A protein motif mediates targeting of proteins to the pyrenoid and appears to link the pyrenoid’s three subcompartments.
Abstract
Approximately one-third of the Earth’s photosynthetic CO2 assimilation occurs in a pyrenoid, an organelle containing the CO2-fixing enzyme Rubisco. How constituent proteins are recruited to the pyrenoid and how the organelle’s subcompartments—membrane tubules, a surrounding phase-separated Rubisco matrix, and a peripheral starch sheath—are held together is unknown. Using the model alga Chlamydomonas reinhardtii, we found that pyrenoid proteins share a sequence motif. We show that the motif is necessary and sufficient to target proteins to the pyrenoid and that the motif binds to Rubisco, suggesting a mechanism for targeting. The presence of the Rubisco-binding motif on proteins that localize to the tubules and on proteins that localize to the matrix–starch sheath interface suggests that the motif holds the pyrenoid’s three subcompartments together. Our findings advance our understanding of pyrenoid biogenesis and illustrate how a single protein motif can underlie the architecture of a complex multilayered phase-separated organelle.
INTRODUCTION
CO2 is the source of carbon for nearly the entire biosphere (1, 2), but its availability is limited in many environments. To overcome this limitation, many photosynthetic organisms use energy to locally concentrate CO2 around the CO2-assimilating enzyme Rubisco (3–6). In eukaryotic algae, which perform a major fraction of primary production in the oceans (7), concentrated CO2 is supplied to Rubisco inside a microcompartment, the pyrenoid (8). The pyrenoid consists of a spheroidal protein matrix containing Rubisco, which in nearly all algae is traversed by membranous tubules through which CO2 is delivered (9, 10). In green algae, the Rubisco matrix is additionally surrounded by a starch sheath (11) that limits CO2 leakage out of the pyrenoid (Fig. 1A) (12).
Fig. 1. A polyclonal antibody raised against the pyrenoid protein SAGA1 interacts with at least five other pyrenoid proteins.
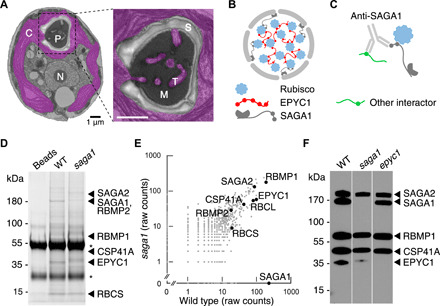
(A) Electron micrograph of a median plane section through an air-acclimated Chlamydomonas cell. N, nucleus; C, chloroplast; P, pyrenoid; M, Rubisco matrix; T, tubules; S, starch sheath. Scale bar, 1 μm. (B) Two proteins, the Rubisco linker EPYC1 and the starch sheath–binding protein SAGA1, have been previously characterized and localized to the pyrenoid. (C) An anti-SAGA1 antibody was incubated with cell lysate in an effort to coimmunoprecipitate proteins that bind to SAGA1. (D) Coomassie-stained SDS-PAGE of proteins immunoprecipitated by the anti-SAGA1 antibody from wild-type (WT) and saga1 mutant lysates. Immunoprecipitated proteins were eluted from anti-SAGA1 antibodies on beads by boiling; beads not incubated with lysate were also boiled for reference. Asterisks show heavy and light immunoglobulin chains. (E) Proteins immunoprecipitated by the SAGA1 antibody from wild type and saga1 were identified by mass spectrometry. Raw spectral counts are plotted on a log scale. (F) Anti-SAGA1 Western blot on denatured protein extracted from wild type, saga1, and epyc1.
In the model green alga Chlamydomonas reinhardtii (Chlamydomonas hereafter), two proteins are known to play central roles in pyrenoid assembly, EPYC1 (13) and SAGA1 (Fig. 1B) (14). EPYC1 is a ~35-kDa intrinsically disordered protein that phase-separates with Rubisco to form the pyrenoid matrix (13, 15, 16). SAGA1 is a ~180-kDa protein with a starch-binding domain that localizes to the periphery of the matrix and is required for normal pyrenoid morphology, although the underlying molecular mechanism is unknown (14). Here, we present how a SAGA1 antibody led us to discover that pyrenoid-localized proteins share a common Rubisco-binding protein sequence motif, revealing a mechanism by which proteins are targeted to the pyrenoid and allowing us to propose a model for how the pyrenoid’s three subcompartments are connected to each other.
RESULTS
An anti-SAGA1 antibody recognizes multiple pyrenoid proteome proteins
In an effort to immunoprecipitate SAGA1-interacting proteins, we incubated a polyclonal SAGA1 antibody with clarified cell lysates from wild-type cells (Fig. 1C and fig. S1). In addition to SAGA1, the antibody precipitated multiple proteins found in the pyrenoid proteome (17), suggesting at first that those proteins may interact with SAGA1. The precipitated proteins included EPYC1, SAGA2, RBMP1, RBMP2, and CSP41A (Fig. 1, D and E, and table S1). The protein we named SAGA2 (Cre09.g394621) is 30% identical to SAGA1 (Cre11.g467712) and, like SAGA1, has a predicted starch-binding domain (fig. S2, A to C). The proteins we named Rubisco-binding membrane proteins RBMP1 (Cre06.g261750) and RBMP2 (Cre09.g416850) have predicted transmembrane domains and were previously found to bind to Rubisco (fig. S2, D to G) (18). CSP41A (Cre10.g440050) is a predicted chloroplast epimerase (fig. S2H) (19).
We found that the precipitation of these pyrenoid proteome proteins was not mediated by SAGA1 but rather that the proteins were directly bound by the SAGA1 antibody. Three lines of evidence supported this conclusion. First, the same proteins were immunoprecipitated from a saga1 mutant lysate (Fig. 1, D and E, and table S1). Second, the predicted molecular weights of EPYC1, SAGA2, RBMP1, and CSP41A showed remarkable agreement with the multiple polypeptides recognized by the same SAGA1 antibody in immunoblots (Fig. 1F; please see Materials and Methods for a potential explanation of why other proteins, including RBMP2, were apparently not detected in these immunoblots) (14). Third, the ~35-kDa band was absent in anti-SAGA1 immunoblots of epyc1 mutant cell extracts, strongly suggesting that EPYC1 was directly recognized by the SAGA1 antibody (Fig. 1F). This putative EPYC1 band showed an apparent upward shift and decreased signal in the saga1 mutant, which could be due to a change in the expression and/or posttranslational modification of EPYC1 in the absence of SAGA1. The apparent direct binding of our SAGA1 antibody to these pyrenoid proteins led us to hypothesize that the antibody recognizes a common sequence present on all six proteins, raising questions about the nature and function of this sequence.
SAGA1, SAGA2, RBMP1, RBMP2, EPYC1, and CSP41A share a common protein motif
To identify potential sequences in the proteins that our antibody could bind, we searched their sequences for similarity to the 19 C-terminal amino acids of SAGA1, against which our antibody had been raised (14). Sequence alignment revealed that all six proteins contain a common motif, with sequence [D/N]W[R/K]XX[L/I/V/A], present at their exact C termini (Fig. 2, A and B).
Fig. 2. A motif found on pyrenoid proteins is necessary and sufficient for targeting proteins to the pyrenoid.
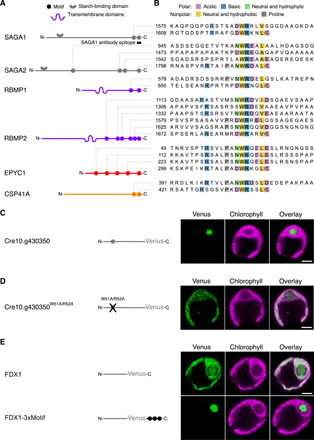
(A) The location of motifs along the primary sequence of each protein is shown (not to scale). The SAGA proteins each have a predicted starch-binding domain. The RBMP proteins each have predicted transmembrane domains (see fig. S2). (B) Sequence alignment of protein regions containing the pyrenoid motif. Motif residues are colored by physicochemical properties. Intensity of coloring is proportional to frequency at a given amino acid position. Peptides with the sequences shown in (B) were synthesized, and their binding to Rubisco was measured by surface plasmon resonance and peptide tiling array (see Fig. 3). (C) The localization of poorly characterized protein Cre10.g430350 was determined by tagging with the fluorescent protein Venus. (D) The localization of the same protein was determined after mutation of the central tryptophan-arginine dipeptide of the motif to a double alanine. (E) Localization of Venus-tagged FDX1 protein without and with the C-terminal addition of three copies of the C-terminal SAGA2 motif. Localization in (C) to (E) was determined by transforming the corresponding constructs into wild-type Chlamydomonas. Scale bar, 2 μm.
We identified an additional 14 variants of this motif at internal positions across all six proteins (Fig. 2, A and B). Most internal occurrences of the motif are immediately followed by an aspartic acid (D) or a glutamic acid (E), both of which contain a carboxyl group. This observation suggests that when the motif is found at the C terminus of the protein, the C-terminal carboxyl group of the protein is functionally important, and when the motif is found internally in the protein, this functionality is provided by the carboxyl group of the D or E side chains that follow the motif.
Further inspection of internal and C-terminal motifs revealed additional characteristics. One of the motif’s X residues is usually a D/E. In most instances of the motif, a proline is found two or three positions upstream of the tryptophan, and one or more positively charged residues (R or K) are found six to eight positions before the tryptophan (Fig. 2B). In summary, all six of the pyrenoid proteins share multiple copies of a common motif, which appears to have been recognized by our SAGA1 antibody.
The motif is necessary and sufficient for targeting a protein to the pyrenoid
The prevalence of a common motif among pyrenoid proteome proteins led us to hypothesize that this motif mediates targeting of proteins to the pyrenoid. To test this hypothesis, we evaluated the impact of disrupting the motif in a pyrenoid-localized protein. We selected Cre10.g430350, an uncharacterized protein present in the pyrenoid proteome (17), which contains a single internal copy of the motif. We validated this protein’s pyrenoid localization by expressing a fluorescently tagged wild-type protein (Fig. 2C and fig. S3A). Disruption of this fluorescently tagged protein’s motif by mutating WR to AA caused the protein to localize homogeneously throughout the chloroplast (Fig. 2D and fig. S3B), indicating that the motif is necessary for targeting this protein to the pyrenoid.
To determine whether the motif is sufficient for targeting a protein to the pyrenoid, we added the motif to ferredoxin 1 (FDX1; Cre14.g626700), a small soluble protein that natively localizes throughout the chloroplast stroma. To increase the chances of observing the motif’s effect, we chose to add three tandem copies of the motif to FDX1. Addition of the motifs relocalized fluorescently tagged FDX1 exclusively to the pyrenoid matrix (Fig. 2E and fig. S4, A and B). We obtained a similar result with another chloroplast protein (fig. S4, C and E). These results demonstrate that the motif is sufficient to localize a chloroplast protein to the pyrenoid matrix.
The motif binds to Rubisco
Two observations led us to hypothesize that proteins bearing the motif are recruited to the pyrenoid via binding to Rubisco. First, the motif is present in each of the regions that were found to mediate EPYC1’s binding to Rubisco in yeast two-hybrid experiments (20) and as short peptides in vitro (21). Second, the other five motif-containing proteins were also previously found to bind to Rubisco: SAGA1 by yeast two-hybrid (14), and SAGA2, RBMP1, RBMP2, and CSP41A by affinity purification–mass spectrometry (18).
To determine whether each variant of the motif can bind to Rubisco, we used surface plasmon resonance to measure the binding of synthetic peptides to Rubisco. With this method, 14 of 20 peptides representing motif variants had a higher affinity to Rubisco than peptides with random sequences. Peptides with C-terminal motifs systematically showed higher affinity to Rubisco than peptides with internal motifs (Fig. 3A). Rubisco bound to all predicted internal motif sites when we incubated purified Rubisco with arrays of peptides tiling across the full-length proteins (Fig. 3, B and C, and fig. S5, A to D; C-terminal sites could not be assayed by this method). These results indicate that all variants of the motif (Fig. 2B) bind to Rubisco in vitro in at least one of our assays. Binding of the proteins to the SAGA1 antibody in the immunoprecipitation experiment (Fig. 1, C and D) suggests that at least one motif on each protein is accessible for Rubisco binding when the proteins adopt their native folding in vivo.
Fig. 3. The motif binds to Rubisco.
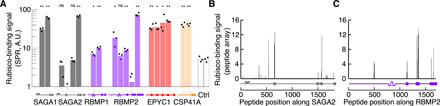
(A) Peptides containing motifs from the indicated proteins were synthesized, and their binding to Rubisco was measured by surface plasmon resonance (SPR). A.U., arbitrary units. Two peptides not containing the motif were included as controls (Ctrl). Each dot shows the binding response of an independent replica. Negative values (when the experimental binding signal was lower than that of the reference cell) are not plotted. The positions of the predicted motifs are indicated below the graph (not to scale). Significance levels of increased binding relative to control peptides were determined using Welch’s t test. *P < 0.05 and **P < 0.01. (B and C) Arrays of 18–amino acid peptides tiling across the sequences of SAGA2 (B) and RBMP2 (C) were synthesized and probed with Rubisco. The binding signal in (B) and (C) is normalized to a control EPYC1 peptide known to bind to Rubisco (one unit of binding) (21). The positions of motifs are indicated below each graph (to scale).
In a parallel study (21), we inadvertently determined where the motif binds on Rubisco. In that study, as part of an effort to understand how EPYC1 clusters Rubisco to form the pyrenoid matrix, we obtained a cryo–electron microscopy structure of Rubisco bound to a peptide from EPYC1. A Rubisco-binding motif is present at positions N62W63R64Q65E66L67E68 on this EPYC1 peptide and plays a central role in the binding interface. One EPYC1 peptide binds to each of the eight Rubisco small subunits of the Rubisco holoenzyme. The motif-containing portion of the peptide adopts an α helix that binds to the Rubisco surface. R64 of the peptide, corresponding to the [R/K] of the motif, forms a salt bridge with a glutamic acid of the Rubisco small subunit. In addition, W63 and L67 of the peptide, corresponding to the W and [L/I/V/A] of the motif, respectively, contribute to a hydrophobic interaction with three hydrophobic residues of the Rubisco small subunit. The central role of the motif residues in the binding interface strongly suggests that all instances of the motif studied here bind to Rubisco using the same mechanism.
The motif is present in proteins other than the ones we studied here
We identified putative instances of the motif in the Chlamydomonas proteome using a simple scoring scheme (table S2; Materials and Methods). Motif scores ranged from 0 (for only a WR or WK dipeptide) to 6 (for sequences that share all canonical features of the motif). High motif scores were modestly enriched among pyrenoid proteome proteins relative to other proteins in the chloroplast proteome (P = 0.047; fig. S6). Twenty-two of 191 pyrenoid proteome proteins have at least one putative motif with a score of 3 or greater, and 100 of these proteins have at least a WR or WK dipeptide. We hypothesize that some of the pyrenoid-localized proteins that do not contain the motif are targeted to the pyrenoid by binding to a motif-containing protein. Some chloroplast proteome proteins that have a putative motif do not localize to the pyrenoid: At least one motif with a score of 3 or greater is present in 52 of 735 chloroplast proteins that are not in the pyrenoid proteome. One potential explanation for this observation is that to mediate targeting to the pyrenoid, a motif must not only be present in the protein sequence but also be accessible on the surface of the folded protein for interaction with Rubisco. Motifs that are present in the internal folds of a protein would not be expected to affect protein localization. We found that the motifs of pyrenoid proteome proteins are more frequently situated in predicted disordered regions, compared with the motifs of chloroplast proteins that are not found in the pyrenoid proteome (Welch’s t test P = 0.007; fig. S7), supporting the idea that motif accessibility is important for pyrenoid targeting.
Together, our findings suggest a mechanism for targeting proteins to the pyrenoid, involving the presence of a common motif that recruits its protein to the pyrenoid via direct binding interactions with Rubisco. It is possible that the mechanism operates via random diffusion of the motif-bearing protein through the chloroplast, followed by capture of the motif by Rubisco when the protein encounters the pyrenoid matrix.
The motif appears to mediate binding between the pyrenoid’s three subcompartments
Beyond providing a mechanism for targeting proteins to the pyrenoid matrix, the motif appears to play a role at the interfaces of the pyrenoid’s three subcompartments. Although Rubisco and EPYC1 are localized in the matrix (Fig. 4A and fig. S8, A and B), we observed that some motif-containing proteins localize to pyrenoid regions other than the matrix. Fluorescently tagged RBMP1 and RBMP2 localized to the tubules (Fig. 4A and fig. S8, C and D). SAGA1 (14) and SAGA2 localized to the interface between the Rubisco matrix and the starch sheath (Fig. 4A and fig. S8, E and F).
Fig. 4. The motif orchestrates the architecture of the pyrenoid’s three subcompartments.
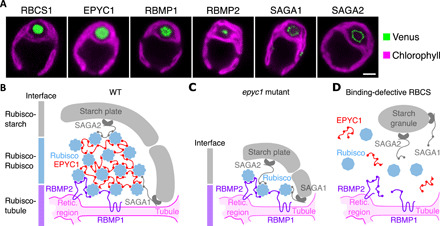
(A) Representative confocal images of Venus-tagged proteins that have the Rubisco-binding motif and Rubisco small subunit (RBCS1). Chlorophyll autofluorescence delimits the chloroplast. Scale bar, 2 μm. (B) Proposed model for how the motif mediates assembly of the pyrenoid’s three subcompartments in wild type. The motif on tubule-localized transmembrane proteins RBMP1 and RBMP2 mediates Rubisco binding to the tubules [Retic. region is the reticulated region of the tubules (9)]. Multiple copies of the motif on EPYC1 link Rubiscos to form the pyrenoid matrix (21). At the periphery of the matrix, the motif on starch-binding proteins SAGA1 and SAGA2 mediates interactions between the matrix and surrounding starch sheath. (C) The model in (B) explains the matrix-less phenotype observed in EPYC1-less mutants. (D) The model also explains the absence of matrix and starch plates in mutants where Rubisco’s binding site for the motif has been disrupted (21).
These observations together with previous work (9, 13–17, 20) are consistent with a model where the Rubisco-binding motif holds together the pyrenoid matrix, the traversing tubules, and the surrounding starch sheath (Fig. 4B). In this model, multiple copies of the motif on EPYC1 mediate cohesion of the matrix by bringing Rubisco holoenzymes together (21). Moreover, the presence of the same motif on the pyrenoid tubule-localized transmembrane proteins, RBMP1 and RBMP2, recruits Rubisco to the tubules and favors assembly of the matrix around them. Last, the presence of the motif on proteins with starch-binding domains, SAGA1 and SAGA2, which localize to the pyrenoid periphery, mediates adherence of the starch sheath to the matrix.
The slightly different localization patterns of RBMP1 and RBMP2 suggest that RBMP1 and RBMP2 could each promote Rubisco matrix binding to a different part of the tubules. Our microscopy data suggest that RBMP2 is confined to the central reticulated region of the tubules (9), whereas RBMP1 appears to localize to the more peripheral tubular regions to the exclusion of the reticulated region. Similarly to the RBMPs, the different localization patterns of the SAGAs also suggest that they may each interface with different features on the starch sheath. As described previously (14), SAGA1 localized to puncta at the periphery of the matrix, likely at the interface between the Rubisco matrix and the starch sheath. We observed that SAGA2 also localized to that interface but appeared to cover the surface of the matrix more homogeneously than SAGA1.
The model explains several previously puzzling observations. First, in a mutant lacking EPYC1, a pyrenoid-like structure still assembles around the tubules, containing some Rubisco enclosed by a starch sheath, although the canonical matrix is absent (13). The presence of both Rubisco and starch in this structure can be explained by a layer of Rubisco that serves as a bridge between motif-containing proteins RBMP1 and RBMP2 on the tubules and motif-containing proteins SAGA1 and SAGA2 on the surrounding starch sheath (Fig. 4C). Second, point mutations that disrupt the EPYC1 binding site on Rubisco not only eliminate the matrix but additionally disrupt the association of the starch sheath with the pyrenoid (21). The additional disruption of the starch sheath can be explained by the same Rubisco-binding site being required not only for binding to EPYC1 but also for binding the motif on other proteins that connect the starch and tubules to Rubisco (Fig. 4D).
DISCUSSION
Our results provide insights into pyrenoid biogenesis and function
Our work reveals a ubiquitous Rubisco-binding motif that is necessary and sufficient for targeting proteins to the pyrenoid and also appears to mediate the overall assembly of the pyrenoid’s three subcompartments. The eightfold symmetry of the Rubisco holoenzyme allows it to interact simultaneously with multiple binding partners via the motif, making Rubisco a central structural hub of the pyrenoid. The valency and binding strengths of the motif to Rubisco vary among the binding partners (Figs. 2A and 3), which could play a role in their relative priority of access to Rubisco, as observed for other phase-separated organelles (22–24).
The Rubisco-EPYC1 condensate can enhance CO2 fixation only if it is anchored around the pyrenoid tubules, which are thought to provide concentrated CO2 (8, 9, 25, 26). Identification in the present work of two tubule-localized proteins that bind to Rubisco provides a plausible explanation for how the Chlamydomonas pyrenoid forms preferentially around tubules rather than anywhere else in the chloroplast. Considering that the tubules can form independently of the matrix (27, 28), whether the motif is required for RBMPs to localize to the tubules remains to be investigated. Our observation that RBMP1 and RBMP2 localize to different subdomains of the tubules suggests that an additional localization mechanism may be at play, such as a preference for a specific membrane curvature.
RBMP1 is predicted to be a Ca2+-activated anion channel of the bestrophin family (fig. S2, D and F), several members of which are thought to supply HCO3− to the lumen of the tubules for conversion to CO2 (29). Although the previously described members localized primarily to membranes outside the pyrenoid, RBMP1 localizes exclusively to the tubules themselves, which may allow it to directly feed HCO3− to the tubules for conversion to CO2.
The principles described here likely apply more broadly
Pyrenoids are thought to have evolved independently multiple times through convergent evolution (30–32). Chlamydomonas belongs to the green algal order Volvocales, a lineage that has been evolving independently from other green algae for the past 70 to 200 million years (33). Homologs of all six pyrenoid proteins studied here are present in other Volvocales, and in each of these homologs, one or more copies of the motif are conserved (fig. S9, A and B to G). The amino acids of the Rubisco small subunit that are essential for binding the motif (21, 34) are also broadly present in the Volvocales (fig. S9H). These results suggest that the motif described here and its functions in pyrenoid biogenesis and protein targeting evolved before the divergence of the Volvocales.
Although the specific sequences and proteins may be different in other algal lineages, we hypothesize that the organizational principles described here apply broadly to pyrenoids across the tree of life. The convergent evolution of pyrenoids may well have been facilitated by the possibility of using a common motif and binding site on Rubisco to perform three functions essential to all pyrenoids: clustering of Rubisco into a matrix, targeting of proteins to the matrix, and connecting the matrix to other structures.
Our findings advance the basic understanding of the biogenesis of the pyrenoid and provide a framework for engineering a pyrenoid into crops for improving yields (8, 35, 36). More broadly, the system presented here provides a remarkable example of how the architecture of a complex phase-separated organelle can be defined by a simple organizing principle.
MATERIALS AND METHODS
Strains and culture conditions
The C. reinhardtii stain CC-4533 (37) was the wild type for all experiments (hereafter WT) and parent for all genetic transformations. The fluorescently tagged strain showing the native localization of SAGA1 was saga1-paroR;SAGA1-Venus-3×FLAG-hygR (14). All strains were maintained at room temperature (RT) (∼22°C) under very low light (<10 μmol photons m−2 s−1), on solidified tris-acetate-phosphate medium (TAP + 1.5% agar) (pH 7.4), using a revised trace elements recipe for increased growth (38). Medium was supplemented with paromomycin at 2 μg ml−1 for all strains (except WT) and, additionally, with hygromycin at 6.25 μg ml−1 for the SAGA1-Venus strain.
All experiments were conducted on photoautotrophically grown cells. Liquid cultures were primed with a loopful of TAP-agar grown cells not older than 2 weeks resuspended into tris-phosphate medium (as TAP above, but without acetate) to a starting concentration less than 105 cells ml−1. Cultures were maintained in an orbital incubator-shaker (Infors) with controlled conditions: 130 rpm, continuous cool white fluorescent light at ∼175 μmol photons m−2 s−1, 22°C, air-enriched with 3% CO2 (v/v) for faster growth, and rescue of saga1 and epyc1. Culture volume for Rubisco extractions was ∼500 ml, for coimmunoprecipitations was ∼250 ml, and for imaging and Western blots was ∼50 ml. Cells were grown in conical flasks with a total capacity at least four times that of the volume of the medium. Culturing time allowed at least six rounds of mitotic division. Cell densities were not allowed to exceed 107 cells ml−1 at any point in time and were subcultured accordingly. Cell densities were measured using a Countess II F automated cell counter (Thermo Fisher Scientific).
For most experiments, cells were acclimated to air-level CO2 concentrations for 6 hours before harvesting to maximize expression of the CO2-concentrating mechanism and packaging of Rubisco into a pyrenoid (39, 40). Cultures destined for confocal imaging were acclimated overnight (∼16 hours). Acclimation to air-level CO2 was performed by pelleting high-CO2 grown cells (1000g, 10 min, RT), followed by gentle resuspension by agitation in fresh air–equilibrated TP medium, before transfer to an air-equilibrated chamber of the same orbital incubator-shaker (agitation, light, and temperature as above). CO2 concentration was periodically monitored with a CO2 sensor (CO2Meter). All experiments aimed for a cell density at the time of harvesting of ∼2 × 106 to 4 × 106 cells ml−1. All strains generated in this work were deposited at the Chlamydomonas Resource Center (https://chlamycollection.org/).
Coimmunoprecipitation and mass spectrometry analysis
Native protein complexes were extracted according to the protocol described by Mackinder et al. (18), with minor modifications. Briefly, all protein extraction steps were performed at 4°C in a cold room, using only fresh algal material. After harvesting (1000g, 5 min, 4°C), cells were washed once in ice-cold TP, repelleted, and suspended in a 1:1 (v/w) ratio of ice-cold 2× immunoprecipitation buffer [400 mM sorbitol, 100 mM Hepes, 100 mM KOAc, 4 mM Mg(OAc)2•4H2O, and 2 mM CaCl2] containing a protease inhibitor cocktail (cOmplete, Roche) and phosphatase inhibitors (2 mM NaF and 0.6 mM Na3VO4). To ease the grinding, the cell slurry was transformed into frozen droplets of ~5 mm diameter by slowly releasing the cell/buffer mixture into liquid N2 through a fine-tipped transfer pipette held ~15 cm above the cryogenic liquid. Releases were timed so as to avoid clumping of not fully frozen material. Each assay used ~1 g of cell/buffer mixture. Mass spectrometry analysis was performed at the Stanford University Mass Spectrometry facility, as previously described (18). Raw spectral counts are given in table S1.
Minor deviations from (18): A 50:50 mixture of Dynabeads protein A and protein G was used; incubation was with an anti-SAGA1 antibody (YenZym); protein complexes bound to magnetic beads were released by boiling for 1 min; denatured protein samples were run on denaturing tris/glycine gradient gel (4 to 15%) and stained with EZBlue (Thermo Fisher Scientific).
Immunoblot analysis
Total proteins were extracted as follows. Cell suspensions (10 ml) were pelleted (3500g, 10 min, 4°C), resuspended in 300 μl of lysis buffer [5 mM Hepes-KOH (pH 7.5), 100 mM dithiothreitol, 100 mM Na2CO3, 2% SDS, 12% sucrose, and cOmplete protease inhibitor cocktail], transferred to a microcentrifuge tube, and heat-denatured in a thermomixer (37°C, 10 min, 750 rpm). Lysate was clarified (16,000g, 5 min, 4°C), aliquoted, flash-frozen in liquid N2, and stored at −80°C until analysis on SDS–polyacrylamide gel electrophoresis (SDS-PAGE).
Gel loading was normalized by total chlorophyll a + b content. Pigments were extracted from 50 μl of cell lysate with 2 ml of 100% methanol. Chlorophylls contained in the clarified extract (16,000g, 2 min) were quantified according to the following equations: chlorophyll a (μg ml−1) = 16.29 A665 − 8.54 A652; chlorophyll b (μg ml−1) = 30.66 A652 − 13.58 A665 (41), after correction for A750. Absorbances were measured in a SmartSpec Plus spectrophotometer (Bio-Rad).
Proteins were separated by size on a denaturing tris/glycine gradient gel (4 to 15%, Criterion TGX, Bio-Rad; 90 V constant, 105 min) and transferred to 0.45-μm polyvinylidene difluoride membrane (Immobilon-P, MilliporeSigma) using a wet electroblotting system (Criterion Blotter, Bio-Rad) and Towbin buffer [20% methanol, 25 mM tris, 192 mM glycine, 20% (v/v) methanol, and 0.05% SDS] at 30 V constant overnight.
For immunoblot analysis, membranes were blocked in tris-buffered saline (TBS) + 0.1% Tween-20 (TBST) containing 5% nonfat dry milk for 1hour at RT or overnight at 4°C, under gentle agitation. Incubations with the primary antibodies were performed in TBST containing 2.5% milk for 1 hour at RT or overnight at 4°C. Membranes were washed in TBST (4×, 10 min, rocking platform) before incubation with the secondary antibody for 1 hour at RT. Membranes were washed again in TBST (4×, 10 min). Immunoreactive proteins were detected using enhanced chemiluminescence (WesternBright ECL, Advansta) followed by x-ray film processing (CL-XPosure Film, Thermo Fisher Scientific; SRX-101A, Konica-Minolta).
Primary antibodies were obtained from YenZym (anti-SAGA1 and anti-EPYC1) and MilliporeSigma (monoclonal anti-FLAG M2 antibody). The polyclonal anti-Rubisco antibody was a gift from H. Griffiths, University of Cambridge, UK. Goat anti-mouse immunoglobulin G (IgG) (H+L) and goat anti-rabbit IgG (H+L) were from Thermo Fisher Scientific. Dilutions: anti-FLAG 1:2500 + secondary 1:10,000; anti-SAGA1 1:2500 + secondary 1:10,000; anti-EPYC1 1:5000 + secondary 1:10,000; anti-Rubisco: 1:10,000 + secondary 1:20,000.
A note on the results from the immunoblot in Fig. 1F: One might ask why only five bands appeared when wild-type lysates were probed by the anti-SAGA1 antibody, considering that variants of the motif are present in hundreds of proteins in the proteome (table S2). One potential explanation for this discrepancy is that the anti-SAGA1 antibodies may be very specific to certain feature variants of the motif that are found only in SAGA1 and in the other four proteins that yield bands in this experiment. For example, because the anti-SAGA1 antibodies were generated against a peptide representing the C terminus of SAGA1, it is possible that the anti-SAGA1 antibodies only recognize the motifs on the C termini of proteins, which would prevent recognition of the vast majority of the motifs we identified in the proteome. Additional specificity to other features such as a C-terminal leucine may explain why SAGA1, SAGA2, RBMP1, EPYC1, and CSP41A were apparently detected in Fig. 1F, whereas RBMP2 (which unlike the other five proteins has a C-terminal valine) was apparently not detected in Fig. 1F (one would otherwise expect RBMP2 to be visible as a band at ~180 kDa in the saga1 mutant) and gave the fewest spectral counts of the six proteins in the anti-SAGA1 immunoprecipitation experiment (Fig. 1E and table S1).
Rubisco purification and quantification
WT Rubisco was extracted as follows. Cell cultures (500 ml) were harvested (~4000g, 15 min, 4°C), resuspended in 1.5 ml of lysis buffer [50 mM bicine (pH 8.0), 10 mM NaHCO3, 10 mM MgCl2, and 1 mM dithiothreitol] containing a protease inhibitor cocktail, and transferred to an ice-cold 50-ml Falcon. Cells were sonicated on ice in 30-s bursts, followed by 30-s pauses with a microprobe set at 60% amplitude (Q125 + CL-18 probe, QSonica), until no intact cells were left. Progress of the lysis was monitored with a light microscope (400×). Total soluble proteins were isolated by centrifugation (16,000g, 30 min, 4°C), and 650 μl of the clarified lysate was loaded on top of a thin-wall ultracentrifugation tube (Ultra-Clear, Beckman Coulter) containing 12 ml of a 10 to 30% sucrose gradient prepared with the lysis buffer. Gradients were made the previous day with a gradient maker (BioComp Instruments) and left to equilibrate at 4°C overnight. Gradients were run at 37,000 rpm for 20 hours in an ultracentrifuge (Optima XE-100 + SW 41 Ti rotor, Beckman Coulter). Fractions (750 μl) were collected either with a piston gradient fractionator (BioComp Instruments) or manually by gravity. Fractions enriched in Rubisco were identified by running 10-μl aliquots in 2:1 Laemmli buffer on SDS-PAGE, followed by staining with EZBlue (same conditions as detailed in the previous section). Fractions with the highest concentration of Rubisco (bands at 55 and 15 kDa for the Rubisco large and small subunits, respectively) were pooled, and buffer was exchanged by dialysis at 4°C overnight (Slide-A-Lyzer 20k MWCO, Thermo Fisher Scientific) using the same buffer as the one for the two Rubisco-peptide binding assays (see the next two sections). Rubisco was concentrated to ~2 mg ml−1 on centrifugal filters (Amicon Ultra-4 100K, MilliporeSigma) before use. Rubisco concentration was determined by Bradford assay (Quick Start Bradford Dye Reagent + BSA Standard Set, Bio-Rad).
Binding of free synthetic peptides to immobilized Rubisco measured by surface plasmon resonance
The surface plasmon resonance experiment was performed on a Biacore 3000 (GE Healthcare) at constant 25°C, using the proprietary Biacore Control Software v.4.1, embedded application wizards “Surface preparation” and “Binding analysis,” and GE’s immobilization kit, buffers, and consumables (no deviation from the manufacturer’s instructions). Optimal pH of 4.5 for amine coupling of purified Rubisco onto a CM5 sensor chip was identified with the aid of the “pH scouting” script. Variable amounts of Rubisco were immobilized in three independent assays (each independent assay used fresh Rubisco from an independent extraction), spanning 2000 to 4000 resonance units (RUs) using the “Aim for immobilized level” script. All peptides (see Fig. 2B) were synthesized by GenScript, with purity ≥85% and nitrogen content validated by analysis on an organic elemental analyzer. Binding assays were run in PBS-P+ buffer [20 mM phosphate buffer (pH 7.4), 2.7 mM KCl, 137 mM NaCl, and 0.05% (v/v) P20 surfactant]. The same buffer was used for peptide solubilization and Rubisco dialysis (see the previous section). Lyophilized peptides were solubilized to a stock concentration of 2.5 mM, aliquoted, flash-frozen in liquid N2, and stored at −80°C until needed. Binding response was measured at a peptide concentration of 1 mM during a single 3-min injection into the sensor’s flow cells at a flow rate of 40 μl min−1. Dissociation was measured while injecting buffer only, at a rate of 40 μl min−1 for 2.5 min. The chip surface was regenerated by flowing buffer for 5 min at a rate of 40 μl min−1. Return to baseline was observed for all peptides except the one corresponding to RBMP2’s fourth instance of the motif, which remained partially insoluble even after addition of dimethyl sulfoxide (DMSO) and was therefore discarded from further analysis (see Fig. 3A). Binding responses were normalized to 1000 RUs of immobilized Rubisco to allow comparison across independent repeats and plotted on a log10 scale. Peptides lacking the predicted Rubisco-binding motif were used as negative controls: GYFAVDHRPNLAILQGELGTKSESMDVRI and SKPAVDLRFYLEIGMQNTA.
Binding of free Rubisco to immobilized synthetic peptides measured by peptide tiling array
Four peptide arrays [30 × 20 spots each on 15 × 10 cm cellulose membranes, SPOT synthesis (42)] were ordered from the Koch Institute for Integrative Cancer Research at Massachusetts Institute of Technology, Biopolymers and Proteomics Laboratory (Cambridge, MA). The arrays were composed of 18–amino acid peptides that tiled across the full length of each of the six Rubisco-binding protein sequences, with a step size of three amino acids. Each peptide was represented by a single spot, except for EPYC1, which included a nonrandomized duplicate in positions 499 to 598 on membrane #1. All other locations of peptides were randomized. EPYC1 (100 spots, two repeats), CSP41A (142 spots), and RBMP1 (217 spots) were arrayed on the same membrane. RBMP2, SAGA1, and SAGA2 each required a separate membrane (558, 537, and 594 spots, respectively). Membranes were incubated with 750 to 2000 μg of purified Rubisco and probed by anti-Rubisco Western blot, as described above. The peptide corresponding to the very C terminus of each protein does not accurately represent the Rubisco-binding motif in this assay, as the peptides are linked to the cellulose via their C termini. This linkage eliminates the carboxyl group, which appears to be important for binding to Rubisco. Binding intensity was quantified in ImageJ (43) by measuring the integrated density of a circle of constant area centered on each blot dot, after background correction on an inverted image (rolling ball radius set to 25 pixels). Binding intensity was normalized to the binding of positive controls to allow comparison across membranes [TRSVLPANWRQELESLRN (21)].
Fluorescent protein tagging and confocal microscopy
All fluorescently tagged proteins in this study were tagged with the fluorescent protein Venus followed by three copies of the FLAG epitope. Open reading frames of the two chloroplast proteins FDX1 and Cre12.g498550 were cloned by Gibson assembly into the vector pLM005-Venus (KX077945.1), as reported by Mackinder et al. (13). A 677–nucleotide (nt)–long GeneArt DNA fragment (Thermo Fisher Scientific) and pLM005-Venus were double digested with Eco RI–HF and Pfl MI (New England Biolabs) and ligated by T4 DNA ligase (16°C). Constructs were validated by Sanger sequencing [759-nt polymerase chain reaction (PCR) product spanning the double digested 643-nt insert] and transformed in WT Chlamydomonas, as described in (13), with two modifications: A four-step pulse electroporator (NEPA21, NEPAGENE) was used, and 50 μg of carrier DNA (MP Biochemicals) was added to each transformation.
RBMP1, RBMP2, and SAGA2 were cloned using homologous recombination based on protocols optimized for Chlamydomonas (29). Several attempts at cloning CSP41A by this and other methods (18) were unsuccessful.
Chlamydomonas transformants were selected on TAP-agar + paromomycin (20 μg ml−1), and high expressors were screened on a fluorescence laser scanner (Typhoon, GE). Expression of full-length fusion proteins was validated by immunoblotting, using an anti-FLAG M2 antibody (figs. S3C and S4F).
Images were captured with a laser scanning microscope (TCS SP5, Leica) using a 100× objective (numerical aperture 1.46). Venus and chlorophyll were excited by argon lasers at 514 and 561 nm, respectively; emission was collected at 525 to 550 nm and 620 to 670 nm, respectively. Zoom-in (3×) acquisition settings were identical for all strains. Two-dimensional median plane cross sections were captured at 200 Hz. Pinhole was set at 1 airy unit. Venus was captured on hybrid detectors, and chlorophyll autofluorescence was captured on photomultiplier tubes. Picture montages were done on ImageJ (43).
Site-directed mutagenesis
Point mutations were generated using a commercial kit (QuikChange II XL, Agilent Technologies). The mutagenic primers were GACTGGCGCAGCTCCGCGGCAACGGAGCTTGAGG and its reverse complement. Mutations were added by PCR to pLM005-Cre10.g430350-Venus-3×FLAG (generated as above) according to the manufacturer’s instructions. The mutated pLM005-Cre10.g430350-Venus-3×FLAG codes for a fluorescently tagged Cre10.g430350 with two substitutions (W51A and R52A).
Bioinformatic search for motifs and motif enrichment analysis
We used a point system to identify motifs in the genome. A potential motif must contain a WR or WK dipeptide and is assigned points for the following additional residues (in positions relative to the W, which is considered the “zero” position): (i) a basic residue (R or K) in position −8 to −6: +1 point (no additional point if multiple instances at those three positions), (ii) a proline (P) in position −3 or − 2: +1 point, (iii) an aspartic acid (D) or an asparagine (N) in position −1: +1 point, (iv) an aliphatic residue (L, I, V, or A) in position +4: +2 points, and (v) an acidic residue (D or E) or a C terminus in position +5: +1 point. Motif scores for all proteins in the proteome are listed in table S2. To test the statistical significance of the motif enrichment in pyrenoid proteins, we used the Mann-Whitney test to evaluate the difference between the two distributions shown in fig. S6, excluding the six proteins in which we originally noticed the motif to avoid our observations biasing the result. The two distributions are different (P = 0.047).
Bioinformatic search of pyrenoid protein homologs
BLAST searches were conducted on publicly accessible repositories and portals: oneKP (44) (https://db.cngb.org/onekp/), Phytozome, and GeneBank/National Center for Biotechnology Information. The Rubisco small subunit sequence alignment and phylogenetic tree were generated with MUSCLE (45).
Supplementary Material
Acknowledgments
We thank C. Dupont (JCVI, La Jolla, CA), as well as present members and alumni of the Jonikas laboratory for discussions and feedback on the manuscript; the Princeton University Confocal Microscopy and Biophysics core facility managers G. Laevsky and V. Gopal Vandavasi for instrumentation support; and R. Leib and C. Adams at the Stanford University Mass Spectrometry facility. Funding: The project was funded by NSF (IOS-1359682 and MCB-1935444), NIH (DP2-GM-119137), and Simons Foundation and Howard Hughes Medical Institute (55108535) grants to M.C.J., and the UK Biotechnology and Biological Sciences Research Council (BB/R001014/1) grant to L.C.M.M. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Author contributions: M.T.M., A.K.I., and M.C.J. conceived the project. A.K.I. performed the coimmunoprecipitation experiment and prepared samples for analysis by mass spectrometry. M.T.M. purified Rubisco and performed Western blots, surface plasmon resonance, protein relocalization experiments, and confocal microscopy. S.H. and M.T.M. performed the peptide tiling array experiment. T.E.-M., J.L., G.Y., L.W., M.T.M., and L.C.M.M. produced fluorescently tagged strains. W.P. conducted the bioinformatic analyses. All authors analyzed the data. M.T.M. and M.C.J. wrote the manuscript with input from all authors. M.C.J. supervised the work. Competing interests: Princeton University, Stanford University, and the University of York have submitted a provisional patent application on aspects of the findings. These applications were submitted to two patent offices: PCT (no. PCT/US2020/044326, filed on 30 July 2020, priority date: 2 August 2019) and Argentina (no. P200102163, filed on 31 July 2020). The authors declare no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/46/eabd2408/DC1
REFERENCES AND NOTES
- 1.Raven J. A., Contributions of anoxygenic and oxygenic phototrophy and chemolithotrophy to carbon and oxygen fluxes in aquatic environments. Aquat. Microb. Ecol. 56, 177–192 (2009). [Google Scholar]
- 2.Bar-Even A., Noor E., Milo R., A survey of carbon fixation pathways through a quantitative lens. J. Exp. Bot. 63, 2325–2342 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Still C. J., Berry J. A., Collatz G. J., DeFries R. S., Global distribution of C3 and C4 vegetation: Carbon cycle implications. Global Biogeochem. Cycles 17, 6-1–6-14 (2003). [Google Scholar]
- 4.Granum E., Raven J. A., Leegood R. C., How do marine diatoms fix 10 billion tonnes of inorganic carbon per year? Can. J. Bot. 83, 898–908 (2005). [Google Scholar]
- 5.Raven J. A., Beardall J., Giordano M., Energy costs of carbon dioxide concentrating mechanisms in aquatic organisms. Photosynth. Res. 121, 111–124 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Maberly S. C., Gontero B., Ecological imperatives for aquatic CO2-concentrating mechanisms. J. Exp. Bot. 68, 3797–3814 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Field C. B., Behrenfeld M. J., Randerson J. T., Falkowski P., Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 281, 237–240 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Hennacy J. H., Jonikas M. C., Prospects for engineering biophysical CO2 concentrating mechanisms into land plants to enhance yields. Annu. Rev. Plant Biol. 71, 461–485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engel B. D., Schaffer M., Kuhn Cuellar L., Villa E., Plitzko J. M., Baumeister W., Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo–electron tomography. eLife 4, e04889 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer M. T., Whittaker C., Griffiths H., The algal pyrenoid: Key unanswered questions. J. Exp. Bot. 68, 3739–3749 (2017). [DOI] [PubMed] [Google Scholar]
- 11.J. D. Pickett-Heaps, Green Algae: Structure, Reproduction and Evolution in Selected Genera (Sinauer Associates, 1975). [Google Scholar]
- 12.Toyokawa C., Yamano T., Fukuzawa H., Pyrenoid starch sheath is required for LCIB localization and the CO2-concentrating mechanism in green algae. Plant Physiol. 182, 1883–1893 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackinder L. C. M., Meyer M. T., Mettler-Altmann T., Chen V. K., Mitchell M. C., Caspari O. D., Freeman Rosenzweig E. S., Pallesen L., Reeves G., Itakura A. K., Roth R., Sommer F., Geimer S., Mühlhaus T., Schroda M., Goodenough U. W., Stitt M., Griffiths H., Jonikas M. C., A repeat protein links rubisco to form the eukaryotic carbon-concentrating organelle. Proc. Natl. Acad. Sci. U.S.A. 113, 5958–5963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itakura A. K., Chan K. X., Atkinson N., Pallesen L., Wang L., Reeves G., Patena W., Caspari O. D., Roth R., Goodenough U. W., McCormick A. J., Griffiths H., Jonikas M. C., A Rubisco-binding protein is required for normal pyrenoid number and starch sheath morphology in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 116, 18445–18454 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freeman Rosenzweig E. S., Xu B., Kuhn Cuellar L., Martinez-Sanchez A., Schaffer M., Strauss M., Cartwright H. N., Ronceray P., Plitzko J. M., Förster F., Wingreen N. S., Engel B. D., Mackinder L. C. M., Jonikas M. C., The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171, 148–162.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wunder T., Cheng S. L. H., Lai S.-K., Li H.-Y., Mueller-Cajar O., The phase separation underlying the pyrenoid-based microalgal Rubisco supercharger. Nat. Commun. 9, 5076 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhan Y., Marchand C. H., Maes A., Mauries A., Sun Y., Dhaliwal J. S., Uniacke J., Arragain S., Jiang H., Gold N. D., Martin V. J. J., Lemaire S. D., Zerges W., Pyrenoid functions revealed by proteomics in Chlamydomonas reinhardtii. PLOS ONE 13, e0185039 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackinder L. C. M., Chen C., Leib R. D., Patena W., Blum S. R., Rodman M., Ramundo S., Adams C. M., Jonikas M. C., A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell 171, 133–147.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker M. E., Grundy W. N., Elkan C. P., Spinach CSP41, an mRNA-binding protein and ribonuclease, is homologous to nucleotide-sugar epimerases and hydroxysteroid dehydrogenases. Biochem. Biophys. Res. Commun. 248, 250–254 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Atkinson N., Velanis C. N., Wunder T., Clarke D. J., Mueller-Cajar O., McCormick A. J., The pyrenoidal linker protein EPYC1 phase separates with hybrid Arabidopsis–Chlamydomonas Rubisco through interactions with the algal rubisco small subunit. J. Exp. Bot. 70, 5271–5285 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.S. He, H. T. Chou, D. Matthies, T. Wunder, M. T. Meyer, N. Atkinson, A. Martinez-Sanchez, P. D. Jeffrey, S. A. Port, W. Patena, G. He, V. K. Chen, F. M. Hughson, A. J. McCormick, O. Mueller-Cajar, B. D. Engel, Z. Yu, M. C. Jonikas, The structural basis of rubisco phase separation in the pyrenoid. bioRxiv 2020.08.16.252809 [Preprint]. 16 August 2020. 10.1101/2020.08.16.252809. [DOI] [PMC free article] [PubMed]
- 22.Banani S. F., Rice A. M., Peeples W. B., Lin Y., Jain S., Parker R., Rosen M. K., Compositional control of phase-separated cellular bodies. Cell 166, 651–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi J.-M., Holehouse A. S., Pappu R. V., Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 49, 107–133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin E. W., Holehouse A. S., Peran I., Farag M., Incicco J. J., Bremer A., Grace C. R., Soranno A., Pappu R. V., Mittag T., Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pronina N. A., Semenenko V. E., Localization of membrane bound and soluble carbonic anhydrase in the Chlorella cell. Fiziol. Rastenii 31, 241–251 (1984). [Google Scholar]
- 26.Raven J. A., CO2-concentrating mechanisms: A direct role for thylakoid lumen acidification? Plant Cell Environ. 20, 147–154 (1997). [Google Scholar]
- 27.Goodenough U. W., Levine R. P., Chloroplast structure and function in ac-20, a mutant strain of Chlamydomonas reinhardtii. 3. Chloroplast ribosomes and membrane organization. J. Cell Biol. 44, 547–562 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caspari O. D., Meyer M. T., Tolleter D., Wittkopp T. M., Cunniffe N. J., Lawson T., Grossman A. R., Griffiths H., Pyrenoid loss in Chlamydomonas reinhardtii causes limitations in CO2 supply, but not thylakoid operating efficiency. J. Exp. Bot. 68, 3903–3913 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee A., Lau C. S., Walker C. E., Rai A. K., Prejean C. I., Yates G., Emrich-Mills T., Lemoine S. G., Vinyard D. J., Mackinder L. C. M., Moroney J. V., Thylakoid localized bestrophin-like proteins are essential for the CO2 concentrating mechanism of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 116, 16915–16920 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villarreal J. C., Renner S. S., Hornwort pyrenoids, carbon-concentrating structures, evolved and were lost at least five times during the last 100 million years. Proc. Natl. Acad. Sci. U.S.A. 109, 18873–18878 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raven J. A., Beardall J., Sánchez-Baracaldo P., The possible evolution and future of CO2-concentrating mechanisms. J. Exp. Bot. 68, 3701–3716 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Griffiths H., Meyer M. T., Rickaby R. E. M., Overcoming adversity through diversity: Aquatic carbon concentrating mechanisms. J. Exp. Bot. 68, 3689–3695 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herron M. D., Hackett J. D., Aylward F. O., Michod R. E., Triassic origin and early radiation of multicellular volvocine algae. Proc. Natl. Acad. Sci. U.S.A. 106, 3254–3258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer M. T., Genkov T., Skepper J. N., Jouhet J., Mitchell M. C., Spreitzer R. J., Griffiths H., Rubisco small-subunit α-helices control pyrenoid formation in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 109, 19474–19479 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rae B. D., Long B. M., Förster B., Nguyen N. D., Velanis C. N., Atkinson N., Hee W. Y., Mukherjee B., Price G. D., Mccormick A. J., Progress and challenges of engineering a biophysical CO2-concentrating mechanism into higher plants. J. Exp. Bot. 68, 3717–3737 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Mackinder L. C. M., The Chlamydomonas CO2-concentrating mechanism and its potential for engineering photosynthesis in plants. New Phytol. 217, 54–61 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Zhang R., Patena W., Armbruster U., Gang S. S., Blum S. R., Jonikas M. C., High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell 26, 1398–1409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S. S., Malasarn D., A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas Reinhardtii. Plant J. 66, 770–780 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell M. C., Meyer M. T., Griffiths H., Dynamics of carbon-concentrating mechanism induction and prot`ein relocalization during the dark-to-light transition in synchronized Chlamydomonas reinhardtii. Plant Physiol. 166, 1073–1082 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strenkert D., Schmollinger S., Gallaher S. D., Salomé P. A., Purvine S. O., Nicora C. D., Mettler-Altmann T., Soubeyrand E., Weber A. P. M., Lipton M. S., Basset G. J., Merchant S. S., Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 116, 2374–2383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porra R. J., Thompson W. A., Kriedemann P. E., Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta Bioenerg. 975, 384–394 (1989). [Google Scholar]
- 42.Hilpert K., Winkler D. F. H., Hancock R. E. W., Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat. Protoc. 2, 1333–1349 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.One Thousand Plant Transcriptomes Initiative , One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madeira F., Park Y. M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A. R. N., Potter S. C., Finn R. D., Lopez R., The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A., ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hofmann K., Stoffel W., TMBase—A database of membrane spanning proteins segments. Biol. Chem. 374, 166 (1993). [Google Scholar]
- 49.Yang T., Liu Q., Kloss B., Bruni R., Kalathur R. C., Guo Y., Kloppmann E., Rost B., Colecraft H. M., Hendrickson W. A., Structure and selectivity in bestrophin ion channels. Science 346, 355–359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terashima M., Specht M., Hippler M., The chloroplast proteome: A survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr. Genet. 57, 151–168 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Mészáros B., Erdõs G., Dosztányi Z., IUPred2A: Context-dependent prediction of protien disorder as a function of redox state and protein binding. Nucleic Acids Res. 46, W329–W337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tardif M., Atteia A., Specht M., Cogne G., Rolland N., Brugiè S., Hippler M., Ferro M., Bruley C., Peltier G., Vallon O., Cournac L., PredAlgo: A new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 29, 3625–3639 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/46/eabd2408/DC1


