Structural and biochemical studies reveal the dual regulation of the ABC transporter OpuA by ionic strength and cyclic-di-AMP.
Abstract
(Micro)organisms are exposed to fluctuating environmental conditions, and adaptation to stress is essential for survival. Increased osmolality (hypertonicity) causes outflow of water and loss of turgor and is dangerous if the cell is not capable of rapidly restoring its volume. The osmoregulatory adenosine triphosphate–binding cassette transporter OpuA restores the cell volume by accumulating large amounts of compatible solute. OpuA is gated by ionic strength and inhibited by the second messenger cyclic-di-AMP, a molecule recently shown to affect many cellular processes. Despite the master regulatory role of cyclic-di-AMP, structural and functional insights into how the second messenger regulates (transport) proteins on the molecular level are lacking. Here, we present high-resolution cryo–electron microscopy structures of OpuA and in vitro activity assays that show how the osmoregulator OpuA is activated by high ionic strength and how cyclic-di-AMP acts as a backstop to prevent unbridled uptake of compatible solutes.
INTRODUCTION
Hypertonic stress of bacteria triggers a rapid uptake of K+ ions and uptake or synthesis of compatible solutes, which restores the turgor (1). The opposite stress, hypotonicity, can lead to cell lysis, which is mitigated by membrane tension–gated mechanosensitive channels that rapidly expel osmolytes and reduce the turgor (2). Compatible solute transporters that respond to hypertonic stress are gated by ionic strength or K+ ions (3–6), but recent studies have shown that the second messenger cyclic-di-AMP can also act as a key regulator of osmoregulatory transport (7–9). OpuA is a type I, ATP (adenosine triphosphate)–binding cassette (ABC) transporter (10) that belongs to a large family of osmoregulatory proteins, named BusA, OpuA, OpuC, OtaA, or ProU, present in bacteria and archaea. OpuA from Lactococcus lactis, the best-studied member, mediates the ATP-driven import of the compatible solute glycine betaine and is gated by ionic strength (11, 12), but mechanistic insights are still elusive.
Cyclic-di-AMP was first discovered as a messenger involved in the initiation of DNA damage checkpoints (13) but is also involved in central metabolism, cell wall metabolism, and, importantly, crucial for the response of bacterial cells to osmotic stress (7, 14–17). While cyclic-di-AMP is essential for survival of bacteria, high concentrations of the molecule are toxic. Higher eukaryotes do not synthesize cyclic-di-AMP, but the ER (endoplasmic reticulum) adapter protein (ERAdP) binds the molecule, which triggers a nuclear factor κB–induced inflammatory cytokine release (18). Here, immune cells detect invading microbes through the presence of second messengers such as cyclic-di-AMP. Cyclic-di-AMP inhibits the transcription of genes of osmoregulatory transporters including those of the opuA operon (19) and binds to a subset of cystathionine-β-synthase (CBS), RCK (regulators of K+ conductance), and USP (universal stress protein) domains of osmoregulatory proteins (8, 9, 16), indicating distinct levels of regulation. Although those studies have shown the importance of cyclic-di-AMP as a signaling molecule and structures of isolated CBS domains have been determined (8, 9), no structure of any transporter in complex with cyclic nucleotides is yet available.
RESULTS AND DISCUSSION
Functional properties of OpuA
OpuA is a homodimeric protein complex, wherein each protomer is composed of two polypeptide chains, with the transmembrane domain (TMD) fused to the substrate-binding domain (SBD) and the nucleotide-binding domain (NBD) fused to a tandem CBS domain (CBS1 and CBS2) (Fig. 1A). To mimic a native-like lipidic environment, OpuA was reconstituted in membrane scaffold protein 1D1 (MSP1D1) nanodiscs for functional characterization and single-particle cryo–electron microscopy (cryo-EM) analysis. Different OpuA/MSP1D1/lipid ratios were tested, and a ratio of 1:10:500 was found optimal, using a lipid composition of 50 mole percent (mol %) 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 38 mol % 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DOPG), and 12 mol % 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC). The adenosine triphosphatase (ATPase) activity of the transporter was determined with a coupled enzyme assay, where the ATP hydrolysis is stoichiometrically coupled to the reduced form of nicotinamide adenine dinucleotide (NADH) oxidation (Fig. 1B). In this lipid composition, OpuA is fully functional and gated by ionic strength (Fig. 1, C to F) (11). The ATPase activity of OpuA is tightly dependent on the presence of glycine betaine (Fig. 1C), indicating that the activities in distant protein domains are mechanistically tightly coupled. The apparent half-maximum effect of glycine betaine on ATPase activity of 5.4 ± 0.7 μM is very similar to the reported Michaelis constant (Km) of 1.9 μM in proteoliposomes (20). The activation by ionic strength (11) is reflected by an increase in ATPase activity at increasing KCl concentration (Fig. 1D). OpuA is also regulated by cyclic-di-AMP (Fig. 1, C to E), with an apparent half-maximum effect for cyclic-di-AMP on the ATPase activity in the low micromolar range (Fig. 1E). Furthermore, the apparent Km for ATP hydrolysis of 4.5 ± 0.5 mM (and cooperativity upon ATP binding with a Hill coefficient of 1.4 ± 0.1) (Fig. 1F) is comparable to the Km of ~3 mM reported for proteoliposomes (21). We conclude that OpuA retains full functionality after reconstitution in lipid nanodiscs.
Fig. 1. Domain organization of OpuA and activity in nanodiscs.
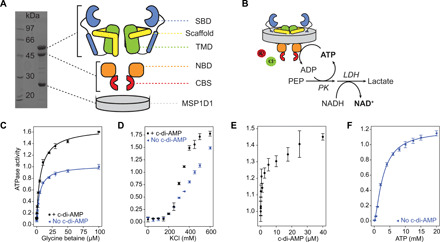
(A) Left, SDS–polyacrylamide gel electrophoresis (PAGE) analysis of OpuA in nanodiscs; right, domain organization of OpuA protomers. (B) Schematic of the coupled enzyme assay to monitor ATPase activity, as shown in (C) to (F). LDH, lactate dehydrogenase; PK, pyruvate kinase. For (C) to (F), the standard assay mixture was composed of 50 mM K-Hepes (pH 7.0), 450 mM KCl, 4 mM phosphoenolpyruvate, 600 μM NADH, 2.1 to 3.5 U of pyruvate kinase, and 3.2 to 4.9 U of lactate dehydrogenase. (C) ATPase activity of OpuA in nanodiscs in the presence of 10 mM ATP with (black circles) and without (blue triangles) 10 μM cyclic-di-AMP. (D) ATPase activity of OpuA in nanodiscs as a function of ionic strength (generated by addition of KCl) in the presence of 100 μM glycine betaine and 10 mM ATP with (black circles) and without (blue triangles) 10 μM cyclic-di-AMP. (E) ATPase activity of OpuA in nanodiscs as a function of the cyclic-di-AMP concentration in the presence of 100 μM glycine betaine and 10 mM ATP. (F) ATPase activity of OpuA as a function of the ATP concentration in the presence of 100 μM glycine betaine. We normalized the ATP hydrolysis activities, as described in Materials and Methods. The error bars represent the SD of three technical replicates. PEP, phosphoenolpyruvate; NADH, reduced form of nicotinamide adenine dinucleotide; NAD+, oxidized form of nicotinamide adenine dinucleotide.
Architecture of OpuA
To gain insight into the transport cycle, we have determined structures of full-length OpuA in nanodiscs in different conformations by single-particle cryo-EM. In the absence of substrate and at high ionic strength, an inward-facing (IF) conformation of wild-type OpuA was captured at 3.3 Å resolution, representing an apo state (Fig. 2A and fig. S1). This IF conformation shows variable spacing between the two NBDs (the different opening angles for different classes are visible in fig. S1D), hinting at a dynamic sampling of distinct degrees of NBDs opening in the apo state. As expected, the SBDs and CBS domains are not resolved in this state, most likely due to their high flexibility in the absence of the substrate glycine betaine and the CBS ligand cyclic-di-AMP.
Fig. 2. Conformational states of OpuA during the transport cycle.
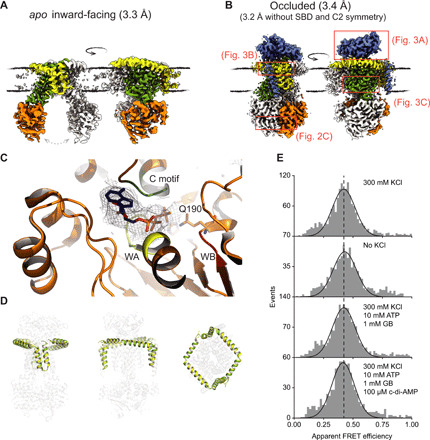
(A) Cryo-EM map of the apo inward-facing conformation of wild-type OpuA at high ionic strength [50 mM KPi (pH 7.0) and 200 mM KCl (pH 7.0)] in the absence of glycine betaine. The NBDs are highly flexible, exposing different opening angles, as emphasized by the gray arrow. The SBD and CBS domains are not resolved. (B) Cryo-EM map of the substrate-loaded occluded conformation of OpuA (E190Q) in the presence of 100 μM glycine betaine, high ionic strength [20 mM Hepes (pH 7.0) and 300 mM KCl], and 10 mM MgATP. The CBS domains are not resolved. (A and B) The color code of the individual domains of one protomer is the same as in Fig. 1, and membrane boundaries are indicated by black lines. (C) ATP (blue sticks with phosphates in orange) bound to the NBD in the occluded conformation. Walker A, walker B, and signature motifs are shown in yellow (WA), red (WB), and green (C motif), respectively (density around ATP is shown as mesh at 6σ). Glutamine-190 is shown as stick. (D) Superposition of the scaffold that is covalently linked to the TMD of OpuA of both the IF (yellow) and occluded (green) conformation. (E) Single-molecule fluorescence resonance energy transfer (smFRET) measurements by alternating laser excitation (ALEX) of Alexa555- and Alexa647-labeled OpuA (S24C) in 20 mM K-Hepes (pH 7.0) at high ionic strength (+300 mM KCl), low ionic strength, turnover conditions [300 mM KCl, 10 mM MgATP, and 1 mM glycine betaine (GB)], and turnover-inhibited conditions [300 mM KCl, 10 mM MgATP, 1 mM glycine betaine, and 100 μM cyclic-di-AMP]. The dashed line marks the peak of the FRET signal.
An occluded conformation, with the transmembrane helices in an outward-oriented fashion, was obtained in the presence of 10 mM MgATP, 100 μM glycine betaine, and at high ionic strength, when the catalytic glutamic acid in the NBD was mutated to a glutamine (E190Q) (Fig. 2B and fig. S2). The structure reveals that one of the SBDs is docked onto the TMD. It disrupts the twofold symmetry of the transporter, resulting in a cryo-EM map at a global resolution of 3.4 Å, but allows an unambiguous modeling of the SBD and the respective anchoring helix. Excluding the SBD and imposing a C2 symmetry during image processing further improved the resolution to 3.2 Å in the remaining regions. The ATP-bound NBDs are arrested in a dimerized closed state, incapable of hydrolyzing the nucleotide because of the E190Q mutation (Fig. 2C). The CBS domains remain flexible and are, therefore, not resolved in the occluded state, which is consistent with the finding that a large part of the CBS domain is natively disordered and connected to the NBD via a linker region (22). The belt protein MSP1D1 wraps tightly around OpuA (fig. S3A), yet OpuA is fully active with comparable kinetics, as in proteoliposomes (Fig. 1, C to F), indicating that the nanodiscs do not perturb transport function.
A unique structural feature found in the cryo-EM maps is a domain that surrounds the TMD, hereafter referred to as the scaffold (Figs. 1A and 2D). It is covalently linked to the TMD and consists of two amphipathic α helices that are lying on top of the outer membrane leaflet and two transmembrane-spanning helices that serve as lipid membrane anchors. By contrast, topology prediction programs located the amphipathic α helices on the intracellular side. To exclude a reconstitution artifact, we labeled OpuA in intact cells by introducing cysteine residues in the amphipathic α helices and used Cys325 in the NBD as negative control. We find that Cys23 and Cys24 in the first amphipathic α helix of the scaffold are accessible for the membrane-impermeable fluorescein-5-maleimide, whereas Cys325 is not, indicating that the amphipathic helices are, as revealed by the structures, indeed present on the outside (fig. S3B). Guided by the sequences of homologous proteins with and without the scaffold, we deleted this domain but found OpuA inactive (fig. S3C). Next, we analyzed by single-molecule fluorescence resonance energy transfer (smFRET) whether the scaffold undergoes conformational changes during the transport cycle. We stochastically labeled Cys24 with Alexa Fluor 555 and Alexa Fluor 647 maleimide dyes and used alternating laser excitation (ALEX) of OpuA nanodiscs in solution (23) to monitor the FRET signal, which provides a measure for the distance between the residues. The FRET signals were virtually identical for all tested conditions: (i) high salt, (ii) low salt, (iii) turnover conditions, and (iv) turnover conditions in the presence of cyclic-di-AMP (Fig. 2E). Further, no notable structural differences in the scaffold were found when comparing the IF and occluded conformation (Fig. 2D). We therefore hypothesize that the scaffold has a static role and might provide stability, be involved in the docking of the SBD, or thin the membrane locally (24) to facilitate wider IF conformations and enable the NBD or CBS domain to interact with the membrane.
Substrate loading of OpuA
From x-ray structures of the isolated SBD domain from OpuA (25) and other proteins, it is known that the SBD changes its conformation upon substrate binding from open to closed (Fig. 3A) and subsequently docks onto the TMD. However, our cryo-EM studies of OpuA in the presence of glycine betaine indicate that the substrate alone is not enough to trap the SBD in a (stable) docked state (fig. S4). Furthermore, in the occluded conformation, we do not find density for glycine betaine in the SBD. We observe a twist-like conformational change in the open-docked SBD, similar to the distortion in MetQ upon docking and substrate release in the methionine ABC transporter MetNI (26). This results in a disruption of the cation-π interactions between the three tryptophan residues in the binding pocket of the SBD and glycine betaine (Fig. 3A), potentially squeezing the substrate out of the SBD into a pocket in the TMD. At the entry to the TMD, we find two highly conserved phenylalanines and two methionines that might interact with glycine betaine during translocation (Fig. 3B). In the IF conformation, these residues are pointing upward, spaced by ~10 Å, potentially allowing glycine betaine to be coordinated in between, whereas in the occluded conformation, they are turned by 90°, closing the putative substrate entry point.
Fig. 3. Substrate delivery and translocation in OpuA.
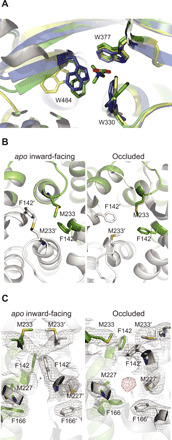
For a global indication of the location with respect to the full map, consult Fig. 2B. (A) Comparison of the x-ray structures of the soluble/isolated OpuA-SBD in an open [yellow; Protein Data Bank (PDB) ID: 3L6G] and a closed substrate-loaded conformation (green; PDB ID: 3L6H) to the docked conformation, as seen in the full-length occluded structure (blue). (B) Comparison of the substrate entry point in the TMD seen from the extracellular side, for the IF conformation (left) and the occluded conformation (right), with residues F142 and M233 presumably closing the substrate entry. (C) G seen from the membrane plane, showing F166 and M227 as well. Unassigned density found in the occluded space of the TMD of the occluded conformation is shown in red (density is shown as mesh at 6σ).
We then examined the occluded conformation for other substrate-binding pockets and found indication for an occluded space in the TMD, just below the extracellular boundary of the membrane. The occluded space is surrounded by phenylalanine, isoleucine, and methionine residues, resulting in a highly hydrophobic environment that coincides with an unassigned density, likely that of glycine betaine (Fig. 3C). We reason that the hydrophobic nature of the binding pocket prevents tight interactions with glycine betaine, similar to what has been proposed for the vitamin B12 importer BtuCD-F (27) but different from the solvent-filled cavity in the membrane domain of the maltose importer MalFGK-E (28). Thereby, substrate release into the cytoplasm is facilitated, allowing the formation of large concentration gradients and the required accumulation of osmolyte to (sub)molar levels under hypertonic stress (29).
Regulation of OpuA by ionic strength
A multiple sequence alignment on different ABC importers, guided by the OpuA structure, shows that the NBDs contain a helix-turn-helix motif (HTH), which is not present in homologs that are not regulated by osmotic stress (Fig. 4A). The motif is located in close proximity to the membrane, consists of two short α helices, and contains a series of positively charged residues that are conserved among osmosensing homologs (Fig. 4, A and B). Moreover, the number of positively charged residues close to the membrane in the ABC transporters ProU and OpuA (Fig. 4A) corresponds qualitatively to the ionic strength activation threshold of the proteins (6, 30). The ionic gating in OpuA requires anionic lipids and is, thus, dependent on the surface charge of the membrane (6, 11, 22, 31). As revealed by the cryo-EM structures, we speculate that the positively charged residues on the HTH interact with the negatively charged lipids. To test this hypothesis, we constructed a mutant in which the positively charged KRIK motif was mutated to AAIA and compared the in vivo glycine betaine uptake activity over a wide range of osmotic (ionic strength) conditions. We find that the mutant has close to maximal activity at low ionic strength and is much less affected by hypertonicity than wild-type OpuA (Fig. 4C).
Fig. 4. Ionic strength sensor and ionic gating of OpuA.
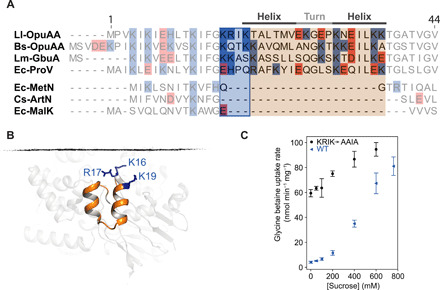
(A) Sequence alignment of part of the NBD with the putative ionic strength sensor. The top three sequences are OpuA homologs that are regulated by ionic strength; the bottom three sequences are nonregulated type 1 ABC importers. KRIK residues are shown in the blue box. Numbering is based on the OpuA sequence. UniProt IDs for the alignment are as follows: Ll-OpuAA, Q9KIF7; Bs-OpuAA, P46920; Lm-GbuA, Q9RR46; Ec-ProV, P14175; Ec-MetN, P30750; Cs-ArtN, Q8RCC2; and Ec-MalK, P68187. (B) HTH region of the NBD in the occluded conformation. The basic residues of KRIK are shown in stick representation in blue. (C) In vivo 14C-glycine betaine uptake by wild-type OpuA (blue triangles) and the KRIK to AAIA mutant (black circles) as a function of osmotic stress (sucrose addition to L. lactis cells), which increases the internal ionic strength. Error bars represent the SD of three independent experiments.
We have previously shown that truncation of the CBS module (11) and mutagenesis of cationic residues on the surface of the CBS domains (22) affect the ionic strength gating of OpuA and, to some extent, the anionic lipid dependence of transport. We have, thus, postulated that the ionic strength sensor would reside in the CBS (11). However, below, we show that the CBS domains with bound cyclic-di-AMP are too distant from the membrane surface to act as an anionic lipid-dependent ionic strength sensor. Yet, we consider it possible that in the absence of cyclic-di-AMP, when a large part of the CBS is natively disordered (22), the CBS may interact with the membrane or with the HTH motif on the NBD. Future smFRET studies may explain the apparent role of the CBS module in ionic strength gating. We now propose that the cationic patch of the HTH in conjunction with an anionic membrane surface forms the osmosensor of OpuA. The interaction between these two partners is modulated by the ambient ionic strength and sets the on/off state of the transporter.
Regulation of OpuA by cyclic-di-AMP
The persistent question on how cyclic-di-AMP regulates the activity of OpuA remains unanswered. We have shown that cyclic-di-AMP stimulates the ATPase activity, while retaining the same dependency on the substrate glycine betaine (Fig. 1C). We also show that cyclic-di-AMP has no impact on the ATPase activity at low ionic strength (less than 250 mM KCl) (Fig. 1D). To elucidate the regulatory effect of cyclic-di-AMP, we reconstituted OpuA in vesicles together with a system for long-term metabolic energy conservation (Fig. 5A) (32). This approach allows us to monitor the impact of cyclic-di-AMP on substrate translocation, instead of ATPase activity alone. Notably, while we show that cyclic-di-AMP stimulates ATPase activity (Fig. 1E), we find that the same messenger inhibits glycine betaine uptake (Fig. 5B).
Fig. 5. Regulation of OpuA by cyclic-di-AMP and proposed mechanism.
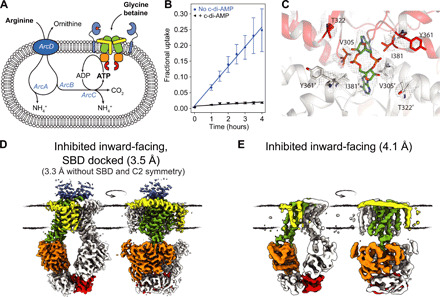
(A) Schematic representation of OpuA reconstituted in vesicles together with the ATP-regenerating system described in (32). (B) Uptake of glycine betaine by OpuA in the vesicles in the presence (black triangles) or absence (blue circles) of 200 μM cyclic-di-AMP. The ATP/ADP (adenosine diphosphate) ratio is kept constant (with ATP ~ 6.5 mM) by the co-reconstituted arginine breakdown pathway (32); error bars represent the SD of three independent experiments. (C) Binding site of cyclic-di-AMP in the substrate-bound cyclic-di-AMP–inhibited IF conformation of OpuA. Density around cyclic-di-AMP is shown as mesh at 6σ. We find one molecule of cyclic-di-AMP coordinated between both CBS domains. (D) Cryo-EM map of the substrate-bound inward-facing cyclic-di-AMP–inhibited conformation of OpuA in 20 mM K-Hepes (pH 7.0) at high ionic strength (+300 mM KCl) and in the presence of 10 μM cyclic-di-AMP, 10 μM MgAMP-PNP, and 100 μM glycine betaine. (E) Cryo-EM map of the IF cyclic-di-AMP–inhibited conformation of OpuA in 50 mM KPi (pH 7.0) at high ionic strength (+200 mM KCl) and in the presence of 10 μM cyclic-di-AMP.
To obtain insights into the transport-inhibited state(s), we determined the structure of the full-length wild-type OpuA under turnover-like conditions (33), namely, at high ionic strength and in the presence of glycine betaine, adenylyl-imidodiphosphate (AMP-PNP) and cyclic-di-AMP (Fig. 5D and fig. S5). We find one of the SBDs docked onto the TMD in a substrate-bound (fig. S5H) closed conformation, different from the open-docked state seen for SBD on the occluded state (Fig. 2B). Although AMP-PNP is bound (fig. S5I), the transporter is stuck in an inward-facing state, rather than the outward-oriented conformation, as found for the occluded state of the E190Q mutant trapped in the presence of ATP. Further, we unambiguously resolve the structure of the CBS domain, with cyclic-di-AMP bound at the interface between the two tandem CBS domains (Fig. 5C) via a V-type interaction (15), which is different from the O-type interaction seen for the isolated CBS domains of the carnitine transporter OpuC (8, 19). The CBS domains are dimerized via a crossover of the polypeptides, whereby the CBS domain of chain 1 interacts with the NBD of chain 2 and vice versa. We now find that the NBDs do not dimerize but are in close proximity, presumably kept in this state by the CBS domain crossover (Fig. 5D).
Last, we determined another structure of OpuA at high ionic strength, in the presence of cyclic-di-AMP alone at 4.1 Å resolution (Fig. 5E and fig. S6). As expected, the SBDs are not resolved, but we find cyclic-di-AMP bound and the two NBDs in the same semiclosed inward-facing state, as observed in the presence of glycine betaine plus AMP-PNP. We propose that the increased ATPase activity in the presence of cyclic-di-AMP (Fig. 1E) is caused by the close proximity of the NBDs. Yet, the CBS domain dimerization does not allow the transporter to open up completely to the intracellular side, which might be required to release the substrate and/or to allow for the SBD to undock and reset the transport cycle, explaining the inhibition of transport by cyclic-di-AMP (Fig. 5B).
Transport cycle of OpuA and conclusions
Using single-particle cryo-EM, complemented with smFRET studies and functional assays, we provide previously unknown insights into the structure, transport cycle, and regulation of OpuA, which is likely representative for other osmoregulatory ABC transporters (30, 34–36). The transport cycle starts with docking of the substrate-bound SBD to a transient narrow OF (outward-facing) conformation. Binding of ATP and dimerization of the NBDs lead to an outward-open state, where glycine betaine loads into a hydrophobic pocket in the TMD. Subsequently, ATP hydrolysis leads to a separation of the NBDs, which induces the transition to an inward-facing state. Full opening to the IF conformation releases the substrate to the cytoplasm and undocks the SBD, resetting the cycle (Fig. 6). The latter step is blocked by the second messenger cyclic-di-AMP (Fig. 6).
Fig. 6. Mechanism of translocation and regulation of transport.
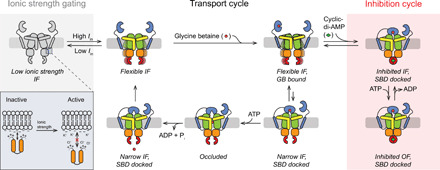
Schematic of the proposed transport cycle of OpuA. At low ionic strength, OpuA is in an inactive state. Ionic strength (~0.2 M) breaks the interaction of the sensor (cationic residues of HTH) with the anionic membrane and activates OpuA (gray shaded area). The transport cycle starts with a flexible IF. The substrate is bound by the SBD (blue), which docks onto the IF conformation. The substrate is translocated into the hydrophobic occluded space inside the outwardly oriented TMD (green). ATP hydrolysis returns the transporter to the IF conformation, and the substrate is pushed into the intracellular environment. The SBD undocks, resetting the transport cycle (unshaded area). In the inhibition cycle, the CBS domains (red) of OpuA dimerize by the binding of cyclic-di-AMP (green). This state leads to the substrate-dependent futile hydrolysis of ATP, as shown in Fig. 1 (C and D), and inhibition of transport, as shown in Fig. 3D.
OpuA activity is controlled by a double brake on different sites, that is, ionic strength and cyclic-di-AMP. Hypertonicity increases the ionic strength inside the cell, which weakens the electrostatic interaction of the cationic HTH with the anionic membrane surface. Under these conditions, OpuA is stimulated, which can lead to a massive accumulation of osmolytes, restoring the turgor and allowing the cell to cope with severe hypertonicity. However, once isotonic conditions have been reached, further uptake of osmolytes needs to be stopped to avoid the internal osmotic pressure reaching lytic values. It has been shown that uncontrolled OpuA can be lethal under low-osmolality conditions (7), indicating that regulation by ionic strength might not be precisely fine-tuned or too slow to stop uptake to (sub)molar levels. Here, cyclic-di-AMP acts as a second gate to prevent unbridled uptake of glycine betaine and rescues the cell from lysis.
MATERIALS AND METHODS
Materials
Common chemicals were of analytical grade and ordered from Sigma-Aldrich Corporation, Carl Roth GmbH & Co. KG, or Merck KGaA. The lipids were obtained from Avanti Polar Lipids Inc. (>99% pure, in chloroform): DOPE (850725C), DOPC (850375C), and DOPG (840475C). n-Dodecyl-β-d-maltoside (DDM) (D97002) was purchased from Glycon Biochemicals GmbH and Triton X-100 (T9284) from Sigma-Aldrich Corporation. 14C-glycine betaine was prepared enzymatically from 14C-choline-chloride [American Radiolabeled Chemicals Inc. (ARC 0208; 55 mCi mmol−1)], as described previously (6).
Expression of OpuA and preparation of membrane vesicles
L. lactis Opu401 containing pNZOpuAhis was cultivated semi-anaerobically at 30°C in a rich medium containing 2% (w/v) gistex LS (Strik BV, Eemnes, NL), 65 mM potassium phosphate (pH 7.0), supplemented with 1% (w/v) glucose, and chloroamphenicol (5 μg ml−1) in a 10-liter bioreactor. The pH was kept at 6.5 with 4 M potassium hydroxide. At an OD600 (optical density at 600 nm) of 2, the nisA promotor was activated by adding 0.05% (v/v) of the supernatant of a nisin-producing strain (NZ9700) (37). After 2 hours of induction, the cells were harvested by centrifugation (15 min, 6000g, 4°C) and resuspended in ice-cold 100 mM KPi (pH 7.0). The cells were centrifuged again and resuspended in 50 mM KPi (pH 7.0) to a final OD of 100 and stored at −80°C.
The cells were broken by two passes at 29,000 psi through a high-pressure device (Constant Systems) in the presence of 2 mM MgSO4 and deoxyribonuclease (DNase) (100 μg ml−1), followed immediately by the addition of 5 mM Na2-EDTA (pH 8.0) and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cell debris was removed by centrifugation (15 min, 22,000g, 4°C), after which the membranes were spun down in an ultracentrifugation step (90 min, 125,000g, 4°C). The membranes were resuspended to a total protein concentration of 10 mg/ml in 50 mM KPi (pH 7.0) with 20% (w/v) glycerol, flash-frozen in liquid nitrogen, and kept at −80°C.
Purification of OpuA
Membrane vesicles containing OpuA were thawed and diluted to a total protein concentration of 3 mg/ml. OpuA was solubilized at 4°C with 0.5% DDM for 60 min, after which the insoluble material was spun down (20 min, 270,000g, 4°C); 0.5 ml column volume of Ni2+-Sepharose resin was equilibrated with 12 column volumes of water and 2 column volumes of buffer [50 mM potassium phosphate (pH 7.0), 200 mM KCl, 15 mM imidazole, and 0.02% DDM]. The supernatant was diluted 2.5 times and incubated with the equilibrated resin for 1 hour at 4°C. The column was then poured, drained, and sequentially washed with 20 column volumes [50 mM potassium phosphate (pH 7.0), 200 mM KCl, 20% glycerol, 50 mM imidazole, and 0.02% DDM]. The protein was eluted in 0.6 column volume for the first and 0.4 column volume for the sequential fractions with elution buffer [50 mM potassium phosphate (pH 7.0), 200 mM KCl, 20% (w/v) glycerol, 500 mM imidazole, and 0.02% DDM]. The purified protein was directly used for reconstitution in nanodiscs or liposomes.
Labeling of OpuA and accessibility of scaffold domain
A small 30-ml M17 culture [with 2% (w/v) glucose and chloroamphenicol (5 μg/ml)] was inoculated with L. lactis Opu401 containing pNZOpuAhis with one of the following cysteine mutations in the OpuA gene: Cys325 in the NBD, or Cys23, Cys24, or Cys25 in the first amphipathic helix of the scaffold domain. The cells were induced at an OD600 of 0.5 to 0.6 with 0.05% (v/v) of the supernatant of a nisin A–producing strain (NZ9700) (37). The culture was grown for another hour and collected by centrifugation (15 min, 6000g, 4°C); washed with 100 mM KPi (pH 7.5), 150 mM KCl, and 20 mM dithiothreitol (DTT); and centrifuged again (15 min, 6000g, 4°C). The cells were resuspended in 1 ml of the same buffer. For labeling, the cells were transferred to 2-ml Eppendorf tubes and centrifuged (3 min, 10,000g, 4°C) and washed twice in the same buffer without DTT. Fluorescein-5-maleimide (2 mM) in DMSO (dimethyl sulfoxide) was added (final DMSO concentration of 5%) and incubated at room temperature for 30 min. The reaction was stopped by addition of 20 mM DTT, and cells were collected by centrifugation (3 min, 10,000g, 4°C) and resuspended in 100 mM KPi (pH 7.5), 150 mM KCl, and 20 mM DTT. Cells were lysed with glass beads in a TissueLyser (Qiagen) for 5 min at 50 Hz. Lysed cells were diluted 4× with 100 mM KPi (pH 7.5), 150 mM KCl, and 20 mM DTT, and membrane fractions were collected by ultracentrifugation (15 min, 264,000g, 4°C), resuspended in 1 ml of the same buffer, and loaded on a 12.5% SDS–polyacrylamide gel electrophoresis (PAGE) gel. The gel was imaged on a LAS-3000 imager.
Purification of MSP1D1
The pMSP1D1 vector was freshly transformed into Escherichia coli BL21 DE3. The cells were grown aerobically in 1-liter baffled flasks using LB broth [1% (w/v) NaCl, 1% (w/v) bacto tryptone, and 0.5% (w/v) bacto-yeast extract with kanamycin (30 μg/ml)]. At an OD600 of 0.8, the cells were induced with 1 mM IPTG (isopropyl-β-d-1-thiogalactopyranoside). After induction, the cells were allowed to grow for 3.5 hours, after which they were harvested by centrifugation (15 min, 6000g, 4°C). The pellet was resuspended in 100 mM KPi (pH 7.8) and centrifuged again (15 min, 6000g, 4°C). After resuspending in the same buffer, the cells were stored at −80°C.
Cells were thawed on ice and supplemented with DNase (100 μg/ml) and 1 mM PMSF. The cells were broken by sonication (70% amplitude, total on time: 6 min; 5 s on, 5 s off). Triton X-100 [1% (w/v)] was added to the lysate and stirred at room temperature for 10 min. Unsolubilized material was spun down by centrifugation (30 min, 30,000g, 4°C). Column volume (7.5 ml) of Ni2+-Sepharose resin was equilibrated with 4 column volumes of water and 4 column volumes of buffer [50 mM potassium phosphate (pH 7.8)]. The resin was incubated together with the supernatant for 1 hour at 4°C. The column was poured, drained, and washed sequentially with 4 column volumes of the following buffers: (i) 40 mM tris-HCl (pH 8.0), 300 mM NaCl, and 1% Triton X-100; (ii) 40 mM tris-HCl (pH 8.0), 300 mM NaCl, 50 mM Na-cholate, and 20 mM imidazole; and (iii) 40 mM tris-HCl (pH 8.0), 300 mM NaCl, and 50 mM imidazole. MSP1D1 was eluted in 12 fractions of 2 ml with elution buffer [40 mM tris-HCl (pH 8.0), 300 mM NaCl, and 500 mM imidazole] and dialyzed overnight at 4°C against 20 mM tris-HCl, 100 mM NaCl, and 0.5 mM EDTA. MSP1D1 was then aliquoted, flash-frozen, and stored at −80°C.
Reconstitution of OpuA in MSP1D1 nanodiscs
For reconstitution in nanodiscs, different OpuA/MSP/lipid ratios were tested. For optimal conditions, 8.6 μM purified OpuA was mixed with 86 μM purified MSP1D1 scaffold protein and 4.3 mM lipids (with a composition of 50% DOPE, 12% DOPC, and 38% DOPG) in 75 mM potassium phosphate (pH 7.0) with 7% (v/v) glycerol and 12 mM DDM to a total volume of 700 μl. After nutating the mixture for an hour at 4°C, 500 mg of SM-2 Bio-Beads (Bio-Rad) was added to adsorb the detergent. The mixture was then allowed to incubate overnight. In the morning, the supernatant was separated from the Bio-Beads and the OpuA nanodiscs were purified by size exclusion chromatography using a Superdex 200 increase 10/300 GL column in 50 mM potassium phosphate, 200 mM KCl, or 20 mM K-Hepes (pH 7.0) plus 300 mM KCl.
ATPase activity assays
As detailed in (12), the ATPase activity of the transporter was determined by a coupled enzyme assay where the NADH absorbance decrease at 340 nm is stoichiometrically coupled to the ATPase activity. The NADH absorbance is coupled to the ATP hydrolysis by the reactions catalyzed by pyruvate kinase and lactate dehydrogenase (Fig. 1B); the enzymes in terms of activity were present in excess of OpuA. The assay was performed in 96-well plates. A Tecan Spark 10-m plate reader was used to measure the NADH absorbance over time. Each well contains 50 mM Hepes (pH 7), 4 mM phosphoenolpyruvate, 600 μM NADH, 2.1 to 3.5 U of pyruvate kinase, and 3.2 to 4.9 U of lactate dehydrogenase. The wells were then supplemented with 100 μM glycine betaine, 300 mM KCl, 10 μM cyclic-di-AMP, and 10 mM MgATP unless specified differently. The 10 mM MgATP was used to start the reaction. In the case of variable ATP concentrations, the assay was started by titrating in 100 μM glycine betaine. We normalized the ATP hydrolysis activities, and a value of 1 corresponds to 200 to 1200 nmol of ATP hydrolyzed per minute × nanomoles of OpuA, depending on the efficiency of nanodisc reconstitution; for each set of experiments, the ATPase activity was normalized to one condition that was identical in all biological replicates.
Cryo-EM sample preparation and data acquisition
Freshly prepared OpuA nanodiscs were concentrated to at least 1 mg/ml using a Vivaspin 500 (50 kDa cutoff) concentrator. Before freezing, OpuA was diluted to 1 mg/ml and respective additives were added (e.g., 100 μM glycine betaine, 10 μM cyclic-di-AMP, 10 μM MgAMP-PNP, or 10 mM MgATP). Holey carbon grids (Quantifoil; Au R1.2/1.3, 300 mesh) were glow-discharged for 20 s at 5 mA. Sample (2.8 μl) was applied on the grids, after which they were blotted for 3 to 5 s in a Vitrobot Mark IV (Thermo Fisher Scientific) at 15° to 22°C and 100% humidity and plunge-frozen into a liquid ethane/propane mixture. The grids were then stored in liquid nitrogen until further use.
Data collection was performed in-house on a 200-keV Talos Arctica microscope (Thermo Fisher Scientific) equipped with a K2 and a post-column BioQuantum energy filter (Gatan) operated in zero-loss mode, with a 20-eV slit, and a 100-μm objective aperture. Automatic collection was done at a calibrated magnification of 49,407× (1.012 Å pixel size) and a nominal defocus range from −0.8 to −1.9 μm using EPU (Thermo Fisher Scientific). Holes were selected with the help of an in-house–written script that calculates ice thickness within DigitalMicrograph (Gatan). Each movie consisted of 60 frames with a total exposure time of 9 s and a dose of 53 electrons/Å2 (0.883 e−/Å2 per frame). FOCUS software (38) was used for on-the-fly quality control, and settings were adjusted if necessary.
Image processing
See figs. S1, S2, and S4 to S6 for a graphical overview on respective datasets. The following detailed description applies for the cryo-EM dataset acquired for the OpuA apo inward-facing sample, but a similar workflow was used for the other datasets. A total of 2861 movies were recorded, and on-the-fly data assessment was conducted using FOCUS 1.1.0 (38). MotionCor2_1.2.1 (39) was used to correct for beam-induced motion, and ctffind4.1.8 (40) was used for estimation of the contrast transfer function (CTF) parameters. Micrographs containing ice or aggregates, micrographs with a low-resolution estimation of the CTF fit (>4 Å), and micrographs out of the defocus range of 0.5 to 2 μm were discarded. The remaining 2196 micrographs were used for further processing. First, crYOLO 1.3.1 (41) was used to automatically pick 1,383,502 particles using a loose threshold. Particle coordinates were imported in RELION 3.0.8 (42), and the particles were extracted with a box size of 256 pixels and binned 2×, yielding a pixel size of 2.024. A total of 786,018 particles were selected after several rounds of two-dimensional (2D) classification, to exclude false positives. For this dataset, an initial reference was generated in RELION. The particle set was further cleaned by several rounds of 3D classifications using the then current best maps, low-pass–filtered to 40 Å, as references in an iterative fashion. Both 2D and 3D classifications were performed, ignoring the CTF until the first peak. The resulting 434,607 particles from the best class(es) were re-extracted with a box size of 256 pixels and a pixel size of 1.012 and used for 3D autorefinement with the unbinned 3D class as reference and, for later refinements, the refined map as reference, yielding a 4.0-Å map. A final 3D classification without image alignment was performed to separate different opening angles, now with the reference low-pass–filtered to 20 Å. A total of 78,021 high-quality particles were used for 3D autorefinement. This refinement was continued as a focused refinement with a mask, excluding most of the lipid bilayer and the MSP1D1 belt protein, improving the resolution to 3.5 Å at convergence. C2 symmetry was imposed during the latter autorefinement, and the obtained maps were used as subsequent references (low-pass–filtered to 40 Å) in an iterative fashion. Postprocessing was done to mask and sharpen the final map, yielding a resolution of 3.5 Å. Sequentially, several rounds of CTF refinement (42) were performed, using per-particle CTF estimation, increasing the resolution to 3.3 Å. Local resolutions were determined using the RELION local resolution algorithm. The 0.143 cutoff criterion (43) was applied for all resolution estimations, using gold-standard FSC (Fourier shell correlation) between both independently refined half-maps (44). The 3DFSC web server (45) was used to estimate directional resolution anisotropy of the density maps.
Similar approaches were taken for the other datasets. In short, for the occluded dataset, 2759 movies were recorded and 2608 were used to select 572,496 particles. Several rounds of 2D and 3D classifications yielded 479,186 and 90,870 high-quality particles, respectively. After sequential 3D autorefinement, CTF refinement, and postprocessing without imposed symmetry, the resolution was 3.4 Å. A 3D classification without image alignment was performed to separate two different orientations of the SBD that were otherwise merged together. Both classes were independently refined (3.5 and 3.6 Å, respectively), after which the nanodisc density was removed using particle subtraction. Both sets were recombined and used for a final refinement and postprocessing with a mask (3.4 Å). Notably, the docking of a single SBD disrupts the twofold symmetry of the transporter. While the unsymmetrized cryo-EM map at a global resolution of 3.4 Å allows an unambiguous modeling of the SBD and the respective anchoring helix, excluding the SBD by focused refinement and imposing a C2 symmetry during image processing further improved the resolution to 3.2 Å in the remaining regions.
For the glycine betaine–only dataset, 1713 movies were recorded and 901 were used to select 615,299 particles. Several rounds of 2D and 3D classifications yielded 172,260 and 30,077 high-quality particles, respectively. After sequential 3D autorefinement and postprocessing with imposed C2 symmetry, the resolution was 4.5 Å. CTF refinement did not improve the resolution further.
For the cyclic-di-AMP–inhibited inward-facing, SBD-docked dataset, 8804 movies were recorded and 7788 were used to select 1,344,075 particles. Several rounds of 2D and 3D classifications yielded 303,810 and 110,161 high-quality particles, respectively. After sequential 3D autorefinement, CTF refinement, and postprocessing without imposed symmetry, the resolution was 3.5 Å; processing without the SBD and C2 symmetry applied yielded a 3.3-Å map.
For the cyclic-di-AMP–inhibited inward-facing dataset, 1517 movies were recorded and 1066 were used to select 657,116 particles. Several rounds of 2D and 3D classifications yielded 133,761 and 13,520 high-quality particles, respectively. After sequential 3D autorefinement, CTF refinement, and postprocessing with imposed C2 symmetry, the resolution was 4.1 Å.
Model building
The model building was done in COOT (46). The resolutions of the maps were of sufficient quality to unambiguously build the model (figs. S1, S2, and S4 to S6 and table S1). The TMD was de novo built, while for the SBD, known crystal structures [Protein Data Bank (PDB): 3L6G and 3L6H] were used as reference (25). For the NBD, the structure of the homolog methionine transporter MetNI (PDB: 6CVL) was used as a starting point (26). The CBS domain was built with the carnitine transporter OpuCA CBS domain (PDB: 5KS7) as reference (8). The models were optimized by iterative rounds of Phenix real-space refinement (47) and manual inspection and corrections of the output models against the map in COOT.
Fourier shell cross-correlation (FSCsum) between model and map was used to determine the quality of the fit. To prevent potential overfitting, random shifts (up to 0.5 Å) were applied to the coordinates of the final model, followed by refinement against the first unfiltered half-map. This provided FSCwork, and the FSC against the second half-map, which was not used at any point during refinement, FSCfree, should have a marginal gap between them, indicating no overfitting of the model. MolProbity (48) was used to validate the chemo-physical properties of the final model. Images were prepared with the PyMOL Molecular Graphics System, version 2.0 Schrödinger LLC, Chimera (49), and ChimeraX (50). In addition, the SBGrid (51) software package tool was used to manage the software packages.
Labeling of OpuA for smFRET
Stochastic labeling was performed with the dyes Alexa555 and Alexa647 maleimide (Thermo Fisher Scientific). OpuA (5 to 10 nmol) was first treated with 10 mM DTT for 10 min to fully reduce oxidized cysteines. After dilution of the protein sample to a DTT concentration of 1 mM, the reduced protein was immobilized on 200 μl of Ni2+-Sepharose resin and washed with 2 ml of buffer A [20 mM K-Hepes (pH 7.0) and 300 mM KCl] to remove the DTT. The resin was incubated in 1 ml of buffer A supplemented with 50 nmol of Alexa555 and 50 nmol of Alexa647 for 2 to 4 hours at 4°C. Subsequently, unbound dyes were removed by washing the column with 3 to 4 ml of buffer A. Elution of the proteins was done by supplementing buffer A with 200 mM imidazole. The labeled protein was further purified by size exclusion chromatography (Superdex 200, GE Healthcare) using buffer A. Sample composition was assessed by recording the absorbance at 280 nm (protein), 559 nm (Alexa555), and 645 nm (Alexa647) to estimate the labeling efficiency. The labeling efficiency was typically about 70%.
smFRET
Solution-based smFRET and ALEX (52) experiments were carried out at 5 to 25 pM labeled protein at room temperature in buffer A supplemented with additional reagents, as stated in the text and previously described (53). Microscope cover slides (no. 1.5H precision cover slides, VWR Marienfeld) were coated with bovine serum albumin (BSA; 1 mg/ml) for 30 to 60 s to prevent fluorophore and/or protein interactions with the glass material. Excess BSA was subsequently removed by washing and exchanged with buffer A. All smFRET experiments were performed using a home-built confocal microscope. Briefly, two laser diodes (Coherent OBIS) with emission wavelengths of 532 and 637 nm were directly modulated for alternating periods of 50 μs and used for confocal excitation. The laser beams were coupled into a single-mode fiber (PM-S405-XP, Thorlabs) and collimated (MB06, Q-Optics/Linos) before entering an oil immersion objective [60×; NA (numerical aperture) of 1.35; UPlanSAPO 60XO, Olympus]. The fluorescence was collected by excitation at a depth of 20 μm. Average laser powers were 30 μW at 532 nm (~30 kW/cm2) and 15 μW at 637 nm (~15 kW/cm2). Excitation and emission light were separated by a dichroic beam splitter (zt532/642rpc, AHF Analysentechnik), which is mounted in an inverse microscope body (IX71, Olympus). Emitted light was focused onto a 50-μm pinhole and spectrally separated (640DCXR, AHF Analysentechnik) onto two single-photon avalanche diodes (TAU-SPADs-100, PicoQuant) with appropriate spectral filtering (donor channel, HC582/75; acceptor channel, Edge Basic 647LP; AHF Analysentechnik). Registration of photon arrival times and alternation of the lasers were controlled by an NI card (PXI-6602, National Instruments).
Analysis of the photon arrival times was done as described in (54). Briefly, to identify fluorescence bursts, a “dual-channel burst search” (55) was used, as done previously (54). Three photon counts per burst were measured: NDA (acceptor emission upon donor excitation), NDD (donor emission upon donor excitation), and NAA (acceptor emission upon acceptor excitation) and assignment are based on the excitation period and detection channel (52). The photon counts were corrected for background, as done previously (56). The apparent FRET efficiency was calculated as NDA/(NDA + NDD), and the stoichiometry S was calculated as (NDA + NDD)/(NDA + NDD + NAA) (52). Binning the detected bursts into 2D apparent FRET versus stoichiometry histograms allowed the selection of the donor- and acceptor-labeled molecules and reduce artifacts arising from fluorophore bleaching (52). The selected 1D apparent FRET histograms were fitted with a Gaussian distribution, using the method of least squares, to obtain a 95% confidence interval for the mean.
Co-reconstitution of ArcD2 and OpuA in liposomes
On the basis of the method described in (57), synthetic lipids were mixed from chloroform stocks in the ratio of 50 mol % DOPE, 12 mol % DOPC, and 38 mol % DOPG, after which they were dried in a rotary vacuum setup (Büchi Labortechnik AG). The dried lipids were dissolved in diethylether, dried again, and rehydrated in 50 mM KPi (pH 7.0) with 200 mM KCl. Last, the mixture was sonicated using a tip sonicator (Sonics and Materials Inc.) (16 cycles, 70% amplitude, 15 s on, 45 s off) and frozen-thawed three times in liquid nitrogen. The liposomes were diluted five times to a final lipid concentration of 4 mg/ml after which they were destabilized by titrating with 10% Triton, until 60% of the initial absorbance after Rsat was reached (57). The proteins and destabilized liposomes were mixed to obtain a ArcD2:OpuA:lipid ratio of 1:2:400 (w/w). After 15 min of mixing at 4°C, the detergent was removed by adding four portions of 200 mg of SM-2 Bio-Beads (10 mg of SM-2 Bio-Beads/mg of lipids) after 15-, 30-, 45-min, and overnight incubation. The last addition was followed by 2 hours of incubation at 4°C, after which the proteoliposomes were collected by ultracentrifugation (2 hours, 125,000g, 4°C). The proteoliposomes were resuspended to a final lipid concentration of 100 mg/ml in 50 mM KPi (pH 7.0).
Encapsulation of the arginine breakdown pathway
The enzymes needed for arginine breakdown and ATP (re)generation were incorporated in the vesicles as previously described (32). In short, the proteoliposomes (66 μl, 6.6 mg of lipid) containing ArcD2 and OpuA were mixed in 50 mM KPi (pH 7.0) with 1 μM ArcA, 2 μM ArcB, 5 μM ArcC1, 5 mM ADP (adenosine diphosphate), 5 mM MgSO4, 0.5 mM ornithine, and, optionally, 10 μM cyclic-di-AMP in a total volume of 200 μl; the final liposome concentration was 33 mg of lipid per milliliter. The final internal medium was composed of 50 mM KPi (pH 7.0) (plus 25 mM NaCl carried over with the purified ArcA, ArcB, and ArcC1) (32). The enzymes and metabolites were encapsulated by five freeze-thaw cycles in liquid nitrogen. After that, the vesicles were extruded 13 times through a 400-nm pore size polycarbonate filter with a prewashed extruder in 100 mM KPi (pH 7.0) supplemented with 0.5 mM ornithine. The vesicles were collected by centrifugation (20 min, 325,000g, 4°C), resuspended in 100 mM KPi (pH 7.0) supplemented with 0.5 mM ornithine, and centrifuged again (20 min, 325,000g, 4°C). Last, the vesicles were resuspended to a final concentration of 165 mg of lipid per milliliter and kept on ice before use in the transport assays.
In vitro transport assays
As described in (32), the vesicles with encapsulated enzymes and metabolites were diluted to a final concentration of 1.67 mg ml−1 in 100 mM KPi (pH 7.0) supplemented with 250 mM KCl and 180 μM glycine betaine, of which 2% (mol/mol) was 14C-radiolabeled. The mixture was incubated for 30 min at 30°C, and a time zero point was taken. The internal ATP production was then started by the addition of 20 mM arginine, and samples of 50 to 100 μl were taken at given time intervals (0 to 5 hours). Samples were immediately diluted in 2 ml of ice-cold quenching buffer [100 mM KPi (pH 7.0)] and filtered over 0.45-μm pore size cellulose nitrate filters to stop the reaction. The filters were then washed with another 2 ml of the same buffer. Radioactivity on the filter was quantified by liquid scintillation counting using Ultima Gold MV scintillation fluid (PerkinElmer) and a Tri-Carb 2800TR scintillation counter (PerkinElmer). Although the pore size of the filters is larger than the diameter of the vesicles, more than 99% of the vesicles are retained (11).
In vivo transport assays
For in vivo uptake assays, cells from strain L. lactis Opu401 carrying the pNZOpuAHis or pNZOpuAhis_K16R17K19A plasmid were grown in M17 supplemented with 1% glucose and chloramphenicol (5 μg/ml). Moderate OpuA expression was induced for 1 hour with 1 10−3% (v/v) of the supernatant of a nisin-producing strain (NZ9700) [Kuipers et al. (37)]. Afterward, the cells were washed twice with, and subsequently diluted to an OD600 of 50 in, ice-cold 50 mM Hepes (pH 7.3). To get linear uptake curves, cells diluted to 0.4 mg of total protein per milliliter were pre-energized for 5 min at 30°C in buffer supplemented with 10 mM glucose. 14C-glycine betaine uptake was initiated with prewarmed 50 mM Hepes (pH 7.3), supplemented with 10 mM glucose, 14C-glycine betaine (1 mM end concentration), and the required sucrose concentration. Acquisition of data points was done similar to what is described for the liposome uptake measurements. Note that quenching of each reaction is done in an ice-cold isotonic buffer. Afterward, leftover cells were broken by shaking in the presence of glass beads (0.1 mm Ø). Approximately 20 μg of the total protein fraction was loaded on gel. To get the membrane fraction, the broken cells were spun down for 12 min at 14,202g at 4°C. The membrane fraction of 0.45 mg of total protein was loaded on gel.
Supplementary Material
Acknowledgments
We thank D.-J. Slotboom for critical reading of the manuscript and M. Punter for IT support. Structural biology applications used in this project were compiled and configured by SBGrid (51). Funding: The work was funded by the ERC Advanced Grant (ABCvolume; no. 670578) to B.P., the NWO Veni grant (722.017.001) to C.P., and the NWO Start-Up grant (740.018.016) to C.P. Author contributions: H.R.S., C.P., and B.P. designed the research; H.R.S. performed the majority of the experiments; H.R.S. and J.R. performed cryo-EM analysis; M.v.d.N. and G.K.S.-W. performed in vivo transport assays; M.d.B. performed smFRET measurements; S.T.K. performed labeling studies; H.R.S., J.R., C.P., and B.P. performed data analysis; and H.R.S., C.P., and B.P. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All data, code, and materials used in the analysis are available upon request to the lead author. The five cryo-EM density maps, OpuA apo inward-facing, OpuA (E190Q) occluded, OpuA in the presence of glycine betaine inward-facing, OpuA-inhibited inward-facing, and OpuA-inhibited inward-facing SBD docked, have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-11782, EMD-11783, EMD-11785, EMD-11784, and EMD-11786, respectively. The deposition includes the cryo-EM map, both half-maps, and the mask used for final FSC calculation. Raw cryo-EM data will be deposited in in the Electron Microscopy Public Image Archive (EMPIAR). Coordinates of four models, OpuA apo inward-facing, OpuA (E190Q) occluded, OpuA-inhibited inward-facing, and OpuA-inhibited inward-facing SBD docked, have been deposited in the Protein Data Bank with accession numbers 7AHC, 7AHD, 7AHE, and 7AHH, respectively.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/47/eabd7697/DC1
REFERENCES AND NOTES
- 1.Wood J. M., Bacterial osmoregulation: A paradigm for the study of cellular homeostasis. Annu. Rev. Microbiol. 65, 215–238 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Blount P., Iscla I., Life with bacterial mechanosensitive channels, from discovery to physiology to pharmacological target. Microbiol. Mol. Biol. Rev. 84, e00055-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Culham D. E., Shkel I. A., Record M. T., Wood J. M., Contributions of coulombic and hofmeister effects to the osmotic activation of Escherichia coli transporter ProP. Biochemistry 55, 1301–1313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krämer R., Morbach S., BetP of Corynebacterium glutamicum, a transporter with three different functions: Betaine transport, osmosensing, and osmoregulation. Biochim. Biophys. Acta 1658, 31–36 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Ressl S., Terwisscha van Scheltinga A. C., Vonrhein C., Ott V., Ziegler C., Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature 458, 47–52 (2009). [DOI] [PubMed] [Google Scholar]
- 6.van der Heide T., Stuart M. C., Poolman B., On the osmotic signal and osmosensing mechanism of an ABC transport system for glycine betaine. EMBO J. 20, 7022–7032 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devaux L., Sleiman D., Mazzuoli M.-V., Gominet M., Lanotte P., Trieu-Cuot P., Kaminski P.-A., Firon A., Cyclic di-AMP regulation of osmotic homeostasis is essential in Group B Streptococcus. PLOS Genet. 14, e1007342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huynh T. N., Choi P. H., Sureka K., Ledvina H. E., Campillo J., Tong L., Woodward J. J., Cyclic di-AMP targets the cystathionine beta-synthase domain of the osmolyte transporter OpuC. Mol. Microbiol. 102, 233–243 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuster C. F., Bellows L. E., Tosi T., Campeotto I., Corrigan R. M., Freemont P., Gründling A., The second messenger c-di-AMP inhibits the osmolyte uptake system OpuC in Staphylococcus aureus. Sci. Signal. 9, ra81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locher K. P., Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Biemans-Oldehinkel E., Mahmood N. A. B. N., Poolman B., A sensor for intracellular ionic strength. Proc. Natl. Acad. Sci. U.S.A. 103, 10624–10629 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karasawa A., Swier L. J. Y. M., Stuart M. C. A., Brouwers J., Helms B., Poolman B., Physicochemical factors controlling the activity and energy coupling of an ionic strength-gated ATP-binding cassette (ABC) transporter. J. Biol. Chem. 288, 29862–29871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witte G., Hartung S., Büttner K., Hopfner K.-P., Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell 30, 167–178 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Commichau F. M., Gibhardt J., Halbedel S., Gundlach J., Stülke J., A delicate connection: c-di-AMP affects cell integrity by controlling osmolyte transport. Trends Microbiol. 26, 175–185 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Corrigan R. M., Campeotto I., Jeganathan T., Roelofs K. G., Lee V. T., Gründling A., Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 9084–9089 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He J., Yin W., Galperin M. Y., Chou S.-H., Cyclic di-AMP, a second messenger of primary importance: Tertiary structures and binding mechanisms. Nucleic Acids Res. 48, 2807–2829 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeden M. S., Schuster C. F., Bowman L., Zhong Q., Williams H. D., Gründling A., Cyclic di-adenosine monophosphate (c-di-AMP) is required for osmotic regulation in Staphylococcus aureus but dispensable for viability in anaerobic conditions. J. Biol. Chem. 293, 3180–3200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia P., Wang S., Xiong Z., Zhu X., Ye B., Du Y., Meng S., Qu Y., Liu J., Gao G., Tian Y., Fan Z., The ER membrane adaptor ERAdP senses the bacterial second messenger c-di-AMP and initiates anti-bacterial immunity. Nat. Immunol. 19, 141–150 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Pham H. T., Nhiep N. T. H., Vu T. N. M., Huynh T. N., Zhu Y., Huynh A. L. D., Chakrabortti A., Marcellin E., Lo R., Howard C. B., Bansal N., Woodward J. J., Liang Z.-X., Turner M. S., Enhanced uptake of potassium or glycine betaine or export of cyclic-di-AMP restores osmoresistance in a high cyclic-di-AMP Lactococcus lactis mutant. PLOS Genet. 14, e1007574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biemans-Oldehinkel E., Poolman B., On the role of the two extracytoplasmic substrate-binding domains in the ABC transporter OpuA. EMBO J. 22, 5983–5993 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patzlaff J. S., van der Heide T., Poolman B., The ATP/substrate stoichiometry of the ATP-binding cassette (ABC) transporter OpuA. J. Biol. Chem. 278, 29546–29551 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Karasawa A., Erkens G. B., Berntsson R. P.-A., Otten R., Schuurman-Wolters G. K., Mulder F. A. A., Poolman B., Cystathionine β-synthase (CBS) domains 1 and 2 fulfill different roles in ionic strength sensing of the ATP-binding cassette (ABC) transporter OpuA. J. Biol. Chem. 286, 37280–37291 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee N. K., Kapanidis A. N., Wang Y., Michalet X., Mukhopadhyay J., Ebright R. H., Weiss S., Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys. J. 88, 2939–2953 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grage S. L., Afonin S., Kara S., Buth G., Ulrich A. S., Membrane thinning and thickening induced by membrane-active amphipathic peptides. Front. Cell Dev. Biol. 4, 46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolters J. C., Berntsson R. P.-A., Gul N., Karasawa A., Thunnissen A.-M. W. H., Slotboom D.-J., Poolman B., Ligand binding and crystal structures of the substrate-binding domain of the ABC transporter OpuA. PLOS ONE 5, e10361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen P. T., Lai J. Y., Lee A. T., Kaiser J. T., Rees D. C., Noncanonical role for the binding protein in substrate uptake by the MetNI methionine ATP binding cassette (ABC) transporter. Proc. Natl. Acad. Sci. U.S.A. 115, E10596–E10604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korkhov V. M., Mireku S. A., Locher K. P., Structure of AMP-PNP-bound vitamin B12 transporter BtuCD–F. Nature 490, 367–372 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Oldham M. L., Khare D., Quiocho F. A., Davidson A. L., Chen J., Crystal structure of a catalytic intermediate of the maltose transporter. Nature 450, 515–521 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Glaasker E., Konings W. N., Poolman B., Glycine betaine fluxes in Lactobacillus plantarum during osmostasis and hyper- and hypo-osmotic shock. J. Biol. Chem. 271, 10060–10065 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Gul N., Poolman B., Functional reconstitution and osmoregulatory properties of the ProU ABC transporter from Escherichia coli. Mol. Membr. Biol. 30, 138–148 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Poolman B., Spitzer J. J., Wood J. M., Bacterial osmosensing: Roles of membrane structure and electrostatics in lipid–protein and protein–protein interactions. Biochim. Biophys. Acta 1666, 88–104 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Pols T., Sikkema H. R., Gaastra B. F., Frallicciardi J., Śmigiel W. M., Singh S., Poolman B., A synthetic metabolic network for physicochemical homeostasis. Nat. Commun. 10, 4239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofmann S., Januliene D., Mehdipour A. R., Thomas C., Stefan E., Brüchert S., Kuhn B. T., Geertsma E. R., Hummer G., Tampé R., Moeller A., Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 571, 580–583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teichmann L., Kümmel H., Warmbold B., Bremer E., OpuF, a new Bacillus compatible solute ABC transporter with a substrate-binding protein fused to the transmembrane domain. Appl. Environ. Microbiol. 84, e01728-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teichmann L., Chen C., Hoffmann T., Smits S. H. J., Schmitt L., Bremer E., From substrate specificity to promiscuity: Hybrid ABC transporters for osmoprotectants. Mol. Microbiol. 104, 761–780 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Whitley P., Zander T., Ehrmann M., Haardt M., Bremer E., von Heijne G., Sec-independent translocation of a 100-residue periplasmic N-terminal tail in the E. coli inner membrane protein proW. EMBO J. 13, 4653–4661 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuipers O. P., de Ruyter P. G. G. A., Kleerebezem M., de Vos W. M., Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64, 15–21 (1998). [Google Scholar]
- 38.Biyani N., Righetto R. D., McLeod R., Caujolle-Bert D., Castano-Diez D., Goldie K. N., Stahlberg H., Focus: The interface between data collection and data processing in cryo-EM. J. Struct. Biol. 198, 124–133 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D., Quentin D., Roderer D., Tacke S., Siebolds B., Schubert E., Shaikh T. R., Lill P., Gatsogiannis C., Raunser S., SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J., Lindahl E., Scheres S. H., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenthal P. B., Henderson R., Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Scheres S. H. W., Chen S., Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan Y. Z., Baldwin P. R., Davis J. H., Williamson J. R., Potter C. S., Carragher B., Lyumkis D., Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T. E., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morin A., Eisenbraun B., Key J., Sanschagrin P. C., Timony M. A., Ottaviano M., Sliz P., Cutting edge: Collaboration gets the most out of software. eLife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kapanidis A. N., Lee N. K., Laurence T. A., Doose S., Margeat E., Weiss S., Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. U.S.A. 101, 8936–8941 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gouridis G., Schuurman-Wolters G. K., Ploetz E., Husada F., Vietrov R., de Boer M., Cordes T., Poolman B., Conformational dynamics in substrate-binding domains influences transport in the ABC importer GlnPQ. Nat. Struct. Mol. Biol. 22, 57–64 (2015). [DOI] [PubMed] [Google Scholar]
- 54.de Boer M., Gouridis G., Vietrov R., Begg S. L., Schuurman-Wolters G. K., Husada F., Eleftheriadis N., Poolman B., McDevitt C. A., Cordes T., Conformational and dynamic plasticity in substrate-binding proteins underlies selective transport in ABC importers. eLife 8, e44652 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nir E., Michalet X., Hamadani K. M., Laurence T. A., Neuhauser D., Kovchegov Y., Weiss S., Shot-noise limited single-molecule FRET histograms: Comparison between theory and experiments. J. Phys. Chem. B 110, 22103–22124 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Boer M., Gouridis G., Muthahari Y. A., Cordes T., Single-molecule observation of ligand binding and conformational changes in FeuA. Biophys. J. 117, 1642–1654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geertsma E. R., Nik Mahmood N. A. B., Schuurman-Wolters G. K., Poolman B., Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc. 3, 256–266 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/47/eabd7697/DC1


