Conserved circBoule RNAs protect male fertility from thermal insults in animals.
Abstract
Circular RNAs (circRNAs) are a large family of newly identified transcripts, and their physiological roles and evolutionary significance require further characterization. Here, we identify circRNAs generated from a conserved reproductive gene, Boule, in species from Drosophila to humans. Flies missing circular Boule (circBoule) RNAs display decreased male fertility, and sperm of circBoule knockout mice exhibit decreased fertilization capacity, when under heat stress conditions. During spermatogenesis, fly circBoule RNAs interact with heat shock proteins (HSPs) Hsc4 and Hsp60C, and mouse circBoule RNAs in sperm interact with HSPA2. circBoule RNAs regulate levels of HSPs by promoting their ubiquitination. The interaction between HSPA2 and circBoule RNAs is conserved in human sperm, and lower levels of the human circBoule RNAs circEx3-6 and circEx2-7 are found in asthenozoospermic sperm. Our findings reveal conserved physiological functions of circBoule RNAs in metazoans and suggest that specific circRNAs may be critical modulators of male reproductive function against stresses in animals.
INTRODUCTION
Circular RNAs (circRNAs) are single-stranded covalently closed RNA molecules (1–4). In recent years, thousands of circRNAs have been identified in animal cells (5–7). In studies of cultured cells or tumors, circRNAs have been found to function as microRNA “sponges,” transcriptional regulators, protein binding partners, and templates for protein translation (5, 6, 8–11). Researchers have just begun to explore the physiological roles of circRNAs in animals (12–15). For example, deletion of the murine circRNA Cdr1as was found to cause abnormalities in excitatory synaptic transmission (8, 12, 13).
While circRNAs have been found among various eukaryotes (16), it remains undetermined whether homologous genes in different species give rise to functionally conserved circRNAs. In mammals, the brain and testis have the largest number of tissue-specific circRNAs (17, 18). One of the first identified circRNAs, circSry, is transcribed from the murine testis determining factor gene Sry (19). While Sry is a mammalian gene, circSry is absent in humans. No back-splicing junction site in the corresponding human genomic region of SRY has been identified in circRNA-sequencing profiles of human testes and sperm or in circRNA databases (20, 21). Another gene, BOULE, a member of the human DAZ (Deleted in AZoospermia) family, functions in the testis during spermatogenesis and is highly conserved in animals; the human BOULE transgene can rescue meiotic defects in infertile boule mutant flies (22–26). These findings stimulated us to investigate whether conserved testis genes such as Boule give rise to circRNAs, and—if present—to investigate their physiological roles in testes. Given the pervasiveness of circRNAs and their regulatory roles in animals, we hypothesized that conserved circRNAs may play critical roles in physiological responses to intrinsic or environmental signals. To test this possibility, we determined the presence of circular Boule (circBoule) RNAs in various animals and examined their physiological function and mechanism of action in flies, mice, and humans.
RESULTS
Considering the facts that Boule is such an essential and conserved gene in male fertility and the testes have abundant and complex profiles of circRNAs in both mice and human (18, 21), we first asked whether any circRNAs were generated from human BOULE. We not only detected expression of circRNA from the human BOULE gene, but also identified circBoule RNA from four other species spanning both vertebrates and invertebrates (Drosophila melanogaster, Gallus gallus domesticus, Sus scrofa domesticus, Mus musculus, and Homo sapiens; Fig. 1A). In the four warm-blooded vertebrates, both Boule mRNA and circBoule RNAs showed testis-specific expression, whereas boule mRNA and circBoule RNAs were also detectable in somatic tissues in the fruit fly. The presence of two fly circBoule RNAs, likely formed by back-splicing of exon 2 with exon 3 (circEx2-3) and exon 4 with exon 7 (circEx4-7), was confirmed by Northern blot and reverse transcription polymerase chain reaction (RT-PCR) (Fig. 1, A and B, and fig. S1, A to C). The widespread distribution of circRNAs and their testicular expression from orthologous loci among metazoan species from flies to human support the hypothesis that circBoule RNAs are highly conserved and argue for the functional significance of these molecules in male reproductive processes.
Fig. 1. Boule circRNAs are conserved and required for male fecundity under heat stress in fruit flies.
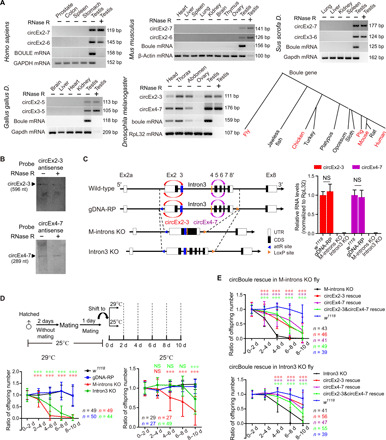
(A) RT-PCR [with or without ribonuclease (RNase) R treatment] of Boule circRNAs in human, mouse, pig, chicken, and fly. A phylogenetic diagram of Boule homologs in animals (animals examined for circBoule labeled in red). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; bp, base pairs. (B) Northern blot of circEx2-3 and circEx4-7 from wild-type (w1118) testes. nt, nucleotide. (C) Left: Scheme for generation of boule genomic DNA replacement (gDNA-RP), knockout (KO) of boule middle introns (introns 2, 3, 4, 5, 6, and 7; M-introns KO), and KO of boule intron 3 (Intron3 KO) flies. Right: Levels of circBoule RNAs in testes of 2-day-old w1118, gDNA-RP, M-introns KO, and Intron3 KO flies reared at 25°C. Ribosomal protein L32 (RpL32) mRNA was used as endogenous control. UTR, untranslated region. (D) Ratio of offspring numbers (normalized to progeny from 0 to 2 days) in w1118, gDNA-RP, M-introns KO, and Intron3 KO male flies mating at 29° or 25°C. Scheme of experimental setup is shown. Details of shifting male flies from 25° to 29°C are described in Materials and Methods. There was no significant difference in fertility between w1118 and gDNA-RP male flies at 29° or 25°C. Red or green asterisks and NS (not significant) indicate statistical analyses between gDNA-RP and M-introns KO or Intron3 KO flies. (E) Ratio of offspring numbers in circEx2-3 rescue, circEx4-7 rescue, or circEx2-3&circEx4-7 rescue male flies (29°C). CircEx2-3 rescue, circEx4-7 rescue, or circEx2-3&circEx4-7 rescue is overexpression of boule circEx2-3, circEx4-7, or circEx2-3&circEx4-7 in M-introns KO or Intron3 KO flies by β2-tubulin promoter. Red, purple, or green asterisks indicate statistical analyses of fertility between circEx2-3, circEx4-7, or circEx2-3&circEx4-7 rescue and the corresponding control (M-introns KO or Intron3 KO). In (D) and (E), n = number of male flies tested. Data are shown as means ± SD. P values from unpaired Student’s t test, ***P < 0.001.
Flies missing circBoule RNAs exhibit reduced male fertility under thermal stress
To investigate the physiological roles of these conserved circRNAs, we generated a D. melanogaster circBoule RNA-specific knockout (KO) that did not seem to affect linear boule mRNA or Boule protein expression. We constructed a fly strain missing all introns between exon 2 and exon 7 of the boule gene [Middle-introns KO (M-introns KO), with the first and last introns present] and another strain, in which intron 3 was deleted (Intron3 KO) (Fig. 1C). As a control, we constructed a fly strain, in which the coding region of boule was replaced with the genomic region of boule (gDNA-RP), using the same recombinational strategy (Fig. 1C). The two major fly circBoule RNAs, circEx2-3 and circEx4-7, were absent in the M-introns KO and Intron3 KO lines (Fig. 1C). Wild-type (w1118), M-introns KO, Intron3 KO, and gDNA-RP lines expressed similar levels of boule mRNA and BOULE protein (fig. S1D).
While both the M-introns KO and Intron3 KO lines appeared normal in morphology and viability, both lines displayed a marked decrease in male fecundity when young adults raised under 25°C were shifted to 29°C (Fig. 1D and fig. S1, E and F). When kept continuously under 25°C, M-introns KO males and, to a lesser degree, Intron3 KO males showed a moderate decrease in fertility over time (Fig. 1D). This reduction in male fertility did not occur when both lines raised under 25°C were shifted to 18°C (fig. S1G). The expression levels of boule mRNA and protein showed no notable difference among w1118, gDNA-RP, M-introns KO, and Intron3 KO testes at 29°C (fig. S1H). To further validate that this heat-induced fertility decline was caused by missing circBoule RNAs, we constructed transgenic flies, with the expression of circEx2-3 or circEx4-7 driven by a male germ cell-specific promoter (β2-tubulin) (24). The expression of either circEx2-3 or circEx4-7 in the M-introns KO or Intron3 KO line partially rescued the fertility phenotype at 29°C (Fig. 1E and fig. S1I). The expression of both circEx2-3 and circEx4-7 together (circEx2-3&circEx4-7 rescue) in the M-introns KO or Intron3 KO line also showed partial rescue and had no obvious difference in male fertility when comparing to that of circEx2-3 or circEx4-7 rescue alone (Fig. 1E). The fact that the two circRNAs together did not show better rescue ability than each circRNA alone might indicate that circEx2-3 and circEx4-7 played redundant roles. It was also possible that partial rescue by the two circBoule RNAs either separately or together might be due to the potential difference (in expression level and/or tempospatial pattern) between the transgenic and the endogenous circBoule RNAs. Together, these data suggest that circBoule RNAs protect the fecundity of fruit fly males in the presence of heat stress.
circBoule RNA KO strains have abnormalities in sperm
We quantified the number of sperm in adult male flies after 10 days of heat stress (29°C). Sperm number was significantly lower in the M-introns KO and Intron3 KO lines compared to controls (Fig. 2A and fig. S2, A to C). A significant portion of sperm in the M-introns KO and Intron3 KO lines under heat stress also exhibited defective nuclear morphology and were positive for the cell death marker TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) (Fig. 2, B and C). The heat stress–related reduction in sperm number in both the M-introns KO and Intron3 KO lines could be partially rescued by male germ cell–specific expression of either circEx2-3 or circEx4-7. Abnormal sperm nuclear morphology could also be partially rescued by circEx2-3 or circEx4-7 in the Intron3 KO line, while only circEx2-3 expression, but not circEx4-7, showed statistically significant rescue effect on sperm nuclear morphology in the M-introns KO line (Fig. 2, D and E). It was possible that circEx2-3 might be more potent (or nonredundant in some way) than circEx4-7 in the M-introns KO background, or it might again be due to expression difference between the transgenic (with a β2-tubulin promoter in M-introns KO background) and the endogenous expression of circEx2-3 or circEx4-7. The expression of both circEx2-3 and circEx4-7 together (circEx2-3&circEx4-7 rescue) in the M-introns KO or Intron3 KO line also showed partial rescue in sperm count and sperm nuclear morphology, although the rescue effects with both circRNAs together did not seem to be the summation of rescue by each circRNA alone (Fig. 2, D and E), and this was consistent with rescue effect in male fertility by the two circRNAs together (Fig. 1E).
Fig. 2. Absence of circBoule RNAs causes abnormalities in fly sperm at 29°C.
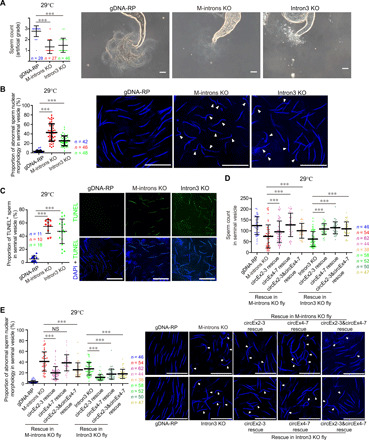
(A) Sperm counts of gDNA-RP (with circBoule RNAs), M-introns KO (missing circBoule RNAs), and Intron3 KO (missing circBoule RNAs) male flies. Representative contrast images of seminal vesicles for sperm counting in gDNA-RP, M-introns KO, and Intron3 KO flies are shown. Sperm count was graded into three artificial grades of low (1), medium (2), and high (3), which is described in fig. S2A and in Materials and Methods. (B) Proportion of sperm with abnormal nuclear morphology in gDNA-RP, M-introns KO, and Intron3 KO males. Representative images are shown [sperm nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI)], and white triangles indicate sperm with abnormally shaped nuclei (sperm nuclei exhibiting curved and not smooth bent morphology). (C) TUNEL assay of gDNA-RP, M-introns KO, and Intron3 KO sperm. Representative images are shown (DAPI, blue; TUNEL, green). (D and E) Sperm count (D) and sperm with defective nuclear morphology (E) in gDNA-RP, M-introns KO, Intron3 KO, circEx2-3 rescue, circEx4-7 rescue, and circEx2-3&circEx4-7 rescue males. Representative images are shown (DAPI, blue), and white triangles indicate mature sperm with abnormally shaped nuclei. All males were continuously mated with females for 10 days at 29°C, followed by another 2 days without mating, and then testes were dissected for further analyses. In all graphs, n = fly testis number. Scale bars, 50 μm (A) and 20 μm (B, C, and E). Data are shown as means ± SD. P values from unpaired Student’s t test, ***P < 0.001.
circBoule RNAs interact with heat shock proteins
Animals encounter various environmental and intrinsic insults (27, 28), and heat shock proteins (HSPs) and noncoding RNAs (both long and short) are critical factors that allow cells to respond to thermal stress (29–31). To identify proteins interacting with circBoule RNAs, we performed RNA pull-down assays in fly testis extracts with circEx2-3 and circEx4-7, respectively. Among a few proteins pulled down, only Hsc4 and Hsp60C, two HSPs, were immunoprecipitated by both circEx2-3 and circEx4-7 (Fig. 3, A and B). Then, we verified their interactions by coimmunoprecipitation (IP) of both circEx2-3 and circEx4-7 in fly testes with either FLAG-tagged Hsc4 or Hsp60C (with an in-frame 3xFLAG tag sequence inserted into the 3′ of the genomic locus of the corresponding HSP gene) (Fig. 3, C and D, and fig. S3, A to C). RNA pull-down of circEx4-7 also pulled down MIP15742p, a protein with unknown function (Fig. 3B). Given the importance of HSPs in thermal stress response and both Hsc4 and Hsp60C pulled down, we focused on the interaction between circBoule RNAs and HSPs and did not further examine the circEx4-7/MIP15742p interaction. The interactions between circBoule RNAs and HSPs were further validated in Drosophila S2 cells (fig. S3D).
Fig. 3. Fly circBoule RNAs interact with Hsp60C and Hsc4 in testis.
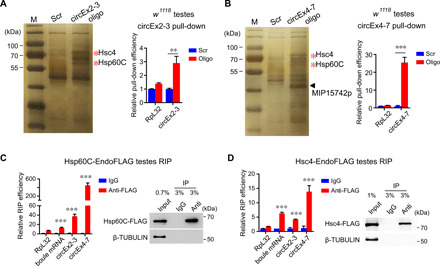
(A and B) RNA pull-down using biotin-labeled junction-antisense oligos against circEx2-3 or circEx4-7 with w1118 testes. Proteins pulled down were analyzed by silver staining, and bands indicated by red asterisks were identified as Hsp60C and Hsc4 proteins through mass spectrometry (MS). The triangle indicates circEx4-7-specific pull-down band (MIP15742p, a protein with unknown function). Bar graph shows the pull-down efficiency of circEx2-3 or circEx4-7. Enrichment was normalized to Scr control of each group. Scr, negative control oligos with scrambled sequences. (C and D) Hsp60C-EndoFLAG and Hsc4-EndoFLAG (3xFlag DNA sequences were inserted into the C terminus of the endogenous Hsp60C or Hsc4 genomic sequence, respectively) fly testes were subjected to RNA IP (RIP) with antibody against FLAG. Precipitated RNAs were analyzed by reverse transcription quantitative PCR (RT-qPCR), and the enrichment was normalized to the immunoglobulin G (IgG) control. Testis lysates and RIP materials were immunoblotted with the indicated antibodies to show the successful pull-down of FLAG-tagged proteins. Data are shown as means ± SD. P values from unpaired Student’s t test, **P < 0.01 and ***P < 0.001.
A specific RNA motif is required for HSP interactions and function of circBoule
We identified two conserved motifs (motif 1 and motif 2) in circBoule RNAs from the five species examined (Figs. 1A and 4A and fig. S3E) (32). IP of Hsp60C and Hsc4 showed that mutation in motif 2 of either circEx2-3 or circEx4-7 significantly decreased the binding of the circBoule RNAs to HSPs (Fig. 4A). Mutation of motif 1 decreased the binding of circEx4-7 but not circEx2-3 to HSPs (Fig. 4A). When motif 2 was mutated, both circEx2-3 and circEx4-7 also lost the ability to rescue the heat-related infertility phenotype, reduction in sperm number, and abnormal sperm morphology (Fig. 4, B and C, and fig. S3F). These data suggest that circBoule RNAs interact with HSPs via conserved RNA motifs, and these interactions are essential for the protective function of circBoule RNAs against heat stress–related male infertility.
Fig. 4. Fly circBoule RNAs interact with HSPs via conserved RNA motifs.
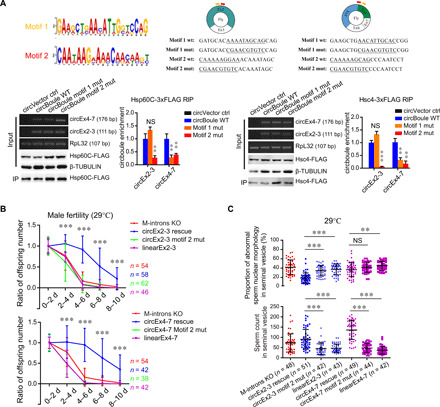
(A) Binding of circBoule RNAs containing mutations in motif 1 or motif 2 sequences to Hsp60C or Hsc4 protein in fly S2 cells. The conserved motifs (motif 1 and motif 2) in circBoule RNAs among species (human, mouse, pig, chicken, and fly) predicted by MEME (32) are shown. Sequence alignment of conserved motifs among species is shown in fig. S3E. Positions of motifs (yellow, motif 1; red, motif 2) in fly circEx2-3 and circEx4-7 and the sequences of mutated motif 1 and motif 2 (underlined) are shown. Bottom: RIP assay of Hsp60C-3xFLAG or Hsc4-3xFLAG. Bar graphs show the relative enrichment of circBoule RNAs [wild-type (WT) or motif-mutated], gel images indicate RT-PCR products of the corresponding RNAs, and representative immunoblots of the corresponding proteins are shown. Mut, mutation. (B) Ratio of offspring numbers (relative to progeny from 0 to 2 days) of circEx2-3 motif 2 mut or circEx4-7 motif 2 mut males (flies mated at 29°C). LinearEx2-3 or linearEx4-7 indicates the expression of fly boule Exon2-3 or Exon4-7 linear sequences without producing circEx2-3 or circEx4-7. Asterisks indicate statistical significance of the difference between circEx2-3 (or circEx4-7) rescue and circEx2-3 (or circEx4-7) motif 2 mut flies. There was no significant difference in fertility among M-introns KO, linearEx2-3 (or linearEx4-7), circEx2-3 motif 2 mut, and circEx4-7 motif 2 mut male flies. Data for M-introns KO in top and bottom panels are the same. n = number of male flies tested. (C) Sperm count and sperm with defective nuclear morphology (tested at 29°C). n = fly testes analyzed. Data are shown as means ± SD. P values from unpaired Student’s t test, **P < 0.01 and ***P < 0.001.
circBoule RNAs regulate Hsp60C and Hsc4 protein levels
In the testes of w1118 and M-introns KO flies exposed to 29°C for 8 days, both Hsp60C and Hsc4 protein levels gradually decreased over time, although both HSPs were significantly elevated in the M-introns KO line than wild type at each time point (Fig. 5, A and B, and fig. S4, A and B). These data suggest that both HSPs are down-regulated in the presence of circBoule RNAs and that this down-regulation may be important for male fecundity under heat stress conditions. We hypothesized that failure to down-regulate HSPs in the absence of circBoule RNAs might underlie the heat stress–related male fertility defect in the circBoule KO mutants. Consistent with this possibility, heat-stressed flies with overexpression of either Hsp60C or Hsc4 (driven by their own promoter and inserted at the attP40 site in chromosome 2) showed decreased male fertility, fewer sperm, and abnormal sperm morphology (Fig. 5, C to E, and fig. S4, C and D). Furthermore, we examined the effects of lower dosage of HSPs in male fertility. First, we generated Hsp60C and Hsc4 mutants using CRISPR-Cas9. Hsp60C homozygous mutant males were infertile with sperm individualization defects (fig. S4E), while Hsc4 homozygous mutants were completely lethal before the pupal stage. Removing one copy of either Hsp60C or Hsc4 in circBoule RNA KO flies demonstrated slower decline in male fertility as compared to circBoule RNA KO flies under heat stress (fig. S4F), supporting that circBoule KO–induced higher level of HSP proteins may be responsible for the accelerated male fertility decline.
Fig. 5. Fly circBoule RNAs regulate protein levels of Hsp60C and Hsc4.
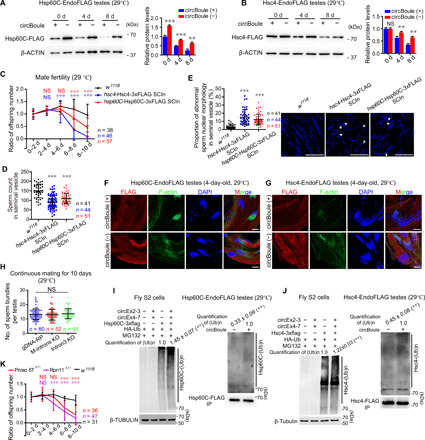
(A and B) Protein levels of Hsp60C and Hsc4 in testis with (w1118) or without (M-introns KO) circBoule at different time points at 29°C. β-ACTIN was used as endogenous control. (C) Ratio of offspring numbers of w1118, hsp60C-Hsp60C-3xFLAG SCIn (single copy insertion), and hsc4-Hsc4-3xFLAG SCIn males at 29°C. Red (or blue) asterisks and NS indicate statistical significance of difference between w1118 and hsp60C-Hsp60C-3xFLAG SCIn (or hsc4-Hsc4-3xFLAG SCIn) flies. n = males tested. (D and E) Sperm count and sperm with defective nuclear morphology in w1118, hsp60C-Hsp60C-3xFLAG SCIn, and hsc4-Hsc4-3xFLAG SCIn males. Representative images of abnormal sperm nuclear morphology are shown. White triangles indicate mature sperm with abnormally shaped nuclei. n = fly testes analyzed. Scale bars, 20 μm. (F and G) IF of Hsp60C and Hsc4 protein in spermatid bundles using FLAG antibody. circBoule (+), w1118 background; circBoule (−), M-introns KO background. Scale bars, 10 μm. (H) Quantification of spermatid bundles in gDNA-RP, M-introns KO, and Intron3 KO fly testes. Males reared at 29°C were mated with females for 10 days. (I and J) circBoule RNAs promoted ubiquitination of Hsp60C and Hsc4 proteins in fly S2 cells and fly testes, respectively. For S2 cells, circBoule RNAs, HSPs, and ubiquitin were expressed with plasmids. For testes, HSPs were first immunoprecipitated and then analyzed for ubiquitination. Ub, ubiquitin. Flies were 4 days old at 29°C. (K) Ratio of offspring numbers of w1118, Prosα6T+/−, and Rpn11+/− males at 29°C. Red (or purple) asterisks and NS indicate statistical significance of difference between Prosα6T+/− (or Rpn11+/−) and w1118 flies. n = males tested. Data are shown as means ± SD. P values from unpaired Student’s t test, **P < 0.01 and ***P < 0.001.
Immunofluorescence (IF) staining of Hsp60C and Hsc4 proteins in fly testes showed that both HSPs are mainly localized in spermatids, specifically in early and late canoe sperm bundles (before individualization) (Fig. 5, F and G, and fig. S4, G and H). Hsp60C protein was mainly localized below spermatid nuclei and at the tail of early and late canoe stage sperm bundles, whereas Hsc4 protein was mainly localized above nuclei and at the tail of early and late canoe stage sperm bundles (Fig. 5, F and G, and fig. S4, G and H). Sperm head nuclei in sperm bundles appeared to be embedded between the two HSPs. Both proteins were undetectable by IF in individualized sperm or at later stages of sperm development. In fly testes missing circBoule RNAs at 29°C, IF signals of Hsp60C and Hsc4 protein in sperm bundles were stronger compared to those in w1118 fly testes (Fig. 5, F and G). Spermatid bundles per testis were not significantly different in flies (at 29°C) with or without circBoule RNAs, indicating that the loss of circBoule RNAs likely affects late spermiogenesis or sperm individualization and maturation (Fig. 5H). These results suggest that, under heat stress conditions, higher levels of Hsp60C and Hsc4 protein due to the lack of circBoule RNAs might lead to abnormalities in spermatid bundles and cause defects during sperm individualization and maturation, eventually resulting in abnormal and fewer sperm.
We further explored the mechanism by which circBoule RNAs down-regulate HSPs. Overexpression of circBoule RNAs led to a decrease in HSP protein levels in fly S2 cells by promoting ubiquitination of both HSP proteins (Fig. 5, I and J, and fig. S4I). Ubiquitination levels of both Hsp60C and Hsc4 were lower in fly testes missing circBoule RNAs (Fig. 5, I and J). Two proteasome genes, Prosα6T (a testis-specific core subunit of 20S proteasome) and Rpn11 (a subunit of 26S proteasome) were further examined for the involvement of the ubiquitin-proteasome pathway in male fertility decline under heat stress. Prosα6T homozygous mutants were defective in sperm individualization and nuclear maturation (fig. S4E), while Rpn11 homozygous mutation led to lethality (at least no adult fly with homozygous genotype). Flies missing one copy of either Prosα6T or Rpn11 showed significant decline in male fertility under 29°C (Fig. 5K). Together, these data indicate a possibility that fly circBoule RNAs interacts with Hsp60C and Hsc4 to facilitate their ubiquitination and degradation and that the resulting lower levels of both HSPs protect spermatogenesis and fertility under conditions of heat stress.
Mice missing circBoule RNAs exhibit male fertility defect under heat stress
The physiological role of circBoule RNAs in Drosophila fertility prompted us to investigate the extent of functional conservation by studying conserved circBoule RNAs in mice (Fig. 6A and fig. S5, A and B). We used CRISPR to generate circBoule RNA KO mice, in which the repeat sequences in the second intron of Boule were deleted (Intron2RPΔ/RPΔ), but functional intronic elements such as the splicing sites and pyrimidine tract remained unchanged (Fig. 6B and fig. S5C). circBoule RNAs generated by back-splicing of exon 3 with other exons, including circEx3-5, circEx3-6, and circEx3-7, were missing from the testes and cauda sperm of the Intron2RPΔ/RPΔ mice (Fig. 6C and fig. S5D). The expression levels of circBoule RNAs formed by other exons, such as circEx2-6 and circEx2-7, and levels of Boule mRNAs were unaltered in the Intron2RPΔ/RPΔ mice compared to wild-type (Fig. 6C). BOULE protein levels and distribution in the testis were not noticeably different in wild-type and Intron2RPΔ/RPΔ mice, and BOULE protein was not detected in cauda sperm (fig. S5E).
Fig. 6. Mice circBoule RNAs and male fertility defect of circBoule KO under heat stress.
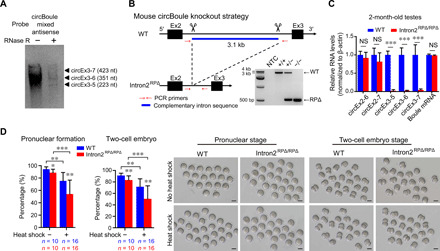
(A) Northern blotting of murine circBoule RNAs. Mix antisense probes against the junctions of circEx3-5, circEx3-6, and circEx3-7 were used to blot RNAs from wild-type mouse testes. (B) Strategy of KO reverse-complementary repeats (RP) in mouse Boule intron2 using CRISPR-Cas9. Gel image of PCR products from mouse genotyping is shown. NTC, no template control; RPΔ, repeat sequences deleted. (C) RNA levels of circBoule RNAs and Boule mRNA were examined in 2-month-old wild-type and Intron2RPΔ/RPΔ mouse testes via RT-qPCR. (D) Quantification of pronuclear and two-cell embryo formation in IVF with mature sperm (cauda sperm) from wild-type and Intron2RPΔ/RPΔ mice. Representative images of pronuclear and two-cell embryo stage are also shown. Heat shock was performed to cauda sperm in epididymis at 42°C for 30 min. Scale bars, 50 μm. Data are shown as means ± SD. P values from unpaired Student’s t test, *P < 0.05, **P < 0.01, and ***P < 0.001.
In adult male Intron2RPΔ/RPΔ and wild-type mice (2 months old), there was no statistical difference in body weight, testis weight, testis size, sperm counts in epididymis, gross histology of testis [hematoxylin and eosin (H&E) staining], sperm motility, or sperm morphology (fig. S5, F to H). Intron2RPΔ/RPΔ males had a slightly smaller litter size on average than age-matched wild-type controls (fig. S5I). We then characterized the potential fertility defect of circBoule KO sperm using in vitro fertilization (IVF) assays in presence or absence of heat stress. There was a decrease in the pronuclear and two-cell embryo formation rate with Intron2RPΔ/RPΔ sperm compared to wild-type sperm (Fig. 6D). Consistent with our finding in fly, mouse Intron2RPΔ/RPΔ sperm exposed to heat stress (30-min exposure of the epididymis to 42°C) displayed more severe defects than wild-type sperm in IVF assays (Fig. 6D and fig. S5J).
Mouse circBoule RNAs interact with HSPA2
circBoule RNAs were present in mouse cauda sperm, and an RNA pull-down assay with circEx3-6 in sperm pulled down a mouse HSP, HSPA2 (Fig. 7A and fig. S5D). Furthermore, HSPA2 also immunoprecipitated circBoule RNAs via RNA IP (RIP), extending molecular interaction of circBoule-HSP from fly to mice (Fig. 7B). Mouse HSPA2 interacted with all three circBoule RNAs, circEx3-5, circEx3-6, and circEx3-7 in the testis (fig. S6, A to D). We also found that mutation of either motif 1 or motif 2 of mouse circBoule decreased binding between HSPA2 and circBoule RNAs (Fig. 7C and fig. S3E) and that HSPA2 protein and circBoule RNAs colocalized in murine sperm (Fig. 7D). Hence, we conclude that interaction between circBoule RNAs and HSP is conserved in mice.
Fig. 7. Mouse circBoule RNAs interact with HSPA2.
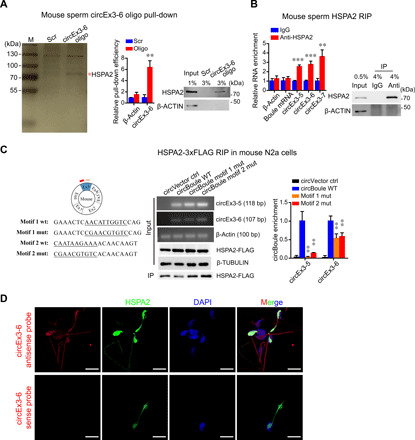
(A) CircEx3-6 co-pulled down HSPA2 in mouse sperm. RNA pull-down using biotin-labeled antisense oligos against the junction of circEx3-6. Proteins pulled down were analyzed by silver staining, and the band indicated by red asterisk was identified to be HSPA2 through MS. The pull-down efficiency of circEx3-6 is shown in the bar graph, and the enrichment is normalized to Scr control. Immunoblot analysis of the HSPA2 co-pulled down is also shown. Scr, negative control oligo with scrambled sequences. (B) IP of HSPA2 in mouse sperm co-pulled down circBoule RNAs (circEx3-5, circEx3-6, and circEx3-7). The RIP enrichment of circBoule RNAs is shown in the bar graph and normalized to IgG control. Immunoblot analysis of HSPA2 is shown. (C) Mutation in motif 1 and motif 2 sequences of circBoule RNAs (circEx3-5 and circEx3-6) decreased circBoule RNAs binding to HSPA2 in mouse N2a cells. Left: The motif positions in mouse circBoule and the wild-type and mutant sequences (underlined) of motif 1 and motif 2 are shown. Enrichment of circBoule RNAs (wild type or motif mutants) from HSPA2-3xFLAG RIP assay is shown in the bar graph, and the circRNAs and proteins overexpressed in cells, as well as the immunoprecipitated HSPA2, are shown in representative gel images of RT-PCR products and immunoblots. (D) In situ hybridization of circEx3-6 (red) using antisense probe or sense probe (negative control) against the junction along with IF of HSPA2 (green) in mouse sperm and counterstaining of nuclei with DAPI (blue). Scale bars, 10 μm. Data are shown as means ± SD. P values from unpaired Student’s t test, **P < 0.01 and ***P < 0.001.
circBoule RNAs regulate HSPA2 protein levels
We next determined the impact of circBoule KO on HSPA2 levels in the testis and in sperm maturation. HSPA2 protein levels and expression pattern in testes of Intron2RPΔ/RPΔ mice did not show significant differences from those of wild-type mice (fig. S6, E and F). However, we observed a faster decline of HSPA2 protein levels during sperm maturation in the absence of circBoule RNAs (Fig. 8A). Mouse sperm mature and gain fertilization potential as they transverse inside the epididymis from caput to cauda segment. HSPA2 protein levels of maturing sperm from caput and cauda decreased significantly in wild-type mice, consistent with previous reports (33), yet the decrease was less marked in Intron2RPΔ/RPΔ sperm (Fig. 8, A and B). During in vitro sperm capacitation, HSPA2 protein levels further decreased in wild-type sperm and again to a lesser degree in Intron2RPΔ/RPΔ sperm (Fig. 8B). During the capacitation of heat-stressed sperm (30-min exposure of the epididymis to 42°C), HSPA2 protein levels in both wild-type and Intron2RPΔ/RPΔ cauda sperm decreased, with Intron2RPΔ/RPΔ sperm consistently to a lesser degree (Fig. 8C). As it was reported that human HSPA2 forms a complex with ARSA (arylsulfatase A; a sperm surface receptor) and was proposed to play important roles in sperm-egg recognition in humans (34, 35), we investigated the impact of loss of mouse circBoule RNAs on HSPA2 and ARSA complex in sperm. In mice sperm, we found that HSPA2 protein coimmunoprecipitated with mouse ARSA (Fig. 8D). In capacitated cauda sperm, both HSPA2 and ARSA protein levels in Intron2RPΔ/RPΔ sperm were higher than those of wild-type sperm; under heat shock, the difference in HSPA2 and ARSA protein levels between capacitated wild-type and Intron2RPΔ/RPΔ sperm was more marked (Fig. 8E).
Fig. 8. Mouse circBoule RNAs down-regulate HSPA2 and can rescue fly phenotypes.
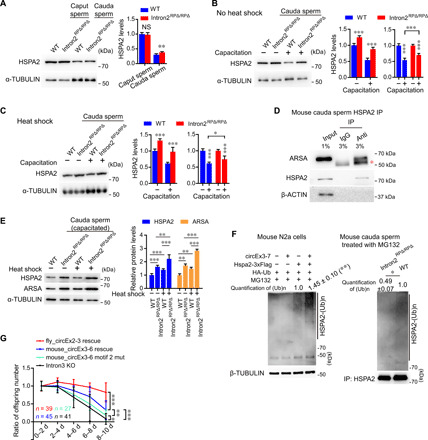
(A) Immunoblotting of HSPA2 protein in caput and cauda sperm of wild-type and Intron2RPΔ/RPΔ mice. Protein levels of HSPA2 are shown in the bar graph. (B and C) Immunoblotting of HSPA2 in cauda sperm (capacitated or not capacitated) of wild-type and Intron2RPΔ/RPΔ mice without or with heat shock pretreatment. The protein levels of HSPA2 are shown in the bar graph. Heat shock pretreatment (C) was performed to cauda sperm in epididymis at 42°C for 30 min. (D) IP of HSPA2 in mouse cauda sperm with co-IP of ARSA. Red asterisk indicates heavy chain of antibodies. (E) Immunoblotting of HSPA2 and ARSA proteins in capacitated cauda sperm of wild-type and Intron2RPΔ/RPΔ mice with or without heat shock pretreatment. The protein levels of HSPA2 and ARSA are shown in the bar graph. (F) circBoule RNAs promote HSPA2 ubiquitination in N2a cells and mouse cauda sperm. For N2a cells, circBoule RNAs, HSPA2 protein, and ubiquitin were expressed with plasmids. Cauda sperm from wild-type and Intron2RPΔ/RPΔ mice were capacitated in buffer containing 100 μM MG132. (G) Ratio of offspring numbers (relative to progeny of 0 to 2 days) of fly_circEx2-3 rescue, mouse_circEx3-6 rescue, mouse_circEx3-6 motif 2 mut, and Intron3 KO males at 29°C. Data for Intron3 KO are the same as in Fig. 1E. n = males tested. For (A) to (F), data are from three independent experiments and shown as means ± SD. P values from unpaired Student’s t test, *P < 0.05, **P < 0.01, and ***P < 0.001.
We further found that the overexpression of circBoule RNAs led to a decrease in HSPA2 protein levels in murine N2a cells via increased HSPA2 ubiquitination (Fig. 8F and fig. S6G). Ubiquitination levels of HSPA2 were lower in Intron2RPΔ/RPΔ cauda sperm than wild-type cauda sperm (Fig. 8F).
Together, these data suggest that HSPA2 proteins are gradually degraded during the process of sperm maturation in the epididymis and the capacitation of cauda sperm, and the absence of circBoule RNAs in Intron2RPΔ/RPΔ sperm leads to inadequate degradation of HSPA2. When exposed to heat stress, the effect of circBoule RNAs loss on HSPA2 protein level is more pronounced, with a bigger difference in the levels of HSPA2 and its partner protein ARSA in capacitated sperm of wild-type and Intron2RPΔ/RPΔ mice. The loss of circBoule RNAs and persistence of HSPA2 may contribute to the compromised sperm fertilization ability under conditions of heat stress.
Mice circEx3-6 partially rescues fertility defects in circBoule RNA KO fly
We then determine the extent of functional conservation of mouse circBoule RNA by expressing mouse circEx3-6 in flies. The expression of mouse circEx3-6 (mouse_circEx3-6) in circBoule RNA KO (Intron3 KO) flies partially rescued the fertility decline phenotype at 29°C, although the rescue ability of mouse circEx3-6 was weaker than that of fly circEx2-3 (Fig. 8G and fig. S6H). When motif 2 of mouse circEx3-6 was mutated (mouse_circEx3-6 motif 2 mut), the rescuing capability of this murine circBoule RNA significantly decreased in comparison to wild-type circEx3-6, albeit still detectable (Fig. 8G and fig. S6H). Fly Hsc4 but not Hsp60C protein interacted with mouse circEx3-6 expressed in fly testis, and Hsc4 had a decreased interaction with mouse_circEx3-6 motif 2 mut (fig. S6I). Given that fly Hsc4 and mouse HSPA2 are members of the HSP70 subfamily, we propose that molecular interaction of circBoule RNAs and HSPs (at least HSP70) underlying male fertility protection are conserved during sperm development from fly to mouse.
circBoule RNAs interact with human HSPA2 and may be affected in human asthenozoospermia
Last, we turned our investigation back to human circBoule as circBoule RNAs were initially identified in human testes and mature sperm (Figs. 1A and 9A). Given the observed roles of circBoule RNAs in fertility in both fly and mouse, we explore whether the expression of human circBoule RNAs might be affected in sperm of infertile men. We found that levels of human circBoule RNAs, circEx3-6, and circEx2-7, but not several other circBoule RNAs, were significantly lower in sperm from patients with asthenozoospermia, a type of infertility due to reduced sperm motility (Fig. 9B). We next examined the interaction between circBoule and HSPA2 in human sperm. The conserved HSP binding sites in human circBoule RNAs are present in exon 5, which is part of the circEx3-6 and circEx2-7 (fig. S3E). In human sperm, we could consistently pull down circEx3-6 and circEx2-7 via RIP, and conversely, RNA pull-down of either circEx3-6 or circEx2-7 co-pulled down HSPA2 (Fig. 9, C and D). As circBoule RNAs down-regulate HSP levels in fly testis and mouse sperm, we examined the correlation of HSPA2 and circEx3-6 and found that HSPA2 protein levels were negatively correlated with circEx3-6 RNA levels in individual normozoospermic sperm samples, but not in asthenozoospermic sperm (Fig. 9E). Since aberrant levels of HSPA2 have been associated with human asthenozoospermia (36), the negative correlation between circEx3-6 and HSPA2 and reduced overall levels of circEx3-6 and circEx2-7 in asthenozoospermia raised the possibility that circBoule RNAs may be important for human fertility. While more detailed investigation of various human infertility sperm samples and larger sample size are warranted to establish the roles of circBoule RNAs in human infertility and their potential as the fertility biomarkers, our combined data argue for their conserved roles in animal sperm development and maturation (Fig. 10), supporting functional roles of circRNAs in human fertility (37).
Fig. 9. Human circBoule RNAs interact with HSPA2 and are associated with human asthenozoospermia.
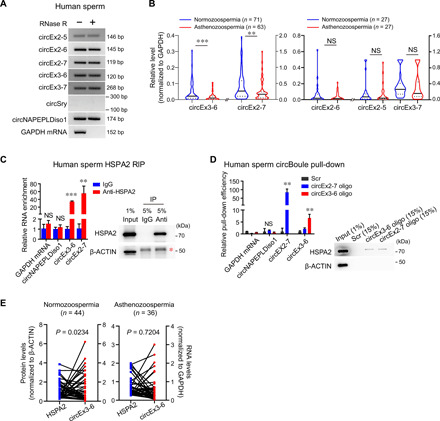
(A) circBoule RNAs are present in human sperm, while circSry is not detected. circNAPEPLDiso1 is a known human sperm circRNA (positive control). (B) Levels of circBoule RNAs (circEx2-5, circEx2-6, circEx2-7, circEx3-6, and circEx3-7) were examined in sperm of normozoospermic and asthenozoospermic patients via RT-qPCR. (C) IP of HSPA2 in human sperm co-pulled down circBoule RNAs (circEx3-6 and circEx2-7). The RIP enrichment of circBoule RNAs is shown in the bar graph and normalized to IgG control. Immunoblot analysis of HSPA2 is shown. Red asterisk indicates heavy chain of antibodies. (D) Pull-down of circBoule RNAs (circEx3-6 and circEx2-7) co-pulled down HSPA2 in human sperm. RNA pull-down using biotin-labeled antisense oligos against the junction of circEx3-6 and circEx2-7, respectively. The pull-down efficiency of circBoule RNAs was showed by bar graph, and the enrichment was normalized to Scr control. Western blot of the HSPA2 co-pulled down was also shown. Scr, negative control oligo with scrambled sequences. (E) HSPA2 protein levels were negatively correlated with circEx3-6 levels in normozoospermic sperm but not in asthenozoospermic sperm. The blue (HSPA2 protein) and red (circEx3-6) dots linked by each line indicate data from individual samples. HSPA2 protein levels were examined by enzyme-linked immunosorbent assay (ELISA); circEx3-6 levels were examined by RT-qPCR. For (B), data are shown in a violin plot; P values from Mann-Whitney test. For (C) and (D), data are from three independent experiments and shown as means ± SD; P values from unpaired Student’s t test. For (E), Spearman correlation test was used to calculate correlation between HSPA2 levels and circEx3-6 levels. **P < 0.01 and ***P < 0.001.
Fig. 10. A working model of the conserved role of circBoule RNAs in maintaining male fertility.
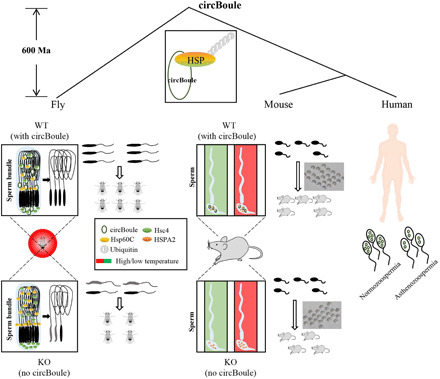
In both flies and mice, circBoule RNAs are required for regulating levels of HSPs when animals are under heat shock. In heat-stressed flies, the absence of circBoule RNAs leads to excessive levels of Hsp60C and Hsc4 proteins in testes, resulting in abnormal sperm nuclear morphology and reduced sperm number. When mature sperm of mice is heat shocked, lack of circBoule RNAs leads to excessive HSPA2 levels and lower fertilization ability. circBoule RNAs appear to regulate levels of HSPs through facilitating their ubiquitination. Levels of human circBoule RNAs (circEx3-6 and circEx2-7) were significantly lower in sperm of asthenozoospermic patients. The interaction and regulation between circBoule RNAs and HSPA2 protein are conserved in human sperm. Such conserved circBoule-HSP function and mechanism might be present in the common ancestor of fly and mammals at least 600 million years (Ma) ago.
DISCUSSION
CircRNAs have been found to be present across eukaryotic species including humans, and some human circRNAs are conserved within mammals. So far, few human circRNAs have been demonstrated to be conserved outside mammals (38), let alone outside vertebrates. Here, we have shown that human circBoule RNAs are present not only in mice, pigs, and chickens, but also in fruit flies, extending conservation of specific human circRNAs to invertebrates and supporting metazoan origin of circBoule RNAs.
BOULE proteins translated from linear mRNAs are known to play essential roles in germline meiosis and spermatid differentiation across multiple organisms (22–26). Boule is the sole DAZ family member in fruit fly, there are several DAZ gene family members, including DAZL and BOULE in both mice and humans, and DAZ 1 to 4 are expressed in human testes; among them, only BOULE produces circRNAs (18, 20). Mice have two DAZ gene family members DAZL and BOULE, and DAZL does not give rise to circRNAs (18, 20). Both human and mouse Boule genes have higher sequence similarity to fly boule than to mammalian DAZ or DAZL (23). There appears to be high evolutionary pressure on Boule to preserve the sequences and functions of both linear and circRNA transcripts. By contrast, the mammalian testis determining factor gene Sry gives rise to circRNA in mice but not in human (Fig. 9A) (20, 21). circBoule RNAs in different species may have roles that are not conserved, and different circBoule RNAs in the same species may have distinct functions, which may or may not be conserved (e.g., fly circEx2-3 and circEx4-7; Fig. 3, A and B). It is clear that dissecting the conserved and nonconserved functions of circRNAs, including circBoule, may shed new light on animal evolution.
In this study, we found that circBoule RNAs interact with conserved HSPs, consistent with the idea that recruitment of proteins may underpin the molecular mechanisms of circRNAs. For example, in a recent study, deletion of mammalian circRNA cia-cGAS resulted in higher expression levels of type I interferons and decreased numbers of dormant long-term HSCs, and cia-cGAS (circular RNA antagonist for cGAS) was found to bind to the DNA sensor cGAS to block its synthase activity (14). HSPs are essential players in various organisms and allow them to respond to an array of internal and external stresses; many HSPs are also important regulators under nonstress conditions (27–29, 36, 39, 40). HSPs can act as protein folding catalysts and mediate protein complex formation (27–29, 36, 39, 40). The ability of HSPs to act as RNA binding proteins is documented but less appreciated in the literature (41, 42). Specific members of all subclasses of HSPs have been found to interact with mRNAs or noncoding RNAs (31, 42).
When under heat stress, dynamic expression of Hsc4 and Hsp60C in fruit flies and HSPA2 in mice may be part of the regulatory response network, although too high levels of these proteins would be harmful to spermatogenesis in flies and sperm in mammals. Hsp60C is a member of the HSPD (HSP60) family, and Hsc4 is a member of the HSPA (HSP70) family (39, 40). Hsp60 might be a testis-specific gene, whereas Hsc4 was found to be expressed in multiple tissues (fig. S3, A and C) (40, 43). Hsp60C was shown to be essential for the development of fly testes and spermatogenesis, as an Hsp60C loss-of-function mutant was completely sterile with significantly smaller testes and fewer spermatocytes (40). Hsc4 was previously shown to play roles in neurotransmitter exocytosis (43); roles of Hsc4 in fly spermatogenesis require further study. Unexpectedly, circBoule RNAs are expressed in multiple tissues of flies but only in testes and sperm of mammals (Figs. 1A and 9A and fig. S5D). For mammals, testes and sperm are not located inside the body cavity and thus are more susceptible to heat stress from the environment. As a cold-blooded animal, all tissues of the fruit fly are subjected to frequent thermal stress from the environment, and presumably, all tissues require protective mechanisms from heat shock. It remains to be determined whether fly circBoule RNAs play similar roles in other tissues. HSPA2 is a member of the HSPA (HSP70) family, and it is critical for germ cell differentiation during mouse spermatogenesis, after testicular sperm maturation, and sperm-egg recognition in mice and human (35, 36, 44, 45). Conservation of interaction of circBoule with HSP proteins from flies to humans and their roles in male fertility support a common metazoan origin of circBoule RNAs and their ancient reproductive function.
Appropriate levels of HSPA2 protein in human sperm may be important, as either too high or too low levels are associated with male infertility diseases such as asthenozoospermia, oligozoospermia, and azoospermia (36). Consistent with the hypothesis of critical and conserved roles of circBoule-HSPs interaction in male fertility, human circBoule RNA (circEx3-6 and circEx2-7) levels are lower in asthenozoospermic sperm, and the negative correlation between levels of circBoule RNA and HSPA2 in normozoospermic sperm is abolished in asthenozoospermic sperm (Fig. 9, B and E). Mammalian sperm are naturally subjected to “heat shock” during fertilization when ejaculated from the lower temperature epididymis into the female reproductive tract (generally, the temperature difference can be 3° to 5°C in mammals) (46). Our study demonstrates the roles of circBoule RNAs in mammalian sperm in heat shock conditions. Mature sperm are of particular interest for studying the roles of circBoule RNAs in coping with heat stress, because BOULE proteins translated from linear mRNAs are not present in mature sperm (fig. S5E). Mouse HSPA2 levels decreased with elevation of testicular temperature (36). HSPA2 and BOULE proteins (thus may be circBoule RNAs) were expressed in essentially same types of cells in murine testes (figs. S5E and S6E). Dosage and tempospatial expression of circBoule RNAs may be critical in the precise tuning of HSP levels. The partial rescue in flies, either by circEx2-3 and circEx4-7 alone or by the two together, could be due to expression differences between the transgenic and endogenous genes. Likewise in Intron2RPΔ/RPΔ mice, where the expression of circEx2-6 and circEx2-7 is not affected, the reduction in exon 5 containing circBoule RNAs was sufficient to cause changes in HSPA2 levels. It will be crucial to characterize the potential involvement of circBoule RNAs in mammalian fertilization, in particular, after physiological heat stresses, and to explore the potential of circBoule RNAs as biomarkers of human male infertility. More broadly, the roles of circBoule RNAs in sperm in coping with heat stress provide another example of functional sperm non-coding RNA and support the importance of the functions of sperm noncoding RNAs in regulating embryo development or even offspring phenotypes (47, 48); as circRNAs have unique features and are supposed to have longer half-life than linear RNAs, their roles in this context are worth further investigation.
As we demonstrated in this study, circBoule RNAs are essential to the fly for fertility protection when under heat stress. CircRNAs, especially ones encoded by coding genes, are often viewed as side products of gene expression; in the case of Boule gene in animals including humans, circRNAs may play key roles in modulating the physiological and cellular response to environmental stress. In conclusion, we illustrated a mechanism by which conserved circBoule RNAs regulate levels of HSPs under heat stress to protect male fertility in animals from flies to mice. Future studies should investigate other potential roles of circRNAs in animal physiology and human health, stress responses, and evolution.
MATERIALS AND METHODS
Human tissue samples
All human studies were approved by the Institutional Ethics Committee of Nanjing Medical University and performed after obtaining written informed consent. Study subjects were from the Department of Urology, First People’s Hospital of Lianyungang (Lianyungang, China) and Department of General Surgery, Jiangsu Provincial Hospital (Nanjing, China). Tissue biopsies intended for RNA isolation were stored in RNAlater RNA stabilization solution (AM7020, Thermo Fisher Scientific, Life Sciences, USA).
Animals
Animal care, breeding, and surgical procedures were performed in accordance with the standard protocols of the Institutional Animal Care and Use Committee, Nanjing Medical University. A portion of each tissue was snap-frozen in liquid nitrogen for later RNA extraction. Adult White Leghorn chickens (G. gallus) were purchased from Beijing Laboratory Animal Research Center (Beijing, China). Adult landrace pigs (S. scrofa) were housed in Nanjing Medical University animal facilities, and standard animal husbandry protocols were followed. Mouse was bred at Animal Core Facility of Nanjing Medical University. Fly (D. melanogaster) stocks were maintained on standard Drosophila food medium at 25°C in a 12-hour light/12-hour dark cycle. Transgenic or genetically manipulated animal models used in this study: Boule Intron2RPΔ/RPΔ mice, boule gDNA-RP fly, boule M-introns KO fly, boule Intron3 KO fly, boule circEx2-3 overexpression fly, boule circEx4-7 overexpression fly, boule circEx2-3 motif 2 mut overexpression fly, boule circEx4-7 motif 2 mut overexpression fly, boule linearEx2-3 overexpression fly, boule linearEx4-7 overexpression fly, mouse circEx3-6 overexpression fly, mouse circEx3-6 motif 2 mut overexpression fly, Hsp60C-EndoFLAG fly, Hsc4-EndoFLAG fly, hsp60C-Hsp60C-3xFLAG overexpression fly, and hsc4-Hsc4-3xFLAG overexpression fly (see below for details).
Construction of boule KO, gDNA-RP, M-introns KO, and Intron3 KO fly strains
The detailed method and vector skeleton were described in the reference (49). Briefly, for generation of boule gDNA-RP, M-introns KO, and Intron3 KO lines, we first generated boule KO “founder” line with ΦC31 attP site using CRISPR-dependent homologous recombination. The fly boule genomic sequence (dm6, chr3L: 9106038-9113540) was targeted by single guide RNAs (sgRNAs) with the following sequences: 5′- CATTAAAATAATCAGCCACATGG-3′ for the upstream site and 5′- GGGGAAGGGACTATAACCATAGG-3′ for the downstream site. In boule KO founder line, the genomic DNA between these two sgRNAs was replaced by an attP site and the white+ (w+) transgenic marker that was flanked by loxP sites and removed by Cre recombinase later. The deleted genomic DNA of boule gene was engineered in vitro to incorporate desired modifications (gDNA-RP, M-introns KO, or Intron3 KO sequences) on an integration vector (pGE-attB) that carries an attB site together with a w+ marker. Then boule gDNA-RP, M-introns KO, or Intron3 KO sequences were integrated into the deletion locus of the founder line through ΦC31-mediated DNA integration. Extra vector sequences, together with w+, were removed by Cre recombinase to generate a final engineered-mutant allele. The gDNA-RP sequence was the deletion part of genomic DNA sequence in founder line. The Intron3 KO sequence was the deletion part of genomic DNA sequence without intron 3 sequence in founder line. The M-introns KO sequence was exon 2 to exon 8 sequence of fly boule gene transcript (FBtr0076538).
Testis-specific expression of circRNAs in Drosophila
To express fly boule circRNAs (circEx2-3 or circEx4-7) specifically in Drosophila testis, we generated transgenic fly alleles by cloning the boule genomic DNA chr3L: 9108074-9114826 (without intron 2) for expression of circEx2-3 and the boule genomic DNA chr3L: 9106551-9112787 (without intron 4, intron 5, and intron 6) for expression of circEx4-7, which both driven by β2-tubulin (betaTub85D) promoter (24), into an integration vector (pGE-attB) individually and integrating them into the attP40 site at chr2L with the help of ΦC31 integrase. Extra vector sequences, together with w+, were removed by Cre recombinase. Motif 2 sequences in vectors for the expression of circEx2-3 or circEx4-7 were mutated (see Fig. 4A) to generate circEx2-3 motif 2 mut or circEx4-7 motif 2 mut fly strains, respectively. Intron 3 sequence in vectors for the expression of circEx2-3 or circEx4-7 was deleted for generating boule linearEx2-3 or linearEx4-7 fly strains, respectively. To obtain mouse Boule circEx3-6 (or circEx3-6 motif 2 mut) expression vectors for the overexpression of mouse circBoule RNAs in fly, sequences of fly Exon2-3 in vector for the expression of fly boule circEx2-3 were replaced by sequences of mouse Boule Exon3-6 (with or without motif 2 mutation).
Generation of Hsp60C-EndoFLAG and Hsc4-EndoFLAG fly strains
To generate Hsp60C-EndoFLAG fly line, 3xFlag sequences were inserted into C terminus of endogenous hsp60C genomic locus using CRISPR-Cas9. Then, the sgRNA vector (sgRNA sequence, 5′- AGAATATAATGACATAGGGA -3′) and donor vector (containing 3xFlag sequences) were injected into fly fertilized eggs expressing Cas9 protein using CRISPR-dependent homologous recombination. Similarly, to generate Hsc4-EndoFLAG fly line, 3xFlag sequences were inserted into C terminus of endogenous hsc4 genomic locus using CRISPR-Cas9. The sgRNAs vector (sgRNAs sequence, 5′- GGTTTAGTCGACCTCCTCGA -3′ and 5′- ATCAGAAATCGGCACACACG -3′) and donor vector (containing 3xFlag sequences) were injected into fly fertilized eggs expressing Cas9 protein using CRISPR-dependent homologous recombination.
Construction of hsc4-Hsc4-3xFLAG SCIn and hsp60C-Hsp60C-3xFLAG SCIn flies
We generated the transgenic fly alleles by cloning the hsp60C genomic DNA chr2L: 5659849-5670838 (without introns) or hsc4 genomic DNA chr3R: 15241187-15246614 (without introns) into an integration vector (pGE-attB) and integrating them into the attP40 site at chr2L with the help of ΦC31 integrase. The 3xFlag sequences were inserted into the C terminus of Hsp60C and Hsc4. Extra vector sequences, together with w+, were removed by Cre recombinase.
Generation of Hsp60C, Hsc4, Prosα6T, and Rpn11 mutant flies
The Hsp60C, Hsc4, Prosα6T, and Rpn11 mutants were generated by nonhomologous end-joining strategy with the help of CRISPR-Cas9 system. Hsp60C coding sequences (CDSs) were targeted by sgRNAs with the following sequences: 5′-GTGGGCCCGTAACTATGCAA-3′ and 5′-GGCCGTGACCATGGGACCAA-3′, to generate a deletion of 97 base pairs (bp), which result in the formation of a premature stop codon. Hsc4 CDS was targeted by sgRNA with the following sequences: 5′-GTGCTTCATGTCAGACTGCA-3′, to generate a small deletion (5 bp), which led to the formation of a premature stop codon. Prosα6T (CG5648) CDS was targeted by sgRNAs with the following sequences: 5′-GCAATGGAGGCGGTGAAACA-3′ and 5’-GCGTGGTGTGCCAGTACATG-3′, to generate a deletion of 186 bp, which would result in a truncated protein without full-function. Rpn11 CDS was targeted by sgRNAs with the following sequences: 5′-GCAGGTGTACATCTCGTCCT-3′ and 5’-GCCATGCCGCAAACGGGTAC-3′, to generate a deletion of 191 bp, which led to the formation of a premature stop codon.
Generation of Boule intron2 repeat sequences deletion (Intron2RPΔ/RPΔ) mice
For the generation of Boule circEx3-5–, circEx3-6–, and circEx3-7–deficient mice, part of the intronic sequences of Boule intron2 (mm10, chr1: 55356985-55360085) was targeted by sgRNAs with the following sequences: 5′-GATTCACAGTATTCCATACA-3′ and 5′-TCCATACAGGGAAGGTATAC-3′ for the upstream site and 5′-ATCTGTTACCTTAATACCAT-3′ and 5′-TCTGCACACCCATGGTATTA-3′ for the downstream site. Cas9 and sgRNA construction and in vitro transcription were performed according to the supplier’s protocol. After confirming the cleavage efficiency of these sgRNAs in mouse NIH3T3 cells [CRL-1658, American Type Culture Collection (ATCC)], in vitro transcribed RNAs containing sgRNAs (10 ng/μl) and Cas9 mRNA (20 ng/μl) were injected into 150 C57BL/6 zygotes, followed by implantation into five ICR (Institute of Cancer Research) strain surrogate mice, and eventually generated 22 F0 founders. This mice strain was generated in the Model Animal Research Center of State Key Lab of Reproductive Medicine, Nanjing Medical University, China. F0 founders were genotyped by PCR primers with the following sequences: 5′-CTTTCCTATACAGCACTCCAAT-3′ and 5′-AAGAAGAATTACTAACTCAGCCT-3′. Wild-type allele had a PCR length of about 3.7 kb, and deficient allele had a PCR length of about 500 bp. Positive F0 founders with target sequence deletion were backcrossed to C57BL/6 mice to obtain deficient allele heritable heterozygous F1 mice. Then, F1 mice were intercrossed to generate Boule Intron2RPΔ/RPΔ homozygous mice.
Cell culture and transfection of plasmids
Mouse N2a cells (CCL-131, ATCC) were maintained under standard culture conditions with Dulbecco’s modified Eagle’s medium plus 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37°C and 5% CO2. Drosophila S2 cells (CRL-1963, ATCC) were maintained under standard culture conditions with Schneider’s Drosophila medium plus 10% FBS and 1% penicillin/streptomycin at room temperature. Plasmid transfection was conducted with Lipofectamine 2000 (11668-027, Invitrogen) according to the supplier’s protocol. To induce the expression of plasmid genes in S2 cells, 0.5 mM CuSO4 was added 24 hours after transfection, and cells were harvested 24 hours after the induction for downstream experiments.
TUNEL assay of fly testes
About 30 to 50 fly testes were dissected out in 1× phosphate-buffered saline (PBS) solution and fixed in 4% paraformaldehyde (PFA) for 20 min at room temperature. Fixed testes were washed with 1× PBS (containing 0.2% Triton X-100) for 30 min, and then TUNEL analysis was performed using TUNEL BrightGreen Apoptosis Detection Kit (A112-02, Vazyme) according to the manufacturer’s manual. Stained slides were imaged on a ZEISS LSM 700 confocal microscope.
Mating set for testing fly fertility
One male and three w1118 females were used. All assays were performed at least 30 replicates per genotype. Briefly, newly hatched unmated males were picked out and kept for 2 days at 25°C and then mated with 2- to 3-day-old virgin females for 1 day at 25°C to allow them “premate” (offspring from the first day mating was not counted in the fertility test). The flies were then transferred to new vials to examine the fertility by counting adult progeny numbers at different time points, and if to examine the effect of thermal changes to male fertility, then premated flies were transferred to new vials under 18°, 25°, or 29°C.
Nuclear morphology and sperm count assessment of mature sperm in seminal vesicle
Fly testes from fertility test were dissected out in 1× PBS solution and fixed in 4% PFA for 20 min at room temperature in dissection dish. Fixed testes were washed with 1× PBS (containing 0.2% Triton X-100) for 30 min and then transferred to the slides, following by staining with 4′,6-diamidino-2-phenylindole (DAPI) for 15 min at room temperature. Stained slides were imaged on a ZEISS LSM 700 confocal microscope. The largest sections of seminal vesicles were imaged for statistical analysis of sperm count and proportion of abnormal nuclear morphology.
Sperm count assessment of mature sperm in seminal vesicle
Fly testes from fertility test were dissected out in 1× PBS and transferred to a drop of 1× PBS on a glass slide. To let the sperm flow out as much as possible, seminal vesicle was torn open and squashed under the coverslip for about 30 s. We then assessed amount of mature sperm based on the volume of the sperm flowed out from the seminal vesicle using three arbitrary grades (fig. S2A) under a ZEISS Axioskop 2 plus microscope equipped with phase-contrast optical lenses. For grade 3, a large volume of sperm flows out and spread rapidly when seminal vesicle was torn open and squashed. The sperm volume of grade 3 includes those of wild-type fly with lots of sperm spreading. Sperm volume flowed out from the seminal vesicles of grade 2 is about 30 to 50% to that of grade 3. Sperm volume flowed out from the seminal vesicles of grade 1 is about <20% to that of grade 3. As this method is subjective, the final grade evaluation of sperm count in seminal vesicles is from three independent researchers.
IF of fly testes
Fly testes were dissected out in 1× PBS solution and fixed in 4% PFA for 20 min at room temperature. Fixed testes were washed with 1× PBS (containing 1% Triton X-100) for 30 min, then transferred to 200-μl tube, and incubated with anti-FLAG primary antibody (F1804, Sigma-Aldrich) overnight at 4°C. Next morning, testes were washed with 1× PBS (containing 1% Triton X-100) for 30 min, incubated with anti-mouse second antibody (ab60316, Abcam) for 1.5 hours, and followed by phalloidin (P5282, Sigma-Aldrich) staining for 30 min at room temperature. Then, testes were transferred to slides and stained with DAPI for 15 min at room temperature. Stained slides were imaged on a ZEISS LSM 700 confocal microscope.
Mouse fertility test
Two-month-old wild-type (n = 10) and Intron2RPΔ/RPΔ (n = 10) male individuals were mated to two ICR virgin females (6 to 8 weeks old), respectively. All mating cages lasted for 4 months, and the litter size was recorded during this period.
Mouse sperm count and mature sperm motility test
To count sperm number, one whole epididymis was removed and minced into small pieces in prewarmed (37°C) 1× PBS in 24-well plates. For each epididymis, 1 ml of 1× PBS were used. The plates were then incubated at 37°C for 30 min to allow mature sperm to swim out. The sperm number was counted in a hemocytometer. To examine mature sperm motility, one caudal epididymis was removed from each mouse and cut into 2 to 3 pieces in prewarmed (37°C) 500 μl of human tubal fluid (HTF; 90125, Irvine Scientific) and then incubated at 37°C for 5 min to allow mature sperm to swim out. Well-mixed sperm suspension was siphoned into Rectangle Boro Tubing (HTR1099, VitroTubes), and mature sperm motility was examined by IVOS II Animal & Research CASA systems (Hamilton Thorne) according to the instructions.
Heat shock treatment of sperm in epididymis
Wild-type and Intron2RPΔ/RPΔ adult male mice (2 months old) were anesthetized with 2,2,2-tribromoethanol (T48402, Sigma-Aldrich) at a dose of 240 mg/kg. The lower third of the body was placed in a water bath at 42°C for 30 min (heat shock). After heat shock treatment, mice were immediately euthanized, and the epididymis was isolated for further analyses.
IVF in mice
IVF was performed according to the standard procedures. Briefly, after the pregnant mare serum gonadotropin injection for 48 hours, oocyte donors (3- to 4-week-old ICR females) were injected with human chorionic gonadotropin (hCG), and the oocyte clutches from donors 13 to 14 hours after hCG were collected into 2 ml of modified 1× PBS prewarmed to 37°C. Sperm from cauda epididymis was incubated for an additional hour in HTF for capacitation. The oocyte clutches with minimal media (PBS) were transferred to the 500-μl drop of fertilization media [HTF with 1% bovine serum albumin (BSA)] containing sperm at 37°C under mixed gas (5% CO2, 5% O2, and 90% N2) for 4 to 6 hours. Then, the excess sperm and debris were removed, the presence of pronuclei and the extruded second polar body in fertilized oocytes were examined, and then the fertilized oocytes were incubated overnight in KSOM (potassium simplex optimization medium) supplemented with amino acids at 37°C under mixed gas (5% CO2, 5% O2, and 90% N2); the two-cell-stage embryos were counted in the next morning.
Histological and IF of mouse testis
Mouse testes were fixed in Hartman’s fixative (H0290, Sigma-Aldrich) for 24 hours at room temperature. The testes were embedded in paraffin and sectioned into 5-μm-thick slides. H&E staining was performed using hematoxylin solution (HHS16, Sigma-Aldrich) and eosin Y (318906, Sigma-Aldrich). The slides were treated with citrate buffer (Beyotime) for antigen retrieval and then incubated in rabbit anti-BOULE (Boule antiserum 101) primary antibody (26), followed by horseradish peroxidase (HRP) conjunctive second antibody detection with the Biotin-Streptavidin HRP Detection Systems (ZSGB-BIO). Slides were then imaged using a ZEISS Axioscope A1 microscope. For immunofluorescence of HSPA2 protein, the testis sections were incubated in rabbit anti-HSPA2 (12797-1-AP, Proteintech) primary antibodies and followed by Rhod Red-X-AffiniPure Donkey Anti-Rabbit immunoglobulin G (IgG) (H + L) (711-295-152, Jackson ImmunoResearch). Slides were then imaged using a ZEISS LSM700 confocal microscope.
Protein mass spectrometry
Specific silver-stained bands were cut, digested, and extracted. The masses of the peptides in the extract were then measured by mass spectrometry (MS) to obtain the peptide mass fingerprints. Next, peptides were selected to undergo fragmentation via tandem MS. Both the MS and tandem MS data were searched against protein sequence databases to determine the proteins present in the gel.
Plasmid construction
All plasmids were constructed with restriction-enzyme digestion and recombinant ligation (C113-02, Vazyme). All plasmids were sequenced for confirmation. To express fly circBoule RNAs (circEx2-3 and circEx4-7), hemagglutinin (HA)–tagged ubiquitin protein, FLAG-tagged Hsc4 protein, and FLAG-tagged Hsp60C protein in S2 cell, pMT/V5-His A vector (gift from Q. Wu) was used. For the expression of circRNAs, circEx2-3 and circEx4-7 were plasmids with inserts corresponding to the exon forming the circRNA and the 5′ and 3′ flanking genomic sequences. To express mouse circBoule RNAs (circEx3-5, circEx3-6, and circEx3-7) in N2a cells, the promoter in the vector was cytomegalovirus (CMV) promoter. CircEx3-5, circEx3-6, and circEx3-7 with the exon forming the circRNA were inserted to circular green fluorescent protein (gift from Z. Wang) flanking sequence using recombinant method. The motifs (motif 1 and motif 2) mutation vectors were constructed on the basis of circEx2-3, circEx4-7, circEx3-5, and circEx3-6 using recombinant method. For the expression of FLAG-tagged HSPA2 and hnRNP M (heterogeneous nuclear ribonucleoprotein M), the vector was p3xFLAG-Myc-CMV-24. For the expression of HA-tagged ubiquitin, the vector was pKH3.
RNA IP
RIP was carried out as previously described with some modifications (6). Briefly, mouse testes were digested with collagenase (1 mg/ml) and 0.25% trypsin for 20 min before the filtration with a 40-μm filter to obtain mouse testes cells. For S2 cells, N2a cells, mouse testes cells, mouse sperm, human sperm, or fly whole testes were fixed and cross-linked in 1% formaldehyde with gentle rotation at 4°C for 20 min and then treated with 0.125 M glycine (to quench formaldehyde cross-linking) for 20 min at 4°C. The samples were resuspended after the centrifuge at 500g for 5 min at 4°C in ice-cold lysis buffer with gentle rotation for 30 min at 4°C and then sonicated with a Sonics Vibra-Cell (3-s on, 6-s off, 5 min, 30%). The samples were centrifuged at 12,000g for 15 min at 4°C, and the supernatant was collected. Protein G Dynabeads (10004D, Thermo Fisher Scientific) were preincubated with antibodies (IgG, anti-FLAG, or anti-HSPA2, 2 μg in the whole system) at room temperature for 2 hours. The supernatants were added to antibodies coupled with Protein G Dynabeads for at least 4 hours at 4°C with rotation. The antibody–Protein G bead complexes were washed five times with lysis buffer. After the last wash, one-fifth of the bead volume was saved for Western blotting. The remaining antibody–Protein G bead complexes were resuspended in 50 μl of lysis buffer and were digested with deoxyribonuclease (DNase) at 37°C for 20 min and 30 μg of proteinase K (C500021-0010, Sangon Biotech) at 56°C for 20 min; this was followed by extraction with TRIzol (15596026, Thermo Fisher Scientific) to obtain RNAs. The lysis buffer was [50 mM tris-Cl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.1% SDS, 1 mM dithiothreitol (DTT), cOmplete protease inhibitor, and ribonuclease (RNase) inhibitor (0.1 U/μl)], except for mouse sperm using lysis buffer [10 mM Hepes, (pH 7.0), 50 mM KCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.5% Triton X-100, transfer RNA (tRNA) (0.1 μg/μl), heparin (0.5 μg/μl), cOmplete protease inhibitor, and RNase inhibitor (0.1 U/μl)].
RNA pull-down
Mouse testes cells were harvested as detailed in RIP. Mouse testes cells were resuspended in lysis buffer [20 mM tris-Cl (pH 7.4), 137 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1% NP-40, cOmplete protease inhibitor, and RNase inhibitor (0.1 U/μl)]. Mouse sperm and human sperm were resuspended in lysis buffer [10 mM Hepes (pH 7.0), 50 mM KCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.5% Triton X-100, tRNA (0.1 μg/μl), heparin (0.5 μg/μl), cOmplete protease inhibitor, and RNase inhibitor (0.1 U/μl)]. Fly whole testes were resuspended in lysis buffer [125 mM tris-Cl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM EDTA, 1 mM DTT, cOmplete protease inhibitor, and RNase inhibitor (0.1 U/μl)]. The samples were rotated for 30 min at 4°C and sonicated for 15 min. The lysate was centrifuged at 17,000g for 15 min. The supernatant was collected and precleared with M-280 Streptavidin Dynabeads (11206D, Thermo Fisher Scientific) at 4°C for 2 hours. Then, the supernatant was collected and added 200 pmol of biotin-DNA oligonucleotides (table S1) at 4°C for 4 hours. M-280 Streptavidin Dynabeads were pretreated three times in the lysis buffer, blocked with yeast total RNA (500 ng/μl) and BSA (1 mg/ml) for 2 hours at room temperature, and then washed three times with lysis buffer. The treated beads were then added into the samples with rotation for 4 hours at 4°C. Beads then were captured with magnets, washed two times with lysis buffer, and three times with lysis buffer supplemented with 500 mM NaCl. RNAs and proteins were analyzed with the same methods described in RIP.
Examination of ubiquitination
To examine ubiquitination of HSPs in cells, N2a cells or S2 cells were transfected with the plasmids for 20 or 44 hours and then treated with 100 μM MG132 (A2585, APExBIO) for 4 hours. To examine ubiquitination of HSPs in mouse cauda sperm and fly whole testes, mouse cauda sperm were treated with 100 μM MG132 for 4 hours in HTF at 37°C, and fly testes were from 4-day-old Hsp60C-EndoFLAG and Hsc4-EndoFLAG flies raised at 29°C. Then, the N2a cells were washed with 1× PBS and followed by ultraviolet (UV)–irradiated for 2 min. The S2 cells, sperm, and fly testes were fixed with 1% formaldehyde for 20 min at 4°C and then quenched with 0.125 M glycine for 20 min at 4°C. The samples were resuspended after the centrifuge at 500g for 5 min at 4°C in ice-cold lysis buffer with gentle rotation for 30 min at 4°C and then sonicated with a Sonics Vibra-Cell (3-s on, 6-s off, 5 min, 30%). The samples were centrifuged at 12,000g for 15 min at 4°C. Protein G Dynabeads (10004D, Thermo Fisher Scientific) were preincubated with antibodies (IgG, anti-FLAG and anti-HSPA2, 2 μg in the whole system) at room temperature for 2 hours. The supernatants were added to antibodies coupled with Protein G Dynabeads for at least 4 hours at 4°C with rotation. The immunoprecipitants and input were then subjected to Western blot analysis to test ubiquitination of HSPA2-FLAG, HSP60C-FLAG, HSC4-FLAG, and HSPA2 proteins. Bead-bound proteins were then analyzed by Western blotting with anti-HA antibody (3724, Cell Signaling Technology) or ubiquitin antibody (sc-8017, Santa Cruz Biotechnology). The N2a and S2 cells were lysed in lysis buffer [50 mM tris-Cl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.1% SDS, 1 mM DTT, cOmplete protease inhibitor, and RNase inhibitor (0.1 U/μl)]. Mouse sperm were lysed in lysis buffer [20 mM tris-Cl (pH 7.4), 137 mM NaCl,1 mM CaCl2, 1 mM MgCl2, 1% NP-40, cOmplete protease inhibitor, and RNase inhibitor (0.1 U/μl)], and fly whole testes were lysed in lysis buffer [125 mM tris-Cl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM EDTA, 1 mM DTT, cOmplete protease inhibitor, and RNase inhibitor (0.1 U/μl)].
Isolation of cytoplasm
The transfected N2a cells were washed with 1× PBS, UV-irradiated, followed by 2 min, resuspended in cytoplasmic lysis buffer [0.5% NP-40, 2 mM MgCl2, 20 mM Hepes (pH 7.4), 10 mM KCl, cOmplete protease inhibitor, and RNase inhibitor (0.1 U/μl)] and incubated on ice for 45 min. Meanwhile, the lysate was mixed by pipette tip every 5 min. The lysate was centrifuged at 5000g for 5 min. The supernatant was collected for the cytoplasmic fraction.
Total RNA extraction, RNase R treatment, and reverse transcription
Total RNA was extracted from collected tissues or cells using TRIzol reagent, according to the manufacturer’s recommendations, and purified by phenol-chloroform extraction after DNase treatment. The RNA concentration was measured by NanoDrop 2000c (Thermo Fisher Scientific, Life Sciences). DNase-treated total RNA (10 μg) were incubated at 37°C for 30 min with or without RNase R (3 U/μg; RNR07250, Epicentre Biotechnologies) and then was reverse-transcribed with PrimeScript RT Master Mix with the supplied protocol (RR036A, TaKaRa Biosystems).
Northern blot
Using the DIG Northern Starter Kit (12039672910, Roche), the corresponding RT-PCR products from circRNA junction was used as templates for T7 transcription. Then, the obtained RNA probes labeled by Digoxin were used for hybridization. Total RNA was extracted from mouse or Drosophila testes with standard TRIzol methods. A total of 8% tris-borate EDTA (TBE)–urea polyacrylamide gel electrophoresis with 8 M urea was prerun for 2 hours. Then, 15 μg of RNA with or without RNase R digestion and RiboRuler High Range RNA Ladder (Thermo Fisher Scientific) were loaded on prerun polyacrylamide gel and run for another 2 hours in 0.5× TBE buffer. RNA in polyacrylamide gel was transferred onto Hybond-N+ membranes (GE Healthcare) by capillary transfer. Hybridization was performed at 60°C overnight. Detection was performed according to the manual (DIG Northern Starter Kit, Roche). Images were taken with an ImageQuant LAS4000 biomolecular imager (GE Healthcare).
Immunofluorescence in situ hybridization coupled with IF of protein
The mouse cauda epididymis was isolated and transferred into 1.5-ml centrifuge tube containing 200-μl prewarmed 1× PBS at 37°C and then minced into two to three small pieces. After 30 min of incubation, the undissolved tissues were removed using 40-mesh strainers. Then, the pure sperm were fixed in 4% paraformaldehyde at room temperature for 30 min and centrifuged at 1000g for 5 min. The sperm deposit was washed twice and resuspended in 1× PBS. After washing, the sperm were resuspended in solution composed of 1× PBS, 20% BSA, and 4% gelatin (2:1:1) and dropped onto the poly-l-lysine–coated slide and covered with a coverslip. Then, the slides were frozen at −80°C for 7 min and peeled off the coverslip quickly. Slides were incubated with HSPA2 primary antibody for 4 hours at room temperature and then followed by incubation with Alexa Fluor 546–labeled secondary antibody (ab150074, Life Technologies) for another 4 hours. RNA probes (table S1) were generated with a TranscriptAid T7 High Yield Transcription Kit (K0441, Thermo Fisher Scientific), using the corresponding insertion in the T vector as the template and then labeled with Alexa Fluor 488 using a ULYSIS Nucleic Acid Labeling Kit (Invitrogen). This kit added a fluor on every G in the probe to amplify the fluorescence intensity. RNA probes were denatured at 80°C for 10 min. Slides were washed with 2× saline sodium citrate (SSC) at 45°C for 10 min and then incubated with the probes at 37°C overnight with and yeast total RNA (500 ng/μl) (Ambion). Slides were then washed with 2× SSC at 45°C for 10 min. The slides were lastly stained with DAPI and observed with confocal microscope.
Human sperm fractioning and purification
Semen samples were selected from men undergoing standard spermiogram analysis during clinical evaluation of infertile couples (couples were diagnosed with infertility if they had been trying to conceive for 12 months or more, and generally, these couples have either female or male infertility). Men were selected for the normozoospermia and asthenozoospermia group according to the guidelines of semen analysis according to the World Health Organization Laboratory Manual for the examination and processing of human semen, fifth edition in 2010. Individual spermiogram parameters were shown in table S2. All men provided written informed consent and agreed to the analysis of genetic material as approved by the Ethics Committee of University of Science and Technology of China. Spermatozoa were then purified by centrifugation in 40% Percoll (GE Healthcare). This procedure results in formation of a pellet fraction consisting mostly of sperm with normal morphology and defective sperm (sperm with morphological abnormalities and/or reduced sperm motility). The separated spermatozoa were subsequently used to isolate RNA and protein.
Enzyme-linked immunosorbent assay of human HSPA2
Human sperm lysates were diluted in coating buffer [0.015 M sodium carbonate and 0.035 M sodium bicarbonate (pH 9.6)] to a final volume of 100 μl and coated onto enzyme-linked immunosorbent assay (ELISA) plate wells (F605031, Sangon) for about 24 hours at 4°C. The coated wells were washed with 1× PBS at room temperature and blocked with 5% BSA at 37°C for 1 hour. Then, the wells were incubated with HSPA2 antibody overnight at 4°C. The wells were washed thrice with 1× PBS (containing 0.1% Tween 20) and incubated with HRP-labeled secondary antibody (1:1000; no. L3012, Signalway Antibody) for 1 hour at room temperature. Then, the wells were washed thrice with 1× PBS (containing 0.1% Tween 20). The bound peroxidase was then subjected to color reaction using EL-TMB Chromogenic Reagent kit (C520026, Sangon). Colorimetric readings were taken at 450 nm in a Multiskan reader (Thermo Fisher Scientific). For normalization, ELISA of β-actin was also performed for each sperm protein lysate.
Statistical analysis
Student’s t tests were used to calculate P values, as indicated in the figure legends. The values reported in the graphs represent averages of actual number of independent experiments, with error bars showing SD. After analysis of variance with F tests, the statistical significance and P values were evaluated with Student’s t tests.
Supplementary Material
Acknowledgments
We would like to thank R. Carthew and T. Kurita for discussion and/or comments on our manuscript. We also like to thank D. Chen for the fly Boule antibody; Q. Wu and Z. Wang for providing plasmids; and B. Xu for help and assistance during the collection of human semen samples. Funding: This work was supported by funding from the National Key R&D Program of China (2019YFA0802600 and 2018YFC1004500), the National Natural Science Foundation of China (31930019, 31725016, 91940303, 31771652, and 31970792), and the Strategic Priority Research Program “Biological basis of aging and therapeutic strategies” of the Chinese Academy of Sciences (XDB39010400). Author contributions: The E.Y.X. lab initiated this research. E.Y.X. and G.S. designed and initiated this project, provided the major funding, and supervised the experiments. G.S., E.Y.X., and J.H. provided the experimental facilities. L.G., S.C., W.X., X.W., C.Z., L. Cheng, and X.L. performed the experiments. L.G., S.C., X.W., L. Chen, Q.S., G.S., E.Y.X., and J.H. analyzed the data. L.G., S.C., G.S., and E.Y.X. wrote the manuscript. All authors have discussed the results and made comments on the manuscript. All authors read and approved the final manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/46/eabb7426/DC1
REFERENCES AND NOTES
- 1.Sanger H. L., Klotz G., Riesner D., Gross H. J., Kleinschmidt A. K., Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 73, 3852–3856 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salzman J., Gawad C., Wang P. L., Lacayo N., Brown P. O., Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLOS ONE 7, e30733 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L., Huang C., Wang X., Shan G., Circular RNAs in eukaryotic cells. Curr. Genomics 16, 312–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X., Wang X., Li J., Hu S., Deng Y., Yin H., Bao X., Zhang Q. C., Wang G., Wang B., Shi Q., Shan G., Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China Life Sci. 63, 1429–1449 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S. D., Gregersen L. H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., Ie Noble F., Rajewsky N., Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., Zhu P., Chang Z., Wu Q., Zhao Y., Jia Y., Xu P., Liu H., Shan G., Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Salzman J., Circular RNA expression: Its potential regulation and function. Trends Genet. 32, 309–316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., Kjems J., Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., Laneve P., Rajewshy N., Bozzoni I., Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66, 22–37.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pamudurti N. R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., Shenzis S., Samson M., Dittmar G., Landthaler M., Chekulaeva M., Rajewshy N., Kadener S., Translation of circRNAs. Mol. Cell 66, 9–21.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L. L., Wang Y., Wong C. C., Xiao X., Wang Z., Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27, 626–641 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piwecka M., Glazar P., Hernandez-Miranda L. R., Memczak S., Wolf S. A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C. A., Fenske P., Trimbuch T., Zywitza V., Plass M., Schreyer L., Ayoub S., Kocks C., Kuhn R., Rosenmund C., Birchmeier C., Rajewsky N., Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Kleaveland B., Shi C. Y., Stefano J., Bartel D. P., A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174, 350–362.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia P., Wang S., Ye B., Du Y., Li C., Xiong Z., Qu Y., Fan Z., A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 48, 688–701.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Zhu P., Zhu X., Wu J., He L., Lu T., Wang Y., Liu B., Ye B., Sun L., Fan D., Wang J., Yang L., Qin X., Du Y., Li C., He L., Ren W., Wu X., Tian Y., Fan Z., IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat. Immunol. 20, 183–194 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Wang P. L., Bao Y., Yee M. C., Barrett S. P., Hogan G. J., Olsen M. N., Dinneny J. R., Brown P. O., Salzman J., Circular RNA is expressed across the eukaryotic tree of life. PLOS ONE 9, e90859 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., Akbalik G., Wang M., Glock C., Quedenau C., Wang X., Hou J., Liu H., Sun W., Sambandan S., Chen T., Schuman E. M., Chen W., Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 18, 603–610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin X., Han M., Cheng L., Chen J., Zhang Z., Shen T., Wang M., Wen B., Ni T., Han C., Expression dynamics, relationships, and transcriptional regulations of diverse transcripts in mouse spermatogenic cells. RNA Biol. 13, 1011–1024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R., Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73, 1019–1030 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Glazar P., Papavasileiou P., Rajewsky N., circBase: A database for circular RNAs. RNA 20, 1666–1670 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong W. W., Li H. M., Qing X. R., Huang D. H., Li H. G., Identification and characterization of human testis derived circular RNAs and their existence in seminal plasma. Sci. Rep. 6, 39080 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eberhart C. G., Maines J. Z., Wasserman S. A., Meiotic cell cycle requirement for a fly homologue of human deleted in Azoospermia. Nature 381, 783–785 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Xu E. Y., Moore F. L., Pera R. A., A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc. Natl. Acad. Sci. U.S.A. 98, 7414–7419 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu E. Y., Lee D. F., Klebes A., Turek P. J., Kornberg T. B., Pera R. A., Human BOULE gene rescues meiotic defects in infertile flies. Hum. Mol. Genet. 12, 169–175 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Shah C., VanGompel M. J., Naeem V., Chen Y., Lee T., Angeloni N., Wang Y., Xu E. Y., Widespread presence of human BOULE homologs among animals and conservation of their ancient reproductive function. PLOS Genet. 6, e1001022 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.VanGompel M. J. W., Xu E. Y., A novel requirement in mammalian spermatid differentiation for the DAZ-family protein Boule. Hum. Mol. Genet. 19, 2360–2369 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akerfelt M., Morimoto R. I., Sistonen L., Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vihervaara A., Duarte F. M., Lis J. T., Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 19, 385–397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feder M. E., Hofmann G. E., Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Liu H., Wang X., Wang H. D., Wu J., Ren J., Meng L., Wu Q., Dong H., Wu J., Kao T. Y., Ge Q., Wu Z. X., Yuh C. H., Shan G., Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat. Commun. 3, 1073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Place R. F., Noonan E. J., Non-coding RNAs turn up the heat: An emerging layer of novel regulators in the mammalian heat shock response. Cell Stress Chaperones 19, 159–172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., Noble W. S., MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ijiri T. W., Merdiushev T., Cao W., Gerton G. L., Identification and validation of mouse sperm proteins correlated with epididymal maturation. Proteomics 11, 4047–4062 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redgrove K. A., Nixon B., Baker M. A., Hetherington L., Baker G., Liu D. Y., Aitken R. J., The molecular chaperone HSPA2 plays a key role in regulating the expression of sperm surface receptors that mediate sperm-egg recognition. PLOS ONE 7, e50851 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redgrove K. A., Anderson A. L., McLaughlin E. A., O’Bryan M. K., Aitken R. J., Nixon B., Investigation of the mechanisms by which the molecular chaperone HSPA2 regulates the expression of sperm surface receptors involved in human sperm-oocyte recognition. Mol. Hum. Reprod. 19, 120–135 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Nixon B., Bromfield E. G., Cui J., De Iuliis G. N., Heat shock protein A2 (HSPA2): Regulatory roles in germ cell development and sperm function. Adv. Anat. Embryol. Cell Biol. 222, 67–93 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Tang C., Xie Y., Yu T., Liu N., Wang Z., Woolsey R. J., Tang Y., Zhang X., Qin W., Zhang Y., Song G., Zheng W., Wang J., Chen W., Wei X., Xie Z., Klukovich R., Zheng H., Quilici D. R., Yan W., m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 30, 211–228 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R., Herzog M., Schreyer L., Papavasileiou P., Lvanov A., Ohman M., Refojo D., Kadener S., Rajewsky N., Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Perkins L. A., Doctor J. S., Zhang K., Stinson L., Perrimon N., Craig E. A., Molecular and developmental characterization of the heat shock cognate 4 gene of Drosophila melanogaster. Mol. Cell. Biol. 10, 3232–3238 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkar S., Lakhotia S. C., The Hsp60C gene in the 25F cytogenetic region in Drosophila melanogaster is essential for tracheal development and fertility. J. Genet. 84, 265–281 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Richter K., Haslbeck M., Buchner J., The heat shock response: Life on the verge of death. Mol. Cell 40, 253–266 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Albihlal W. S., Gerber A. P., Unconventional RNA-binding proteins: An uncharted zone in RNA biology. FEBS Lett. 592, 2917–2931 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Bronk P., Wenniger J. J., Dawson-Scully K., Guo X., Hong S., Atwood H. L., Zinsmaier K. E., Drosophila Hsc70-4 is critical for neurotransmitter exocytosis in vivo. Neuron 30, 475–488 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Govin J., Caron C., Escoffier E., Ferro M., Kuhn L., Rousseaux S., Eddy E. M., Garin J., Khochbin S., Post-meiotic shifts in HSPA2/HSP70.2 chaperone activity during mouse spermatogenesis. J. Biol. Chem. 281, 37888–37892 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nixon B., Bromfield E. G., Dun M. D., Redgrove K. A., McLaughlin E. A., Aitken R. J., The role of the molecular chaperone heat shock protein A2 (HSPA2) in regulating human sperm-egg recognition. Asian J. Androl. 17, 568–573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter R. H., Temperature gradients in female reproductive tissues. Reprod. BioMed. Online 24, 377–380 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y., Shi J., Rassoulzadegan M., Tuorto F., Chen Q., Sperm RNA code programmes the metabolic health of offspring. Nat. Rev. Endocrinol. 15, 489–498 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Chen Q., Human sperm RNA code senses dietary sugar. Nat. Rev. Endocrinol. 16, 200–201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang J., Zhou W., Dong W., Watson A. M., Hong Y., Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. U.S.A. 106, 8284–8289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/46/eabb7426/DC1


