Abstract
Aging is a complex biological process closely linked with the occurrence and development of age-related diseases. Despite recent advances in lifestyle management and drug therapy, the late diagnosis of these diseases causes severe complications, usually resulting in death and consequently impacting social economies. Therefore, the identification of reliable biomarkers and the creation of effective treatment alternatives for age-related diseases are needed. Circular RNAs (circRNAs) are a novel class of RNA molecules that form covalently closed loops capable of regulating gene expression at multiple levels. Several studies have reported the emerging functional roles of circRNAs in various conditions, providing new perspectives regarding cellular physiology and disease pathology. Notably, accumulating evidence demonstrates the involvement of circRNAs in the regulation of age-related pathologies, including cardio-cerebrovascular disease, neurodegenerative disease, cancer, diabetes, rheumatoid arthritis, and osteoporosis. Therefore, the association of circRNAs with these age-related pathologies highlights their potential as diagnostic biomarkers and therapeutic targets for better disease management. Here, we review the biogenesis and function of circRNAs, with a special focus on their regulatory roles in aging-related pathologies, as well as discuss their potential as biological biomarkers and therapeutic targets for these diseases.
Keywords: circRNAs, biomarker, aging, age-related diseases
1. Introduction
Aging is an ineluctable biological process primarily regulated by several evolutionary conserved mechanisms [1, 2]. It is characterized by a progressive loss of physiological integrity caused by the cellular and molecular damage accumulation, resulting in impaired bodily functions and increased susceptibility to diseases [3, 4]. Despite accumulating evidence demonstrating that aging is a major risk factor for human diseases, the molecular mechanisms underlying this process and its link to these diseases are unknown [2, 5]. Therefore, it is important to identify the molecules that play key roles in the aging process, as well as their function in the development of age-related diseases.
Circular RNAs (circRNAs), a novel type of universal and diverse endogenous transcripts that has been a recent focus in the transcriptomics field, were first identified in viroid in 1976 [6] and first observed through an electron microscope in the cytoplasm of eukaryotic cell in 1979 [7]. CircRNAs form covalently closed loop structures with neither 5′-3′ polarities nor polyadenylated tails, and therefore, are more stable than linear RNAs and insusceptible to degradation by RNA exonuclease or RNase R [8-10]. Subsequent reports revealed that circRNAs can act as miRNA sponges, transcriptional regulators, binding partners of proteins, or even translated into functional proteins [11-15]. Furthermore, circRNAs are abundant, relatively stable, specifically expressed in tissues, and evolutionary conserved among species, affording them the potential to be biomarkers for human diseases [8, 16, 17].
Recent studies have identified several circRNAs as regulators of various pathways that are involved in aging and cellular senescence [18-21]. In particular, dysregulated circRNAs were implicated in the pathophysiology of age-related diseases, including cardio-cerebrovascular disease, neurodegenerative disease, cancer, diabetes, rheumatoid arthritis, and osteoporosis [22-24]. Here, we review the biogenesis and function of circRNAs and their potential as biomarkers of age-related diseases.
2. Biogenesis and function of circRNAs
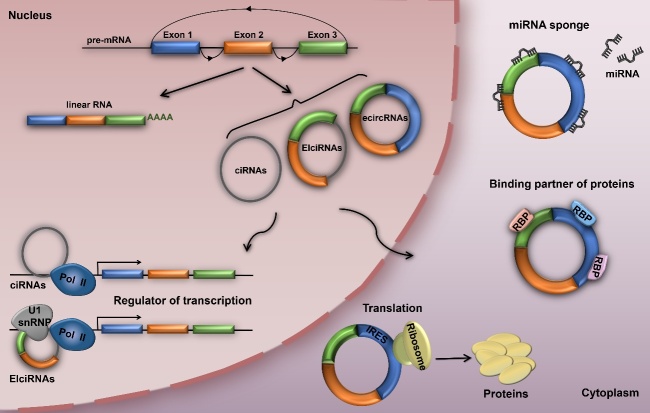
CircRNAs are derived from precursor mRNAs (pre-mRNAs), which are ubiquitous in eukaryotic cells, transcribed by RNA polymerase II (RNA Pol II) [13, 25-27]. Recent studies have revealed that circRNA biogenesis is different from the canonical splicing of linear RNAs [27]. CircRNAs are mainly generated by a process called back-splicing, where downstream exons are spliced to upstream exons in reverse order [28-31], and can be classified into three types - exonic circRNAs (ecircRNAs), retained-intron circRNAs or EIciRNAs and intronic circRNAs (ciRNAs) (Fig. 1) [11, 12, 27, 29]. In addition, circRNAs biogenesis of characteristic back-splicing reaction can be facilitated by complementary flanking Alu elements, specific splicing factors (e.g., Quaking and MBL), or exon skipping [13, 17, 27, 32-34]. Studies have also revealed that ecircRNAs, primarily localized in the cytoplasm, can mediate miRNA function, while ciRNAs, mainly found in the nucleus, can function as transcriptional regulators of their parental gene by interacting with RNA Pol II [12, 14].
Figure 1.
The biogenesis and function of circular RNAs (circRNAs). CircRNAs can be classified into three types - exonic circRNAs (ecircRNAs), retained-intron circRNAs or EIciRNAs and intronic circRNAs (ciRNAs). Nuclear circRNAs can regulate parental gene transcription, while cytoplasmic circRNAs can act as miRNA sponges, transcriptional regulators, binding partners of proteins, or even translated into functional proteins.
CircRNAs play important roles in regulating gene expression at multiple levels via neutralization of endogenous miRNAs, protein binding, regulation of parental gene expression, or translation into functional proteins (Fig. 1) [12-14, 18, 35, 36]. As competitive endogenous RNAs (ceRNAs), circRNAs can negatively regulate miRNAs by competing for miRNA-binding sites, thereby indirectly regulating the expression of the miRNA targets [14, 37]. The most representative is a circRNA named ciRS-7 or CDR1as, which contains over 70 selectively conserved miRNA target sites, serving as miR-7 sponge [8, 14]. Likewise, cSRY that is specifically expressed in mice testes harbors 16 binding sites for miR-138 [14, 38]. In addition, circRNAs can also bind to, sequester, and transport RBPs, suggesting another process for circRNA-mediated gene regulation [39, 40]. It has been reported that circFoxo3 can suppress cell cycle by interacting with CDK2 and p21 [39]. Another study also found that circFoxo3 could regulate cardiac senescence through interacting with senescence-associated proteins (e.g., ID1 and E2F1) and stress-related proteins (e.g., HIF1a and FAK) in the cytoplasm [18]. Several studies also found that EIciRNAs and ciRNAs can regulate transcription of their parental genes [11, 12]. The circEIF3J and circPAIP2 EIciRNAs that are mainly localized in the nucleus can bind to U1 small nuclear ribonucleoproteins (snRNPs), form the EIciRNA-U1 snRNP complex, and interact with RNA Pol II in the promoter region, consequently enhancing the transcription of their parental genes [11]. On the other hand, the ci-ankrd52 ciRNA can directly interact with RNA Pol II and promote the transcription of its parental gene, ANKRD52 [12]. Several studies also revealed that circRNAs containing internal ribosome entry site elements (IRES) or prokaryotic ribosome-binding sites can encode proteins in vivo or in vitro [30, 35, 41]. It has been found that circMbl3 could translate protein in fly heads [42]. More recently, a novel circRNA named circPINTexon2 was reported to contain short open reading frames (sORFs) that can be translated to proteins when driven by IRES in glioblastoma [43]. This discovery provides a new direction for future circRNA studies.
3. CircRNAs as potential biomarkers for age-related diseases
Based on their characteristics and biological functions, circRNAs have a great potential to act as essential biomarkers to predict disease progression and prognosis. The circRNAs previously identified that are potential biomarkers in age-related diseases, specifically cardio-cerebrovascular disease, neurodegenerative disease, cancer, and diabetes, are summarized in Table 1.
Table 1.
CircRNAs as potential biomarkers for age-related diseases.
| Diseases | Type of diseases | circRNAs | Expression | Biological function | Refs. |
|---|---|---|---|---|---|
| Cardio-cerebrovascular disease | Heart failure | circHRCR | Downregulated | Inhibits cardiac hypertrophy and heart failure by sponging miR-223 | [44] |
| Myocardial infarction | circMFACR | Upregulated | Mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression | [47] | |
| Cardiac senescence | circFoxo3 | Upregulated | Promotes cardiac senescence by arrest ID-1, E2F1, FAK, and HIF1a in the cytoplasm | [18] | |
| Atherosclerosis | circANRIL | Downregulated | Controls ribosome biogenesis through binding to PES1 and modulates pathways of atherogenesis | [45] | |
| Coronary artery disease | hsa_circ_0124644 | Upregulated | Potential diagnostic biomarker of CAD in the peripheral blood | [46] | |
| hsa_circ_0001879 | Upregulated | A novel biomarker to diagnose CAD | [48] | ||
| hsa_circ_0004104 | Upregulated | A novel biomarker to diagnose CAD | [48] | ||
| Stroke | circDLGAP | Downregulated | Ameliorates ischemic stroke outcomes by targeting miR-143 | [49] | |
| circHECTD1 | Upregulated | Contributes to astrocyte activation and cerebral infarction by targeting miR-142-TIPARP | [52] | ||
| circFUNDC1 | Upregulated | Biomarker for AIS diagnosis and prediction of outcomes | [50] | ||
| circPDS5B | Upregulated | Biomarker for AIS diagnosis and prediction of outcomes | [50] | ||
| circCDC14A | Upregulated | Biomarker for AIS diagnosis and prediction of outcomes | [50] | ||
| Neurodegenerative | Alzheimer's disease | ciRS-7 | Downregulated | Regulates the expression of UBE2A by sponging miR-7 | [57] |
| disease | Parkinson’s disease | ciRS-7 | Upregulated | Modulates the α-synuclein aggregation pattern in PD by targeting miR-7 | [59] |
| Multiple system atrophy | circIQCK | Upregulated | Potential biomarker for MSA | [60] | |
| circMAP4K3 | Upregulated | Potential biomarker for MSA | [60] | ||
| circEFCAB11 | Upregulated | Potential biomarker for MSA | [60] | ||
| circDTNA | Upregulated | Potential biomarker for MSA | [60] | ||
| circMCTP1 | Upregulated | Potential biomarker for MSA | [60] | ||
| Amyotrophic lateral sclerosis | hsa_circ_0023919 | Downregulated | Blood biomarker for ALS | [62] | |
| hsa_circ_0063411 | Upregulated | Blood biomarker for ALS | [62] | ||
| hsa_circ_0088036 | Upregulated | Blood biomarker for ALS | [62] | ||
| Cancer | Prostate cancer | circCSNK1G3 | Upregulated | Promotes cell growth by interacting with miR-181 | [65] |
| circAMOTL1L | Downregulated | Facilitates cell migration and invasion through binding miR-193a-5p | [66] | ||
| circ_0044516 | Upregulated | Promotes prostate cancer cell proliferation and metastasis and serves as a potential biomarker | [67] | ||
| Breast cancer | circCNOT2 | Upregulated | A useful biomarker to choose the right type of therapy or to monitor breast cancer | [68] | |
| circEPSTI1 | Upregulated | Regulates cell proliferation and apoptosis of TNBC by targeting BCL11A via miR-4753/6809 | [69] | ||
| hsa_circ_001783 | Upregulated | Correlates with tumor burden and serves as a novel prognostic and therapeutic target for breast cancer | [70] | ||
| Colorectal cancer | circKLDHC10 | Upregulated | Potential circulating biomarker for CRC diagnosis | [56] | |
| circ_0001178 | Upregulated | Promising biomarker for liver metastases from CRC | [71] | ||
| circ_0000826 | Upregulated | Promising biomarker for liver metastases from CRC | [71] | ||
| circCCDC66 | Upregulated | Predictive biomarker for CRC detection and prognosis | [72] | ||
| Diabetes | Diabetes | hsa_circ_0054633 | Upregulated | Circulating diagnostic biomarker for pre-diabetes and T2DM | [77] |
| hsa_circ_11783-2 | Downregulated | Closely related to T2DM and might be novel therapeutic targets in diabetes | [78] | ||
| Diabetic retinopathy | circ_0005015 | Upregulated | Facilitates retinal endothelial angiogenic function via sponging miR-519d-3p | [80] | |
| circHIPK3 | Upregulated | Regulates retinal endothelial cell function and vascular dysfunction by sponging miR-30a |
[81] | ||
| Diabetic cataract | circKMT2E | Upregulated | Involves in the pathogenesis of diabetic cataract | [82] | |
| Other diseases | Rheumatoid arthritis | hsa_circ_0044235 | Downregulated | Potential diagnostic biomarker of RA patients | [86] |
| Osteoporosis | circRUNX2 | Downregulated | Promotes the expression of osteogenic differentiation-related proteins by sponging miR-203 | [90] | |
| circ_0002060 | Upregulated | Potential diagnostic biomarker and therapeutic target in osteoporosis | [91] |
3.1 CircRNAs in cardio-cerebrovascular disease
Cardio-cerebrovascular disease (CVD) is an important age-related disease that is the most prominent cause of human mortality and morbidity worldwide. Despite the extensive amount of research for CVD, the survival of CVD patients has not significantly improved due to the lack of rapid and accurate diagnostic procedure and effective treatment. Thus, early diagnosis and proper therapeutic intervention are critical for improving the survival rates of CVD patients.
Recent studies have suggested that circRNAs may play important roles in the initiation and development of CVD [18, 44-50]. The first evidence confirming that circRNAs are involved in the regulation of heart physiology and pathology was reported by Wang et al. in 2016, who discovered a circRNA termed heart-related circRNA (HRCR) that can act as an endogenous miR-223 sponge and inhibit cardiac hypertrophy, decreasing the probability of heart failure [44]. Subsequently, the circMFACR/miR-652-3p/MTP18 axis was discovered to regulate cardiomyocyte apoptosis, mitochondrial fission, and myocardial infarction (MI), indicating that circMFACR may be a potential therapeutic target for cardiovascular disease [47]. Another group investigated the relationship between circRNAs and aging and found that circFoxo3, which was highly expressed in the aged hearts of both humans and mice, interacted with ID-1, E2F1, FAK, and HIF1a and retained them in the cytoplasm, limiting their anti-senescent and anti-stress roles and consequently promoting cardiac senescence [18]. On the other hand, a group from Germany and the USA focused on the role of circRNAs in atherosclerosis and found that circANRIL confers atheroprotection by controlling ribosomal RNA (rRNA) maturation and modulating the associated pathways during atherogenesis, suggesting that circRNAs can alter RNA function and ultimately influence atherosclerosis development [45]. In addition, Zhao et al. reported that the hsa_circ_0124644 in the peripheral blood may be a sensitive and specific biomarker for diagnosing coronary artery disease (CAD) [46], while another study presented a transcriptome-wide overview of aberrantly expressed circRNAs in CAD patients and identified hsa_circ_0001879 and hsa_circ_0004104 as novel CAD biomarkers [48].
Stroke is the leading cause of disability and death worldwide, with approximately 80% of the cases attributed to ischemia [51]. Bai et al. found that circDLGAP4 levels were significantly decreased in the plasma of acute ischemic stroke (AIS) patients [49]. On the other hand, Han et al. reported that the circHECTD1-MIR142-TIPARP axis was involved in ischemic stroke, providing translational evidence that circHECTD1 can serve as a novel biomarker and therapeutic target for stroke [52]. In addition, Zuo et al. found that the significantly increased levels of circFUNDC1, circPDS5B, and circCDC14A in the plasma of AIS patients were positively correlated with infarct volume, suggesting these circRNAs may be potential biomarkers for AIS diagnosis [50].
3.2 CircRNAs in neurodegenerative disease
CircRNAs were also found to be highly expressed in the brain compared to other tissues, prompting several researchers to investigate their association with nervous system diseases [53, 54]. In one study, the total circRNA expression significantly increased in the aging central nervous system of Drosophila, suggesting that circRNAs may serve as aging biomarkers [55]. In addition, a recent study reported that exosomal circRNAs are capable of traversing the blood-brain barrier (BBB), making them perfect candidates as potential diagnostic tools for neurodegenerative disease [56].
One well-known example is the ciRS-7 (or CDR1as), a circRNA sponge and an inhibitor of miR-7 [57-59]. Dysregulated ciRS-7-miR-7 interaction was discovered in the hippocampus of Alzheimer's disease (AD) patients [57]. In addition, Lukiw et al. found that ciRS-7 deficiency resulted in the decreased expression of selective miR-7 targets, such as the AD-associated target, UBE2A [57]. Subsequently, this conjecture was confirmed by another study, implying that ciRS-7 may serve as an effective target for AD treatment [58]. Interestingly, another miR-7 target called α-synuclein is implicated in the pathophysiology of Parkinson’s disease (PD), suggesting that ciRS-7 also plays a role in modulating the α-synuclein aggregation pattern in PD [59]. Another research group performed circRNA sequencing of the brain samples from multiple system atrophy (MSA) patients and identified five circRNAs, namely IQCK, MAP4K3, EFCAB11, DTNA, and MCTP1, that were overexpressed in the white matter of the cortical tissue [60]. Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease mainly characterized by muscle atrophy, speech difficulties, and respiratory insufficiency [61]. To identify biomarkers for ALS, Dolinar et al. compared the circRNA alterations in the leukocyte samples of ALS patients and healthy controls, and identified three circRNAs, has circ 0023919, hsa_circ_0063411, and has circ 0088036, as potential blood-based biomarkers of ALS [62].
3.3 CircRNAs in cancer
Age is the single most important determinant for risk of cancer [63]. Although malignant tumors can occur at all ages, cancer disproportionately strikes individuals aged 65 years and older [64]. However, the primary cause for major age-related cancers (e.g., prostate, breast, colorectal) remains unknown [63]. Therefore, it is important to emphasize the need for research focusing on the prevention of age-related cancer and the planning of treatment and care for elderly patients.
There is mounting evidence that circRNAs play critical roles in the regulation of cancer development and progression [56, 65-72]. Prostate cancer is one of the most common aggressive tumors in elderly men [73]. Recently, Chen et al. found that circCSNK1G3 can promote prostate cancer cell proliferation at least partially through interaction with miR-181b/d [65]. Another study revealed that circAMOTL1L was downregulated in human prostate cancer and circAMOTL1L-miR-193a-5p interaction facilitated cancer cell migration and invasion [66]. In addition, the exosomal circ_0044516 in the blood of prostate cancer patients was found to play an important role in cell survival and metastasis, suggesting its significance as a potential biomarker for prostate cancer [67].
On the other hand, breast cancer is the most frequently diagnosed cancer in elderly women [74, 75]. CircCNOT2 was proven to be a useful biomarker for choosing the right type of cancer therapy and monitoring breast cancer in a minimally invasive manner [68]. Another study reported that circEPSTI1 can function as a positive regulator of cell proliferation and apoptosis in triple-negative breast cancer (TNBC) by targeting BCL11A via miR-4753/6809 and act as an independent prognostic biomarker for survival in TNBC patients [69]. In addition, a higher level of hsa_circ_001783, which was significantly correlated with heavier tumor burden in breast cancer patients, may serve as a novel prognostic and therapeutic target for breast cancer [70].
The involvement of circRNAs in colorectal cancer (CRC), has a high incidence in both elderly men and women, has also been reported [56, 63, 71, 72]. Li et al. demonstrated that circKLDHC10, which was enriched and stable in exosomes, may serve as a potential circulating biomarker for CRC diagnosis [56]. In a circRNA sequence study, Xu et al. discovered that circ_0001178 and circ_0000826 were significantly upregulated in CRC metastatic tissues and may be used as biomarkers for liver metastases from CRC [71]. Furthermore, the findings of Hsiao et al. indicated that the elevated expression level of circCCDC66 was a good predictive biomarker for CRC detection and prognosis [72]. Collectively, these studies suggest that circRNAs can be potential biomarkers for cancer diagnosis and novel targets for cancer treatment.
3.4 CircRNAs in diabetes
Diabetes is an age-related metabolic disorder involving insulin secretion abnormalities and defects due to the action of insulin against its target tissues [76]. There were nearly 410 million diabetic patients around the world in 2015, of which approximately 46.5% have not yet been diagnosed [77]. Furthermore, the high morbidity and mortality rates in diabetic patients impose a huge social and economic burden on human society, therefore, early diagnosis and intervention are urgently needed. In a recent study, the level of hsa_circ_0054633 in the peripheral blood was associated with diabetes and may serve as a circulating diagnostic biomarker for pre-diabetes and type 2 diabetes mellitus (T2DM) [77]. Another research group verified that hsa_circ_11783-2 was closely related to T2DM, indicating that this circRNA may be a novel therapeutic target for diabetes, indicating that this circRNA might serve as a potential diagnostic biomarker of RA [78].
In the advanced stages of diabetes, patients often experience various vascular complications, which are the major causes of disability and high mortality among diabetic patients [79]. In a recent study, circ_0005015 was verified to be upregulated in the plasma, vitreous sample, and fibrovascular membranes of diabetic retinopathy (DR) patients [80]. Likewise, circHIPK3 expression was also significantly upregulated in diabetic retinas; silencing of circHIPK3 in vivo alleviated retinal vascular dysfunction, suggesting that this circRNA is a potential target for controlling proliferative DR [81]. Diabetic cataract (DC) is the leading cause of non-traumatic visual impairment and blindness worldwide. Fan et al. demonstrated that circKMT2E was involved in the pathogenesis of DC, providing a new target for the non-surgical treatment strategies [82].
3.5 CircRNAs in other age-related diseases
Rheumatoid arthritis (RA), which affects approximately 1% of the world’s population, is a chronic systemic autoimmune disease characterized by the debilitating in?ammation and destruction of the joints [83-85]. Early diagnosis and proper treatment can effectively relieve the pathogenetic condition in RA patients. Growing evidence has shown that dysregulated expression of circRNAs was associated with RA [86, 87]. For example, Luo et al. found that the hsa_circ_0044235 in the peripheral blood was significantly downregulated in RA patients, indicating that this circRNA might serve as a potential diagnostic biomarker of RA [86].
The relationship between circRNAs and osteoporosis, a multifactorial and common bone disease characterized by an increased risk of bone fracture (BF) due to the fragility and reduction of bone mass, has also been explored [88, 89]. One study reported that circRUNX2 can sponge miR-203 and promote the expression of osteogenic differentiation-related proteins, such as RUNX2, OCN, OPN, and BSP, consequently preventing osteoporosis [90]. Another study explored the expression of circRNAs using microarray analysis and identified circ_0002060 as a potential diagnostic biomarker and therapeutic target for osteoporosis [91].
4. Conclusion and perspectives
In recent years, circRNAs have gradually become one of the most prominent targets in the field of transcriptomics because of their critical roles in the regulation of gene expression and development of several diseases. The characteristic stability, abundance, and tissue-specific expression of circRNAs confer them great potential for use as biomarkers of various diseases [8, 16, 17]. Notably, circRNAs can exist in the exosomes and plasma due to their excellent stability, thus, providing a more convenient way for diagnosing pathologies [56, 92, 93]. Here, we presented some examples of circRNAs that are involved in age-related diseases that may serve as potential biomarkers. However, further studies specifically aimed at elucidating the function of circRNAs in the aging process are still required.
Studies investigating the diagnostic implications and significance of circRNAs are limited, since many primarily focus on the differential expression of circRNAs. Potential biomarker applications will require an in-depth knowledge of the mechanism regarding how circRNAs are changing in relation to disease development and progression. Furthermore, the reliability and sensitivity of using circRNAs as biomarkers requires thorough validation. Finally, detecting circRNAs in the blood cells or exosomes is more expensive and time-consuming than existing checks, thus, the improvement of existing clinical methods for RNA detection is urgently needed.
In conclusion, further investigations regarding the function and mechanism underlying the associations between circRNAs and age-related diseases are required. In addition, future studies may identify additional promising circRNA biomarkers for potential clinical use.
Acknowledgments
This study was supported by the grants from the National Natural Science Foundation of China (81871120), Natural Science Foundation of Guangdong Province (2019A1515010334, 2017KZDXM039), and Yangfan Training Program of Guangdong Province (4YF16006G).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- [1].Zarrouk A, Vejux A, Mackrill J, O'Callaghan Y, Hammami M, O'Brien N, et al. (2014). Involvement of oxysterols in age-related diseases and ageing processes. Ageing Res Rev, 18:148-162. [DOI] [PubMed] [Google Scholar]
- [2].Dimmeler S, Nicotera P (2013). MicroRNAs in age-related diseases. EMBO Mol Med, 5:180-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bao Q, Pan J, Qi H, Wang L, Qian H, Jiang F, et al. (2014). Aging and age-related diseases--from endocrine therapy to target therapy. Mol Cell Endocrinol, 394:115-118. [DOI] [PubMed] [Google Scholar]
- [4].Noren Hooten N, Fitzpatrick M, Wood WH 3rd, De S, Ejiogu N, Zhang Y, et al. (2013). Age-related changes in microRNA levels in serum. Aging (Albany NY), 5:725-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].De A, Ghosh C (2017). Basics of aging theories and disease related aging-an overview. Pharma Tutor, 5:16-23. [Google Scholar]
- [6].Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A, 73:3852-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hsu MT, Coca-Prados M (1979). Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature, 280:339-340. [DOI] [PubMed] [Google Scholar]
- [8].Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495:333-338. [DOI] [PubMed] [Google Scholar]
- [9].Suzuki H, Tsukahara T (2014). A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci, 15:9331-9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen LL, Yang L (2015). Regulation of circRNA biogenesis. RNA Biol, 12:381-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol, 22:256-264. [DOI] [PubMed] [Google Scholar]
- [12].Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. (2013). Circular intronic long noncoding RNAs. Mol Cell, 51:792-806. [DOI] [PubMed] [Google Scholar]
- [13].Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol Cell, 56:55-66. [DOI] [PubMed] [Google Scholar]
- [14].Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature, 495:384-388. [DOI] [PubMed] [Google Scholar]
- [15].Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. (2017). Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell, 66:22-37 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013). Cell-type specific features of circular RNA expression. PLoS Genet, 9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 19:141-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, et al. (2017). Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J, 38:1402-1412. [DOI] [PubMed] [Google Scholar]
- [19].Yu AQ, Wang ZX, Wu W, Chen KY, Yan SR, Mao ZB (2019). Circular RNA CircCCNB1 sponges micro RNA-449a to inhibit cellular senescence by targeting CCNE2. Aging (Albany NY), 11:10220-10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Panda AC, Grammatikakis I, Kim KM, De S, Martindale JL, Munk R, et al. (2017). Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res, 45:4021-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Haque S, Ames RM, Moore K, Pilling LC, Peters LL, Bandinelli S, et al. (2019). circRNAs expressed in human peripheral blood are associated with human aging phenotypes, cellular senescence and mouse lifespan. Geroscience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kumar L, Shamsuzzama, Jadiya P, Haque R, Shukla S, Nazir A (2018). Functional Characterization of Novel Circular RNA Molecule, circzip-2 and Its Synthesizing Gene zip-2 in C. elegans Model of Parkinson's Disease. Mol Neurobiol, 55:6914-6926. [DOI] [PubMed] [Google Scholar]
- [23].Huang G, Li S, Yang N, Zou Y, Zheng D, Xiao T (2017). Recent progress in circular RNAs in human cancers. Cancer Lett, 404:8-18. [DOI] [PubMed] [Google Scholar]
- [24].Floris G, Zhang L, Follesa P, Sun T (2017). Regulatory Role of Circular RNAs and Neurological Disorders. Mol Neurobiol, 54:5156-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liang D, Wilusz JE (2014). Short intronic repeat sequences facilitate circular RNA production. Genes Dev, 28:2233-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, et al. (2015). Exon circularization requires canonical splice signals. Cell Rep, 10:103-111. [DOI] [PubMed] [Google Scholar]
- [27].Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014). Complementary sequence-mediated exon circularization. Cell, 159:134-147. [DOI] [PubMed] [Google Scholar]
- [28].Cocquerelle C, Mascrez B, Hetuin D, Bailleul B (1993). Mis-splicing yields circular RNA molecules. Faseb j, 7:155-160. [DOI] [PubMed] [Google Scholar]
- [29].Barrett SP, Wang PL, Salzman J (2015). Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife, 4:e07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang Y, Wang Z (2015). Efficient backsplicing produces translatable circular mRNAs. RNA, 21:172-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. (2016). Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res, 26:1277-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell, 160:1125-1134. [DOI] [PubMed] [Google Scholar]
- [33].Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. (2015). Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep, 10:170-177. [DOI] [PubMed] [Google Scholar]
- [34].Zaphiropoulos PG (1996). Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A, 93:6536-6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. (2017). Translation of CircRNAs. Mol Cell, 66:9-21 e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. (2018). Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Thomas LF, Saetrom P (2014). Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics, 30:2243-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. (1993). Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell, 73:1019-1030. [DOI] [PubMed] [Google Scholar]
- [39].Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res, 44:2846-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu, et al. (2017). The Circular RNA Interacts with STAT3, Increasing Its Nuclear Translocation and Wound Repair by Modulating Dnmt3a and miR-17 Function. Mol Ther, 25:2062-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Perriman R, Ares M Jr, (1998). Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA, 4:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. (2017). Translation of CircRNAs. Mol Cell, 66:9- 21.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. (2018). A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun, 9:4475-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. (2016). A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J, 37:2602-2611. [DOI] [PubMed] [Google Scholar]
- [45].Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun, 7:12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L, et al. (2017). Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep, 7:39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, et al. (2017). Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ, 24:1111-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang L, Shen C, Wang Y, Zou T, Zhu H, Lu X, et al. (2019). Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis, 286:88-96. [DOI] [PubMed] [Google Scholar]
- [49].Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R, et al. (2018). Circular RNA DLGAP4 Ameliorates Ischemic Stroke Outcomes by Targeting miR-143 to Regulate Endothelial-Mesenchymal Transition Associated with Blood-Brain Barrier Integrity. J Neurosci, 38:32-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zuo L, Zhang L, Zu J, Wang Z, Han B, Chen B, et al. (2019). Circulating Circular RNAs as Biomarkers for the Diagnosis and Prediction of Outcomes in Acute Ischemic Stroke. Stroke:STROKEAHA119027348. [DOI] [PubMed] [Google Scholar]
- [51].Adams HP Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, et al. (1993). Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke, 24:35-41. [DOI] [PubMed] [Google Scholar]
- [52].Han B, Zhang Y, Zhang Y, Bai Y, Chen X, Huang R, et al. (2018). Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy, 14:1164-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, et al. (2015). Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell, 58:870-885. [DOI] [PubMed] [Google Scholar]
- [54].You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, et al. (2015). Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci, 18:603-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. (2014). Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep, 9:1966-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. (2015). Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res, 25:981-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lukiw WJ (2013). Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet, 4:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ (2016). Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer's Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7). Genes(Basel), 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM (2009). Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A, 106:13052-13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chen BJ, Mills JD, Takenaka K, Bliim N, Halliday GM, Janitz M (2016). Characterization of circular RNAs landscape in multiple system atrophy brain. J Neurochem, 139:485-496. [DOI] [PubMed] [Google Scholar]
- [61].Hobson EV, Harwood CA, McDermott CJ, Shaw PJ (2016). Clinical aspects of motor neurone disease. Medicine, 44:552-556. [Google Scholar]
- [62].Dolinar A, Koritnik B, Glavac D, Ravnik-Glavac M (2019). Circular RNAs as Potential Blood Biomarkers in Amyotrophic Lateral Sclerosis. Mol Neurobiol, 56:8052-8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hansen J (1998). Common cancers in the elderly. Drugs Aging, 13:467-478. [DOI] [PubMed] [Google Scholar]
- [64].Yancik R (2005). Population aging and cancer: a cross-national concern. Cancer J, 11:437-441. [DOI] [PubMed] [Google Scholar]
- [65].Chen S, Huang V, Xu X, Livingstone J, Soares F, Jeon J, et al. (2019). Widespread and Functional RNA Circularization in Localized Prostate Cancer. Cell, 176:831-843 e822. [DOI] [PubMed] [Google Scholar]
- [66].Yang Z, Qu CB, Zhang Y, Zhang WF, Wang DD, Gao CC, et al. (2019). Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway. Oncogene, 38:2516-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Li T, Sun X, Chen L (2019). Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. [DOI] [PubMed] [Google Scholar]
- [68].Smid M, Wilting SM, Uhr K, Rodriguez-Gonzalez FG, de Weerd V, Prager-Van der Smissen WJC, et al. (2019). The circular RNome of primary breast cancer. Genome Res, 29:356-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, et al. (2018). circEPSTI1 as a Prognostic Marker and Mediator of Triple-Negative Breast Cancer Progression. Theranostics, 8:4003-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, et al. (2019). Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis, 10:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Xu H, Wang C, Song H, Xu Y, Ji G (2019). RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol Cancer, 18:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, et al. (2017). Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res, 77:2339-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sebesta EM, Anderson CB (2017). The Surgical Management of Prostate Cancer. Semin Oncol, 44:347-357. [DOI] [PubMed] [Google Scholar]
- [74].Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. (2016). Cancer statistics in China, 2015. CA Cancer J Clin, 66:115-132. [DOI] [PubMed] [Google Scholar]
- [75].DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A (2017). Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin, 67:439-448. [DOI] [PubMed] [Google Scholar]
- [76].Boitard C (2002). Insulin secretion in type 2 diabetes: clinical aspects. Diabetes Metab, 28:4s33-38. [PubMed] [Google Scholar]
- [77].Zhao Z, Li X, Jian D, Hao P, Rao L, Li M (2016). Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetologica, 54:237-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Li X, Zhao Z, Jian D, Li W, Tang H, Li M (2017). Hsa-circRNA11783-2 in peripheral blood is correlated with coronary artery disease and type 2 diabetes mellitus. Diabetes Vasc Dis Res, 14:510-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Sena CM, Pereira AM, Seica R (2013). Endothelial dysfunction - a major mediator of diabetic vascular disease. Biochim Biophys Acta, 1832:2216-2231. [DOI] [PubMed] [Google Scholar]
- [80].Zhang SJ, Chen X, Li CP, Li XM, Liu C, Liu BH, et al. (2017). Identification and Characterization of Circular RNAs as a New Class of Putative Biomarkers in Diabetes Retinopathy. Invest Ophthalmol Vis Sci, 58:6500-6509. [DOI] [PubMed] [Google Scholar]
- [81].Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, et al. (2017). Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation, 136:1629-1642. [DOI] [PubMed] [Google Scholar]
- [82].Fan C, Liu X, Li W, Wang H, Teng Y, Ren J, et al. (2019). Circular RNA circ KMT2E is up-regulated in diabetic cataract lenses and is associated with miR-204-5p sponge function. Gene, 710:170-177. [DOI] [PubMed] [Google Scholar]
- [83].Wang W, Zhang Y, Zhu B, Duan T, Xu Q, Wang R, et al. (2015). Plasma microRNA expression profiles in Chinese patients with rheumatoid arthritis. Oncotarget, 6:42557-42568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Hruskova V, Jandova R, Vernerova L, Mann H, Pecha O, Prajzlerova K, et al. (2016). MicroRNA-125b: association with disease activity and the treatment response of patients with early rheumatoid arthritis. Arthritis Res Ther, 18:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Goekoop-Ruiterman YP, Huizinga TW (2010). Rheumatoid arthritis: can we achieve true drug-free remission in patients with RA? Nat Rev Rheumatol, 6:68-70. [DOI] [PubMed] [Google Scholar]
- [86].Luo Q, Zhang L, Li X, Fu B, Deng Z, Qing C, et al. (2018). Identification of circular RNAs hsa_circ_0044235 in peripheral blood as novel biomarkers for rheumatoid arthritis. Clin Exp Immunol, 194:118-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Zheng F, Yu X, Huang J, Dai Y (2017). Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol Med Rep, 16:8029-8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Teitelbaum SL (2000). Bone resorption by osteoclasts. Science, 289:1504-1508. [DOI] [PubMed] [Google Scholar]
- [89].Coleman RE (2011). Bone cancer in 2011: Prevention and treatment of bone metastases. Nat Rev Clin Oncol, 9:76-78. [DOI] [PubMed] [Google Scholar]
- [90].Yin Q, Wang J, Fu Q, Gu S, Rui Y (2018). CircRUNX2 through has-miR-203 regulates RUNX2 to prevent osteoporosis. J Cell Mol Med, 22:6112-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Huang Y, Xie J, Li E (2019). Comprehensive circular RNA profiling reveals circ_0002060 as a potential diagnostic biomarkers for osteoporosis. J Cell Biochem, 120:15688-15694. [DOI] [PubMed] [Google Scholar]
- [92].Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C (2005). Exosomal-like vesicles are present in human blood plasma. Int Immunol, 17:879-887. [DOI] [PubMed] [Google Scholar]
- [93].Hoorn EJ, Pisitkun T, Zietse R, Gross P, Frokiaer J, Wang NS, et al. (2005). Prospects for urinary proteomics: exosomes as a source of urinary biomarkers. Nephrology (Carlton), 10:283-290. [DOI] [PubMed] [Google Scholar]