered by association with the Musca domestica larvae gut microbiome. ISME J. 11, 100–111. 10.1038/ismej.2016.103, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Yang X., Jiao S., Zhang J., Ye B., Gao S. (2014). Sulfonamide-resistant bacteria and their resistance genes in soils fertilized with manures from Jiangsu Province, Southeastern China. PLoS One 9:e112626. 10.1371/journal.pone.0112626, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte W. (1998). Medical consequences of antibiotic use in agriculture. Science 279, 996–997. 10.1126/science.279.5353.996, PMID: [DOI] [PubMed] [Google Scholar]
- Wu X., Tian Z., Lv Z., Chen Z., Liu Y., Yong X., et al. (2020). Effects of copper salts on performance, antibiotic resistance genes, and microbial community during thermophilic anaerobic digestion of swine manure. Bioresour. Technol. 300:122728. 10.1016/j.biortech.2019.122728, PMID: [DOI] [PubMed] [Google Scholar]
- Yi J., Zheng R., Li F. E., Chao Z., Deng C. Y., Wu J. (2012). Temporal and spatial distribution of Bacillus and Clostridium histolyticum in swine manure composting by fluorescent in situ hybridization (FISH). Appl. Microbiol. Biotechnol. 93, 2625–2632. 10.1007/s00253-011-3558-y, PMID: [DOI] [PubMed] [Google Scholar]
- Zeng G., Yu M., Chen Y., Huang D., Zhang J., Huang H., et al. (2010). Effects of inoculation with Phanerochaete chrysosporium at various time points on enzyme activities during agricultural waste composting. Bioresour. Technol. 101, 222–227. 10.1016/j.biortech.2009.08.013, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang J., Chen M., Sui Q., Tong J., Jiang C., Lu X., et al. (2016). Impacts of addition of natural zeolite or a nitrification inhibitor on antibiotic resistance genes during sludge composting. Water Res. 91, 339–349. 10.1016/j.watres.2016.01.010, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang R., Gu J., Wang X., Li Y., Liu J., Lu C., et al. (2019). Response of antibiotic resistance genes abundance by graphene oxide during the anaerobic digestion of swine manure with copper pollution. Sci. Total Environ. 654, 292–299. 10.1016/j.scitotenv.2018.11.094, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang M., He L. Y., Liu Y. S., Zhao J. L., Zhang J. N., Chen J., et al. (2020). Variation of antibiotic resistome during commercial livestock manure composting. Environ. Int. 136:105458. 10.1016/j.envint.2020.105458, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences data reported in this study have been deposited in NCBI SRA with the accession number PRJNA663659 (https://www.ncbi.nlm.nih.gov/sra/PRJNA663659).