Abstract
Pineoblastoma (PB) are rare, aggressive pediatric brain tumors of the pineal gland with modest overall survival despite intensive therapy. We sought to define the clinical and molecular spectra of PB to inform new treatment approaches for this orphan cancer. Tumor, blood, and clinical data from 91 patients with PB or supratentorial primitive neuroectodermal tumor (sPNETs/CNS-PNETs), and 2 pineal parenchymal tumors of intermediate differentiation (PPTIDs) were collected from 29 centres in the Rare Brain Tumor Consortium. We used global DNA methylation profiling to define a core group of PB from 72/93 cases, which were delineated into five molecular subgroups. Copy number, whole exome and targeted sequencing, and miRNA expression analyses were used to evaluate the clinico-pathologic significance of each subgroup. Tumors designated as group 1 and 2 almost exclusively exhibited deleterious homozygous loss of function alterations in miRNA biogenesis genes (DICER1, DROSHA, and DGCR8) in 62 and 100% of group 1 and 2 tumors respectively. Recurrent alterations of the oncogenic MYC-miR-17/92-RB1 pathway were observed in the RB and MYC subgroup, respectively characterized by RB1 loss with gain of miR-17/92, and recurrent gain or amplification of MYC. PB sub-groups exhibited distinct clinical features: group 1-3 arose in older children (median ages 5.2-14.0 years) and had intermediate to excellent survival (5-year OS of 68.0-100%), while Group RB and MYC PB patients were much younger (median age 1.3-1.4 years) with dismal survival (5-year OS 37.5% and 28.6%, respectively). We identified age <3 years at diagnosis, metastatic disease, omission of upfront radiation, and chr 16q loss as significant negative prognostic factors across all PBs. Our findings demonstrate that PB exhibit substantial molecular heterogeneity with sub-group associated clinical phenotypes and survival. In addition to revealing novel biology and therapeutics, molecular sub-grouping of PB can be exploited to reduce treatment intensity for patients with favorable biology tumors.
Keywords: pineoblastoma, PNET, PPTID, miRNA, RB, MYC
Introduction
Malignant brain tumors are the leading cause of pediatric cancer-related death and disability. Embryonal brain tumors (EBTs) are the largest group of brain tumors diagnosed in children 0-14 years old and comprise 20% of all pediatric brain neoplasms [47]. Although historically classified based on tumor location and similar primitive neuroectodermal tumor (PNET) histology [24], EBTs are known to comprise a spectrum of molecular diseases with distinct clinico-pathologic features [8]. Medulloblastoma (MB) which represents 60% of childhood EBTs has been most studied, while rare EBTs, which comprise ~40% of EBTs are understudied and poorly understood. These include atypical rhabdoid/teratoid tumors (ATRTs), embryonal tumors with multilayered rosettes (ETMRs), as well as pineoblastoma (PB) - all historically treated as high-risk brain tumors with intensified regimens [30,29].
PBs comprise 30% of all pineal region tumors and may be difficult to distinguish from other tumors including germ cell tumors, high-grade gliomas, ATRTs, ETMRs and lower-grade pineal parenchymal tumors of intermediate differentiation (PPTIDs) [32]. PB have been grouped in clinical and biological studies with other EBTs arising in cerebral locations, called supratentorial primitive neuro-ectodermal tumors (sPNETs or CNS-PNETs) [42]. As there are few dedicated PB studies, the clinical and molecular spectra, and best treatment approach for these highly malignant tumors remains to be established. A recent large clinical retrospective study indicated radiotherapy (RT) but not high-dose chemotherapy (HDC) improved survival of PB patients ≥4 years old [45], although prospective consortia studies show improved survival for older children with intensified multi-modal approaches [29,27,11]. Historical sPNET studies also reported 5-yr OS of 50-65% for older children with pineal region EBTs, while patients < 3-5 years old had poorer 5-yr OS of 15-40% [30,20]. Whether these observations reflect age-related treatment biases or biological differences remain unknown.
Limited animal modeling data [57] and clinical studies of heritable “tri-lateral” retinoblastoma [14,6] suggest a role for RB1 and related tumor suppressor pathways in PB. In addition, miRNA biogenesis gene defects have also been recently implicated in PBs [15,58]. MiRNAs which are critical post-transcriptional regulators, undergo complex processing by endonucleases (DROSHA, DGCR8, and DICER1) into mature miRNAs that function in RNA-induced silencing complexes (RISC) [37]. Although, several small studies have reported DICER1 and DROSHA alterations in PB, the spectrum of RB and miRNA biogenesis alterations and their clinical significance in PBs remains to be fully evaluated. In this study, we integrated global DNA methylation profiling, copy number, and whole exome (WES) and targeted sequencing analyses on a large cohort of PB patients enrolled in a rare brain tumor registry to investigate the molecular and clinic-pathologic spectrum of PB.
Materials and Methods
Tumor, blood, and clinical data
Tumor tissue, blood, and clinical data from 93 patients diagnosed with PB, related sPNETs/CNS-PNETs, or PPTID were collected from 29 centres as part of the global Rare Brain Tumor Consortium biorepository and clinical registry (rarebraintumorconsortium.ca) using procedure approved by Research Ethic Board at the Hospital for Sick Children and participating institutions (Supplementary Table 1, online resource) . All cases were diagnosed at their referring institutions. Available pathology reports and prepared slides were all reviewed by an experienced pediatric neuropathologist. Six of these cases have previously been analyzed by Affymetrix 100K single-nucleotide polymorphism (SNP) array and reported by Miller et al.[42]. DNA from frozen tissue or formalin-fixed, paraffin-embedded materials, and blood were extracted using the Qiagen AllPrep DNA/RNA Mini kit (Qiagen, Germany), and total RNA from 6 tumors was prepared with nCounter miRNA prep kit according to standard protocol.
Molecular and bioinformatic analyses
Tumor DNA was analysed on the Illumina HumanMethylation450 or MethylationEPIC methylation arrays (Illumina, San Diego, CA) as described previously (www.tcag.ca) [64,59] and 5000-15,000 most variable probes (standard deviation >0.3) were used for all downstream analyses (R v3.3.1). Tumor types were determined using unsupervised cluster analyses of methylation data against 1200 reference tumor profiles [59].
For t-distributed stochastic neighbor embedding, default parameters were used, except for perplexity = 10 (Rtsne v0.15, R v3.5.3). For hierarchal clustering, 1-Pearson correlation was used for distance measuring, with average linkage (pheatmap R package, R v3.61). k-means clustering was performed with Euclidean for distance measuring, and average linkage (ConsensusClusterPlus R package). Non-matrix factorization (NMF) analysis was performed with ranks (k) 2-10 at 100 runs (NMF v0.20.6). Tumor copy number profiles were determined using Conumee (version 1.8.0) and GISTIC2 (v2.0.23) [41] analyses on methylation and Illumina Omni SNP array.
WES analysis was performed on the Illumina HiSeq 4000 platform (Genome Quebec, TCAG), with variant calling using the Mutek2 pipeline (Ontario Institute for Cancer Research). Targeted sequencing was performed on the Ion Torrent platform using custom primers (Thermo Fisher Scientific) and the Ion Reporter variant calling pipeline (Genome Quebec, ResourcePath) [41]. Mutations were called deleterious or potentially deleterious based respectively on calls by both or one of the Sorting Intolerant From Tolerant (SIFT) (<0.05) or Polyphen-2 (>0.909) tool scores [66,1]. MiRNA expression was determined based on the NanoString miRNA panel (NanoString Technologies Inc.) [59] for available tumor-derived miRNA.
Statistical analyses
Event-free survival (EFS) was defined as interval between time of diagnosis to first event: tumor recurrence or progression, death from any cause, or last follow-up for those without events. Overall survival (OS) was defined as interval between time of diagnosis and death from any cause or last follow-up. Survival and prognostic factor analyses were performed on cases treated with curative intent and for which complete treatment and outcome information were available. Survival estimates were performed using Kaplan-Meier method with 95% CI, with log-rank testing used for comparisons. Fisher exact and Kruskal-Wallis analyses were used to evaluate association of specific clinical features (age, tumor location, stage) with PB sub-groups, while univariate Cox proportional hazards regression modelling was used to identify clinical and treatment prognostic factors. All statistical analyses were performed in R v3.6.1.
Results
PB segregates into five molecular subgroups with distinct copy number profiles
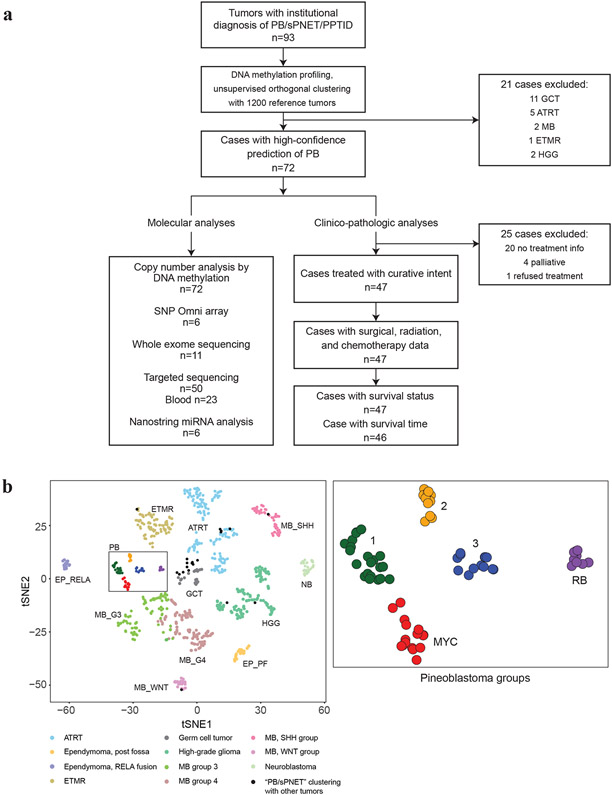
Global methylation data from 93 tumors diagnosed as PB, sPNETs/CNS-PNETs, or PPTID were analysed against a reference cohort of 1200 pediatric brain tumors [59] using unsupervised orthogonal clustering (t-distributed stochastic neighbour embedding, NMF, K-means and hierarchal clustering) analyses (Fig. 1a, b). Attributable to the difficulty in diagnosing PB, 21/93 cases clustered with other tumor entities (11 germ cell tumor, 5 ATRTs, 2 MB, 2 high-grade glioma, 1 ETMR), and were excluded from further analysis. The remaining 72 tumors which segregated in one distinct cluster were further characterized using NMF, hierarchal clustering, and K-means clustering which revealed 5 robust sub-groups with highest co-phenetic co-efficient score at k=5 (Supplementary Fig. 1, online resource). We designated these as group 1, 2, 3, RB, and MYC PB sub-groups, respectively consisting of 21, 11, 13, 9, and 18 tumors, based on specific copy number and mutational features described below.
Figure 1. PB comprise five molecular sub-groups.
a. Flow diagram of analyses performed: 93 primary tumors with institutional diagnosis of pineoblastoma (PB) or supratentorial PNET (sPNET) were analysed using global methylation profiling and compared against a reference cohort of 1200 pediatric brain tumors to identify and exclude samples that segregated with other brain tumors. A cluster of robust, molecularly confirmed 72 PBs were further characterized using methylation and SNP arrays for copy number alterations, mutational analyses using WES and targeted sequencing, and Nanostring analyses for miRNA expression. Clinical, treatment, and molecular sub-group data available for 46 PB patients treated with curative intent were integrated for clinic-pathologic analyses.
b. t-Distributed stochastic neighbour embedding (tSNE) plots of DNA methylation clustering patterns of 93 presumed PB samples relative to 951/1200 representative pediatric brain tumor entities demonstrate PB clusters separately from other tumor entities. Plots using the top 12,500 most varying methylation probes by standard deviation (SD) are shown. Tumors are shown as colored spheres which include atypical teratoid rhabdoid tumor (ATRT), ependymoma posterior-fossa (EP_PF) or supratentorial, RELA-fusion (EP_RELA), embryonal tumor multiple rosettes (ETMR), germ cell tumor (GCT), high-grade glioma (HGG), neuroblastoma (NB), medulloblastoma WNT (MB_WNT), SHH (MB_SHH), group 3 (MB_G3), and group 4 (MB_G4). Black spheres indicate tumors with an institutional diagnosis of PB that segregated with other known brain tumor entities are (n=21). A robust cluster of 72 PBs is boxed; blow-up image of PB cluster on right shows five molecular PB sub-groups designated as 1, 2, 3, RB, and MYC.
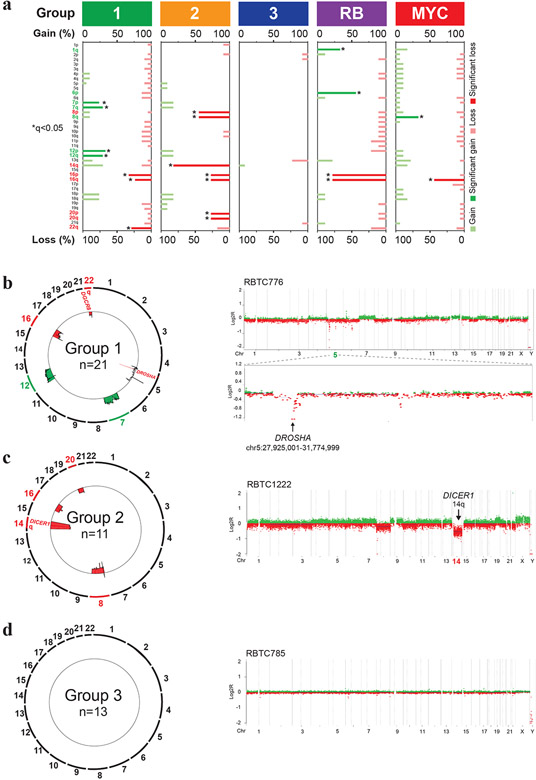
To further investigate PBs sub-groups, we performed copy number analyses using Conumee and GISTIC2 analyses on methylation and SNP array data (Fig. 2a), which revealed few significant overlapping copy number alterations except for chr 16 loss seen in all but group 3 PBs. Group 1 tumors most frequently exhibited broad gains of chr 7 (5/21; 24%) and chr 12 (6/21; 29%) and losses of chr 16 (5/21; 24%) and 22q (6/21; 29%). More detailed analysis revealed 14% (3/21) of group 1 tumors exhibited recurrent loss of a minimal 1.4Mb region on chr 5p13.3 encompassing DROSHA, which mediates primary-miRNA processing (Fig. 2b). In group 2 tumors, DNA methylation (Fig. 2c) and SNP array (Supplementary Fig. 2a, online resource) data showed broad chr 14q (9/11; 82%) losses where miRNA endonuclease gene DICER1 maps, and focal homozygous DROSHA loss in one sample. Additionally, group 2 tumors exhibited loss of chr 8 (5/11; 45%), 16 (3/11; 27%), and 20 (3/11; 27%). In contrast, group 3 PB had no significant recurrent copy number alterations except for chr13q loss in 3/13 (23%) samples (Fig. 2d).
Figure 2. PB molecular subgroups have distinct copy number landscapes.
a. Pattern of copy number alterations across PB molecular sub-groups as determined using GISTIC analyses of global methylation data. Chromosomal regions with recurrent copy number gains (green) or losses (red) significantly enriched within each PB sub-group are highlighted; asterisk indicates false discovery rate of q<0.05.
b, c. Composite circos plots of global methylation profiles showing recurrent copy number gains (green) and losses (red) in 21 group 1 and 11 group 2 PBs. Focal or broad alterations associated with miRNA biogenesis loci DICER1, DROSHA and DGRC8 are highlighted. Higher resolution copy number profiles generated using Conumee, of representative group 1 and group 2 samples with respective focal chr 5p13.3 targeting DROSHA and chr 14q loss associated with DICER1, are shown on the right.
d. Composite circos plot of global methylation profiles in 13 group 3 PBs. Higher resolution copy number profile generated using Conumee of a representative group 3 sample is shown on right.
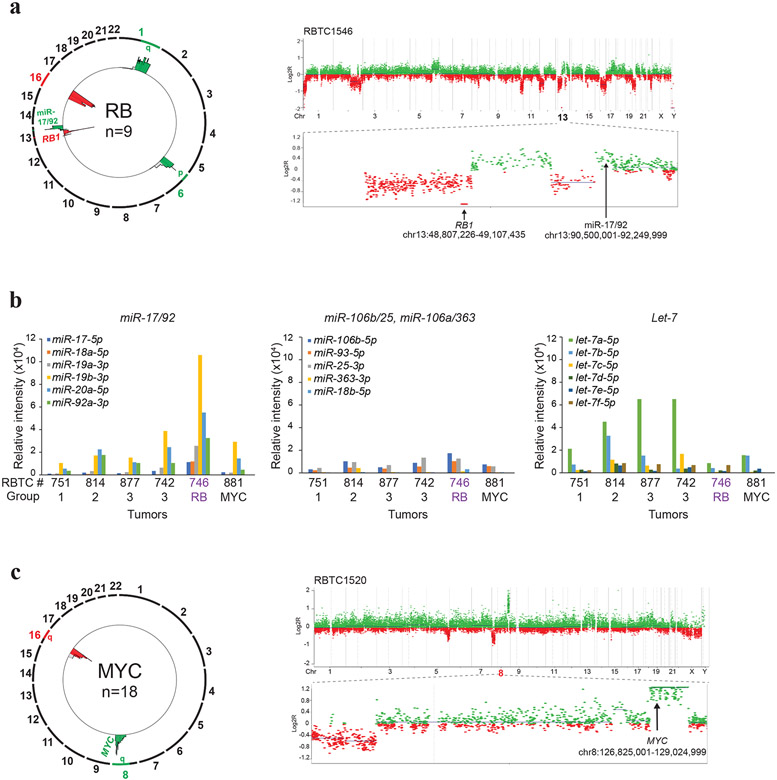
In the fourth designated RB sub-group, methylation and SNP arrays showed recurrent losses of a focal 0.6Mb chr 13q14.2 region spanning RB1 in 56% (5/9) of samples (Fig. 3a; Supplementary Fig. 2b, online resource); 80% (4/5) of these also harbored focal gains of a 1.9Mb chr13q13.3 region spanning the miR-17/92 oncogene previously implicated in retinoblastoma [12]. Nanostring expression profiling on a cohort of 6 primary PBs indicated copy number driven miR-17/92 expression in a group RB PB (RBTC746), without significant changes in expression of paralogous loci, miR-106b/25 and miR-106a/363, or the unrelated let-7 locus (Fig. 3b). The RB sub-group also exhibited broad chr 1q (3/9; 33%) and 6p (5/9; 55%) gains and chr 16 losses (7/9; 78%) (Fig. 3a). The fifth sub-group, designated as the MYC PB, exhibited recurrent focal gains (7/18; 39%) or amplification (2/18; 11%) of a 1.2 Mb chr 8q24.21 segment encompassing MYC and chr 16q losses (8/18; 44%) (Fig. 3c).
Figure 3. Recurrent copy number alterations in RB and MYC sub-group PBs.
a. Composite circos plot of global methylation profiles from 9 RB subgroup PBs depicting recurrent copy number gains (green) and losses (red); recurrent copy number alterations associated with miR-17/92 and RB1 are highlighted. Higher resolution copy number profile of a representative tumor RBTC 1546 with homozygous loss of RB1 at chr13q14.2 and copy number gain encompassing miR-17/92 at chr 13q31.3 is shown on right.
b. Copy number driven expression of miR-17/92 in RB sub-group PB. MiRNA expression levels for the miR-17-92, paralogous miR106a-363, miR-106b-25 and unrelated let-7 loci was determined from NanoString(v.3) miRNA expression data from 6 PBs. Plots show relative, normalized probe intensities of miRNAs in PB sub-groups; miRNA expression levels of RBTC746 with focal chr13q13.3 copy number gains targeting miR-17-92 shown in Figure A, is highlighted.
c. Composite circos plot of global methylation profiles from 18 MYC subgroup PBs. Recurrent focal chr 8q amplification/gains (green) and chr 16q losses (red) are highlighted. Higher resolution copy number profile of a representative tumor, RBTC 1520, with focal MYC amplification is shown on right.
miRNA biogenesis defects, RB1 loss, and MYC activation characterize PB sub-groups
To extend our copy number analyses we performed WES for 11 samples and targeted sequencing of DICER1, DROSHA, DGCR8, XPO5, TARBP2, RB1, and TP53 for 48 tumor and 21 matched blood samples with limited materials; 2 additional tumors and 2 blood samples only had materials sufficient for targeted DICER1 and TP53 sequencing only.
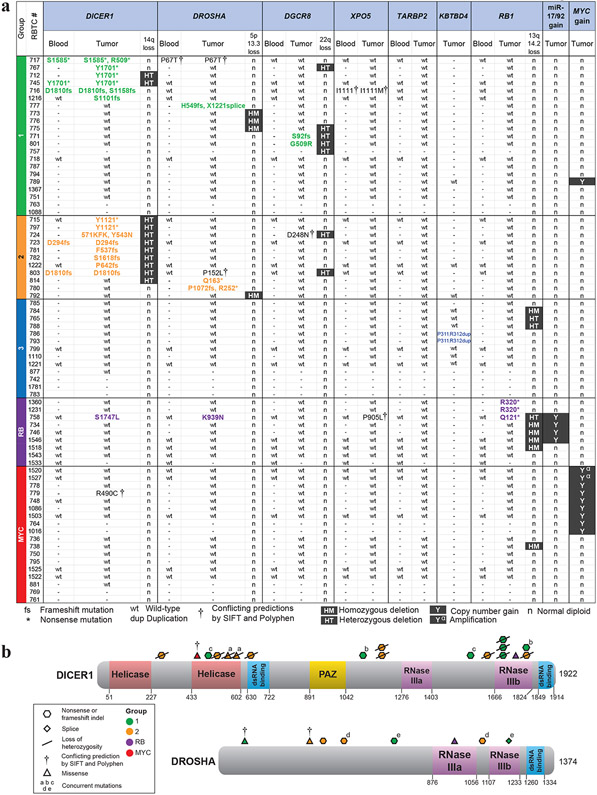
Sequencing analyses revealed mutually exclusive recurrent, deleterious, loss of function mutations of DICER1, DROSHA, or DGCR8 almost exclusively in group 1 and 2 PBs (Fig. 4a). Significantly 11/15 DICER1, 6/8 DROSHA and 3/3 DGCR8 alterations were novel cancer mutations not reported in COSMIC (https://cancer.sanger.ac.uk/cosmic) (Table 1). Of 15 unique DICER1 mutations in 16 PBs, 11 were nonsense/frameshift and 4 were missense mutations. Truncating DICER1 mutations were located within or prior to the RNase IIIb domain, while missense mutations mapped to the RNase IIIb and Helicase domains. DROSHA mutations, which were distributed throughout the gene, were also predominantly truncating (5/8), while only 1/3 DGCR8 mutations was predicted to be truncating. Less common alterations included two novel, potentially deleterious missense mutations of XPO5, which functions in pre-miRNA export. No alterations in TARBP2, a DICER miRNA loading complex gene, mutated in a spectrum of cancers [21,16], were seen in our PB cohort. Significantly, we also identified germline DICER1 mutation in 5 patients and a potential deleterious missense germline DROSHA mutation (RBTC717, c.199C>A; p.P67T) in one patient. Of note, all DICER1 mutations in group 1 (6/6) and 2 (9/9) PBs were accompanied by deleterious somatic DICER1 mutations or heterozygous chr 14q loss, variant allele frequency >96%, or complete chr14q loss. Similarly, three tumors with DGCR8 mutations, both in groups 1 and 2 PBs, also exhibited chr 22q loss. Collectively our data shows critical miRNA biogenesis genes are targeted by copy number alterations and/or mutations in 62 (13/21) and 100% (11/11) respectively of group 1 and 2 PBs (Fig. 4).
Figure 4. Recurrent mutations/alterations of miRNA biogenesis genes, RB1 and MYC characterize PB sub-groups.
a. Summary of mutations and copy number alterations associated with miRNA biogenesis gene (DICER1, DROSHA, DGCR8, XPO5, TARBP2), KBTBD2, RB1, miR-17/92, and MYC determined using a combination of targeted sequencing, WES, methylation and SNP array based copy number analyses in individual PBs of different sub-groups with tumor and matched blood DNA available for study. Samples lacking materials for specific assay are indicated by (-); broad copy number alterations determined by methylation or SNP-based copy number analyses are indicated by HT (heterozygous), HM (homozygous), n (normal diploid) or presence (Y) of MYC focal gains or amplification (α) is indicated. Status or specific gene alterations determined by targeted sequencing or WES is indicated as wt (wild-type); * (stop-gain mutation), fs (frameshift insertion or deletion), † (deleterious missense mutation predicted by SIFT and Polyphen2). All predicted truncating gene mutations are highlighted.
b. Schema of DICER1 and DROSHA mutations relative to maps of corresponding proteins. Type and location of mutations are shown as colored symbols relative to amino acid sequence numbers and known or predicted functional domains; colors of mutation symbols correspond to tumor sub-group.
Table 1.
Summary of PB mutations identified in this study
| Gene | Mutation | Type | Predicted deleterious effect of mutation |
Observed in PB cohort |
Observations in COSMIC |
Cancer types in COSMIC |
|---|---|---|---|---|---|---|
| DICER1 | Y1701* | Nonsense | Truncating | 3 | 1 | Liver |
| S1585* | Nonsense | Truncating | 1 | Novel | ||
| R509* | Nonsense | Truncating | 1 | 3 | Melanoma | |
| Y1121* | Nonsense | Truncating | 2 | Novel | ||
| D1810fs | Frameshift indel | Truncating | 2 | Novel | ||
| S1158fs | Frameshift indel | Truncating | 1 | Novel | ||
| S1101fs | Frameshift indel | Truncating | 1 | Novel | ||
| D294fs | Frameshift indel | Truncating | 1 | Novel | ||
| F537fs | Frameshift indel | Truncating | 1 | Novel | ||
| S1618fs | Frameshift indel | Truncating | 1 | Novel | ||
| P642fs | Frameshift indel | Truncating | 1 | Novel | ||
| Y543N | Missense | Altered helicase domain | 1 | Novel | ||
| S1747L | Missense | Altered RNase IIIb domain | 1 | 2 | Breast | |
| R490C | Missense | Helicase Domain † | 1 | 1 | Bladder | |
| 571_573delinsKFK | Missense | Helicase Domain - unknown | 1 | Novel | ||
| DROSHA | Q163* | Nonsense | Truncating | 1 | Novel | |
| R252* | Nonsense | Truncating | 1 | Novel | ||
| H549fs | Frameshift indel | Truncating | 1 | Novel | ||
| P1072fs | Frameshift indel | Truncating | 1 | Novel | ||
| X1221_splice | Splice site | Truncating | 1 | Novel | ||
| P152L | Missense | Not in functional domain † | 1 | Novel | ||
| P67T | Missense | Not in functional domain † | 1 | 1 | Large intestine | |
| K939N | Missense | Altered RNase IIIa domain | 1 | 1 | Breast | |
| DGCR8 | S92fs | Frameshift indel | Truncating | 1 | Novel | |
| G509R | Missense | Immediately adjacent to DRBM1 domain | 1 | Novel | ||
| D248N | Missense | Not in functional domain † | 1 | Novel | ||
| XPO5 | I1111M | Missense | Unknown † | 1 | Novel | |
| P905L | Missense | Unknown † | 1 | Novel | ||
| RB1 | R320* | Nonsense | Truncating | 2 | 21 | Retinoblastoma, endometrial, breast |
| Q121* | Nonsense | Truncating | 1 | 2 | Lung, thyroid | |
| KBTBD4 | p.P311_R312dup | In-frame insertion | Altered Kelch binding domain | 2 | 0 | PPTID a, MB b |
Targeted sequencing of ten group 3 PB did not reveal any miRNA biogenesis genes, RB1 or TP53 alterations. Interestingly, additional WES analyses of group 3 PB samples revealed 2/8 (RBTC786 and −793) harbored similar in-frame insertions (c.935_936insCGTGGG and c.937_938insGCCGTG, respectively) in KBTBD4, which encodes a Cul3 E3 ubiquitin ligase adaptor, resulting in a p.P311_R312dup affecting the Kel substrate binding domain (Supplementary Fig. 3, online resource) [7]. While the c.937_938insGCCGTG mutation has recently been proposed to be a marker for PPTIDs [35], both cases of PPTID in our cohort, which were group 3 tumors, did not have this alteration on WES [18]. While both our tumors with this mutation were institutionally diagnosed as PB, they had lower Ki67 labeling indices (Supplementary Table 2, online resource) more consistent with PPTID based on values reported by Fèvre-Montange et al. [18]. Examining all group 3 tumors diagnosed as PB, Ki67 scores were at the threshold between PPTID and PB (mean 19.2%, range 10-40%). In total, group 3 tumors (mean 16.6%, range 3-40%) had significantly lower Ki67 scores than tumors belonging to other groups (mean 39.6%, range 10-75%) (p=0.003 by Kruskal-Wallis test).
In contrast to group 1 and 2 tumors, sequencing of 22 RB and MYC sub-group PBs revealed only two potentially deleterious DICER1 mutations without evident LOH, one each in a MYC (RBTC779, c.1468N>T; p.R490C) and a RB subgroup (RBTC758, c.5240C>T; p.S1747L) tumor. Consistent with copy number analyses, sequencing revealed 3/8 (38%) RB sub-group tumors had recurrent stop-gain RB1 (p.R320* and p.Q121*) mutations previously reported in other cancers including retinoblastoma [39,22]. Amongst the RB subgroup patients, one (RBTC1231) presented in the context of tri-lateral retinoblastoma for which confirmatory germline testing could not be performed. Targeted sequencing of 21 blood samples, including 5 from RB subgroup patients, did not reveal any additional RB1 germline mutations. Notably, we did not identify somatic or germline TP53 mutations in 48 PBs and 10 matched blood DNA samples sequenced.
PB subgroups have distinct clinico-pathologic features
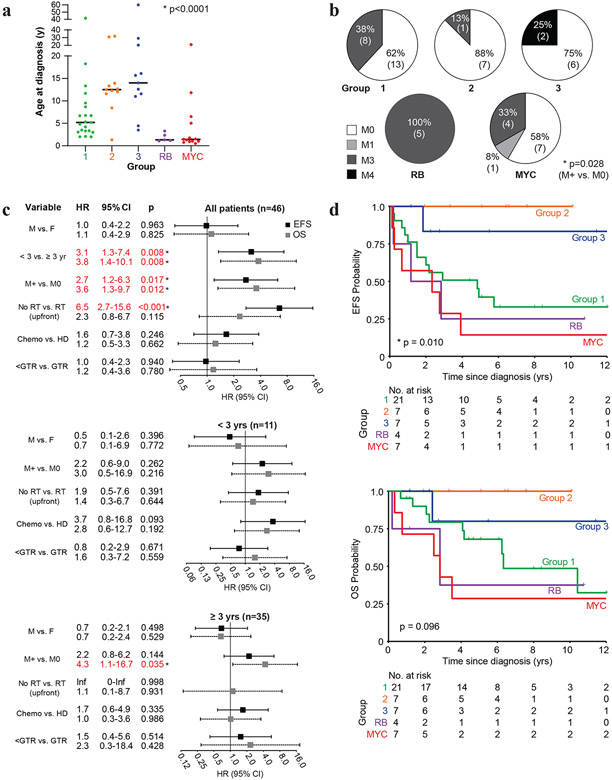
Although PB predominantly arises in children, we observed a wide range of ages from six months to 60 years among 61/72 patients with available data, with 87% of patients ≤18 yrs of age, and children <3yrs comprising 28% of all patients (Supplementary Table 3, online resource). Comparison of clinical features showed no gender bias in the entire cohort (p=0.127) although, there was a predominance of females and males respectively in the RB (1 male:3.5 female) and MYC (2.6 male: 1 female) group of patients (Table 2). While children with group 1-3 PBs had respective median ages of 5.2, 12.5, and 14.0 years at diagnosis, the RB and MYC group patients were much younger with median ages of 1.3 and 1.4 years respectively (p<0.0001) (Fig. 5a). Staging data available for 54 patients indicated 39% (21/54) of primary PB were metastatic; 20/21 patients presented with M3/M4 disease, and only one with M1 disease. Incidence of metastases at diagnosis differed significantly across PB groups (p= 0.028) with group 2 and RB group patients respectively exhibiting the lowest (13% M2; 1/8 patients) and highest incidence (100% M3; 5/5 patients) of metastases (Fig. 5b).
Table 2.
Summary of patient features and treatment across PB sub-groups
| Total | Group 1 | Group 2 | Group 3 | RB | MYC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | n | % | |||||
| Clincial features | Number of patients | 72 | 21 | 29 | 11 | 15 | 13 | 18 | 9 | 13 | 18 | 25 | p-value | |||
| Gender | 72 | 21 | 11 | 13 | 9 | 18 | 0.127 | |||||||||
| Male | 36 | 50 | 10 | 48 | 4 | 36 | 7 | 54 | 2 | 22 | 13 | 72 | ||||
| Female | 36 | 50 | 11 | 52 | 7 | 64 | 6 | 46 | 7 | 78 | 5 | 28 | ||||
| Age | 61 | 21 | 11 | 11 | 5 | 13 | <0.0001 * | |||||||||
| Median (yrs) (range) | 6.5 | (0.5-60) | 5.2 | (2.0-41.5) | 12.5 | (1.3-31.9) | 14.0 | (3.5-60) | 1.3 | (1.1-3.3) | 1.4 | (0.5-21.0) | ||||
| <1 | 3 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 23 | ||||
| 1 to <3 | 14 | 23 | 3 | 14 | 1 | 9 | 0 | 0 | 4 | 80 | 6 | 46 | ||||
| 3 to <10 | 20 | 33 | 14 | 67 | 1 | 9 | 2 | 18 | 1 | 20 | 2 | 15 | ||||
| 10 to <18 | 16 | 26 | 2 | 10 | 7 | 64 | 6 | 55 | 0 | 0 | 1 | 8 | ||||
| ≥18 | 8 | 13 | 2 | 10 | 2 | 18 | 3 | 27 | 0 | 0 | 1 | 8 | ||||
| Stage | 54 | 21 | 8 | 8 | 5 | 12 | 0.028 * | |||||||||
| M0 | 33 | 61 | 13 | 62 | 7 | 88 | 6 | 75 | 0 | 0 | 7 | 58 | ||||
| M+ | 21 | 39 | 8 | 38 | 1 | 12 | 2 | 25 | 5 | 100 | 5 | 42 | ||||
| Treatment features: intent to tre at group | Surgery | 47 | 21 | 7 | 7 | 4 | 8 | 0.431 | ||||||||
| GTR | 17 | 36 | 5 | 24 | 3 | 43 | 4 | 57 | 1 | 25 | 4 | 50 | ||||
| <GTR | 30 | 64 | 16 | 76 | 4 | 57 | 3 | 43 | 3 | 75 | 4 | 50 | ||||
| Radiotherapy | 47 | 21 | 7 | 7 | 4 | 8 | 0.041 * | |||||||||
| Yes | 37 | 79 | 17 | 81 | 7 | 100 | 7 | 100 | 2 | 50 | 4 | 50 | ||||
| No | 10 | 21 | 4 | 19 | 0 | 0 | 0 | 0 | 2 | 50 | 4 | 50 | ||||
| Chemo | 46 | 21 | 7 | 6 | 4 | 8 | 0.749 | |||||||||
| HDC | 29 | 63 | 15 | 71 | 4 | 57 | 3 | 50 | 3 | 75 | 4 | 50 | ||||
| Conventional | 17 | 37 | 6 | 29 | 3 | 43 | 3 | 50 | 1 | 25 | 4 | 50 | ||||
| Survival analysis | Status | 47 | 21 | 7 | 7 | 4 | 8 | 0.019 * | ||||||||
| Dead | 18 | 38 | 9 | 43 | 0 | 0 | 1 | 14 | 2 | 50 | 6 | 75 | ||||
| Alive | 29 | 62 | 12 | 57 | 7 | 100 | 6 | 86 | 2 | 50 | 2 | 25 | ||||
| Recurrence | 46 | 21 | 7 | 7 | 4 | 7 | 0.003 * | |||||||||
| Yes | 21 | 46 | 12 | 57 | 0 | 0 | 1 | 14 | 2 | 50 | 6 | 86 | ||||
| No | 25 | 54 | 9 | 43 | 7 | 100 | 6 | 86 | 2 | 50 | 1 | 14 | ||||
| Median follow-up time (yrs) (range) | 4.2 (0.2-20.3) | 4.7 (0.7-20.3) | 6.4 (1.9-10.1) | 2.8 (1.2-13.9) | 2.0 (0.2-10.8) | 3.2 (0.3-17) | ||||||||||
| 5-yr survival (%) | ||||||||||||||||
| EFS (95% CI) | 48.1 (32.2-62.3) | 39.5 (18.5-60.0) | 100 (n/a) | 83.3 (27.3-97.5) | 25 (0.9-66.5) | 14.3 (0.7-46.5) | 0.009 * | |||||||||
| OS (95% CI) | 65.0 (47.8-77.7) | 68.0 (42.0-84.2) | 100 (n/a) | 80 (20.4-96.9) | 37.5 (1.1-80.8) | 28.6 (4.11-61.2) | 0.096 | |||||||||
Figure 5. Molecular sub-groups of PB have distinct clinicopathologic features.
a. Scatterplot of age at diagnosis for PB patients relative to tumor molecular sub-group. Bar indicates median age as determined using Kruskal-Wallis test.
b. Frequency of metastatic (M+; M1, −3, −4) and non-metastatic (M0) disease determined as per the Chang staging system is shown relative to PB sub-groups; significance in distribution of M+ versus M0 patients across all PB sub-groups was determined using Fisher exact test.
c. Forest plot of Hazard ratio (HR) from univariate Cox proportional hazards regression model of gender (male/M vs. female/F), age, metastatic status (M+ vs M0), radiotherapy (no upfront RT vs. upfront RT), conventional chemotherapy only (chemo) vs. high-dose chemotherapy (HD), and extent of tumor removal (less than gross total resection/GTR vs GTR) on EFS (black) and OS (gray) was performed on data from 46 patients treated with curative intent. Whiskers denote 95% confidence interval.
d. Kaplan-Meier survival analyses of event free (EFS) and overall survival (OS) for 46 patients treated with curative intent stratified by PB sub-groups. Plots abbreviated to maximum of 12 years from diagnosis. For patients with group 1-3, RB, and MYC PBs EFS were respectively, 39.5%, 100%, 83.3%, 25.0%, and 14.3%; 5-year OS were 68.0%, 100%, 80%, 37.5%, and 28.6%.
46/72 PB patients were treated with curative intent and had complete treatment and outcome information available (Table 2). Univariate analyses revealed age <3yrs as a significant negative prognostic factor for EFS (HR 3.1, CI 1.3-7.4, p=0.008) and OS (HR 3.8, CI 1.4-10.1, p=0.008). EFS (HR 2.7, CI 1.2-6.3; p=0.017) and OS (HR 3.6, CI 1.3-9.7, p=0.012) were also significantly inferior in patients with metastatic disease at diagnosis. Patients who did not receive upfront RT also had inferior EFS (HR 6.5, CI 2.7-15.6, p<0.001) but not OS (HR 2.3, CI 0.8-6.7, p=0.115), while receipt of conventional chemotherapy only vs. HDC, and extent of surgery were not significantly associated with EFS or OS. As PB patients <3yrs are often treated without RT or with delayed RT regimens, we also examined prognostic factors stratified by age <3 and ≥3yrs at diagnosis. These analyses showed no significant prognostic factors except a trend toward poorer EFS with conventional dose chemotherapy compared to HDC among 11 children <3yrs, while metastatic disease remained a significant negative prognostic factor for OS (HR 4.3; 95% CI 1.1-16.7; p=0.035) but not for EFS in children ≥3yrs of age at diagnosis.
Kaplan-Meier survival analyses for all PB patients treated with curative intent revealed 5-yr EFS and OS respectively of 48.1 and 65.0%. Consistent with our Cox proportional hazards regression model, patients < 3 yrs, metastatic disease at diagnosis, and who were not treated with upfront RT had significantly poorer survival. The 5-yr EFS and OS for patients stratified by ≥ 3 vs. < 3 yrs of age were 58.2% vs. 18.2% (p=0.005) and 77.0% vs. 24.2%, (p=0.005) respectively (Supplementary Fig. 4a, online resource), while patients with localized and metastatic disease had 5-yr EFS and OS of 60.5% vs. 29.4% (p=0.012) and 78.7% vs. 44.8% (p=0.008) respectively. Patients treated with and without upfront RT had respective 5yr EFS of 58.8% vs 10% (p<0.0001), while upfront RT was also associated with a trend towards improved survival: 5-yr OS for patients who received upfront RT was 71% vs. 40% for patients who did not receive upfront RT (p=0.106) (Supplementary Fig. 4b-c, online resource).
In addition to clinical risk factors, our analyses indicated PB survival also correlated with molecular features of tumors. Notably, EFS differed significantly across the five molecular subgroups of PB (p=0.009) while OS trended toward significance (p=0.096), with group 2 PB patients exhibiting a striking 100% 5-yr EFS/OS (Fig. 5d). In contrast, the RB and MYC sub-groups of PB, which correlated with youngest age at diagnosis and highest frequency of chr 16q loss, had the lowest 5-yr EFS/OS of only 25%/37.5% and 14.3%/28.6%, respectively (Table 2). Because chr 16q loss was associated with these high-risk groups but also seen recurrently in groups 1 and 2, we analyzed whether it was independently associated with poorer outcomes. Indeed, across all cases, those with chr 16q loss compared to unaltered chr 16q were associated respectively with 16.1% vs. 63.0% 5yr-EFS (p=0.015) while OS were respectively 47.0% vs. 72.4% (p=0.139) (Supplementary Fig. 5, online resource). Collectively our data suggest distinct tumor biology are associated with different clinical risk features and may contribute significantly to disparate treatment-related outcomes in PBs.
Discussion
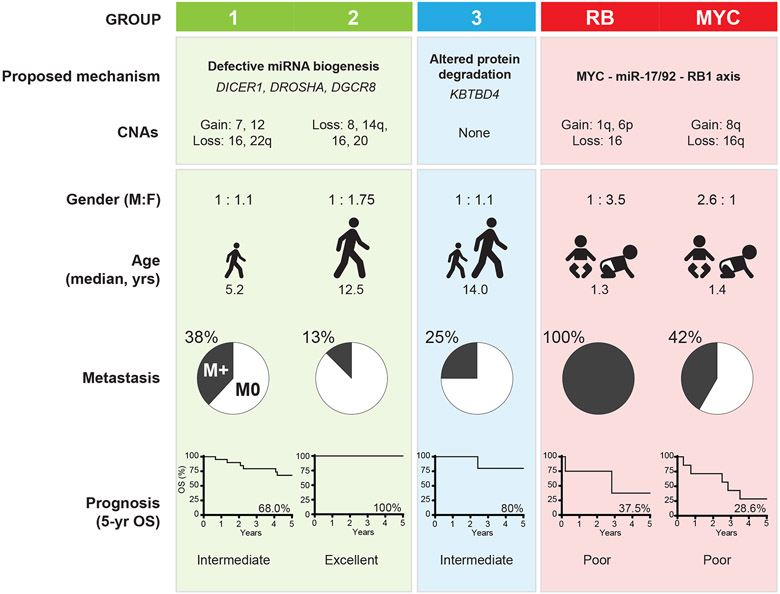
PBs are high-risk brain tumors with only modest long-term survival despite multi-modal intensive regimens and for which there remains limited data to inform novel therapeutic approaches [20,29,45,27,11]. Here we performed an integrated molecular and clinic-pathologic analyses of a large cohort and demonstrate PBs comprise 5 molecular sub-groups with distinct clinico-pathologic and survival features. Group 1 and 2 PB which arise in older children exhibit recurrent miRNA biogenesis gene defects; group 3 PB which affects adolescents and adults exhibit few alterations, while the RB and MYC sub-groups affecting children age <18 months harbor RB1 and MYC alterations. Our data indicate age <3yrs, metastases at diagnosis and tumor molecular features as important determinants of survival in PB patients (summarized in Fig. 6) and provide an important framework for prospective studies.
Figure 6.
Schematic summary of molecular and clinical features across PB sub-groups
Strikingly, we identified deleterious mutations in multiple components of the miRNA processing machinery almost exclusively in group 1 and 2 PBs. Consistent with association of PBs with DICER1 predisposition syndrome [15,58], we identified germline and somatic DICER1 mutations in addition to somatic DROSHA and DGCR8 mutations, which have not been reported in PBs to date. With the exception of reported nonsense mutations [15] in RBTC717 and −745, all of the DICER1 mutations identified in our study were novel and those in group 1 PB most commonly affected the RNase IIIb domain which selectively processes 5p miRNA [26]. Interestingly, imbalanced abundance of 5p versus 3p miRNAs due to RNase IIIb domain mutations have been implicated as important oncogenic mechanisms [28,3,53]. In contrast, mutations in group 2 tumors affecting both RNase domains were predicted to completely impair miRNA maturation, as reported in Wilm’s tumors [54]. Group 1 and 2 PBs with DICER1 mutations exhibited LOH as reported in smaller PB studies [15,58]. This is in stark contrast to DICER1 mutations in other tumors, where LOH is rare and truncating germline mutations are associated with hotspot missense mutation of the RNase IIIb domain [19]. In PBs, the second hit appears to be either chr 14q loss or a second truncating mutation of both RNase domains. Interestingly, murine tumors with bi-allelic Dicer1 knockout appear to be selected against [33] suggesting PB tumors likely retain some residual DICER1 activity, either through conserved RNase IIIa domain (in group 1) or other aberrant functions not involving the RNase domains (in group 2). The unique pattern of DICER1 somatic and germline mutations observed in our cohort suggest specificity of the second hit in the formation of this tumor.
We identified truncating and damaging missense mutations of both DROSHA and DGCR8, but at much lower frequency than DICER1. As these mutations were seen only in group 1 and 2 PBs, and in only 10% (6/59) of tumors in our study, it is perhaps not surprising that DROSHA and DGCR8 mutations were not reported in recent WES or whole genome sequencing studies of 19 PBs [60,35]. DROSHA mutations are frequent in Wilm’s tumors, where > 70% are missense mutations at E1147 in the RNase IIIb domain [68,67]. Although the IIIa and b domains respectively processes the 3p and 5p arms of pri-miRNA, the reported missense mutations do not appear to cause an imbalance in 5p and 3p mature miRNAs, but may act via dominant-negative mechanisms to globally downregulate miRNA production [54,65]. Our findings suggest miRNA maturation may also be globally downregulated in a subset of group 1 and 2 PBs via homozygous loss or biallelic truncating mutations of several critical miRNA endonucleases. All DGCR8 mutations in our PB samples were accompanied by chr 22 loss or LOH, similar to LOH in Wilms tumors with hotspot DGCR8 dsRBD mutations that impair mature miRNA expression [65,67,68]. Interestingly, one group 1 PB (RBTC757) exhibited loss of chr 22 copy in the context of the chr 22q11.2 deletion syndrome (22q11.2DS, DiGeorge syndrome). The minimal chr 22q11.2DS region which encompasses DGCR8 has also been linked to two prior cases of PB [46,61,34], and suggest DGCR8 loss may predispose to PB.
Although heritable retinoblastoma is associated with increased risk for PB [43], RB1 alterations have not been described in sporadic PB. In the RB subgroup, we observed recurrent RB1 homozygous loss or inactivating stop-gain mutation with LOH consistent with a classic two-hit mechanism. Associated with RB1 loss, we observed recurrent copy number gains of chr 13q31.3 which encompasses the oncogenic miR-17/92 cluster. In Rb/p107-deficient mice, miR-17/92 overexpression drives retinoblastoma formation by targeting Cdkn1a (p21/Cip1) to increase retinal cell proliferation [12], an oncogenic process that requires intact Dicer1 function [48] and may explain the paucity of miRNA biogenesis gene mutations in the RB sub-group of PBs. Of note we observed that MYC, which is also known to drive neoplastic growth by upregulating miR-17/92 [36], was recurrently gained/amplified in the MYC PB sub-group. These observations suggest common oncogenic mechanisms driven by a MYC-miR-17/92-RB1 axis [55,56] may underlie the aggressive biological features seen in these PB subgroups
KBTBD4 is a member of a large family of Bric-a-brac/Tramtrack/Broad (BTB) complex-containing adaptor proteins that complex with CUL3 E3 ubiquitin ligase and serve as a bridge between CUL3 and its substrate via a kelch interaction domain [10,7]. Substrates are then ubiquitinated and marked for degradation in the ubiquitin proteasome pathway. Hotspot mutations affecting the kelch domain have been reported in group 3 and 4 MB [49], and three cases of PPTID [35], and have been proposed as an oncogenic driver. While the targeted substrate of KBTBD4 has not been demonstrated, similar mutations in other BTB proteins that affect the substrate-binding domain or cause loss-of-function have been reported in a variety of cancers [10]. For example, in prostate cancer, androgen receptor signaling is implicated in tumor initiation and progression, as well as development of resistance to anti-androgen therapy [9]. Mutations affecting the androgen receptor-binding domain of BTB protein SPOP [5] leads to the failure of ubiquitination by CUL3 and thus, enhanced androgen receptor signaling [2]. Our WES analyses identified hotspot kelch domain mutations in 2/8 sequenced group 3 tumors. Although both these cases, diagnosed as PB, had lower Ki67/MIB-1 proliferation indices more consistent with PPTIDs [18], we did not observe this alteration in our two cases of PPTID or other group 3 PBs. While the hotspot KBTBD4 mutation have been proposed to be a marker for PPTID [35], our data suggests this mutation is characteristic for at least some group 3 tumors rather than exclusively all PPTIDs. With the caveat that Ki67/MIB-1 scores can be subjective and variable depending on tumor sample size, our review of scores in our cohort suggest group 3 is mainly composed of PBs with lower Ki67 indices in the range of PPTIDs, and tumors diagnosed as PPTIDs. Thus, tumors diagnosed as PPTID may be biologically similar to a proportion of PBs based on their shared global DNA methylation profile and silent chromosomal copy number landscape. Alternatively, some PPTIDs may be mis-diagnosed as PBs.
We observed on univariate analysis that loss of chr 16q was a significant negative prognostic marker for EFS and trending toward significance for OS. Interestingly, in another childhood embryonal cancer, Wilm’s tumor, chr 16q loss is also an established independent negative prognostic marker for relapse and death, and is being used to risk stratify patients with favorable histology tumors for intensified therapy [25]. Whether the loss-of-heterozygosity (LOH) of chr 16q disrupts a putative tumor suppressor or is a result of greater genomic instability remains to be elucidated for Wilm’s tumor. Some groups have proposed that LOH 16q may involve the effects of tumor-associated genes E2F4, COX4 [50], and CTCF [44], which all reside on chr 16q. We did not see mutations affecting these genes in our limited WES.
However, RB family tumor suppressor RBL2 (p130) also resides on 16q and is inactivated or lost in multiple cancers, including retinoblastoma [4,62,13,69,52]. We found that group RB and MYC tumors are characterized by chr 16q loss and an oncogenic MYC-miR-17/92-RB1 axis. Interestingly, in pancreatic adenocarcinoma, high expression of one member of the miR-17/92 cluster, miR-17-5p, directly targets RBL2 to inhibit RBL2-mediated repression of E2F4 target genes (MYC, CCND1, JUN), thereby enhancing proliferation [69]. RBL2 targeting is also seen in ovarian carcinoma via overexpression of miR-17/92 paralog miR-106a [38]. RBL2 could be similarly targeted by loss of chr 16q in PB. However, we did not observe RBL2 mutations in our limited WES, nor that chr 16q loss and miR-17/92 gain/amplification were mutually exclusive. Further studies will have to be completed to fully characterize the MYC-miR-17/92-RB axis and the role of RBL2 in PB.
Clinical studies of PBs to date have been limited by its rarity and lack of large, disease specific prospective cohorts. The recently completed Children’s Oncology Group high-risk EBT trial ACNS0332, enrolled 34 patients >3yrs, however separate clinical and molecular analyses of the PB cohort has not been reported [29]. Our clinical findings are limited by the retrospective nature of our registry-based cohort, and relatively smaller numbers compared to other studies of far more common childhood EBTs. Indeed, only recently have two clinical analyses with larger numbers, both retrospective, been published: a single institution study of 41 patients from St. Jude Research Hospital [51] and a pooled analysis of 135 patients from SIOP-E and US Head Start trial groups [45]. No previous published study has yet performed a combined molecular and clinical analysis as we have sought to do here. The clinical applicability of our findings will likely require further validation through continued collaboration with other research groups to pool enough data to power subgroup-specific risk stratification and inform therapy.
Nonetheless, consistent with prior studies [31,23,40,63,51,45], we identified young age at diagnosis (<3 yrs), metastatic disease, and omission of upfront RT as negative prognostic factors for PB survival. Also in agreement with published observations [51,45,17] our analysis did not reveal prognostic correlations with HDC or extent of surgery across all PB patients, although there was a trend toward improved EFS in children <3yrs who received HDC.
In contrast to the excellent outcome in group 2 PB (5-yr OS 100%), groups 1 and 3 patients had intermediate outcomes (68.0 and 80%), while groups RB and MYC patients had poorest outcomes (37.5 and 28%). Metastatic disease and chr 16 loss, which correlate with poorer survival across the entire cohort, was also enriched in group 1, RB, and MYC PBs, thus suggesting adverse molecular and clinical risk features may account partly for the poorer outcomes of these patients.
While the difference in EFS and OS between group 2 and 3 is due to just one group 3 patient who recurred then died from disease, another group 3 patient was only treated with palliative chemotherapy and thus not included in our intent-to-treat analysis. Both patients had extra-CNS (M4) metastasis at diagnosis. In contrast, of nine patients with group 2 tumors and clinical data, two were excluded from our intent-to-treat analysis: one who refused treatment, and another who died from intraoperative complications. No treated patients recurred or died. These differences in clinical features between the two groups not captured by intent-to-cure only EFS/OS estimates have led us to assign group 2 a superior prognosis to group 3.
The impact of different age-related therapeutic approaches likely contributes to differences in outcomes across PB patients. Indeed, we observed a significant difference in proportion of patients <3yrs (4/11; 36.4%) vs. those ≥3yrs of age at diagnosis (33/36; 91.7%; p<0.001) who received upfront RT, suggesting RT avoidance may play a role in adverse outcomes seen in younger patients who primarily had group 1, RB and MYC PB. Of note, group 2 and 3 PB had the highest median age at diagnosis, including three patients >20yrs of age who were alive at last follow-up after therapy that included only up-front RT. In contrast, two adult patients >20yrs at diagnosis who had group 1 and MYC tumors, both died despite receiving multimodal therapy including CSI, suggesting intensive therapy may not completely negate adverse tumor biology. Despite the prognostic impact of RT demonstrated by our study and that of others, it is also interesting to note that 5/29 long term survivors in our cohort who never received radiation therapy were young patients with group 1 (2 patients), MYC (2 patients), and RB (1 patient) PBs.
Our integrative molecular and clinico-pathologic analyses in this study which has identified five distinct molecular sub-groups of PB has provided important new insights into the pathogenesis of PB and confirm the importance of cancer predisposition related to miRNA biogenesis and RB1 gene defects in PB patients. Our study indicates groups 1-3 PBs patients treated with contemporary multi-modality regimens have intermediate to excellent outcomes but also highlight critical treatment gaps for younger PB patients most susceptible to radiation-related toxicities. Although our retrospective study has limitations, it represents one of the largest integrated clinical and molecular analyses of PB to date and provides new and critical information to inform therapy reduction in prospective clinical trials for favorable risk patients and development of novel therapies for high risk patients.
Supplementary Material
Supplementary Figure 1 PBs comprise 5 molecular sub-groups
Global methylation data generated from 72 PBs using Illumina 450K or EPIC arrays were analysed using NMF, HCL and K-means clustering methods to identify molecular sub-groups. a. Non-matrix factorization (NMF) analyses was performed on global methylation data using top 5000-15, 000 DNA methylation probes as determined by standard deviation (SD). Highest co-phonetic score was determined at rank (k) = 5 with 5000 probes; corresponding NMF heat map generated with 5000 probes is shown with PB sub-group designation. b. Silhouette plot of NMF analysis indicating best fit of individual PB sample within molecular sub-group c. Hierarchal (HCL) and K-means cluster analyses of global methylation data using the 5000 most variable probes by standard deviation indicating 5 sub-groups of PBs
Supplementary Figure 2 SNP array copy number analyses of PB
Copy number calls were generated using ASCAT (Allele specific copy number analysis of tumor) on Illumina Omni SNP array data generated from PBs a. ASCAT for group 2 tumor RBTC814 showing homozygous chr 14 loss b. ASCAT plot for group RB tumor RBTC746 showing homozygous RB1 loss and gain of miR-17/92.
Supplementary Figure 3 Schematic of hotspot mutations in Kelch domain of KBTBD4
a. IGV screenshot of aligned reads from whole exome sequencing of RBTC786 and −793 demonstrating in-frame insertions in KBTBD4, resulting in identical p.P311_R312 duplication mutation. Comparison is made to the same mutation reported by Lee JC, et al. 2019. b. Mapped in-frame insertions (in yellow) in our cohort, compared to those described by Lee JC, et al. in three PPTID samples, which are identical to that seen in RBTC793. All mutations result in the same p.P311_R312 duplication mutation.
Supplementary Figure 4 Impact of age, metastatic status and radiation treatment on PB survival
Event free (EFS) and overall survival (OS) analyses were determined for 46 patients treated with curative intent, using the Kaplan-Meier method and log rank tests. a. EFS and OS of patients <3 or ≥3 years age at diagnosis. 5-yr EFS: 18.2 vs 58.2%; 5-yr OS: 24.2 vs. 77.0% for <3 yrs vs. ≥3 yrs. b. EFS and OS of patients without (M0) and with metastases (M+) at diagnosis. 5-yr EFS: 29.4 vs. 60.5%; 5-yr OS: 44.9 vs. 78.7% for M+ vs. M0. c. EFS and OS of patients treated with and without upfront radiation therapy. 5-yr EFS: 10.0 vs. 58.8%; 5-yr OS: 40.0 vs. 71.0% for not radiated vs. radiated.
Supplementary Figure 5 Impact of chr 16 loss on event free and overall survival of PB survival
Event-free (EFS) and overall survival (OS) analyses were determined for 46 patients treated with curative intent, stratified by chr 16 loss or no chr 16 q loss in tumor specimens, using the Kaplan-Meier method and log rank tests. 5-yr EFS: 17.6 vs. 61.1%; 5-yr OS: 52.1 vs. 70.0% for chr 16q loss vs. no loss.
Supplementary Table 1 Molecular analysis performed on tumors and blood samples
Supplementary Table 2 Reported Ki67/MIB-1 scores and presence of hotspot KBTBD4 mutation among PB
Supplementary Table 3 Clinical characteristics and treatment details for PB patients
Acknowledgements:
RBTC biorepository and contributing tumor bank including NeuroBioTec Collection (Groupement Hospitalier Est, Bron, FRANCE), J. Loukides for biological specimens; C Hanzen, I Tennevet, O Langlois, D Frappaz for clinical data; M Fèvre-Montange for histopathologic review with A.J.; A Field for targeted panel design. This project was funded by the Canadian Institutes of Health Research (grant no. 137011) and b.r.a.i.n.child to A. H. D.A.H. is supported by the NIH/NCI (grant 2R01CA143167). B.K.L. is a Garron Family Cancer Centre Research Fellow. AH holds a Tier 1 Canada Research Chair.
Footnotes
Past presentations:
Parts of this study have been presented as abstracts at the American Society of Clinical Oncology Annual Meeting (June 1-5, 2018, Chicago, IL) where it was awarded a Conquer Cancer Merit Award, the International Symposium on Pediatric Neuro-Oncology (June 30-July 3, 2018, Denver, CO), and the International Rhabdoid Tumor Meeting and Satellite Rare Tumor Symposium, (April 21-22, 2018, Lake Louise, AB, Canada).
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Adzhubei I, Jordan DM, Sunyaev SR (2013) Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet Chapter 7:Unit 7 20. doi: 10.1002/0471142905.hg0720s76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An J, Wang C, Deng Y, Yu L, Huang H (2014) Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep 6:657–669. doi: 10.1016/j.celrep.2014.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anglesio MS, Wang Y, Yang W, Senz J, Wan A, Heravi-Moussavi A et al. (2013) Cancer-associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse-strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J Pathol 229:400–409. doi: 10.1002/path.4135 [DOI] [PubMed] [Google Scholar]
- 4.Baldi A, Esposito V, De Luca A, Fu Y, Meoli I, Giordano GG, Caputi M, Baldi F, Giordano A (1997) Differential expression of Rb2/p130 and p107 in normal human tissues and in primary lung cancer. Clin Cancer Res 3:1691–1697 [PubMed] [Google Scholar]
- 5.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP et al. (2012) Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 44:685–689. doi: 10.1038/ng.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blach LE, McCormick B, Abramson DH, Ellsworth RM (1994) Trilateral retinoblastoma--incidence and outcome: a decade of experience. Int J Radiat Oncol Biol Phys 29:729–733 [DOI] [PubMed] [Google Scholar]
- 7.Canning P, Cooper CD, Krojer T, Murray JW, Pike AC, Chaikuad A et al. (2013) Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J Biol Chem 288:7803–7814. doi: 10.1074/jbc.M112.437996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D et al. (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. doi: 10.1038/nature26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL (2004) Molecular determinants of resistance to antiandrogen therapy. Nat Med 10:33–39. doi: 10.1038/nm972 [DOI] [PubMed] [Google Scholar]
- 10.Chen HY, Chen RH (2016) Cullin 3 Ubiquitin Ligases in Cancer Biology: Functions and Therapeutic Implications. Front Oncol 6:113. doi: 10.3389/fonc.2016.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chintagumpala M, Hassall T, Palmer S, Ashley D, Wallace D, Kasow K et al. (2009) A pilot study of risk-adapted radiotherapy and chemotherapy in patients with supratentorial PNET. Neuro Oncol 11:33–40. doi: 10.1215/15228517-2008-079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conkrite K, Sundby M, Mukai S, Thomson JM, Mu D, Hammond SM, MacPherson D (2011) miR-17~92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev 25:1734–1745. doi: 10.1101/gad.17027411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Andrilli G, Masciullo V, Bagella L, Tonini T, Minimo C, Zannoni GF et al. (2004) Frequent loss of pRb2/p130 in human ovarian carcinoma. Clin Cancer Res 10:3098–3103. doi: 10.1158/1078-0432.ccr-03-0524 [DOI] [PubMed] [Google Scholar]
- 14.de Jong MC, Kors WA, de Graaf P, Castelijns JA, Kivelä T, Moll AC (2014) Trilateral retinoblastoma: a systematic review and meta-analysis. Lancet Oncol 15:1157–1167. doi: 10.1016/S1470-2045(14)70336-5 [DOI] [PubMed] [Google Scholar]
- 15.de Kock L, Sabbaghian N, Druker H, Weber E, Hamel N, Miller S et al. (2014) Germ-line and somatic DICER1 mutations in pineoblastoma. Acta Neuropathol 128:583–595. doi: 10.1007/s00401-014-1318-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Vito C, Riggi N, Cornaz S, Suva ML, Baumer K, Provero P, Stamenkovic I (2012) A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer cell 21:807–821. doi: 10.1016/j.ccr.2012.04.023 [DOI] [PubMed] [Google Scholar]
- 17.Farnia B, Allen PK, Brown PD, Khatua S, Levine NB, Li J, Penas-Prado M, Mahajan A, Ghia AJ (2014) Clinical outcomes and patterns of failure in pineoblastoma: a 30-year, single-institution retrospective review. World Neurosurg 82:1232–1241. doi: 10.1016/j.wneu.2014.07.010 [DOI] [PubMed] [Google Scholar]
- 18.Fevre-Montange M, Vasiljevic A, Frappaz D, Champier J, Szathmari A, Aubriot Lorton MH et al. (2012) Utility of Ki67 immunostaining in the grading of pineal parenchymal tumours: a multicentre study. Neuropathol Appl Neurobiol 38:87–94. doi: 10.1111/j.1365-2990.2011.01202.x [DOI] [PubMed] [Google Scholar]
- 19.Foulkes WD, Priest JR, Duchaine TF (2014) DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer 14:662–672. doi: 10.1038/nrc3802 [DOI] [PubMed] [Google Scholar]
- 20.Friedrich C, von Bueren AO, von Hoff K, Gerber NU, Ottensmeier H, Deinlein F et al. (2013) Treatment of young children with CNS-primitive neuroectodermal tumors/pineoblastomas in the prospective multicenter trial HIT 2000 using different chemotherapy regimens and radiotherapy. Neuro Oncol 15:224–234. doi: 10.1093/neuonc/nos292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garre P, Perez-Segura P, Diaz-Rubio E, Caldes T, de la Hoya M (2010) Reassessing the TARBP2 mutation rate in hereditary nonpolyposis colorectal cancer. Nat Genet 42:817–818; author reply 818. doi: 10.1038/ng1010-817 [DOI] [PubMed] [Google Scholar]
- 22.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G et al. (2015) Comprehensive genomic profiles of small cell lung cancer. Nature 524:47–53. doi: 10.1038/nature14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilheeney SW, Saad A, Chi S, Turner C, Ullrich NJ, Goumnerova L et al. (2008) Outcome of pediatric pineoblastoma after surgery, radiation and chemotherapy. J Neurooncol 89:89–95. doi: 10.1007/s11060-008-9589-2 [DOI] [PubMed] [Google Scholar]
- 24.Gonzales M (2001) The 2000 World Health Organization classification of tumours of the nervous system. J Clin Neurosci 8:1–3. doi: 10.1054/jocn.2000.0829 [DOI] [PubMed] [Google Scholar]
- 25.Grundy PE, Breslow NE, Li S, Perlman E, Beckwith JB, Ritchey ML et al. (2005) Loss of heterozygosity for chromosomes 1p and 16q is an adverse prognostic factor in favorable-histology Wilms tumor: a report from the National Wilms Tumor Study Group. J Clin Oncol 23:7312–7321. doi: 10.1200/JCO.2005.01.2799 [DOI] [PubMed] [Google Scholar]
- 26.Gurtan AM, Lu V, Bhutkar A, Sharp PA (2012) In vivo structure-function analysis of human Dicer reveals directional processing of precursor miRNAs. RNA 18:1116–1122. doi: 10.1261/rna.032680.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gururangan S, McLaughlin C, Quinn J, Rich J, Reardon D, Halperin EC et al. (2003) High-dose chemotherapy with autologous stem-cell rescue in children and adults with newly diagnosed pineoblastomas. J Clin Oncol 21:2187–2191. doi: 10.1200/JCO.2003.10.096 [DOI] [PubMed] [Google Scholar]
- 28.Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, Prentice L et al. (2012) Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med 366:234–242. doi: 10.1056/NEJMoa1102903 [DOI] [PubMed] [Google Scholar]
- 29.Hwang EI, Kool M, Burger PC, Capper D, Chavez L, Brabetz S et al. (2018) Extensive Molecular and Clinical Heterogeneity in Patients With Histologically Diagnosed CNS-PNET Treated as a Single Entity: A Report From the Children's Oncology Group Randomized ACNS0332 Trial. J Clin Oncol:JCO2017764720. doi: 10.1200/JCO.2017.76.4720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakacki RI, Burger PC, Kocak M, Boyett JM, Goldwein J, Mehta M, Packer RJ, Tarbell NJ, Pollack IF (2015) Outcome and prognostic factors for children with supratentorial primitive neuroectodermal tumors treated with carboplatin during radiotherapy: a report from the Children's Oncology Group. Pediatr Blood Cancer 62:776–783. doi: 10.1002/pbc.25405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jakacki RI, Zeltzer PM, Boyett JM, Albright AL, Allen JC, Geyer JR et al. (1995) Survival and prognostic factors following radiation and/or chemotherapy for primitive neuroectodermal tumors of the pineal region in infants and children: a report of the Childrens Cancer Group. J Clin Oncol 13:1377–1383. doi: 10.1200/JCO.1995.13.6.1377 [DOI] [PubMed] [Google Scholar]
- 32.Jouvet A, Vasiljevic A, Nakazato Y, Tanaka S (2016) Tumours of the pineal region In: Louis D (ed) WHO Classification of Tumours of the Central Nervous System. 4 edn. International Agency for Research on Cancer, pp 170–182 [Google Scholar]
- 33.Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T (2009) Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev 23:2700–2704. doi: 10.1101/gad.1848209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambert MP, Arulselvan A, Schott A, Markham SJ, Crowley TB, Zackai EH, McDonald-McGinn DM (2018) The 22q11.2 deletion syndrome: Cancer predisposition, platelet abnormalities and cytopenias. Am J Med Genet A 176:2121–2127. doi: 10.1002/ajmg.a.38474 [DOI] [PubMed] [Google Scholar]
- 35.Lee JC, Mazor T, Lao R, Wan E, Diallo AB, Hill NS et al. (2019) Recurrent KBTBD4 small in-frame insertions and absence of DROSHA deletion or DICER1 mutation differentiate pineal parenchymal tumor of intermediate differentiation (PPTID) from pineoblastoma. Acta Neuropathol. doi: 10.1007/s00401-019-01990-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Choi PS, Casey SC, Dill DL, Felsher DW (2014) MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer cell 26:262–272. doi: 10.1016/j.ccr.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin S, Gregory RI (2015) MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15:321–333. doi: 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, Gersbach E, Zhang X, Xu X, Dong R, Lee P et al. (2013) miR-106a represses the Rb tumor suppressor p130 to regulate cellular proliferation and differentiation in high-grade serous ovarian carcinoma. Mol Cancer Res 11:1314–1325. doi: 10.1158/1541-7786.MCR-13-0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohmann DR, Gerick M, Brandt B, Oelschlager U, Lorenz B, Passarge E, Horsthemke B (1997) Constitutional RB1-gene mutations in patients with isolated unilateral retinoblastoma. Am J Hum Genet 61:282–294. doi: 10.1086/514845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massimino M, Gandola L, Spreafico F, Luksch R, Collini P, Giangaspero F et al. (2006) Supratentorial primitive neuroectodermal tumors (S-PNET) in children: A prospective experience with adjuvant intensive chemotherapy and hyperfractionated accelerated radiotherapy. Int J Radiat Oncol Biol Phys 64:1031–1037. doi: 10.1016/j.ijrobp.2005.09.026 [DOI] [PubMed] [Google Scholar]
- 41.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G (2011) GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12:R41. doi: 10.1186/gb-2011-12-4-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller S, Rogers HA, Lyon P, Rand V, Adamowicz-Brice M, Clifford SC et al. (2011) Genome-wide molecular characterization of central nervous system primitive neuroectodermal tumor and pineoblastoma. Neuro Oncol 13:866–879. doi: 10.1093/neuonc/nor070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moll AC, Imhof SM, Bouter LM, Kuik DJ, Den Otter W, Bezemer PD, Koten JW, Tan KE (1996) Second primary tumors in patients with hereditary retinoblastoma: a register-based follow-up study, 1945-1994. Int J Cancer 67:515–519. doi: [DOI] [PubMed] [Google Scholar]
- 44.Mummert SK, Lobanenkov VA, Feinberg AP (2005) Association of chromosome arm 16q loss with loss of imprinting of insulin-like growth factor-II in Wilms tumor. Genes Chromosomes Cancer 43:155–161. doi: 10.1002/gcc.20176 [DOI] [PubMed] [Google Scholar]
- 45.Mynarek M, Pizer B, Dufour C, van Vuurden D, Garami M, Massimino M et al. (2017) Evaluation of age-dependent treatment strategies for children and young adults with pineoblastoma: analysis of pooled European Society for Paediatric Oncology (SIOP-E) and US Head Start data. Neuro Oncol 19:576–585. doi: 10.1093/neuonc/now234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen L, Crawford JR (2018) Pineoblastoma in a child with 22q11.2 deletion syndrome. BMJ Case Rep 2018. doi: 10.1136/bcr-2018-226434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.SEER Cancer Statistics Review 1975-2015, Table 29.1 (2019) https://seer.cancer.gov/csr/1975_2015/browse_csr.php. [Google Scholar]
- 48.Nittner D, Lambertz I, Clermont F, Mestdagh P, Kohler C, Nielsen SJ et al. (2012) Synthetic lethality between Rb, p53 and Dicer or miR-17-92 in retinal progenitors suppresses retinoblastoma formation. Nat Cell Biol 14:958–965. doi: 10.1038/ncb2556 [DOI] [PubMed] [Google Scholar]
- 49.Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T et al. (2017) The whole-genome landscape of medulloblastoma subtypes. Nature 547:311–317. doi: 10.1038/nature22973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan Z, He H, Tang L, Bu Q, Cheng H, Wang A, Lyu J, You H (2017) Loss of heterozygosity on chromosome 16q increases relapse risk in Wilms’ tumor: a meta-analysis. Oncotarget 8:66467–66475. doi: 10.18632/oncotarget.20191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parikh KA, Venable GT, Orr BA, Choudhri AF, Boop FA, Gajjar AJ, Klimo P (2017) Pineoblastoma-The Experience at St. Jude Children's Research Hospital. Neurosurgery 81:120–128. doi: 10.1093/neuros/nyx005 [DOI] [PubMed] [Google Scholar]
- 52.Priya K, Jada SR, Quah BL, Quah TC, Lai PS (2009) High incidence of allelic loss at 16q12.2 region spanning RBL2/p130 gene in retinoblastoma. Cancer Biol Ther 8:714–717. doi: 10.4161/cbt.8.8.7921 [DOI] [PubMed] [Google Scholar]
- 53.Pugh TJ, Yu W, Yang J, Field AL, Ambrogio L, Carter SL et al. (2014) Exome sequencing of pleuropulmonary blastoma reveals frequent biallelic loss of TP53 and two hits in DICER1 resulting in retention of 5p-derived miRNA hairpin loop sequences. Oncogene 33:5295–5302. doi: 10.1038/onc.2014.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rakheja D, Chen KS, Liu Y, Shukla AA, Schmid V, Chang TC et al. (2014) Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun 2:4802. doi: 10.1038/ncomms5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramaswamy V, Remke M, Adamski J, Bartels U, Tabori U, Wang X et al. (2016) Medulloblastoma subgroup-specific outcomes in irradiated children: who are the true high-risk patients? Neuro Oncol 18:291–297. doi: 10.1093/neuonc/nou357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramaswamy V, Taylor MD (2017) Medulloblastoma: From Myth to Molecular. J Clin Oncol 35:2355–2363. doi: 10.1200/JCO.2017.72.7842 [DOI] [PubMed] [Google Scholar]
- 57.Saab R, Rodriguez-Galindo C, Matmati K, Rehg JE, Baumer SH, Khoury JD et al. (2009) p18Ink4c and p53 Act as tumor suppressors in cyclin D1-driven primitive neuroectodermal tumor. Cancer Res 69:440–448. doi: 10.1158/0008-5472.CAN-08-1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sabbaghian N, Hamel N, Srivastava A, Albrecht S, Priest JR, Foulkes WD (2012) Germline DICER1 mutation and associated loss of heterozygosity in a pineoblastoma. J Med Genet 49:417–419. doi: 10.1136/jmedgenet-2012-100898 [DOI] [PubMed] [Google Scholar]
- 59.Sin-Chan P, Mumal I, Suwal T, Ho B, Fan X, Singh I et al. (2019) A C19MC-LIN28A-MYCN Oncogenic Circuit Driven by Hijacked Super-enhancers Is a Distinct Therapeutic Vulnerability in ETMRs: A Lethal Brain Tumor. Cancer cell 36:51–67.e57. doi: 10.1016/j.ccell.2019.06.002 [DOI] [PubMed] [Google Scholar]
- 60.Snuderl M, Kannan K, Pfaff E, Wang S, Stafford JM, Serrano J et al. (2018) Recurrent homozygous deletion of DROSHA and microduplication of PDE4DIP in pineoblastoma. Nat Commun 9:2868. doi: 10.1038/s41467-018-05029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stevens T, van der Werff Ten Bosch J, De Rademaeker M, Van Den Bogaert A, van den Akker M (2017) Risk of malignancy in 22q11.2 deletion syndrome. Clin Case Rep 5:486–490. doi: 10.1002/ccr3.880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Susini T, Massi D, Paglierani M, Masciullo V, Scambia G, Giordano A, Amunni G, Massi G, Taddei GL (2001) Expression of the retinoblastoma-related gene Rb2/p130 is downregulated in atypical endometrial hyperplasia and adenocarcinoma. Hum Pathol 32:360–367. doi: 10.1053/hupa.2001.23514 [DOI] [PubMed] [Google Scholar]
- 63.Timmermann B, Kortmann RD, Kuhl J, Rutkowski S, Meisner C, Pietsch T et al. (2006) Role of radiotherapy in supratentorial primitive neuroectodermal tumor in young children: results of the German HIT-SKK87 and HIT-SKK92 trials. J Clin Oncol 24:1554–1560. doi: 10.1200/JCO.2005.04.8074 [DOI] [PubMed] [Google Scholar]
- 64.Torchia J, Golbourn B, Feng S, Ho KC, Sin-Chan P, Vasiljevic A et al. (2016) Integrated (epi)-Genomic Analyses Identify Subgroup-Specific Therapeutic Targets in CNS Rhabdoid Tumors. Cancer cell 30:891–908. doi: 10.1016/j.ccell.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Torrezan GT, Ferreira EN, Nakahata AM, Barros BD, Castro MT, Correa BR et al. (2014) Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat Commun 5:4039. doi: 10.1038/ncomms5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC (2016) SIFT missense predictions for genomes. Nat Protoc 11:1–9. doi: 10.1038/nprot.2015.123 [DOI] [PubMed] [Google Scholar]
- 67.Walz AL, Ooms A, Gadd S, Gerhard DS, Smith MA, Guidry Auvil JM et al. (2015) Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer cell 27:286–297. doi: 10.1016/j.ccell.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wegert J, Ishaque N, Vardapour R, Georg C, Gu Z, Bieg M et al. (2015) Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors. Cancer cell 27:298–311. doi: 10.1016/j.ccell.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 69.Zhu Y, Gu J, Li Y, Peng C, Shi M, Wang X et al. (2018) MiR-17-5p enhances pancreatic cancer proliferation by altering cell cycle profiles via disruption of RBL2/E2F4-repressing complexes. Cancer Lett 412:59–68. doi: 10.1016/j.canlet.2017.09.044 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 PBs comprise 5 molecular sub-groups
Global methylation data generated from 72 PBs using Illumina 450K or EPIC arrays were analysed using NMF, HCL and K-means clustering methods to identify molecular sub-groups. a. Non-matrix factorization (NMF) analyses was performed on global methylation data using top 5000-15, 000 DNA methylation probes as determined by standard deviation (SD). Highest co-phonetic score was determined at rank (k) = 5 with 5000 probes; corresponding NMF heat map generated with 5000 probes is shown with PB sub-group designation. b. Silhouette plot of NMF analysis indicating best fit of individual PB sample within molecular sub-group c. Hierarchal (HCL) and K-means cluster analyses of global methylation data using the 5000 most variable probes by standard deviation indicating 5 sub-groups of PBs
Supplementary Figure 2 SNP array copy number analyses of PB
Copy number calls were generated using ASCAT (Allele specific copy number analysis of tumor) on Illumina Omni SNP array data generated from PBs a. ASCAT for group 2 tumor RBTC814 showing homozygous chr 14 loss b. ASCAT plot for group RB tumor RBTC746 showing homozygous RB1 loss and gain of miR-17/92.
Supplementary Figure 3 Schematic of hotspot mutations in Kelch domain of KBTBD4
a. IGV screenshot of aligned reads from whole exome sequencing of RBTC786 and −793 demonstrating in-frame insertions in KBTBD4, resulting in identical p.P311_R312 duplication mutation. Comparison is made to the same mutation reported by Lee JC, et al. 2019. b. Mapped in-frame insertions (in yellow) in our cohort, compared to those described by Lee JC, et al. in three PPTID samples, which are identical to that seen in RBTC793. All mutations result in the same p.P311_R312 duplication mutation.
Supplementary Figure 4 Impact of age, metastatic status and radiation treatment on PB survival
Event free (EFS) and overall survival (OS) analyses were determined for 46 patients treated with curative intent, using the Kaplan-Meier method and log rank tests. a. EFS and OS of patients <3 or ≥3 years age at diagnosis. 5-yr EFS: 18.2 vs 58.2%; 5-yr OS: 24.2 vs. 77.0% for <3 yrs vs. ≥3 yrs. b. EFS and OS of patients without (M0) and with metastases (M+) at diagnosis. 5-yr EFS: 29.4 vs. 60.5%; 5-yr OS: 44.9 vs. 78.7% for M+ vs. M0. c. EFS and OS of patients treated with and without upfront radiation therapy. 5-yr EFS: 10.0 vs. 58.8%; 5-yr OS: 40.0 vs. 71.0% for not radiated vs. radiated.
Supplementary Figure 5 Impact of chr 16 loss on event free and overall survival of PB survival
Event-free (EFS) and overall survival (OS) analyses were determined for 46 patients treated with curative intent, stratified by chr 16 loss or no chr 16 q loss in tumor specimens, using the Kaplan-Meier method and log rank tests. 5-yr EFS: 17.6 vs. 61.1%; 5-yr OS: 52.1 vs. 70.0% for chr 16q loss vs. no loss.
Supplementary Table 1 Molecular analysis performed on tumors and blood samples
Supplementary Table 2 Reported Ki67/MIB-1 scores and presence of hotspot KBTBD4 mutation among PB
Supplementary Table 3 Clinical characteristics and treatment details for PB patients