Abstract
The human frontal cortex is unusually large compared with many other species. The expansion of the human frontal cortex is accompanied by both connectivity and transcriptional changes. Yet, the developmental origins generating variation in frontal cortex circuitry across species remain unresolved. Nineteen genes that encode filaments, synapse, and voltage-gated channels are especially enriched in the supragranular layers of the human cerebral cortex, which suggests enhanced corticocortical projections emerging from layer III. We identify species differences in connections with the use of diffusion MR tractography as well as gene expression in adulthood and in development to identify developmental mechanisms generating variation in frontal cortical circuitry. We demonstrate that increased expression of supragranular-enriched genes in frontal cortex layer III is concomitant with an expansion in corticocortical pathways projecting within the frontal cortex in humans relative to mice. We also demonstrate that the growth of the frontal cortex white matter and transcriptional profiles of supragranular-enriched genes are protracted in humans relative to mice. The expansion of projections emerging from the human frontal cortex arises by extending frontal cortical circuitry development. Integrating gene expression with neuroimaging level phenotypes is an effective strategy to assess deviations in developmental programs leading to species differences in connections.
Keywords: cortex, development, human, supragranular-enriched genes
Introduction
The human frontal cortex is unusually large compared with that of many other species such as rodents, though the claim that the frontal, and in particular the prefrontal cortex, is unusually large in humans relative to apes remains controversial (Sherwood and Smaers, 2013; Semendeferi et al. 2002; Barton and Venditi 2013; Carlén 2017; Donahue et al. 2018). The expansion of the frontal cortex in humans versus rodents reflects modifications to its underlying molecular architecture and patterns of connectivity.
Patterns of gene expression across the layers of the cerebral cortex are generally conserved across primates and rodents, but there are some notable differences across species. A study profiling ~1000 genes found that 79% have conserved pattern of expression across layers in humans and in mice (Zeng et al. 2012), but that 21% exhibited species-specific variation. Of particular interest, 19 genes (e.g., NEFH, SYT2, and VAMP1) are especially enriched in the supragranular layers (i.e., layer III) of the cerebral cortex in humans relative to mice (Zeng et al. 2012; Krienen et al. 2016). These genes encode filaments (e.g., NEFH), synapse (e.g., VAMP1), and voltage-gated channels (e.g., SCN4B). The differential expression of these genes across cortical layers suggests major modifications to cross-cortical connectivity patterns between humans and mice. Because layer III pyramidal neurons generally project within the white matter to target various cortical areas, the increased expression of genes encoding filaments and voltage-gated channels in layer III of humans suggest enhanced corticocortical projections in humans relative to mice (Zeng et al. 2012; Krienen et al. 2016).
The notion that humans as other primates possess enhanced cross-cortical projections compared with other mammals is consistent with the observation that primates possess an expansion of layer II–IV neurons (Charvet et al. 2017a), some of which form long-range corticocortical projections (Charvet et al. 2019a). The expansion of layer III neuron numbers in the human frontal cortex is also evident from single-cell sequencing studies that show that excitatory layer III neurons represent ~46.5% of excitatory frontal cortex neurons in humans, but only ~24% in mice (Fig. 1; Luo et al. 2017). Thus, major differences in cell type organization across the depth of the cortex are associated with modifications to frontal cortical circuitry in humans.
Figure 1.
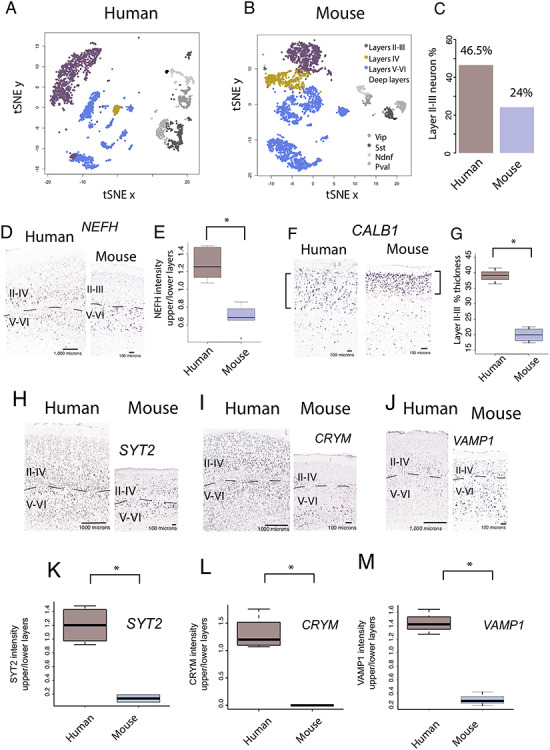
Single-cell methylome sequencing through the frontal cortex of a human (A) and a mouse (B) shows that the human frontal cortex contains relatively more excitatory layer III neurons (C) than mice. Frontal cortical excitatory neurons comprise 46.5% excitatory layer III neurons in humans but only 24% in mice. (D) NEFH expression is relatively and significantly increased in layers II–III of humans compared with mice (E). Moreover, the thickness of layer III as assessed by CALB1 (F) is significantly expanded in humans compared with mice (G). These data together suggest major modifications to long-range cortical pathways emerging from layer III of the frontal cortex in humans compared with mice. Of the supragranular enriched genes, SYT2 (H), CRYM (I), VAMP1 (J) expression is significantly increased in layers II–III versus V–VI in humans relative to mice (K–M). Sections of NEFH, CRYM, SYT2, and VAMP1 mRNA expression are from the Allen Human Brain Atlas 2010 (Allen Institute for Brain Science). Data for humans are available at human.brain-map.org.
We compare the frontal cortex of humans and mice because of their obvious differences in structural organization, which we use to develop a framework to integrate neuroimaging with molecular level phenotypes. Integrating across scales of organization from neuroimaging to molecular level phenotypes is a challenging enterprise to identify evolutionary changes in connections and their underlying developmental programs. Previous methods such as tract-tracers, diffusion MR tractography, or carbocyanine dyes suffer from a number of limitations such that the study of evolutionary changes in connectivity patterns has largely remain unexplored. Accordingly, this study is as much a proof of concept with which to link multiple scales of organization as it is a study to identify deviations in developmental programs giving rise to the emergence of frontal cortical circuitry in humans (Fornito et al. 2019). We select humans and mice as we can build from prior work to identify deviations in developmental programs.
Human development is unusually long compared with that of mice and many other species (Clancy et al. 2001; Workman et al. 2013). Identifying deviations in the timing of developmental programs requires controlling for overall changes in developmental schedules (Charvet et al. 2017b) because developmental processes take longer in humans compared with many other species. Previous work tackling evolutionary changes in developmental programs between humans and model organisms has employed various strategies to compare developmental schedules across species. Those include the use of actual age, birth, or puberty as a basis with which to compare developmental schedules across species (Finlay and Workman 2013; Sakai et al. 2017; Finlay and Charvet 2018). Relying on a few developmental transformations such as birth can be problematic. For example, birth does not align with the timing of most other neurodevelopmental transformations (i.e., events), and should therefore not be used as a basis with which to find corresponding ages across species (Finlay and Darlington 1995; Clancy et al. 2007; Charvet and Finlay, 2018). Rather than relying on the timing of a select few developmental transformations, we use the translating time model that relies on the timing of 271 developmental events gathered across 19 mammalian species to find corresponding ages between humans and mice. We use this resource to infer deviations in developmental programs generating variation across the two species (Workman et al. 2013). Because the translating time model extends up to 2 years of age in humans and their equivalent in other species, we use temporal changes in gene expression from RNA sequencing from the frontal cortex of humans and mice at different postnatal ages to find corresponding time points up to 30 years of age in humans and their equivalent in mice.
We compare transcriptional profiles of supragranular-enriched genes from the frontal cortex of humans versus mice in adulthood and in development because they encode filaments (e.g., NEFH) and voltage-gated channels that are linked to long-range corticocortical pathways (Hof et al. 1995; Zeng et al. 2012; Krienen et al. 2016; Nguyen et al. 2017). We also compare the developmental trajectories in the growth of the frontal cortex white matter between humans and mice because the white matter houses projections coursing to and from the frontal cortex. We use these data to test whether there are deviations in the timing of projection development coursing within, to or from the frontal cortex across these two species. Our findings demonstrate that protracted development of long-range projections accounts for the expansion of frontal cortex cross-cortical circuitry in humans.
Materials and Methods
Comparative Analyses of Layer III of Adult Humans and Mice
We performed a number of analyses to confirm the expansion of layer III neurons and corticocortical pathways in the human frontal cortex compared with mice (Fig. 1). Those include comparative analyses from diffusion MR tractography (Fig. 2), as well as gene expression from bulk and single cells to identify whether humans possess an expansion of long-range corticocortical projections in the frontal cortex compared with mice (Fig. 1; Zeng et al. 2012; Charvet et al. 2017a; Charvet et al. 2019a). We compared 1) the relative thickness of layer III across the frontal cortex of humans and mice with use of CALB1 expression because it is strongly expressed in layers II–III but not in other layers (Supplementary Fig. 1; Nieto et al. 2004), 2) the relative number of layer III neurons in humans and in mice from a previously published single-cell methylome sequencing study (Luo et al. 2017), and 3) the relative expression of supragranular-enriched genes across the layers of the human and mouse frontal cortex. We also compared corticocortical pathways with the use of diffusion MR tractography to test whether corticocortical pathways are expanded in humans compared with mice.
Figure 2.
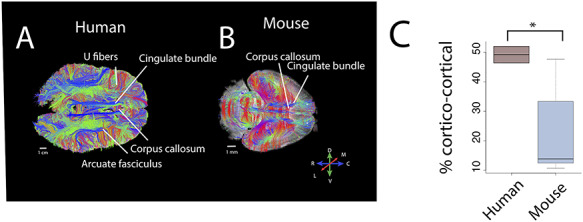
There are major differences in corticocortical pathways between humans (A) and mice (B) as assessed with high-angular resolution diffusion MR tractography. Horizontal slice filters through a human (A) and a mouse (B) show some of the pathways coursing through the white matter of the cortex. Those include the cingulate bundle and corpus callosum as well as the presumptive corticospinal tract in mice (*). (C) We quantified the relative percentage of pathways connecting cortical areas in humans and in mice. Such an analysis shows that the percentage of identified corticocortical pathways is significantly higher in humans than mice. Abbreviations: A: anterior; P: posterior; M: medial; L: lateral, D: dorsal; V: ventral.
We first measured the relative thickness of supragranular layers of adult human and mice frontal cortices. Supragranular layers were defined by the expression of CALB1 in humans and mice because CALB1 is a marker that can be used to define layers II–III (Nieto et al. 2004). Thickness measurements were taken from CALB1 in situ hybridization (ISH) images of humans from 28 to 31 years of age (n = 3) and mice at postnatal day 56 (n = 4) made available as part of the Allen Brain Atlas (2010 Allen Institute for Brain Science; Fig. 1). A grid was placed on sections of either human or mouse frontal cortices, and we randomly selected sites to measure the thickness of layer III from CALB1 ISH sections across the depth of the cortex. We used adjacent Nissl stains to measure the thickness of layers I–VI. Layer VI was bounded by the white matter. The relative thickness of CALB1 is computed as layer III versus layer I–VI thickness, and we performed a t-test on these data. We applied the same approach to measure the relative thickness of layer III from Nissl-stained sections (Supplementary Fig. 1).
We measured the intensity of supragranular-enriched expression of some genes across layers II–IV versus layers V–VI in the frontal cortex of humans and mice (i.e., CRYM, SYT2, VAMP1, NEFH; Supplementary Table 1). Previously, a select number of genes were shown to be preferentially expressed in layers III of the human primary visual cortex relative to mice, but it is not clear whether these observed species differences extend to the frontal cortex. To quantify differences in mRNA expression across layers of the frontal cortex, we downloaded ISH images for CRYM, SYT2, and VAMP1 made available by the Allen Brain Atlas. We placed a rectangular grid with 4–6 sections per individual as we had done previously to quantify the relative intensity of these genes across layers II–IV and layers V–VI (Charvet et al. 2019a). Randomly selected frames were aligned along the cortical surface. Frame widths were 500 and 1000 μm in mice and humans, respectively. The height of frames varied with the thickness of layers. These analyses were performed in Image J. We used cytoarchitecture from adjacent Nissl-stained section in humans, and in mice, as well as atlases to define cortical areas, as well as layers II–III and layers V–VI. Layers II–III were distinguished from layers IV–VI based on a small cell-dense layer IV evident from Nissl-stained sections. We compare the relative expression of these genes in layers II–III versus V–VI in humans and in mice. Considering the relative expression of these genes permits correcting for variation in intensity across brain samples. This approach is similar to that used previously to quantify densities in NEFH expression across layers II–III and V–VI (Charvet et al., 2015, Charvet et al. 2019b). We also include these comparisons of NEFH expression in the present study (Charvet et al. 2019a).
Comparative Analyses of Diffusion MR Tractography in Humans and Mice in Adulthood
We used high-angular resolution diffusion MR imaging tractography to quantify the relative number of pathways emerging to and from the frontal cortex of humans and mice (Fig. 2). A total of two postmortem human and five mouse brains were placed in a perfluoropolyether (Fomblin; Solvay, Brussels, Belgium) solution (invisible in magnetic resonance imaging [MRI]). The human brains were scanned with a 3T clinical Siemens MRI scanner (Erlangen, Germany) at the A. A. Martinos Center for Biomedical Imaging. We selected a steady-state free precession sequence with a repetition time (TR) of approximately 38 ms and an echo time (TE) of approximately 25 ms for diffusion acquisition (McNab et al. 2009). Forty-four diffusion-weighted images (b = 1000 s/mm2) and four nondiffusion-weighted images were acquired. The b value with the steady-state free precession sequence is T1 dependent. The other postmortem brain of a 34-year-old female brain is made available via the Allen Brain Institute (Ding et al. 2016), and details of procedures have been described previously (Ding et al. 2016). Briefly, diffusion-weighted data were acquired with a 3D steady-state free precession sequence (TR = 29.9 ms, α = 60°, TE = 24.96 ms, 900 μm; two averages). Diffusion weighting was applied along 44 directions distributed over the unit sphere (effective b value = 3686 s/mm2; 8 b = 0 images; Ding et al. 2016). We consider both of these human diffusion MR scans when comparing the relative proportion of corticocortical pathways in humans versus mice.
The mouse brains were scanned with a 4.7 T Bruker Biospec MRI system equipped with a high-performance gradient and a radiofrequency coil that best fit the small brains at the A. A. Martinos Center for Biomedical Imaging. A 3D diffusion-weighted spin-echo echo planar imaging sequence was used with a TR of approximately 1000 ms and TE of 40 ms to image the mouse brain. Sixty diffusion-weighted measurements (b = 8000 s/mm2) and one nondiffusion-weighted measurement (b = 0) were acquired. Details of spatial resolution are listed in Supplementary Table 2. These brains were used in previous studies, and details of scanning procedures have been described in more detail (Charvet et al. 2017a; Charvet et al. 2019b).
Diffusion MRI data were processed with Diffusion Toolkit (www.trackvis.org). All orientation distribution functions (ODFs) were normalized by the maximum ODF length within each voxel, fractional anisotropy (FA) was calculated from orientation vectors by fitting the data to the usual tensor model (e.g., Takahashi et al. 2010). We used a fiber assessment by continuous tracking algorithm to reconstruct pathways. No FA threshold was applied in reconstructing tracts, which is an approach consistent with that of other studies (e.g., Schmahmann et al. 2007; Takahashi et al. 2010, 2011; Kanamaru et al. 2017). We used TrackVis (http://trackvis.org) to visualize pathways. We set a horizontal slice filter (3-mm thick) through the brain of the mouse at the level of the frontal cortex white matter to capture pathways for display purposes in Figure 2. We set an region of interest (ROI) through the white matter of the frontal cortex in humans to capture pathways coursing to and from the frontal cortex. No minimum length fiber was applied to the human brain but thresholds applied to the mouse for visualization purposes in Figure 2 (minimum fiber length = 5 mm; max fiber length = 20 mm).
To quantify the relative number of pathway types coursing through the frontal cortex white matter, we randomly selected an ROI consisting of a voxel in the white matter of the frontal cortex of the human and the mouse. We identified whether the fibers coursing through the randomly selected ROI consist of the corpus callosum, the cingulate bundle, other corticocortical pathways, or pathways connecting cortical and subcortical structures (Supplementary Figs 2 and 3 and Supplementary Table 3). If a pathway was observed coursing through the dorsal midline, it was considered to consist of the corpus callosum. If the tractography neither clearly targeted subcortical, or cortical structures, we did not classify the pathway. Pathways connecting cortical and lateral limbic structures were considered to be cortico-subcortical pathways. Other corticocortical pathways consisted of U fibers as well as long-range projection pathways (e.g., arcuate fasciculus). We randomly selected 6–13 sites and 12–31 sites across serial coronal planes through the unilateral frontal cortex in mice and humans, respectively. More ROIs were selected through the human brain than in mice because the frontal cortex of humans is larger than in mice. We then computed the relative number of identified pathway types (i.e., callosal, cingulate, other corticocortical, or cortico-subcortical) per individual, and performed a t-test to identify whether the relative number of observed other corticocortical pathways is significantly greater in humans than in mice.
Subsampling Procedures
We subsampled the number of voxels through the white matter frontal cortex in the two species to ensure that the relative proportion of corticocortical pathways were robust to variation in sampling procedures. To that end, we randomly selected 28–34 voxels through the white matter of the frontal cortex from diffusion MR scans of mice (n = 4) and a human. From this large sample, we randomly subsampled the number of voxels and computed the relative percentage of corticocortical pathways in both species. For each number of selected voxels, we randomly subsampled 10 sites, and we averaged the percentage of corticocortical pathways in each individual (Supplementary Fig. 4). Such an approach shows that regardless of the number of randomly selected voxels, the percentage of identified corticocortical pathways is approximately 50% in the human and 10–20% in mice. The percentage of corticocortical pathways in humans falls outside the 95% confidence intervals (CIs) generated from mice. These analyses demonstrate that our comparative analyses of pathway types are robust to variation in sample size.
Developmental Trajectories in Supragranular-Enriched Gene Expression in Human and Mouse Frontal Cortex
We use a previously published RNA sequencing dataset from human (midfrontal gyrus; n = 7; around birth to 55 years of age) and mouse frontal cortical areas. Ages ranged from embryonic day 11 to 22 months of age in mice (n = 21). Ages ranged from around birth to 53 years of age in humans (n = 7; Lister et al. 2013). Expression values are in fragments per kilobase million (FPKM; GEO accession number: GSE47966; Lister et al. 2013). To correct for variation in library size across samples, the mean number of reads across all collected samples was divided by the number of reads from the sample of interest. Only reads in coding regions, which could be lifted over (with the liftOver tool) across species, were considered. We compare the expression of select genes from two different sources to ensure consistency across datasets (Supplementary Fig. 5). Replicates and number of mapped reads are shown in Supplementary Figure 6. All statistical analyses were performed with the programming language R.
We filtered the dataset to consider orthologous expressed genes with an “OK” status. If a gene has several copies, we selected the gene with the greatest expression. We identified orthologous genes as defined by the mouse genome database (Smith et al. 2018). Only genes with an average expression threshold above 0.5 FPKM across the examined ages for each species were considered (mice: postnatal day: 3–8 months after birth; humans: 35 days after birth to 52 years of age). We consider only expressed genes (FPKM > 0.5 averaged across age ranges per species), which produced 18 supragranular-enriched genes for consideration. Of these supragranular-enriched genes, NEFH, VAMP1, and SYT2 reach a plateau of expression in both species. Notably, NEFH (a marker of long-range projecting neurons), SYT2 expression (synaptic related), and VAMP1 appear to plateau in their expression later in humans than in mice once variation in developmental schedules between these two species are controlled for (Fig. 3; Charvet et al. 2019b). That is, the expression of supragranular-enriched genes such as NEFH, SYT2, and VAMP1 steadily increases to plateau in humans and in mice but appears to plateau later in humans than in mice once variation in developmental schedules is controlled for. To capture temporal variation in supragranular-enriched genes between these two species, we compared an index of variation (i.e., the coefficient of variation) at multiple age ranges between the two species.
Figure 3.
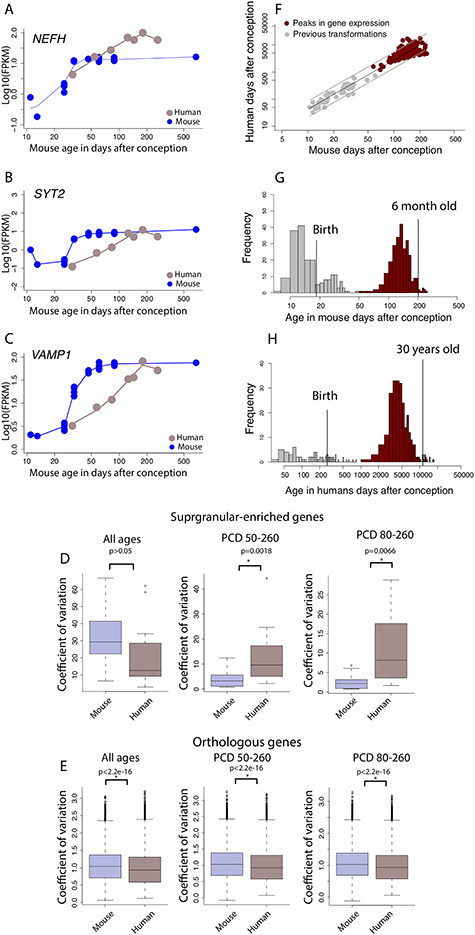
Developmental time courses of (A) NEFH, (B) SYT2, and (C) VAMP1 show protracted developmental time course in humans relative to mice. (D) The coefficient of variation in the expression of supragranular-enriched genes across all examined age ranges is not significantly different between humans and mice. However, the coefficient of variation in the expression of supragranular-enriched genes is significantly increased in humans relative to mice if only later ages are considered, either from PCD 50 to 260 or PCD 80 to 260 in mice and their equivalent in humans. (E) The increased coefficient of variation is not general to all expressed orthologous genes because the coefficient of variation is significantly less in humans compared with mice whether all age ranges are tested or whether only late stages are considered. (F) The timing in peak expression of genes is conserved enough to find corresponding ages across species at late stages of development. The timing of transformations and peaks in gene expression in the log10 days after conception in mice is regressed against those in humans and extends the age range of identified corresponding time points in humans and mice to span postnatal ages. Histograms of corresponding time points from transcriptional variation versus those extracted from anatomical and behavioral transformations demonstrate that we extend the age range of identified corresponding time points to adulthood. The inclusion of transcriptional variation provides a basis on which to find corresponding time points up to 30 years of age in humans (G) and 6 months of age in mice (H).
We test whether the coefficient of variation (i.e., ratio of the standard deviation to the mean) in the expression of supragranular-enriched genes is greater at late stages of development in humans compared with mice. Protracted variation in gene expression linked to the development of long-range projections suggests ongoing modifications to the development of long-range projections. That is, our analysis assumes that genes that vary more in one species relative to another at late stages of development reflect protracted developmental of long-range projections in one species relative to another.
To that end, we fitted a smooth spline through the log10 of FPKM versus the log10 of age expressed in days after conception for each gene in each species. We then extrapolated corresponding ages across the two species to compare equivalent developmental time points, but we retained fetal stages in mice as it appeared that, at least, some of these genes vary during fetal development in mice. Across all analyses, we added a value of 1 to all logged10 FPKM values to consider genes that may not be expressed at a particular time point. We computed the coefficient of variation for each gene across various age ranges of interest. We then performed a t-test on the coefficient of variation of supragranular-enriched genes and all orthologous genes in humans and mice to identify whether the variation in the expression of supragranular-enriched genes is significantly increased at late stages of development in humans compared with mice (Fig. 3). We repeated the same analyses across all orthologous genes to ensure that gene expression is not simply more variable in humans than in mice. To ensure that the temporal patterns of at least some of these supragranular-enriched genes are consistent with other RNA sequencing datasets, we compare the temporal patterns of NEFH expression and VAMP1 from the frontal cortex of humans and mice with that from the Allen Brain Atlas (Supplementary Fig. 5; Lein et al. 2007; Lister et al. 2013; Miller et al. 2014; Hawrylycz et al. 2012; Allen Developing Mouse Brain Atlas 2008; BrainSpan Atlas of the Developing Human Brain 2010). A qualitative comparison suggests strong concordance across datasets.
Temporal Changes in Gene Expression to Find Corresponding Ages Across Species
We next test whether temporal changes in gene expression in the frontal cortex are conserved enough to find corresponding ages across species. To that end, we selected genes with an average FPKM expression value above 0.5. To find corresponding ages at late stages of development, we fit a smooth spline through the logged values of FPKM versus the log-transformed values for age in mice to predict FPKM values at ages that are equivalent to those in humans. Because fitting a smooth spline should reduce variance around the mean, we fit a smooth spline when regressing logged values of FPKM versus the log-transformed values for age in humans as well. These smoothing steps resulted in a total of seven samples of humans and mice to be tested for nonlinear associations (Supplementary Fig. 6).
For each species, we tested which genes exhibit a significant peak in their expression with age with easynls package in R (model 2), examples are shown in Supplementary Figure 7. The age in which a plateau of expression was observed was constrained to span the same age ranges in both species (Supplementary Fig. 6). We corrected for multiple testing with a false discovery rate (FDR) BH threshold P value set to 0.05. We filtered the number of associations further to only include expressed genes that significantly correlate in both species. To that end, we correlated (cor) the log10-based FPKM values in humans (n = 7) and in mice (n = 7) matched for age. We then selected genes that correlate positively and significantly between the two species, and we corrected for multiple testing with an FDR threshold set to P < 0.05. This resulted in 261 genes with a significant peak in expression in both species. We then tested whether these data can be used to extrapolate corresponding ages to later time points in both species. To that end, we included prior work capturing the timing of developmental transformations in humans and mice (Charvet and Finlay 2018). We aimed to increase the number of developmental transformations for consideration in mice. Considering strong conservation in the timing of developmental transformations in rats and mice (Workman et al. 2013), we used previously collected transformations in rats and mice (Workman et al. 2013; Finlay and Charvet 2018) and extrapolated corresponding ages between rats and mice by linear regression through the log-transformed values in the timing of developmental transformations for rats versus those in mice (y = 1.05x-0.11, R2 = 0.96, df = 137). We then extrapolated corresponding ages from rats to mice, which resulted in 58 developmental transformations in mice (Fig. 3F). This approach served to increase the number of transformations for comparison between humans and mice.
Developmental Trajectories of Frontal Cortex White Matter Growth in Humans and Mice
To assess whether there are deviations in the development of white matter pathways between humans and mice, we measured the volume of the frontal cortex white matter in the left hemisphere at successive stages of development in 36 humans (gestational week 36–18 years old) and 47 mice (P2–P60; Supplementary Table 4 and Fig. 4). We used in vivo and ex vivo structural and diffusion MRI scans to capture the growth of the frontal cortex white matter in humans and in mice (Shi et al. 2011; Chuang et al. 2011; Khan et al. 2018b; Khan et al. 2018a; Richardson et al. 2018). In humans as in mice, we used atlases to demarcate the white matter of the frontal cortex from the remaining cortical areas (Ding et al. 2016). The anterior cingulate cortex was included as part of the frontal cortex if the frontal cortex was observed in the same plane of view. In mice, the caudal or posterior boundary between the frontal cortex and remaining cortex was bounded by the presence of the hippocampus from coronal planes. We used atlases (Paxinos and Franklin 2019) to define the lateral boundary between the frontal cortex and somatosensory cortex, which was the most challenging to delineate in mice. We applied this definition of the frontal cortex systematically in humans and in mice.
Figure 4.
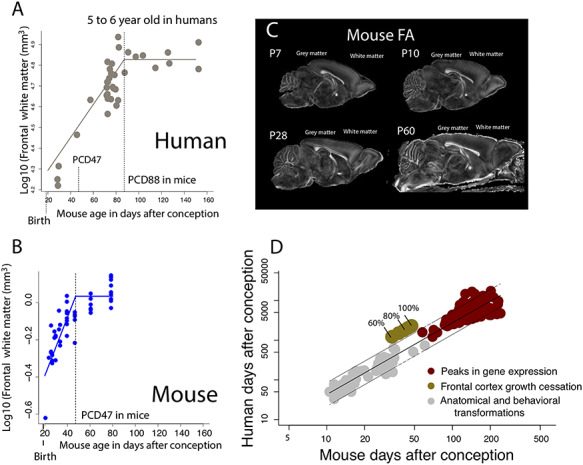
We measured the growth of the frontal cortex white matter in humans (A) and in mice (B) from structural MR scans in humans or FA images from diffusion-weighted scans in mice (C). In mice, the frontal cortex white matter was visualized from FA images of diffusion-weighted scans at different ages, which can be used to distinguish the gray from the white matter. (A) The frontal cortex white matter growth in humans is mapped onto mouse age according to the translating time model (Workman et al. 2013). After controlling for variation in developmental schedules across the two species, the frontal cortex white matter growth is extended in humans (A) compared with mice (B). According to the nonlinear regression used to capture frontal cortex white matter cessation, the frontal cortex white matter ceases to grow at about 5.5 years of age in humans, which corresponds to PCD 89 in mice. In contrast the frontal cortex white matter ceases to grow relatively earlier in mice (PCD 47). Vertical dashed lines represent the timing of frontal cortex white matter growth cessation in both species. (D) To further ensure that the frontal cortex white matter is indeed protracted in humans compared with the timing of other transformations, we compare the timing of frontal cortex white matter cessation growth in humans and mice relative to the timing of transformations assessed from prior work as well as from peaks in gene expression. We used these nonlinear regressions to extract when the frontal cortex white matter reaches 100%, 90%, 80%, 70%, and 60% of the white matter frontal cortex adult volume (A, B) according to the nonlinear regressions. We assess whether the timing of frontal cortex white matter growth occurs later than expected relative to the timing of other transformations. To that end, we regress the log10-based values of the timing of developmental transformations in humans and in mice and we generate 95% prediction intervals from these data. Arbitrary epochs captured from frontal cortex white matter growth falls outside the 95% prediction intervals from these data. That is, the growth of the frontal cortex white matter is protracted in humans relative to the timing of other transformations.
We used FA images from diffusion-weighted images in mice because FA permitted demarcating the frontal cortex white matter at successive stages of development and differences between gray and white matter were clearly observed (Fig. 4B). In humans, we selected structural MR scans because structural MR scans yielded better resolution than FA images. A minimum of five serial planes were used to reconstruct the volume of the frontal cortex in humans and in mice. We multiplied the area by the section spacing to compare volumes of frontal cortex white matter over the course of development and across species. We compare the timing of frontal cortex white matter growth cessation between the two species to test whether the frontal cortex white matter grows for longer than expected in humans compared with mice. In mice as in humans, we averaged frontal cortex white matter volumes for each age prior to testing for a linear plateau with easynls (model 3) implemented with the programming language R. We fit a nonlinear regression with the log10-transformed values for the frontal cortex versus age expressed in days after conception. Age in humans was mapped onto mouse age according to the translating time model (Workman et al. 2013; http://translatingtime.org). Because the observed age ranges span later time points than those predicted by the translating time model, we extrapolated corresponding ages to later ages in humans and mice as we have done previously (Charvet et al. 2017b; Charvet et al. 2019a; Charvet et al. 2019b). We extracted multiple time points from the nonlinear regression of the log10-transformed values of the frontal cortex white matter volume versus age in humans and mice. We fitted epochs through these growth trajectories. We extracted when the frontal cortex white matter reaches 100%, 90%, 80%, 70%, and 60% of its adult volume. Dividing these growth trajectories into epochs permits determining when the frontal cortex white matter reaches a certain percentage of adult brain structure and whether the growth of frontal cortex white matter is protracted in one species relative to the other.
Results
We first test whether layer III is expanded in the frontal cortex of humans compared with mice. We also use high-angular resolution diffusion MR tractography to assess whether humans possess relatively more corticocortical pathways compared with mice coursing through the frontal cortex. We then identify the developmental basis of such variation in frontal cortical connections. We compare temporal changes in supragranular-enriched gene expression as well as the growth of the frontal cortex white matter between humans and in mice.
Expansion of Layer III and Long-Range Projections in Humans in Adulthood
We perform a number of analyses to identify differences in layer III neuron organization in the frontal cortex of humans versus mice in adulthood. Single-cell methylome sequencing from the frontal cortex of humans and mice demonstrate that the relative number of excitatory layer III versus excitatory layer IV–VI neurons in humans (layer III neurons: 873, layer IV–VI neurons: 999) is greater than expected by chance compared with mice (excitatory layer III neurons: 690; excitatory layer IV–VI neurons: 2160; χ2 = 256.56, P < 0.05; Fig. 1A,B). That is, 46.5% of excitatory frontal cortical neurons are located in layer III in humans but only 24% of excitatory layer III are observed in mice (Fig. 1C; Luo et al. 2017). If layer III neurons projecting over long distance are indeed expanded in the frontal cortex of humans, we would expect genes expressed by large neurons in layers III to be increased in humans relative to mice. NEFH is typically expressed by large neurons that often project over long distances. In the frontal cortex, we previously showed that NEFH expression in layers II–IV versus layers V–VI in humans is significantly greater than in mice (n = 6 humans, n = 6 mice, t = 6.5318; P < 0.05; Fig. 1D,E), further demonstrating an expansion of long-range projecting neurons in the frontal cortex of humans relative to mice (Charvet et al. 2019b). The relative thickness of layer III as assessed from CALB1 expression is significantly expanded in the frontal cortex of humans compared with mice (t = −10.185, P < 0.05, n = 4 mice 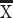 = 20%, 3 humans
= 20%, 3 humans 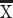 = 39%; Fig. 1F,G). The relative thickness of layers II–III as defined cytoarchitecturally is likewise relatively expanded in humans compared with mice (Supplementary Fig. 1). Together, these findings suggest an expansion and major modifications to layer III neurons in the human frontal cortex compared with mice.
= 39%; Fig. 1F,G). The relative thickness of layers II–III as defined cytoarchitecturally is likewise relatively expanded in humans compared with mice (Supplementary Fig. 1). Together, these findings suggest an expansion and major modifications to layer III neurons in the human frontal cortex compared with mice.
The human frontal cortex also possesses a suite of genes preferentially expressed in layer III of the visual cortex, which encode filaments synapse and voltage-gated channels (Zeng et al. 2012). To confirm the supragranular enrichment of these genes in the frontal cortex of humans relative to mice, we quantified the relative intensity of expression of some supragranular-enriched genes (i.e., SYT2, CRYM, and VAMP1) across layers II–IV versus V–VI across the frontal cortex of humans and mice (Fig. 1H–J). This is because prior work focused on comparing the expression of these genes focused on the primary visual cortex (Zeng et al. 2012), and it is not clear whether species differences in the expression of these supragranular-enriched genes are also evident for the frontal cortex or specific to the primary visual cortex. In the frontal cortex, SYT2, CRYM, and VAMP1 are significantly preferentially expressed in layers II–IV in humans compared with mice (SYT2: t = −7.3234, P < 0.05, n = 4 humans 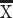 = 1.20, 2 mice
= 1.20, 2 mice 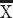 = 0.13; CRYM, t = −8.3528, P < 0.05, n = 4 humans
= 0.13; CRYM, t = −8.3528, P < 0.05, n = 4 humans 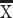 = 1.3, 2 mice
= 1.3, 2 mice 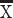 = 0; VAMP1, t = −9.3108, P < 0.05, n = 3 humans
= 0; VAMP1, t = −9.3108, P < 0.05, n = 3 humans 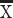 = 1.44, n = 3 mice
= 1.44, n = 3 mice 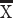 = 0.131; Fig. 1K–M). These findings demonstrate that humans deviate from mice in possessing major modifications in gene expression within layer III, which may be a molecular signature underlying major modifications to long-range corticocortical projections neurons in the frontal cortex.
= 0.131; Fig. 1K–M). These findings demonstrate that humans deviate from mice in possessing major modifications in gene expression within layer III, which may be a molecular signature underlying major modifications to long-range corticocortical projections neurons in the frontal cortex.
Diffusion MR tractography permits visualizing pathways across the entire brain of different species. We use high-angular resolution diffusion MR tractography to test for the expansion of corticocortical pathways in the frontal cortex of humans. An ROI placed through the white matter of the frontal cortex shows a number of corticocortical pathways such as the arcuate fasciculus and uncinate fasciculus in humans (Fig. 2A) that are not observed in mice (Fig. 2B). Horizontal slice filters used to capture fibers coursing to and from the frontal cortex in mice only identify the cingulate bundle, the corpus callosum (Fig. 2B), and cortico-subcortical pathways (i.e., fibers coursing across the dorsal to ventral direction). To identify whether humans possess relatively more corticocortical pathways than mice, we randomly placed ROIs through the frontal cortex white matter to capture fibers, and we classified observed pathways as belonging to the corpus callosum, cortico-subcortical, cingulate bundle, or other corticocortical pathways in both species in humans (n = 2) and mice (n = 5; Fig. 2 and Supplementary Fig. 3). We computed the relative number of identified pathway types (i.e., callosal, cingulate, other corticocortical, or cortico-subcortical) per individual (Supplementary Fig. 3). A t-test on the relative number of corticocortical pathways in two humans and five mice shows that humans possess significantly more corticocortical pathways (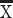 = 49.20%) coursing through the white matter of the frontal cortex than mice (
= 49.20%) coursing through the white matter of the frontal cortex than mice (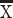 = 23.6%; t = −3.2818, n = 2 humans, n = 5 mice, P < 0.05; Fig. 2C). These species differences are robust to variation in sampling procedures (Supplementary Fig. 4). These findings demonstrate that increased expression of supragranular-enriched genes is concomitant with an expansion in long-range corticocortical projections emerging to or from the frontal cortex in humans.
= 23.6%; t = −3.2818, n = 2 humans, n = 5 mice, P < 0.05; Fig. 2C). These species differences are robust to variation in sampling procedures (Supplementary Fig. 4). These findings demonstrate that increased expression of supragranular-enriched genes is concomitant with an expansion in long-range corticocortical projections emerging to or from the frontal cortex in humans.
Developmental Time Course of Supragranular-Enriched gene Expression in Humans Versus Mouse
Comparative analyses of RNA sequencing data from the frontal cortex at successive ages can reveal differences in developmental trajectories in transcription, and evolutionary modifications to biological pathways. We tested whether the developmental time course of supragranular-enriched genes is protracted in humans compared with mice with the use of RNA sequencing from bulk samples taken from frontal cortical regions of humans and mice. We focus on supragranular-enriched genes that encode axon, voltage-gated sodium, potassium, and calcium channels, extracellular matrix, and synaptic-related genes. To compare the developmental trajectories of these genes, we mapped human age onto mouse age according to the translating time model (Workman et al. 2013). Because the translating time model finds corresponding ages up to 2 years of age in humans and its equivalent across species, we extrapolated corresponding ages to span later times as we had done previously (Finlay and Charvet 2018; Charvet et al. 2019b).
A cursory examination of some of these genes (e.g., NEFH, VAMP1, and SYT2) shows that these genes peak or plateau in both species but that the expression of these genes steadily increase in humans for much longer than in mice once for variation in developmental schedules is controlled for (Fig. 3A–C), which suggests that the developmental profile of at least some of these supragranular genes may be protracted in humans relative to mice. To confirm that the temporal pattern of at least some of these supragranular-enriched genes is indeed protracted in humans compared with mice, we compare the coefficient of variation of supragranular-enriched genes in humans and in mice at all and late stages of development (Fig. 3D). This analysis assumes that increased variation in one species relative to another would reflect evolutionary changes in developmental programs. The coefficient of variation in the expression of supragranular-enriched genes is not significantly different in humans (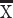 = 20.78%) compared with mice (
= 20.78%) compared with mice (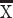 = 21.09%; D = 0.278; P = 0.5, n = 18; Fig. 3D). However, the coefficient of variation is significantly greater in humans than in mice once the examined age ranges span late time points (Fig. 3D). That is, the coefficient of variation is significantly increased in humans relative to mice once the examined age ranges span postconception day (PCD) 50 and beyond (ks test: D = 0.61, P < 0.05, n = 18; Fig. 3D) or PCD80 and beyond in mice (ks test: D = 0.55; P < 0.05, n = 18; Fig. 3D) and their equivalent ages in humans. In other words, humans possess prolonged variation in the expression of at least some supragranular-enriched genes relative to mice, which suggests a prolonged duration of development linked to the emergence and expansion of corticocortical pathways in humans.
= 21.09%; D = 0.278; P = 0.5, n = 18; Fig. 3D). However, the coefficient of variation is significantly greater in humans than in mice once the examined age ranges span late time points (Fig. 3D). That is, the coefficient of variation is significantly increased in humans relative to mice once the examined age ranges span postconception day (PCD) 50 and beyond (ks test: D = 0.61, P < 0.05, n = 18; Fig. 3D) or PCD80 and beyond in mice (ks test: D = 0.55; P < 0.05, n = 18; Fig. 3D) and their equivalent ages in humans. In other words, humans possess prolonged variation in the expression of at least some supragranular-enriched genes relative to mice, which suggests a prolonged duration of development linked to the emergence and expansion of corticocortical pathways in humans.
One caveat with this interpretation is that the expression of genes may simply be more variable in humans than in mice. The coefficient of variation of all expressed orthologous genes (n = 10 815) is actually significantly less in humans it is in mice whether all examined age ranges are considered (D = 0.09; P < 2.2e-16; n = 10 815), or late age ranges are examined (PCD and beyond: ks test: D = 0.08, P < 0.05, n = 10 815; PCD 80 and beyond, ks test: D = 0.08, P < 0.05, n = 10 815; Fig. 3E). Therefore, the increased variation in the expression of supragranular-enriched genes in the human frontal cortex at late stages of development is not a product of overall increased variation in gene expression in the human frontal cortex, but is specific to supragranular-enriched genes. We also use temporal changes in gene expression to extrapolate corresponding ages from humans to mice during postnatal ages (Fig. 3F–H).
Translating Time with Temporal Variation in Transcription
We have extrapolated corresponding ages in humans and mice to span later time points. However, the translating time model only extends up to 2 years of age in humans and its equivalent in other species (Workman et al. 2013). It is an open question as to whether it is actually possible to find corresponding ages at later stages of postnatal development. To ensure the validity of extrapolating the translating time model to later ages in both humans and in mice, we identify corresponding ages from either positive or negative peaks in the expression of orthologous genes from the frontal cortex of humans and mice (Fig. 3F–H). We fit a smooth spline though the log10 FPKM values in humans and in mice versus age expressed in days after conception, and we extrapolate corresponding ages between humans and mice from around birth to 53 years of age, and their equivalent ages in mice. This resulted in seven time points for comparison in the two species. We fit a quadratic model with the log-transformed normalized expression of each gene versus age in each species (Benjamini and Hochberg: BH FDR threshold: P < 0.05) to determine the age in which a gene might peak in its expression in both species. We then only selected genes with a positive correlation between the two species, and we corrected for multiple testing with an FDR threshold set to P < 0.05. Such an analysis yielded 261 genes, which significantly correlate, and peak either positively or negatively in both species. We compare corresponding ages extracted from temporal changes in gene expression from the frontal cortex with previously collected transformations used to find corresponding ages across species (n = 59; Finlay and Charvet 2018). We fit a linear model with the logged values of developmental event timing from previously collected transformations and peaks in gene expression expressed in age in days after conception in mice and humans. This model accounts for 94.31% of the variance (y = 1.80x − 0.17, df = 318, P < 0.05; Fig. 3F), and these analyses find corresponding age up to 30 years of age in humans (Fig. 3G) and 6 months of age in mice (Fig. 3H). Integrating the age of peaks in gene expression with anatomical transformations is an effective strategy to find corresponding ages at later stages of development in humans and in mice.
Growth of Frontal Cortex White Matter in Humans and Mice
We compare the timetable of long-range cortical pathways emerging to and from the frontal cortex in humans as in mice by comparing the growth of unilateral frontal cortex white matter across the two species to assess whether the development of long-range projections emerging to or from the frontal cortex is protracted in humans relative to the timing of other transformations (Fig. 4A–C). In both species, the frontal cortex white matter grows postnatally but eventually plateaus in its growth. Human age is mapped onto mouse age according to the translating time model (Fig. 4A,C). We used a nonlinear model (easynls, model 3) to identify when the frontal cortex white matter ceases to grow in humans and mice and we use this regression to assess when the frontal cortex white matter achieves 60%, 70%, 80%, 90%, and 100% of its adult volume as a basis to compare the growth of the frontal cortex white matter between the two species.
The frontal cortex white matter ceases to grow on PCD 47 in mice (y = 0.03 + 0.015x(x − 47.22); R2 = 0.70; P(coefficient a) = 0.66, P(coefficient b) < 0.05; P(coefficient c) < 0.05; n = 12) and around 5.5 years of age in humans. In humans, 5.5 years of age is equivalent to PCD 87 in mice (y = 4.83 + 0.01x(x − 87), R2 = 0.81, P(coefficient a) < 0.05, P(coefficient b) < 0.05, P(coefficient c) < 0.05, n = 35). Events that occur between PCD 66 and 80 in mice occur on 1.3–1.5 years of age (PCD 735–833; n = 2) in humans (Workman et al. 2013). The timing of frontal cortex white matter growth cessation falls outside the 95% CI of these data (PCD 828). The age of frontal cortex white matter cessation occurs later than expected in humans once variation in developmental schedules are controlled for (Fig. 4A,C).
The protracted growth of the frontal cortex white matter cessation is further supported by the observation that the timing of frontal cortex white matter growth falls outside the 95% prediction intervals generated from the timing of peaks in gene expression and anatomical transformations in humans and mice (Fig. 4D). Therefore, the growth of the frontal cortex white matter, which houses projections coursing within the white matter of the frontal cortex, is protracted in humans relative to the timing of other transformations.
Discussion
The present study focuses on comparative analyses of pathways from the frontal cortex from diffusion MR imaging and supragranular-enriched genes in order to identify evolutionary changes in connectivity patterns in adulthood and in development. Our findings demonstrate that integrating RNA sequencing data with neuroimaging phenotypes is an effective approach to identify evolutionary changes in connections in adulthood as well as in development. We also demonstrate that considering temporal changes in gene expression is an effective strategy to find corresponding ages between humans and mice during postnatal development and in adulthood.
Integrating comparative analyses from diffusion MR tractography with supragranular-enriched genes converge on the observation that the human frontal cortex is preferentially composed of layer III neurons, concomitant with increased cross-cortical projections through the white matter of the frontal cortex in humans. We use diffusion MR tractography and find an expansion in corticocortical pathways in the frontal cortex of humans, which falls in line with predictions from our comparative analyses of supragranular-enriched genes. We further demonstrate that the developmental trajectories of supragranular-enriched genes and growth of projections emerging to and from the frontal cortex are both protracted in their developmental profile in humans. Thus, the expansion of layer III neurons and concomitant expansion of projections coursing within the white matter of the frontal cortex in humans is achieved by extending the duration of their development. We have yet to identify the extent to which such modifications are unique to humans, shared among primates, and how such modifications to developmental programs vary across mammals.
The integration of diffusion MR imaging with comparative analyses of gene expression permits identifying variation in connectivity patterns across species. Although the rapid acquisition of information offered from diffusion MR tractography provides a high-throughput approach with which to study the evolution and development of connections (McNab et al. 2009; Takahashi et al. 2010, 2012; Song et al. 2015; Mars et al. 2019), diffusion MR imaging suffers from several limitations. Diffusion MR tractography has difficulty resolving crossing fibers as well as the precise terminations of tracts. Diffusion MR imaging may preferentially identify pathways based on certain characteristics such as axon length or myelination at the expense of other features (Reveley et al. 2015; Schilling et al. 2018). These biases become accentuated in brains that vary in size. We therefore integrate observations from diffusion MR tractography with comparative analyses of supragranular-enriched genes to overcome these limitations and to study the evolution and development of connections between humans and mice. Both lines of evidence converge on the finding that humans possess a relative expansion of neurons projecting over long distances within the frontal cortex.
Several hypotheses have been proposed to account for variation in connectivity patterns across species, but were not explicitly tested due to the lack of methods available to quantify variation in connectivity patterns across species in adulthood and in development (Innocenti 1995; Striedter 2005). Tract-tracers, diffusion MR tractography, and carbocyanine dyes on their own suffer from a number of technical limitations (Nudo and Masterton 1989, 1990; Chen et al. 2006; Reveley et al. 2015; Heilingoetter and Jensen 2016). Diffusion MR tractography identifies pathways across the entire brain, but suffers from a number of false positives, especially in in vivo (Thomas et al. 2014; Reveley et al. 2015). As a result, hypotheses focused on the evolution and development have largely gone unexplored. Our work integrates neuroimaging and molecular level phenotypes to identify deviations in connections, and their underlying developmental programs. We demonstrate that such an approach is an effective strategy to address evolutionary modifications to developmental programs leading to connectivity patterns in the human frontal cortex.
The establishment of connections extends postnatally in both humans and mice. Comparing developmental programs underlying variation in connections across species relies on identifying corresponding time points across species. We here use temporal trajectories in gene transcription from the frontal cortex in both species to find corresponding time points up to 30 years of age in humans and their equivalent in mice. Identifying sources of conservation and variation in developmental programs during postnatal ages is an essential enterprise as it will permit better extrapolating findings from model organisms to humans. This is because researchers often use model organisms during postnatal development and in adulthood to understand basic developmental processes and dysfunctions that occur in humans (Charvet and Finlay 2018). Extrapolated ages from the translating time model fall in line with our results extracted from peaks in gene expression from the frontal cortex of both species (Workman et al. 2013). Controlling for variation in developmental schedules reveals that the maturation of pathways emerging to or from the frontal cortex is protracted relative to the timing of other transformations in humans relative to mice.
Modifications to the timing of developmental programs have repeatedly been demonstrated as an important source of variation in brain structure and function across vertebrates. For instance, a combination of comparative analysis from thymidine studies to birth date neuronal types and histological data used to quantify the relative number of neuronal types in adulthood demonstrated that the disproportionate expansion of cortical neuron numbers in human and nonhuman primates is achieved by extending the duration of cortical neurogenesis relative to the timing of most other transformations (Rakic 1974, 1995, 2002; Clancy et al. 2001; Charvet et al. 2017b; Charvet et al. 2019a). The extension in the duration of cortical neurogenesis accounts for the amplification of layer II–IV neuron numbers, many of which participate in local and long-range corticocortical projections (Cahalane et al. 2014; Charvet et al. 2017a; Charvet et al. 2019a). We demonstrate yet another important developmental change in the timing of biological pathways (i.e., heterochrony) giving rise to the emergence of the human brain. That is, extending the development of projections accounts for their expansion within the human frontal cortex, and the overrepresentation of frontal cortex layer III neurons in humans. We have yet to identify whether such variation is specific to the frontal cortex or general to the isocortex. Moreover, it is not clear how much variation exists within mammals and how representative the phenotypes observed in mice are of mammals. We have only selected two species for comparisons. A broad sample of mammalian species is needed to identify which of the observed species differences represent the derived or ancestral condition.
The framework as articulated here may be applied to resolve ongoing debates focused on the evolution of the human brain. One example is whether the human frontal cortex, and in particular the prefrontal cortex white matter, is unusually large compared with apes (Donahue et al. 2018). The lack of consensus across studies suggests the need to move beyond gross measures of white matter, and to focus on integrating additional measures of connectivity at multiple scales of organization across species. The set of supragranular-enriched genes coupled with diffusion MR tractography provides an opportunity with which to investigate the evolution of long-range corticocortical pathways more broadly across mammals and more closely in the human lineage to identify sources of variation in connections across species. Such an approach would resolve whether the expansion of long-range corticocortical pathways within the frontal cortex is unique to humans or shared among primates.
Funding
This work was supported by the National Institute of General Medical Sciences (NIGMS) (grant 5P20GM103653) and the National Institute of Neurological Disorders and Stroke (NINDS) (grant R25NS09537) for research at Delaware State University. This work was also supported by a National Institutes of Health INBRE grant from the National Institute of General Medical Sciences (P20GM103446 to C.J.C) and the Eunice Shriver Kennedy National Institute of Child Health and Development (NICHD) (R01HD078561, R21HD069001 to E.T.) as well as the National Institute Institute of Neurological Disorders and Stroke (NINDS) (R01NS109475 to E.T). The opinions in this article are not necessarily those of the National Institutes of Health.
Notes
We thank Dr Melissa Harrington for her support at Delaware State University. A developmental series of mouse fractional anisotropy scans were obtained as a courtesy of Dr Mori (lbam.med.jhmi) at Johns Hopkins University available at http://cmrm.med.jhmi.edu. In situ hybridization data and associated images from mice and humans were taken from the Allen Institute Website and the BrainSpan Atlas of the developing human brain. These data are available at http://developingmouse.brain-map.org and http://www.brainspan.org, which are supported by the National Institute of Health Contract HHSN-271-2008-00047-C to the Allen Institute for Brain Science.
Conflict of Interest: None declared.
Supplementary Material
References
- Barton RA, Venditi C. 2013. Human frontal lobes are not relatively large. Proc Natl Acad Sci U S A. 110(22):9001–9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahalane DJ, Charvet CJ, Finlay BL. 2014. Modeling local and cross-species neuron number variations in the cerebral cortex as arising from a common mechanism. Proc Natl Acad Sci U S A. 111(49):17642–17647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlén M 2017. What constitutes the prefrontal cortex? Science. 358(6362):478–482. doi: 10.1126/science.aan8868. [DOI] [PubMed] [Google Scholar]
- Charvet CJ, Finlay BL. 2018. Comparing adult hippocampal neurogenesis across species: translating time to predict the tempo in humans. Front Neurosci. 12:706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Hof PR, Raghanti MA, Kouwe AJ, Sherwood CC, Takahashi E. 2017a. Combining diffusion magnetic resonance tractography with stereology highlights increased cross-cortical integration in primates. J Comp Neurol. 525:1075–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Šimić G, Kostović I, Knezović V, Vukšić M, Leko MB, Takahashi E, Sherwood CC, Wolfe MD, Finlay BL. 2017b. Coevolution in the timing of GABAergic and pyramidal neuron maturation in primates. Proc Biol Sci. 284:pii: 20171169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Das A, Song JW, Tindal-Burgess DJ, Kabaria P, Dai G, Kane T, Takahashi E. 2019a. High angular resolution diffusion MRI reveals conserved and deviant programs in the paths that guide human cortical circuitry. Cereb Cortex. 30:1447–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Palani A, Kabaria P, Takahashi E. 2019b. Evolution of brain connections: integrating diffusion MR tractography with gene expression highlights increased corticocortical projections in primates. Cereb Cortex. 29:5150–5165. doi: 10.1093/cercor/bhz054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BK, Miller SM, Mantilla CB, Gross L, Yazemski MJ, Windebank AJ. 2006. Optimizing conditions and avoiding pitfalls for prolonged axonal tracing with carbocyanine dyes in fixed rat spinal cords. J Neurosci Methods. 154:256–263. [DOI] [PubMed] [Google Scholar]
- Chuang N, Mori S, Yamamoto A, Jiang H, Ye X, Xu X, Richards LJ, Nathans J, Miller MI, Toga AW et al. 2011. An MRI-based atlas and database of the developing mouse brain. Neuroimage. 54:80–89. doi: 10.1016/j.neuroimage.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy B, Darlington RB, Finlay BL. 2001. Translating developmental time across mammalian species. Neuroscience. 105:7–17. [DOI] [PubMed] [Google Scholar]
- Clancy B, Finlay BL, Darrington RB, Anand KJ. 2007. Extrapolating brain development from experimental species to humans. Neurotoxicology. 28(5):931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SL, Royall JJ, Sunkin SM, Ng L, Facer BA, Lesnar P, Guillozet-Bongaarts A, McMurray B, Szafer A, Dolbeare TA et al. 2016. Comprehensive cellular-resolution atlas of the adult human brain. J Comp Neurol. 524:3127–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue CJ, Glasser MF, Preuss TM, Rilling JK, Van Essen DC. 2018. Quantitative assessment of prefrontal cortex in humans relative to nonhuman primates. Proc Natl Acad Sci U S A. 115(22):E5183–E519292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BL, Darlington RB. 1995. Linked regularities in the development and evolution of mammalian brains. Science. 268:1578–1584. [DOI] [PubMed] [Google Scholar]
- Finlay BL, Workman AD. 2013. Human exceptionalism. Trends Cogn Sci. 17:199–201. [DOI] [PubMed] [Google Scholar]
- Fornito A, Arnatkevičiūtė A, Fulcher BD. 2019. Bridging the gap between connectome and transcriptome. Trends Cogn Sci. 23(1):34–50. [DOI] [PubMed] [Google Scholar]
- Hawkes K, Finlay BL. 2018. Mammalian brain development and our grandmothering life history. Physiol Behav. 193(Pt A):55–68. doi: 10.1016/j.physbeh.2018.01.013. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Gguillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL et al. 2012. An anatomically comprehensive atlas of the adult human transcriptome. Nature. 489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilingoetter CL, Jensen MB. 2016. Histological methods for ex vivo axon tracing: a systematic review. Neurol Res. 38:561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof PR, Nimchinsky EA, Morrison JH. 1995. Neurochemical phenotype of corticocortical connections in the macaque monkey: quantitative analysis of a subset of neurofilament protein-immunoreactive projection neurons in frontal, parietal, temporal, and cingulate cortices. J Comp Neurol. 362:109–133. [DOI] [PubMed] [Google Scholar]
- Innocenti GM 1995. Exuberant development of connections, and its possible permissive role in cortical evolution. Trends Neurosci. 18:397–402. [DOI] [PubMed] [Google Scholar]
- Kanamaru Y, Li J, Stewart N, Sidma RL, Takahashi E. 2017. Cerebellar pathways in mouse model of Purkinje cell degeneration detected by high-angular resolution diffusion imaging tractography. Cerebellum. 16(3):648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Rollins CK, Ortinau CM, Afacan O, Warfield SK, Gholipour A. 2018a. Tract-specific group analysis in fetal cohorts using in utero diffusion tensor imaging. MICCAI. Cham: Springer; pp. 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Vasung L, Marami B, Rollins CK, Afacan O, Ortinau CM, Yang E, Warfield SK, Gholipour A. 2018b. Fetal brain growth portrayed by a spatiotemporal diffusion tensor MRI atlas computed from in utero images. Neuroimage. 185:593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krienen FM, Yeo BT, Ge T, Buckner RL, Sherwood CC. 2016. Transcriptional profiles of supragranular-enriched genes associate with corticocortical network architecture in the human brain. Proc Natl Acad Sci U S A. 113(4):E469–E478. doi: 10.1073/pnas.1510903113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD et al. 2013. Global epigenomic reconfiguration during mammalian brain development. Science. 341(6146):1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Keown CL, Kurihara L, Zhou J, He Y, Li J, Castanon R, Lucero J, Nery JR, Sandoval JP et al. 2017. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 357(6351):600–604. doi: 10.1126/science.aan3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, O’Muircheartaigh J, Folloni D, Li L, Glasser MF, Jbabdi S, Bryant KL. 2019. Concurrent analysis of white matter bundles and grey matter networks in the chimpanzee. Brain Struct Funct. 224:1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab JA, Jbabdi S, Deoni SC, Douaud G, Behrens TE, Miller KL. 2009. High resolution diffusion-weighted imaging in fixed human brain using diffusion-weighted steady state free precession. Neuroimage. 46:775–785. [DOI] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K et al. 2014. Transcriptional landscape of the prenatal human brain. Nature. 508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MQ, Wu Y, Bonilla LS, von Buchholtz LJ, Ryba NJ. 2017. Diversity amongst trigeminal neurons revealed by high throughput single cell sequencing. PLoS One. 12:e0185543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. 2004. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 479(2):168–180. [DOI] [PubMed] [Google Scholar]
- Nudo RJ, Masterton RB. 1989. Descending pathways to the spinal cord: II. Quantitative study of the tectospinal tract in 23 mammals. J Comp Neurol. 286:96–119. [DOI] [PubMed] [Google Scholar]
- Nudo RJ, Masterton RB. 1990. Descending pathways to the spinal cord, III: sites of origin of the corticospinal tract. J Comp Neurol. 296:559–583. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KB. 2019. Paxinos and Franklin's the mouse brain in stereotaxic coordinates. Sunderlaund, (MA): Academic press. [Google Scholar]
- Rakic P 1974. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 183:425–427. [DOI] [PubMed] [Google Scholar]
- Rakic P 1995. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 18:383–388. [DOI] [PubMed] [Google Scholar]
- Rakic P 2002. Pre- and post-developmental neurogenesis in primates. Clin Neurosci Res. 2:29–39. [Google Scholar]
- Reveley C, Seth AK, Pierpaoli C, Silva AC, Yu D, Saunders RC, Leopold DA, Ye F. 2015. Superficial white matter fiber systems impede detection of long-range cortical connections in diffusion MR tractography. Proc Natl Acad Sci U S A. 112(21):E2820–E2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson H, Lisandrelli G, Riobueno-Naylor A, Saxe R. 2018. Development of the social brain from age three to twelve years. Nat Commun. 9:1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Mikami A, Suzuki J, Miyabe-Nishiwaki T, Matsui M, Tomonaga M, Hamada Y, Matsuzawa T, Okano H, Oishi K. 2017. Developmental trajectory of the corpus callosum from infancy to the juvenile stage: comparative MRI between chimpanzees and humans. PLoS One. 12(6):e0179624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling K, Gao Y, Janve V, Stepniewska I, Landman BA, Anderson AW. 2018. Confirmation of a gyral bias in diffusion MRI fiber tractography. Hum Brain Mapp. 39:1449–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahmann JD, Pandya DN, Wang R, Dai G, D’Arceuil HE, de Crespigny AJ, Wedeen VJ. 2007. Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain. 130:630–65353. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Lu A, Schenker N, Damasio H. 2002. Humans and great apes share a large frontal cortex. Nat Neurosci. 5(3):272–276. [DOI] [PubMed] [Google Scholar]
- Shi F, Yap PT, Wu G, Jia H, Gilmore JH, Lin W, Shen D. 2011. Infant brain atlases from neonates to 1-and 2-year-olds. PloS one. 6:e18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Blake JA, Kadin JA. 2018. Mouse Genome Database (MGD)-2018: knowledgebase for the laboratory mouse. Nucleic Acids Res. 46:D836–D842. doi: 10.1093/nar/gkx1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JW, Mitchell PD, Kolasinski J, Grant EP, Galaburda AM, Takahashi E. 2015. Asymmetry of white matter pathways in developing human brains. Cereb Cortex. 25:2883–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striedter GF 2005. Principles of brain evolution. Sinauer Associates. [Google Scholar]
- Takahashi E, Dai G, Wang R, Ohki K, Rosen GD, Albert M, Grant PE, Wedeen VJ. 2010. Development of cerebral fiber pathways in cats revealed by diffusion spectrum imaging. Neuroimage. 49:1231–124040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E, Dai G, Rosen GD, Wang R, Ohki K, Rebecca D, Galaburda AM, Wedeen VJ, Ellen Grant P. 2011. Developing neocortex organization and connectivity in cats revealed by direct correlation of diffusion tractography and histology. Cereb Cortex. 21:200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E, Folkerth RD, Galaburda AM, Grant PE. 2012. Emerging cerebral connectivity in the human fetal brain: an MR tractography study. Cereb Cortex. 2:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Frank QY, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C. 2014. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci U S A. 111(46):16574–16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman AD, Charvet CJ, Clancy B, Darlington RB, Finlay BL. 2013. Modeling transformations of neurodevelopmental sequences across mammalian species. J Neurosci. 33(17):7368–7383. doi: 10.1523/JNEUROSCI.5746-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh WS, Bernard A, Royall JJ, Glattfelder KJ, Sunkin SM, Morris JA, Guillozet-Bongaarts A, et al. 2012. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 149(2):483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


