Abstract
Animal models recapitulating features of chronic colitis, such as ulcerative colitis, Crohn’s disease, or HIV infection, are critical to study disease pathogenesis and test novel therapeutics. In this study, we used a proteomics approach to explore the molecular intestinal response in two rhesus macaque (RM) animal models of experimentally induced colitis using dextran sulfate sodium (DSS) and simian immunodeficiency virus (SIV) infection. Proteomic analysis detected more than 2500 proteins in colonic tissue collected from 30 RMs. Differential protein expression analysis revealed a protein expression pattern in DSS-treated RMs resembling the proteome of human ulcerative colitis. In a group of 12 DSS-treated RMs compared to 6 with no treatment, decrease in expression of proteins related to mitochondrial energy metabolism, including fatty acid metabolism was noted, while innate immune activation pathways, including complement and coagulation proteins were upregulated. SIV infection of RMs resulted in increased innate immune responses related to viral defense. Proteomic signatures of barrier damage were apparent in both DSS treatment or SIV infection. These results demonstrate that DSS treatment in a non-human primate model resembles features of human ulcerative colitis, making this a promising tool to study important immunological mechanisms in inflammatory bowel disease.
INTRODUCTION
Ulcerative colitis (UC) is a form of inflammatory bowel disease (IBD) that affects millions of individuals, predominantly in Western countries, with reported annual rates as high as 24.3 per 100,000 persons.1 The development and etiology of UC is complex, but involves a combination of environmental, genetic, and immunoregulatory factors.2 The main features of UC are disruption of the gut epithelial barrier leading to neutrophil infiltration, microbial translocation, and chronic relapsing remitting inflammation caused by dysregulation of immunomodulatory pathways. Current treatments are insufficient3,4 and if these therapies fail or if IBD leads to severe complications, surgery is usually required.
The most common animal models to study disease pathogenesis or novel therapies of UC are chemically induced models of colitis in small animals, primarily rodents, including dextran sulfate sodium (DSS), oxazolone, or 2,4,6-trinitro benzene sulfonic acid. Translatability of these models into human disease however is poor5 and may be improved by using animals that are genetically closely related to humans. Recently, DSS-treated rhesus macaques (RMs, Macaca mulatta) were shown to recapitulate many effects of gut barrier damage commonly associated with colitis, including epithelial disruption, neutrophil infiltration, microbial translocation, and systemic inflammation.6,7 These features of DSS-treated RMs suggest that this may be an ideal model to study human IBD, given their highly related physiology, anatomy, and immune systems. However, the molecular alterations and pathways occurring in the gut in this model system have not been fully elucidated.
Previous studies of the gut mucosal barrier dysfunction in IBD have shown increased immune activation and inflammation in the colon of affected individuals. There is evidence showing that neutrophil infiltration,8 complement activation,9 oxidative stress,10 and apoptosis11 may all play a role in barrier disruption and inflammation during IBD. There is also evidence of decreased energy metabolism playing a role in IBD. Studies show that energy metabolism,12 amino acid metabolism,13,14 and mitochondrial and oxidation/reduction reactions15 are all perturbed during UC.
In this study, we utilized a proteomics-based systems biology approach to evaluate molecular alterations in the gut of two colitis models, namely DSS-induced colitis and SIV-infected RMs. Here, we show that these model systems recapitulate many of the molecular dysfunction signatures observed in IBD and HIV infection, including mitochondrial and metabolic proteome dysfunction, immune activation, and barrier disruption.
RESULTS
Proteomic analysis of colon tissue from RMs
In this study, we performed a detailed proteomics-based systems biology approach to evaluate molecular alterations in the gut in RM models of DSS chemically induced colitis and SIV infection. We collected transverse colon tissue from the gut of 31 RMs that consisted of seven different experimental groups as described in the Methods section. Principal component analysis (PCA) showed chronic DSS-treated RM clustering separately from the untreated control animals (Fig. 1a). RMs subjected to an acute DSS treatment were expectedly found between the untreated control and chronic DSS-treated clusters. SIV-infected RMs, whether they received DSS treatment or not, clustered with untreated RMs. CD4+ T cell depletion had no apparent effect on the gut proteome (Supplementary Fig. 1). This is likely because the proteins detected in this study are predominantly from cell types other than CD4+ T cells. Based on the above results, RMs were consolidated into five different experimental groups (untreated: DSS untreated; acute treatment: DSS acute; chronic treatment: DSS chronic; SIV infected: SIV+; DSS treated and SIV infected: DSS SIV+) and the effects of CD4+ T cell depletion were not further explored in downstream analysis.
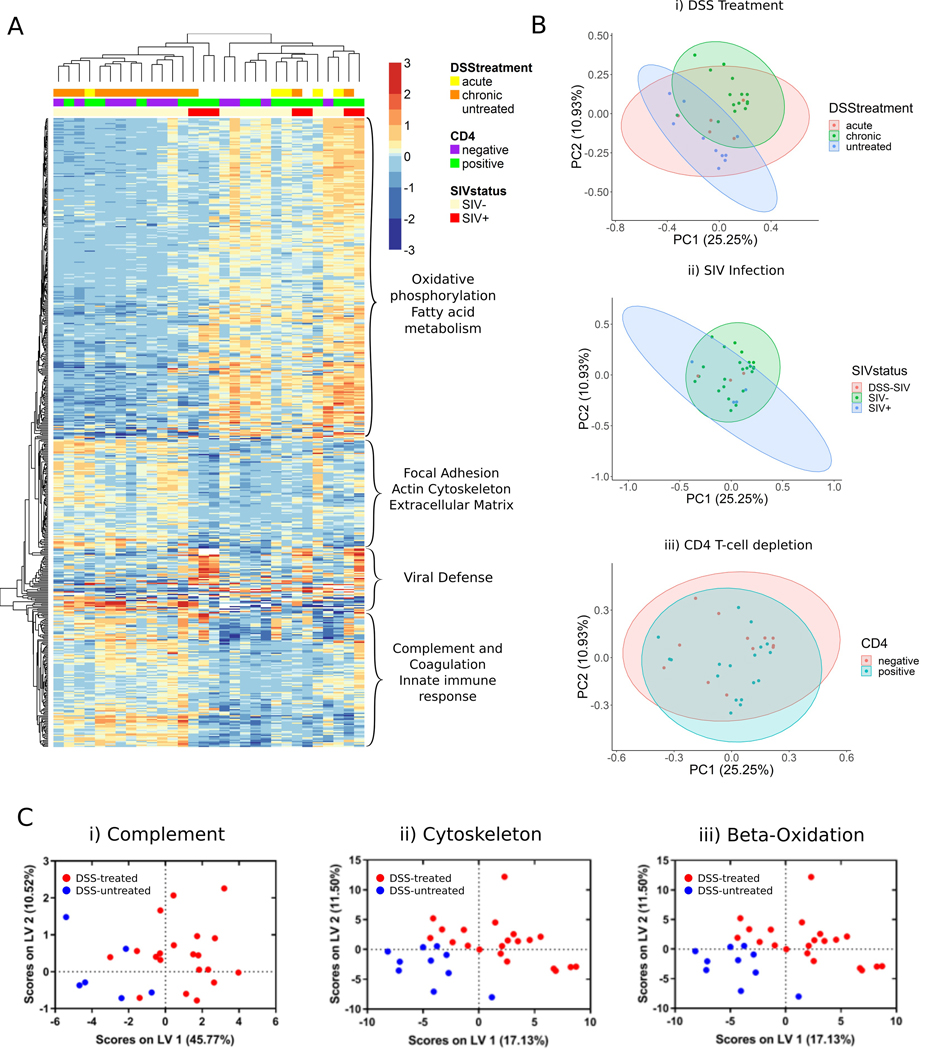
Fig. 1.
Overview of colon protein expression in a chemically induced and SIV rhesus macaque model of ulcerative colitis. a Subject PCA plots demonstrating the effect of (i) DSS treatment and (ii) SIV infection on protein expression. PCA plots show two separate clusters for untreated and DSS-treated RMs. RMs with acute DSS treatment are distributed between untreated and treated clusters. SIV+ RMs tended to cluster with RMs not treated with DSS. RMs treated with both SIV and DSS tended to cluster with RMs without DSS treatment. b (i) Heatmap of 425 differentially expressed proteins between DSS-untreated (n = 6), and DSS chronic RMs (n = 12, adjusted p value<0.05, BH correction). Protein expression is also shown for RMs with acute DSS treatments (n = 5). Main functions based on DAVID analysis (adjusted p value <0.05, BH correction) are listed to the right of each cluster. Major downregulated pathways included oxidative phosphorylation, fatty acid metabolism, and butanoate metabolism. Upregulated pathways included complement and coagulation cascades, ribosomal pathways, and regulation of the actin cytoskeleton. (ii) Heatmap of 37 upregulated proteins in SIV+ RMs against DSS-untreated RMs. DAVID analysis of two clusters (adjusted p value <0.05, BH correction) revealed upregulation of defense response to virus and proteasome complex proteins. (c) Biological pathways were analyzed for their ability to classify DSS-untreated RMs (DSS untreated, SIV+ RMs, n = 10) from DSS-treated RMs (DSS acute, DSS chronic, DSS SIV+ n = 20). (i) Thirteen complement components were detected in the dataset and were able to classify DSS-treated RMs (red) from non-treated RMs (blue) with 90.2% calibration (Cal) accuracy and 67.4% cross-validation (CV) accuracy. (ii) One hundred and twenty eight proteins involved in cytoskeletal processes showed a 97.2% Cal accuracy and 76.9% CV accuracy. (iii) Seventeen proteins involved in β-oxidization of fatty acids were also able to distinguish DSS-treated RMs with 80.7% Cal accuracy and 60.3% CV accuracy
Proteomic analysis of these colon tissue samples quantified 2511 protein groups across 30 replicates. Statistical analysis revealed that 530 unique proteins were differentially expressed across the five treatment groups. The highest number of differentially expressed proteins were detected in the DSS chronic treatment group compared to DSS untreated (187 upregulated, 250 downregulated). Unsupervised hierarchical clustering of differentially expressed proteins between these two groups identified two main protein clusters that associated with DSS treatment (Fig. 1b). Functional annotation of the upregulated cluster revealed proteins involved in complement and coagulation, the innate immune response, ribosomes, the phagosome, actin cytoskeleton regulation, and focal adhesion. Functional annotation of the downregulated cluster revealed these proteins to be involved in oxidative phosphorylation, the citrate cycle, fatty acid metabolism, the spliceosome, and DNA replication. A summary on the effects of DSS treatment in the RM colon can be found in Fig. 2.
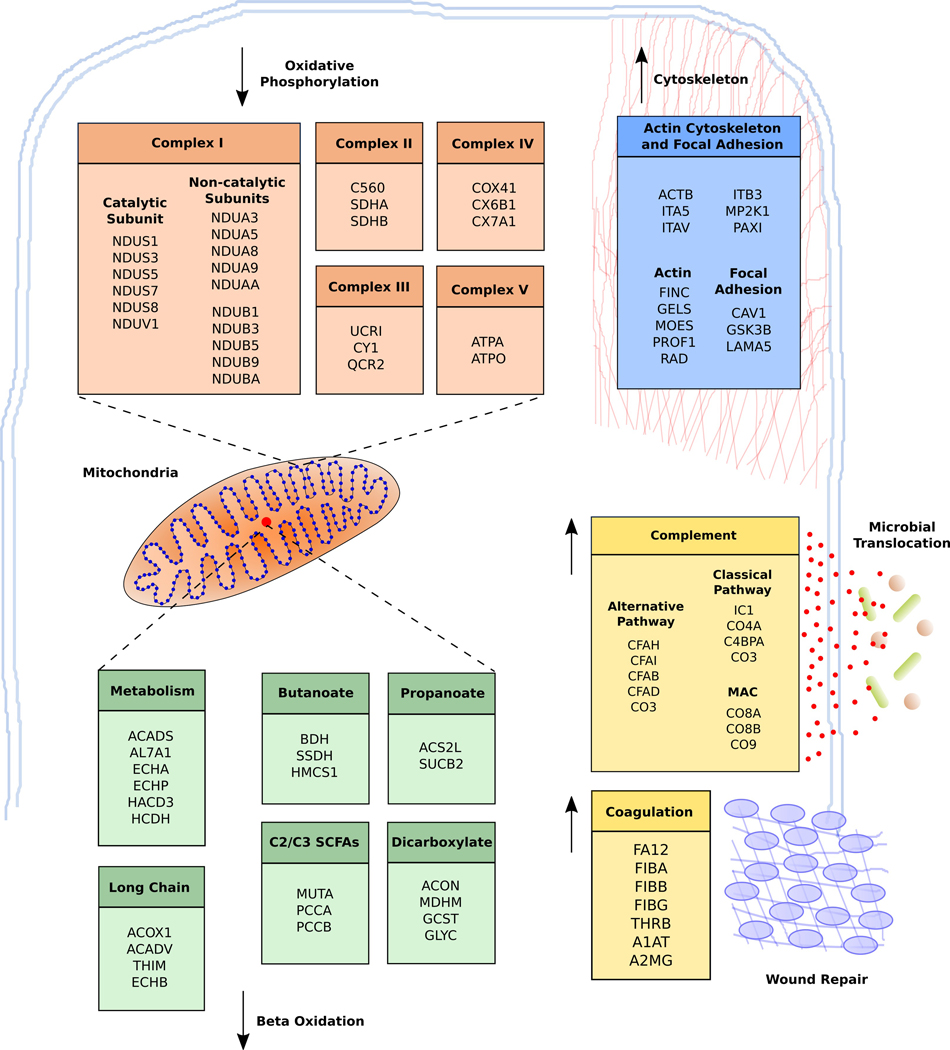
Fig. 2.
Outline of the effects of DSS treatment on the colon tissue proteome of rhesus macaques. List of significantly regulated proteins (adjusted p value <0.05, BH correction) between DSS chronic and DSS-untreated RMs. Multiple components belonging to each of the five complexes important in oxidative phosphorylation were downregulated. Enzymes involved in short- and long-chain fatty acid metabolism inside the mitochondria were also downregulated. Upregulated components include proteins involved in the actin cytoskeleton and focal adhesion. There was also significant upregulation of complement components related to innate immune activation and coagulation. All pathways were determined to be significantly upregulated in DSS chronic RMs as per DAVID analysis (adjusted p value <0.05, BH correction)
Complement, coagulation, and innate immune activation in DSS chronic RMs
Several components of the complement system were upregulated in DSS chronic RMs demonstrating significant signs of innate immune activation (Fig. 2, Supplementary Fig. 2). Complement and innate immune activation has been observed in studies of IBD and in chemically induced models of colitis.9,16 All branches of complement activation were upregulated, including the classical pathway (CO3, CO4A, C4BPA), the alternative/lectin (CFAB, CFAD) pathways, and the membrane attack complex (CO8A, CO8B, CO9), as well as proteins that inhibit the complement pathway (CFAH, CFAI, IC1). The acute-phase response was also activated in DSS chronic RMs. α−1-Antitrypsin, serum amyloid P-component, α−1-acid glycoprotein 1, α−1-acid glycoprotein 2, inter-α-trypsin inhibitor heavy chain H4, haptoglobin, argininosuccinate synthase, prothrombin (THRB), signal transducer and activator of transcription, and fibronectin are all upregulated in DSS chronic RMs providing evidence of innate immune activation as a result of DSS treatment. Finally, several proteins involved in coagulation pathways including fibrinogen chain α, fibrinogen chain β, fibrinogen chain γ, THRB, integrin α−5 (ITA5), integrin β−3 (ITB3), and integrin α-V (ITAV) were all upregulated in DSS chronic RMs.
Ribosomal proteins upregulated in DSS chronic RMs
Ribosomal proteins RS6 and RL6 were both upregulated in DSS-treated RMs. These proteins have previously been implicated in the control of cell growth and proliferation.17,18 To the best of our knowledge, ribosomal proteins have not been implicated in DSS treatment and this may represent a potential pathway that acts as a response to cellular damage caused by DSS.19 Unbiased pathway analysis of ribosomal proteins detected in this study revealed up- and downregulation of several other ribosomal proteins that depend on DSS treatment (Supplementary Fig. 3). There was a total of eight upregulated ribosomal proteins and eight downregulated ribosomal proteins in DSS-treated RMs.
Mitochondrial dysfunction and perturbation of fatty acid metabolism in DSS chronic RMs
Impairment of mitochondrial function and fatty acid metabolism20–23 is common in UC and we observed possible impairment of these pathways in this RM DSS model. Components belonging to the main five electron transport structures in mitochondria were downregulated in DSS-treated RMs (Fig. 2). There were 16 components of the NADH dehydrogenase complex (complex I), 3 components in the succinate dehydrogenase complex (complex II), 3 ubiquinone oxidoreductase proteins (complex III), 3 cytochrome c oxidase proteins (complex IV), and two proteins from ATP synthase (complex V). These results suggest significant impairment of oxidative phosphorylation and energy metabolism in the colon of DSS-treated RMs.
Fatty acid metabolism components were also perturbed with DSS treatment. These proteins have overlapping functions involved in the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways fatty acid degradation, fatty acid metabolism, butanoate metabolism, propanoate metabolism, and dicarboxylate and glyoxylate metabolism (Fig. 2, Supplementary Fig. 4). We identified unique enzymes belonging to each pathway indicating disruption in metabolism of both long- and short-chain fatty acids. Acyl-coenzyme A oxidase 1, acetyl-CoA acyltransferase (THIM), trifunctional enzyme subunit beta (ECHB), very long-chain specific acyl-CoA dehydrogenase are all enzymes important for the degradation and metabolism of long-chain fatty acids. Hydroxymethylglutaryl-CoA synthetase (HMCS1), and D-β-hydroxybutyrate dehydrogenase are enzymes involved specifically in butyrate metabolism. We also identified several enzymes important in C2/C3 SCFA metabolism including propionyl-CoA carboxylase α chain, propionyl-CoA carboxylase β chain, and methylmalonyl-CoA mutase. Finally, we identified two enzymes in the dicarboxylate metabolism pathway that are also important for amino acid metabolism, aminomethyltransferase (GCST), and serine hydroxymethyltransferase (GLYC). Overall, these results indicate several overlapping or cascading effects that lead to decreased energy, fatty acid, and amino acid metabolism potentially related to mitochondrial dysfunction.
Antiviral response pathways in SIV+ and DSS SIV+ RMs
There was also a small cluster of 37 proteins upregulated in SIV+ compared to DSS-untreated RMs (Fig. 1b, Supplementary Fig. 5). The same proteins appear to also be increased in DSS SIV+ compared to DSS-untreated RMs, although they did not meet statistical significance. These proteins were mostly related to natural responses against viral infection. This includes proteins with previously known antiviral activity (MX1, GBP3, IFIT1, IFIT3), proteins that signal the transcription of antiviral factors (DDX58, OAS2, OAS3, E2AK2), and proteins involved in antigen processing and presentation (1A80, TAP1). Many of these proteins, including MX1, IFIT3, OAS2, and OAS3, are known interferon-stimulated genes that showed increased expression in SIV-infected RMs in a previous study.24
Immune activation and mitochondrial dysfunction with DSS treatment and SIV infection
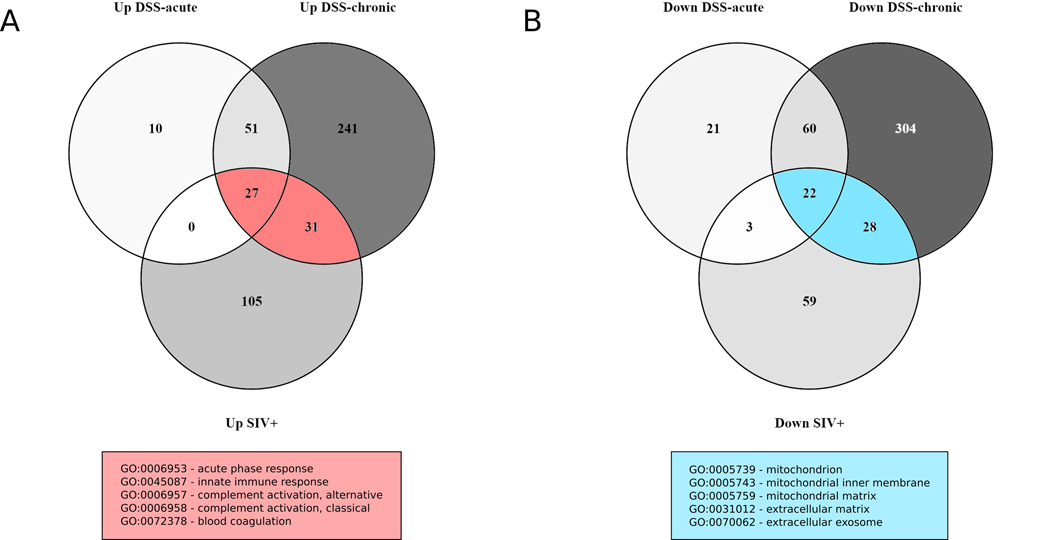
To explore potential overlap in immune activation and inflammatory pathways in DSS-treated and SIV-infected RMs, we decided to conduct an exploratory analysis involving the differentially expressed proteins in these groups. We used a less stringent statistical cut-off (unadjusted P value <0.05) to compare proteins differentially expressed between DSS acute and DSS-untreated, DSS chronic and DSS-untreated, and SIV+ and DSS-untreated groups. There was a total of 949 unique proteins differentially expressed between these groups at this significance level. We created two Venn diagrams each for upand downregulated proteins to show the overlap in differentially expressed proteins in these three treatment groups (Fig. 3). The majority of proteins upregulated in the DSS acute group overlapped with the proteins in the DSS chronic treatment group (78 out of 88 proteins). There were 58 proteins that overlapped between the SIV+ and the DSS acute and DSS chronic treatment groups (12.5% of all upregulated proteins). The majority of these proteins were related to complement activation (13 proteins). There were also eight proteins with serine-type endopeptidase inhibitor activity upregulated in both treatment groups.
Fig. 3.
Venn diagrams showing overlap in differential protein expression in DSS-treated and SIV-infected RMs for upregulated (a) and downregulated (b) proteins (non-adjusted p value <0.05). Representative up- or downregulated biological processes are shown for overlapping proteins in DSS-treated and SIV+ RMs (DAVID analysis, BH-adjusted p value <0.05). The majority of proteins up- and downregulated in the DSS acute treatment groups overlapped with those upregulated in the DSS chronic group. There was a total of 58 and 53 up- and downregulated proteins, respectively, detected in both DSS-treated and SIV+ RMs demonstrating overlap between response to DSS treatment and SIV infection
We then performed a similar analysis for the downregulated proteins in SIV+, DSS acute, and DSS chronic treatment groups. There were 53 downregulated proteins that were in both SIV+ and DSS treatment groups (~10% of all downregulated proteins). Functional analyses showed that most of these proteins were part of the mitochondria, play a role in oxidative phosphorylation, or are part of the extracellular matrix. According to gene ontology, 3 of 4 succinate dehydrogenase (complex II) components, 3 of 10 cytochrome c reductase (complex III) components, and 3 of 17 cytochrome c oxidase (complex IV) components were downregulated. In addition to this, we detected 16 of 45 NADH dehydrogenase components and 2 of 15 ATPase synthase components downregulated in these treatment groups. These results show some potential overlap in DSS treatment and SIV infection related to innate immune activation and mitochondrial dysfunction.
DISCUSSION
We have previously published the extensive characterization of the severity of colitis (in both acute and chronic) in this DSS non-human primate (NHP) model,7 where we histologically demonstrated the severity of disease in the large bowel, including intestinal epithelial damage, inflammatory polymorphonuclear cell infiltration, and increased fibroblast and capillary vessel formation. Lesion grades were confirmed using a modified version of Geboes et al.25 by a pathologist who was blinded to these groupings. Our follow-up proteomic analysis of colon tissues from these animals revealed altered expression of molecular pathways related to complement and coagulation, the innate immune system, coagulation, mitochondrial dysfunction, and fatty acid metabolism. A caveat of this study is that we did not provide direct functional evidence of the consequences of the proteome changes occurring in DSS-treated RMs. However, these results resemble the altered proteome of UC in humans from multiple other proteomic studies conducted in gut tissue providing evidence that our model system would be useful as a clinical model to study UC.12,15,26 Another challenge is the low number of animals used in this study, which is a challenge that all NHP studies face due to extensive care and handling that leads to exceptional costs required to maintain even a small cohort of primates; we were able to demonstrate key molecular pathways alongside previously published histological defects in the gut mucosa of these animals that align with human IBD, supporting the suitability of this model system to explore mechanisms of this disease.
There are several proteomic studies conducted to study IBD in both human and animal models of disease. In a recent study of UC in humans, the colon tissue proteome was compared in patients suffering from UC to that of healthy controls.26 We compared up- and downregulated proteins in each study and found relative concordance to our study, including upregulation of innate immune factors such as dual oxidase 2, neutrophil gelatinase-associated lipocalin, and myeloperoxidase, as well as downregulation of the mitochondrial energy metabolism factors hydroxybutyrate dehydrogenase and NADH dehydrogenase subunit 5. A recent DSS mouse model study using Two-dimensional difference gel electrophoresis (2D-DIGE) examined the gut tissue proteome difference in DSS-treated against untreated mice and found upregulation of the mitochondrial protein HMCS2 and leukocyte elastase inhibitor and downregulation of protein disulfide isomerase A3, periredoxin-6, and vimentin.27 In our study, mitochondrial factors HMCS1 and HMCS2 were downregulated, while protein disulfide isomerase A5 was upregulated in disagreement with their study. We found upregulation of leukocyte elastase inhibitor and downregulation of peroxiredoxin-5, which matched their results. Vimentin was not differentially expressed in this study. In another study of IBD, 2D-DIGE was used to measure protein expression changes in interleukin-10 (IL-10)-deficient mice against healthy controls and reported downregulation of short-chain fatty acid, lipid, carbohydrate, and amino acid metabolism pathways and upregulation of proteins involved in cytoskeleton assembly, immune responses, the acute-phase response, and nucleic acid-binding proteins.28
Immune activation and bacterial translocation appears to be one of the main causes of persistent inflammation during UC and other forms of IBD.29,30 The immune responses observed in patients with UC may include increased numbers of neutrophils, macrophages, and dendritic cells along with increased proinflammatory cytokines such as IL-1β, tumor necrosis factor-α, IL-6, and IL-8.30 This response typically also includes activation of complement.31–34 There were significant signs of innate immune activation in RMs with chronic DSS treatment similar to that found in human UC, including the coagulation and complement cascade pathways. There was no specific branch of the complement activation system that was activated in DSS-treated RMs. Components of the alternative pathway that inhibit complement activation (CFAH), those that contribute to complement activation (CFAB, CFAD), and the classical/lectin pathway (IC1, C4BPA, CO4A) were all upregulated in DSS chronic RMs.
There were also signs of blood coagulation in DSS-treated RMs that may be the result of chemically induced epithelial barrier disruption. Increased fibrinogen and THRB indicate the formation of fibrin in the colon of these RMs. Formation of fibrin is a normal part of the clotting process, but excessive amounts of fibrin are known to contribute to fibrosis such as in the lung35 or liver.36 Increased integrins were also associated with DSS treatment. ITB3, ITA5, and ITAV are known to act as receptors for a multitude of different extracellular matrix components that can contribute to fibrosis. There are numerous examples in the literature on how integrins can contribute to fibrosis in different tissues.37–39 Signs of fibrosis are common in patients with IBD40 and this proteomic analysis reveals that the NHP model may be an important avenue of research into this aspect.
Our study detected decreased expression of oxidative phosphorylation proteins in mitochondrial complex II, III, and IV in RMs treated with DSS compared to untreated controls. In a previous study, activity of mitochondrial respiratory complex II, III, and IV were decreased by 50–60% in the colonic mucosa of UC patients20 supporting our findings. In one recent study of gene expression in patients with Crohn’s disease, patients who had decreased expression of mitochondrial genes were more likely to develop specific complications associated with this type of IBD.41 Disruption of mitochondrial function and energy metabolism is well documented in patients with IBD,20,21,42,43 highlighting the fact that systems biology analysis can inform functional pathways.
It is well known that there is a relationship between fatty acid metabolism and UC in both human and animal models.23,44–46 In another study of gene expression in colonic mucosal biopsies, several genes involved in butyrate metabolism were under-expressed in UC patients compared to controls.47 In our study, proteomic analysis showed decreased expression of proteins in short-chain fatty acid metabolism pathways in the colon of DSS chronic RMs. Mitochondrial short-chain specific acyl-CoA dehydrogenase (ACADS) was downregulated in both of these studies. The authors also identified decreased expression of enoyl-CoA hydratase (ECHM) and 3-hydroxyacyl-CoA dehydrogenase type-2 (HCD2), enzymes that catalyze the next two steps in butyrate metabolism after the action of ACADS. While we did not identify decreased expression of these enzymes specifically in this study, we did detect decreased expression of peroxisomal bifunctional enzyme (ECHP) and hydroxyacyl-coenzyme A dehydrogenase (HCDH) that can alternatively catalyze the same two steps in butyrate metabolism. Overall, DSS treatment in RMs appears to mimic the effect of impaired short-chain fatty acid metabolism in human UC patients. Further study of fatty acid metabolism with direct assays of enzyme activity will be required to confirm impaired fatty acid metabolism in the gut tissue of these RMs.
Gut pathology is well documented in HIV infection and in SIV-infected RMs. CD4+ T cells rapidly deplete from the gut of RMs within the first few days of SIV infection48, making it the primary target for viral replication during the early stages of infection. What follows is breakdown of the gut epithelial barrier that leads to microbial translocation, infiltration of neutrophils, and increased inflammation.6 We detected similarities in mitochondrial dysfunction, decreased extracellular matrix proteins, and complement activation that might contribute to persistent inflammation in both models of SIV+ and DSS chronic RMs. To our knowledge, these effects have not been documented in the colon of SIV-infected RMs. Overall, this analysis provides evidence that some of the gut barrier dysfunction at the molecular level in SIV infection can be recapitulated with DSS treatment.
There are limitations to our study that must be considered. Although we observed similar proteome differences to other proteomic studies of IBD,26,27 we did not confirm these pathway changes by other methods. Furthermore, we did not evaluate how these proteome changes relate to metabolic dysfunction. However, in a study of DSS treatment in mice,49 proton nuclear magnetic resonance analysis of metabolites in the urine of DSS-treated mice showed an increase of citric acid cycle metabolites, including citrate, fumarate, and 2-oxoglutarate, suggesting an impairment of mitochondrial function. Further study will be required to find the impact DSS treatment has in this RM model of colitis.
In this study, we used quantitative proteomics on colon tissue collected from RMs to evaluate molecular alterations in the gut from RM models of colonic inflammation, namely an NHP model of DSS chemically induced colitis and SIV infection. The effects of DSS treatment at the proteome level appeared to be similar to the effects of UC in humans. Notably, decreased mitochondrial function, decreased short-chain fatty acid metabolism proteins, increased complement, and increased innate immune system proteins. Given the similarity between this NHP model and human studies of UC, the DSS model of colitis in RMs may prove useful to not only study the disease pathology of UC in humans but also test novel therapeutic interventions for IBD. Currently, mouse models of chemically induced colitis are the most common animal models of colitis; however, RMs provide a much more genetically, physiologically, and immunologically similar model to humans and should provide better information on the preclinical efficacy of new treatment modalities of colitis. Overall, this model represents a new way to study UC in a system that closely resembles human UC and could potentially lead to new treatment options for patients suffering from this disease.
METHODS
RM DSS treatment
All RMs (Macaca mulatta) in this study were mature and of Indian origin, consisting of both male and female animals ranging in age from ~2 to 7 years, and were housed at the National Institutes of Health (NIH) in accordance with the Association for the Assessment and Accreditation of Laboratory Animal Care standards and all procedures were performed according to the protocols approved by the Institutional Animal Care and Use Committee of the National Cancer Institute (Assurance #A4149–01) as previously reported.50 In total, 31 RMs were used across the various groups in this study: 2 untreated RMs, 4 RMs with CD4 depletion, 6 RMs with acute DSS treatment, 4 RMs with chronic DSS treatment, 8 RMs with chronic DSS treatment and CD4 depletion, 4 SIV-infected RMs, and 3 SIV-infected RMs with chronic DSS treatment. Six of the seven SIV-infected RMs were infected with SIV strain SIVmac251, while the remaining RM was infected with SIVmac239M. SIV infections were allowed to reach the chronic stages of infection and then euthanized in the range of 34–86 weeks post infection. CD4 depletion was achieved with eight subcutaneous injections of anti-CD4R1 antibody (NHP Reagent Resource; 50 mg/kg) every 3 weeks, starting at week 0, and the efficacy was confirmed via flow cytometry of peripheral blood throughout the experiment. Before use, all animals were tested and confirmed seronegative for Macacine herpes virus 1 (herpes B), SIV, simian T-lymphotropic virus, and simian retrovirus, and were negative for simian retrovirus by PCR as well. In addition, before study, all animals were treated with enrofloxacin (10 mg kg−1 once daily for 10 days), paromomycin (25 mg kg−1 twice daily for 10 days), and fenbendazole (50 mg kg−1 once daily for 5 days), followed by weekly fecal culture and parasite exams for 3 weeks to ensure they were free of common enteric pathogens. A post-treatment period of at least 4 weeks was chosen to allow time for stabilization of the microbiome, although not formally tested in this study. After study completion, all animals were euthanized, and a detailed necropsy was performed with extensive tissue collection, including full thickness gastrointestinal pieces that were opened longitudinally, rinsed in phosphate-buffered saline, snap frozen in liquid N2, and stored at −80 °C until further processing.
DSS administration
Mild-to-moderate colitis was induced with DSS (MP Biomedicals), molecular weight 35,000–50,000 Da, resuspended in sterile, and filtered drinking water containing non-colored Kool-Aid to increase palatability.7 For acute colitis, a fresh bottle containing 0.25–1% (w/v) DSS was given to the animals daily for 10 days. For chronic colitis, 0.25% DSS was given for 6–8 alternating cycles, consisting of 14 days DSS, followed by a 14-day interval with plain drinking water. If the bottle of DSS-containing water was consumed prior to the end of the day, fresh plain water was given to the animals. The animals were closely monitored for disease progression throughout the experiment, via measures including weight change, endoscopy, and magnetic resonance imaging.
Colon tissue processing for mass spectrometry analysis
A small ~1 cm2 piece of freeze-dried tissue was cut into smaller pieces with a scalpel and added to a tube containing 5 mL of tissue lysis buffer (7 M urea, 2 M thiourea, 40 mM Tris, 100 mM Dithiothreitol, pH 7.5) containing MS-Safe protease and phosphatase inhibitors (Sigma). The tissue was homogenized with a tissue homogenizer (gentleMACS Octo Dissociator, Miltenyi Biotec) and debris was removed from the supernatants by centrifugation. The tissue lysate was then aliquoted and frozen at −80 °C. The protein concentration of each sample was determined by 2D Quant kit (GE Life Sciences).
Peptide sample preparation for mass spectrometry
A total of 100 μg of protein was mixed with urea buffer (50 mM HEPES, 8 M urea, pH 8.4) to bring the final volume to 600 μL and left at room temperature for 10 min. The lysate was then added to a 10 kDa molecular weight cut-off spin filter and digested as per a modified filter-assisted sample protocol method.51 Peptides were purified by high-performance liquid chromatography and vacuum dried before dissolving in 200 μL of mass spectrometry Buffer A (2% acetonitrile in 0.1% formic acid) for liquid chromatography with tandem mass spectrometry analysis. Hyper reaction monitoring calibration standard peptides (Biognosys) were spiked into each sample as per the manufacturer’s instructions. Sample order was randomized and then analyzed in both data-independent (DIA) and data-dependent modes of acquisition. Details on liquid chromatography and mass spectrometry settings can be found in the Supplementary Information. The DIA windows were set to those found in a previous article using DIA acquisition.52
Proteomic data analysis
Mass spectrometry files were analyzed in directDIA mode with Spectronaut 10 using the default BGS Factory settings. The data were searched against a fasta database downloaded from uniprot.org53 containing all human protein sequences in the SwissProt database. For differential protein expression analysis, data were exported in MSstats format and uploaded into R for modeling and statistical analysis with the MSstats package.54 The MSstats model relies on the lme and lmer functionalities in R to fit linear models for protein quantification and then performs differential protein expression analysis between each treatment group.54 The Benjamini–Hochberg (BH) method was used to correct p values for multiple comparisons. Proteins were considered differentially expressed between groups if the adjusted p value was <0.05. After analysis, one outlier sample was removed because the median signal intensity was 1.5 standard deviations away from the median signal intensity of all other samples. The presence of this outlier was further confirmed by visual inspection of PCA plots and hierarchical clustering (unpublished data, Burgener Lab, 2018). Protein functional analysis was performed using the DAVID functional annotation toolset version 6.855 and KEGG gene ontology.56 A statistical cut-off threshold of p value <0.05 that was adjusted for multiple comparison (BH correction) was employed for differential protein functional analysis. KEGG pathway analysis was performed with the KEGGREST R package to map Uniprot IDs in each pathway.57 Venn diagrams were constructed using Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Biological pathways of interest (complement components, cytoskeletal factors and fatty acid-beta oxidation proteins) identified from functional analysis were assessed for their prediction ability for classifying DSS-treated from untreated RMs using partial least-square discriminant analysis (PLS-DA) implemented in Matlab software (Mathworks, Natick, MA) and the PLS toolbox from Eigenvector research (Eigenvector Research Inc., Manson, WA). Proteins from each pathway were analyzed separately. Protein expression values were log 2 normalized, mean centered, and underwent variance scaling. Cross-validation (CV) of models was performed using iterative exclusions of random subsets during calibration. The calibration and CV accuracy of each model from the first two loading variables are reported.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Specimen Support Core of the AIDS and Cancer Virus Program and the animal husbandry and technical staff of the Laboratory Animal Sciences Program, Frederick National Laboratory for Cancer Research, for expert animal care and handling. We also thank Max Abou for technical support for sample preparation of samples for proteomic analysis. Funding for this research was provided by grants from the National Institute of Research (A.B., R01DK112254) and the Public Health Agency of Canada (A.B.). This work was supported in part by federal funds from the National Cancer Institute, National Institutes of Health (Contract No. HHSN261200800001E) and by the Oregon National Primate Research Center NIH grant award P51OD011092 (J.D.E). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
ADDITIONAL INFORMATION
The online version of this article (https://doi.org/10.1038/s41385-019-0200-2) contains supplementary material, which is available to authorized users.
Competing interests: The authors declare no competing interests.
REFERENCES
- 1.Molodecky NA et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 142, 46–54 e42 (2012). quiz e30. [DOI] [PubMed] [Google Scholar]
- 2.de Souza HS & Fiocchi C Immunopathogenesis of IBD: current state of the art.Nat. Rev. Gastroenterol. Hepatol 13, 13–27 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Baumgart DC & Sandborn WJ Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet 369, 1641–1657 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Kozuch PL & Hanauer SB Treatment of inflammatory bowel disease: a review of medical therapy. World J. Gastroenterol. 14, 354–377 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeVoss J & Diehl L Murine models of inflammatory bowel disease (IBD): challenges of modeling human disease. Toxicol. Pathol. 42, 99–110 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Estes JD et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 6, e1001052 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hao XP et al. Experimental colitis in SIV-uninfected rhesus macaques recapitulates important features of pathogenic SIV infection. Nat. Commun. 6, 8020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fournier BM & Parkos CA The role of neutrophils during intestinal inflammation. Mucosal Immunol. 5, 354–366 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Hodgson HJ, Potter BJ & Jewell DP Humoral immune system in inflammatory bowel disease: I. Complement levels. Gut 18, 749–753 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam G, Apostolopoulos V, Zulli A & Nurgali K NADPH oxidases and inflammatory bowel disease. Curr. Med. Chem 22, 2100–2109 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Iwamoto M, Koji T, Makiyama K, Kobayashi N & Nakane PK Apoptosis of crypt epithelial cells in ulcerative colitis. J. Pathol. 180, 152–159 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Shkoda A et al. Differential protein expression profile in the intestinal epithelium from patients with inflammatory bowel disease. J. Proteome Res. 6, 1114–1125 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Barcelo-Batllori S et al. Proteomic analysis of cytokine induced proteins inhuman intestinal epithelial cells: implications for inflammatory bowel diseases. Proteomics 2, 551–560 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Balasubramanian K et al. Metabolism of the colonic mucosa in patients with inflammatory bowel diseases: an in vitro proton magnetic resonance spectroscopy study. Magn. Reson. Imaging 27, 79–86 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Hsieh SY et al. Comparative proteomic studies on the pathogenesis of human ulcerative colitis. Proteomics 6, 5322–5331 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Fang K et al. Temporal genomewide expression profiling of DSS colitis reveals novel inflammatory and angiogenesis genes similar to ulcerative colitis. Physiol. Genom. 43, 43–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gou Y et al. Ribosomal protein L6 promotes growth and cell cycle progression through upregulating cyclin E in gastric cancer cells. Biochem. Biophys. Res. Commun. 393, 788–793 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Li Y et al. Relationship between ATM and ribosomal protein S6 revealed by the chemical inhibition of Ser/Thr protein phosphatase type 1. Biosci. Biotechnol. Biochem 76, 486–494 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Wirtz S et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 12, 1295–1309 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Sifroni KG et al. Mitochondrial respiratory chain in the colonic mucosal of patients with ulcerative colitis. Mol. Cell. Biochem 342, 111–115 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Santhanam S et al. Mitochondrial electron transport chain complex dysfunction in the colonic mucosa in ulcerative colitis. Inflamm. Bowel Dis. 18, 2158–2168 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Roediger WE The colonic epithelium in ulcerative colitis: an energy-deficiency disease? Lancet 2, 712–715 (1980). [DOI] [PubMed] [Google Scholar]
- 23.Chapman MA et al. Butyrate oxidation is impaired in the colonic mucosa of sufferers of quiescent ulcerative colitis. Gut 35, 73–76 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandler NG et al. Type I interferon responses in rhesus macaques prevent SIVinfection and slow disease progression. Nature 511, 601–605 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geboes K et al. A reproducible grading scale for histological assessment of inflammation in ulcerative colitis. Gut 47, 404–409 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schniers A et al. The proteome of ulcerative colitis in colon biopsies from adults—optimized sample preparation and comparison with healthy controls. Proteom. Clin. Appl 11, 11–12 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Naito Y et al. Identification of inflammation-related proteins in a murine colitis model by 2D fluorescence difference gel electrophoresis and mass spectrometry. J. Gastroenterol. Hepatol 25, S144–S148 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Cooney JM et al. Proteomic analysis of colon tissue from interleukin-10 gene-deficient mice fed polyunsaturated fatty acids with comparison to transcriptomic analysis. J. Proteome Res. 11, 1065–1077 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Johansson ME et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 63, 281–291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sartor RB Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. Nat. Clin. Pract. Gastroenterol. Hepatol 3, 390–407 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Hermanowicz A, Sliwinski Z, Nawarska Z & Banas B Humoral immune system and ulcerative colitis activity. II. Complement level. Arch. Immunol. Ther. Exp. (Warsz.) 32, 509–514 (1984). [PubMed] [Google Scholar]
- 32.Teisberg P & Gjone E Humoral immune system activity in inflammatory bowel disease. Scand. J. Gastroenterol 10, 545–549 (1975). [PubMed] [Google Scholar]
- 33.Sugihara T et al. The increased mucosal mRNA expressions of complement C3 and interleukin-17 in inflammatory bowel disease. Clin. Exp. Immunol 160, 386–393 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halstensen TS, Mollnes TE, Garred P, Fausa O & Brandtzaeg P Surface epithelium related activation of complement differs in Crohn’s disease and ulcerative colitis. Gut 33, 902–908 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imokawa S et al. Tissue factor expression and fibrin deposition in the lungs of patients with idiopathic pulmonary fibrosis and systemic sclerosis. Am. J. Respir. Crit. Care Med 156, 631–636 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Dhar A et al. Thrombin and factor Xa link the coagulation system with liver fibrosis. BMC Gastroenterol. 18, 60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henderson NC et al. Targeting of alphav integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat. Med. 19, 1617–1624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed NI et al. The alphavbeta1 integrin plays a critical in vivo role in tissue fibrosis. Sci. Transl. Med 7, 288ra279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H et al. Mechanosensing by the alpha6-integrin confers an invasive fibroblast phenotype and mediates lung fibrosis. Nat. Commun. 7, 12564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rieder F & Fiocchi C Intestinal fibrosis in IBD—a dynamic, multifactorial process. Nat. Rev. Gastroenterol. Hepatol 6, 228–235 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Kugathasan S et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 389, 1710–1718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novak EA & Mollen KP Mitochondrial dysfunction in inflammatory bowel disease. Front. Cell Dev. Biol 3, 62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Packiriswamy N, Coulson KF, Holcombe SJ & Sordillo LM Oxidative stress-induced mitochondrial dysfunction in a normal colon epithelial cell line. World J. Gastroenterol. 23, 3427–3439 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roediger WE & Nance S Metabolic induction of experimental ulcerative colitisby inhibition of fatty acid oxidation. Br. J. Exp. Pathol 67, 773–782 (1986). [PMC free article] [PubMed] [Google Scholar]
- 45.Hudert CA et al. Transgenic mice rich in endogenous omega-3 fatty acids are protected from colitis. Proc. Natl Acad. Sci. USA 103, 11276–11281 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiese DM et al. Serum fatty acids Are correlated with inflammatory cytokines in ulcerative colitis. PLoS ONE 11, e0156387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Preter V et al. Impaired butyrate oxidation in ulcerative colitis is due to decreased butyrate uptake and a defect in the oxidation pathway. Inflamm. Bowel Dis. 18, 1127–1136 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Veazey RS et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280, 427–431 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Dong F, Zhang L, Hao F, Tang H & Wang Y Systemic responses of mice to dextran sulfate sodium-induced acute ulcerative colitis using 1H NMR spectroscopy. J. Proteome Res. 12, 2958–2966 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Smedley J et al. Tracking the luminal exposure and lymphatic drainage pathways of intravaginal and intrarectal inocula used in nonhuman primate models of HIV transmission. PLoS ONE 9, e92830 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Birse KM et al. Unbiased proteomics analysis demonstrates significant variability in mucosal immune factor expression depending on the site and method of collection. PLoS ONE 8, e79505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bruderer R et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteom. 14, 1400–1410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.UniProt Consortium T UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46, 2699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi M et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Kanehisa M & Goto S KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tenebaum D KEGGREST: Client-Side REST Access to KEGG. R package version 1.24.0 (2019). http://bioconductor.org/packages/release/bioc/html/KEGGREST.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.