Abstract
Gu-tong formula (GTF) has achieved good curative effects in the treatment of cancer-related pain. However, its potential mechanisms have not been explored. We used network pharmacology and molecular docking to investigate the molecular mechanism and the effective compounds of the prescription. Through the analysis and research in this paper, we obtained 74 effective compounds and 125 drug-disease intersection targets to construct a network, indicating that quercetin, kaempferol, and β-sitosterol were possibly the most important compounds in GTF. The key targets of GTF for cancer-related pain were Jun proto-oncogene (JUN), mitogen-activated protein kinase 1 (MAPK1), and RELA proto-oncogene (RELA). 2204 GO entries and 148 pathways were obtained by GO and KEGG enrichment, respectively, which proved that chemokine, MAPK, and transient receptor potential (TRP) channels can be regulated by GTF. The results of molecular docking showed that stigmasterol had strong binding activity with arginine vasopressin receptor 2 (AVPR2) and C-X3-C motif chemokine ligand 1 (CX3CL1) and cholesterol was more stable with p38 MAPK, prostaglandin-endoperoxide synthase 2 (PTGS2), and transient receptor potential vanilloid-1 (TRPV1). In conclusion, the therapeutic effect of GTF on cancer-related pain is based on the comprehensive pharmacological effect of multicomponent, multitarget, and multichannel pathways. This study provides a theoretical basis for further experimental research in the future.
1. Introduction
Cancer-related pain remains common and severe for many patients, especially in the advanced stage, while the prevalence is approximately more than 70% [1]. A large number of people suffer from mild to severe pain before they die, and only few patients with cancer pain are well managed because the pain is difficult to relieve. Cancer-related pain is an aggregation of many different cancer types, which is a mixed pain representing a homogenous pathological process, mainly including acute pain, inflammatory pain, nerve pain, and tumor-induced pain [2]. Currently, the principal strategy for cancer-related pain management has been based on an ordinal three-step analgesic ladder, but its clinical use is usually limited by the side effects of gastrointestinal bleeding, constipation, respiratory depression, and so on [3, 4].
In China, the application of Gu-tong Formula (GTF) in treatment of cancer-related pain has a history of decades, and it has been a patented prescription (Patent No.: 201410415620.1). GTF is prepared from a formula of nine Chinese medicines, including Radix Aconiti Lateralis Preparata (RALP), Cortex Cinnamomi (CC), Rhizoma Curculiginis (RC), Herba Asari (HA), Rhizoma Zingiberis (RZ), Radix Clematidis (CCO), Pseudobulbus Cremastrae seu Pleiones (PCSP), Scorpio (BMK), and Flos Caryophylli (FC). Preliminary clinical trials showed that GTF can observably reduce the frequency of breakthrough cancer pain and the requirements of medication [5]. However, the related mechanisms of its analgesic effect have not been entirely explored.
It is known that the Chinese herbal formula has a characteristic of multicomponent, multitarget, and multipathways; it means traditional experimental methods cannot detect its complex mechanisms absolutely. Network pharmacology, as a combination of pharmacology and pharmacodynamics, emphasizing the integration of disease, gene, target, and drug, has been widely used for exploring the overall effect of drugs on the treatment of diseases from a macroscopic and systematic point of view. Therefore, we adopt the network pharmacology approach to discover potential mechanisms of GTF in the treatment of cancer-related pain and use molecular docking for reverse verification.
2. Methods
2.1. Active Ingredients and Related Targets in GTF
2.1.1. Pharmacokinetic Predictions
The active ingredients in GTF (except for BMK) were acquired from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database, a platform designed for herbs. Selecting oral bioavailability (OB) ≥30% and drug-likeness (DL) ≥0.18 as the screening criteria, the acquired active ingredients were searched for related protein targets from TCMSP and DrugBank databases [6, 7]. The active ingredients in BMK were obtained from the Traditional Chinese Medicine Information Database (TCMID) [8] and BATMAN-TCM [9]. Eventually, six effective compounds of BMK were collected, which were bufotoxin, chlorotoxin, katsutoxin, cholesterol, stearin, and 20-hexadecanoylingenol. Then, the chemical structure formula of the BMK active ingredients were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), followed by importing to the PharmMapper database [10] and selecting the effective targets with high match (norm fit > 0.7).
2.1.2. Potential Target Genes of Cancer-Related Pain
The data for the target genes of cancer-related pain were acquired from the DisGeNET database [11], GeneCards [12], and the Online Mendelian Inheritance in Man (OMIM) database [13]. All the data were standardized through the UniProt database [14].
DisGeNET database: search strategy—Set the disease name as “crushing pain, widespread chronic pain, postoperative pain, nerve pain, and intractable pain” in the gene disease network interface; 64 genes were collected (Table S1).
GeneCards: search strategy—Set the keyword as “cancer related pain” and the score >4 after logging into GeneCards; 1093 genes were collected (Table S2).
OMIM database: search strategy—Set the keyword as “cancer-related pain”; 22 genes were collected (Table S3).
2.1.3. Drug-Disease-Target Network Construction
Intersection genes, which may treat the disease, were obtained by intersecting the drug targets and disease targets. We used Cytoscape v3.7.2 to construct the drug-compound-disease intersection gene network and then carried out topology analysis [15]. Finally, we calculated the degree value of each node in the network by cytoHubba plug-in to screen out key pharmacodynamic molecules [16].
2.1.4. Protein-Protein Interaction (PPI) Data and Hub Gene Screening
We imported the intersection genes into the STRING database [17], and the species were set as Homo sapiens. The confidence score was set ≥0.9 to construct our PPI network [18], and the acquired network was imported into Cytoscape v3.7.2. The degree value of each node in the network was calculated using cytoHubba plug-in, and the top 10% was selected as the hub gene. Then, the biological process of GO was analyzed for the hub gene.
2.2. Enrichment Analysis
2.2.1. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
ClusterProfiler package was used to perform GO and KEGG enrichment analysis of the intersection gene with p < 0.05 [19].
2.2.2. Molecular Docking Verification
AutoDock was used to perform the receptor-ligand docking simulation calculation of key pharmacodynamic molecules and screened core targets. The protein structure, downloaded from the PDB database, was imported into POCASA v1.1 [20]. Meanwhile, the position of the active site was investigated in the PubMed database to verify the predicted position by POCASA v1.1. The docking operation used the Lamarckian genetic algorithm, and the rigid receptor-flexible ligand docking pattern was used in the docking process. The number of runs was 50, and the maximum energy evaluation was 2,500,000 [21]. The ligand corresponding to the target protein was used as a positive control. After all the docking simulations were completed, heat maps were made according to the strongest affinity of key pharmacodynamic molecules and core targets.
3. Results and Discussion
In this study, we obtained 74 active compounds (Table S4) and 269 potential targets of GTF after deleting duplicates. The active compound-related targets in TCMSP and PharmMapper databases are listed in Tables S5 and S6, respectively. 1144 potential target genes of cancer-related pain were collected, and we obtained 125 drug-disease intersection targets (Figure 1(a)).
Figure 1.
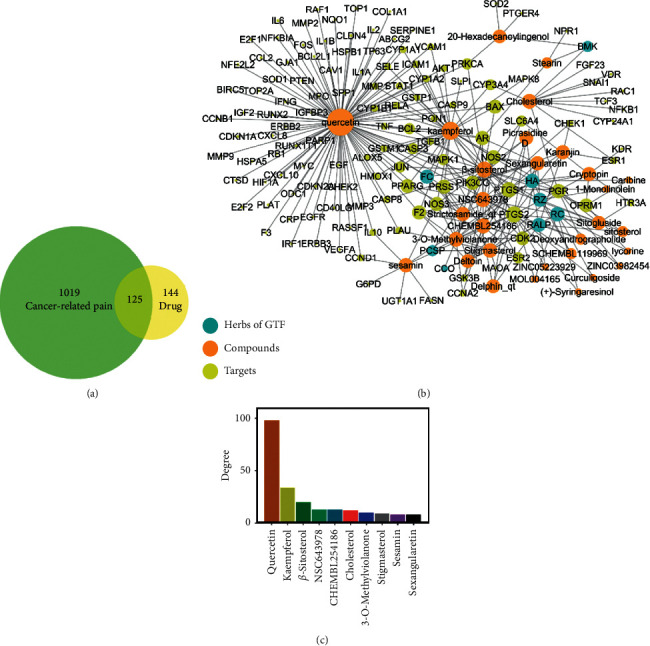
(a) Venn map of disease-related targets and drug targets. (b) Gu-tong formula (GTF) drug-disease-target network. Radix Aconiti Lateralis Preparata (RALP), Cortex Cinnamomi (CC), Rhizoma Curculiginis (RC), Herba Asari (HA), Rhizoma Zingiberis (RZ), Radix Clematidis (CCO), Pseudobulbus Cremastrae seu Pleiones (PCSP), Scorpio (BMK), and Flos Caryophylli (FC). (c) The top ten key pharmacodynamic molecules with the degree value.
3.1. GTF Drug-Disease-Target Network
The intersection targets were imported into Cytoscape v3.7.2 to construct a drug-disease-target network (Figure 1(b)), including 163 nodes and 296 edges, with a network heterogeneity of 2.336 and a network centralization of 0.590. It indicated that some nodes in the network were more concentrated than others. The network showed that compounds with most high connectivity were quercetin (degree = 98), kaempferol (degree = 34), and β-sitosterol (degree = 20), which were sorted by the degree value, suggesting that these three compounds were possibly the most important compounds in GTF (Figure 1(c)). Quercetin and kaempferol are flavonoids, and β-sitosterol is a phytosterol. All of them can regulate the way of cell apoptosis, proliferation, and phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) pathways to achieve the purpose of antitumor [22–24]. Moreover, they also have a strong inhibitory effect on inflammation through the inhibition of lipoxygenase and cyclooxygenase pathways. Among them, kaempferol has been used as a chemosensitizer in clinical research and has the potential of reducing toxicity and enhancing efficacy [25].
We preliminary analyzed the targets of the compounds in the network. NPR1 with a 0.672 disease-specific index (DSI) was targeted by stearin (extracted from BMK) and was important in crushing pain. ORPM1, which was targeted by β-sitosterol (extracted from FC, RZ, PCSP, CCO, and RC), caribine, cryptopine (extracted from HA), and lycorine (extracted from RC), played an important role in nerve pain, and its DSI was 0.479. The DSI of CXCL8 targeted by quercetin (extracted from FC) in intractable pain was 0.342. These three types of pain were crucial in cancer-related pain.
3.2. PPI Network
The intersection targets were imported into the STRING 11.0 database, and then, 104 nodes and 324 edges can be obtained, with an average degree of 5.18 for each node and an average neighbor of 6.231 for each edge (Figure 2(a)). As shown in Figure 2(b), the hub genes in the network screened by cytoHubba plug-in were JUN (degree = 28), MAPK1 (degree = 25), RELA (degree = 22), etc. Among them, RELA, RB1, NFKB1, TNF, and ESR1 had the function of regulating inflammatory response (GO: 0050727) and cellular response to reactive oxygen species (GO: 0034614). JUN, AKT1, FOS, and MAPK8 played the role of response to mechanical stimulus (GO: 0009612).
Figure 2.
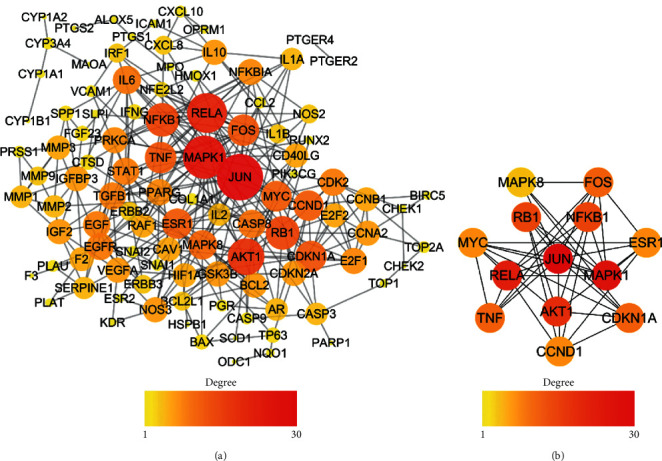
(a) PPI network of the intersection targets. (b) The hub genes in the PPI network.
TNF mainly encoded a multifunctional proinflammatory cytokine, combination with the transient receptor potential vanilloid-1 (TRPV1), playing an important role in inflammatory pain and nerve pain [26, 27]. Downregulating TNF expression can effectively inhibit the occurrence of inflammatory pain and nerve pain [28]. Moreover, MAPK8, JUN, AKT1, and RELA can regulate cell proliferation and cell cycle, which was crucial in tumorigenesis. GTF can regulate the expression of these genes to affect the development of tumors.
3.3. GO Enrichment
To further explore the multiple mechanisms of GTF, GO enrichment analysis (take biological process for example) was performed (Figure 3), and 2204 GO entries were enriched.
Figure 3.
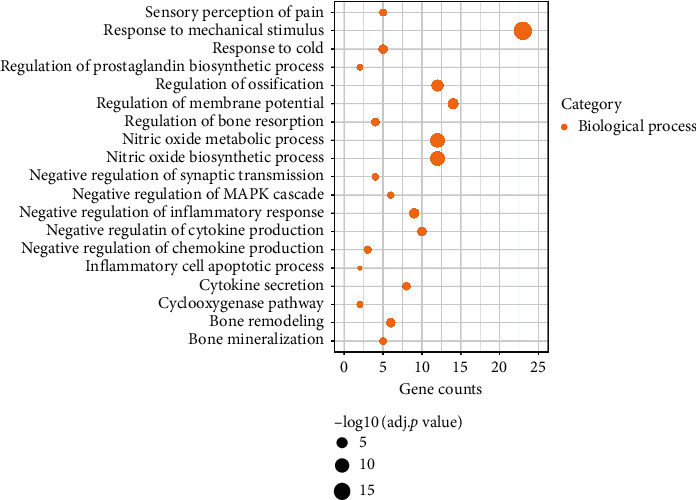
Gene Ontology (GO) enrichment result of GTF in treatment of cancer-related pain. The size of the bubble represents the different adjusted p value.
We found that GTF was involved in regulating the production of neuronal action potential via the regulation of membrane potential (GO: 0042391) and synaptic transmission (GO: 0050805). The activation of neurons and the conduction of electrical signals were the fundamental causes of nerve sensitization and pain [29]. GTF inhibited the conduction of pain by means of controlling the transmission of neurotransmitters to maintain the stability of the postsynaptic membrane.
What is more, GTF inhibited the secretion of inflammatory factors in the inflammatory response by means of regulating the prostaglandin (PG) biosynthetic process (GO: 0031392), cyclooxygenase pathway (GO: 0019371), inflammatory response (GO: 0050728), and cytokine production (GO: 0001818). Among them, PG induced inflammation and hyperalgesia, leading to the occurrence and aggravation of pain. In the first stage of three-step pain relief, it achieved the purpose of pain relief by inhibiting the target and cyclooxygenase pathway [2]. GTF also played a role in this pathway and the target and inhibited the occurrence of inflammatory pain. Moreover, it relieved the pain caused by tumor compression and sensitivity to cold stimulation after chemotherapy via the response to mechanical stimulus (GO: 0009612) and cold (GO: 0009409). Activation of the MAPK pathway was crucial in the development of pain [30], and GTF-negative regulated the MAPK cascade (GO: 0043409) to restrain the occurrence of pain.
Cancer-related pain caused by bone metastasis and bone destruction is usually difficult to relieve [31]. It was believed that bone resorption and bone formation were the key factors of bone destruction caused by tumor [32]. Interestingly, we found that GTF protected bones through regulation of ossification (GO: 0030278), bone resorption (GO: 0045124), and remodeling (GO: 0046849). Detailed GO enrichment information is shown in Table 1. The multitarget and multifunctional characteristics of GTF played a certain role in alleviation of cancer-related pain by regulating nerves, reducing inflammation and mechanical stimulation, and protecting bone.
Table 1.
Gene Ontology (GO) enrichment result.
| ID | Description | p value | Count |
|---|---|---|---|
| GO:0009612 | Response to mechanical stimulus | 2.60 × 10−19 | 23 |
| GO:0006809 | Nitric oxide biosynthetic process | 5.86 × 10−12 | 12 |
| GO:0046209 | Nitric oxide metabolic process | 1.24 × 10−11 | 12 |
| GO:0030278 | Regulation of ossification | 1.55 × 10−7 | 12 |
| GO:0042391 | Regulation of membrane potential | 3.96 × 10−6 | 14 |
| GO:0050728 | Negative regulation of inflammatory response | 1.41 × 10−5 | 9 |
| GO:0009409 | Response to cold | 1.09 × 10−4 | 5 |
| GO:0001818 | Negative regulation of cytokine production | 1.52 × 10−4 | 10 |
| GO:0046849 | Bone remodeling | 1.55 × 10−4 | 6 |
| GO:0045124 | Regulation of bone resorption | 5.98 × 10−4 | 4 |
| GO:0050663 | Cytokine secretion | 8.37 × 10−4 | 8 |
| GO:0032682 | Negative regulation of chemokine production | 1.27 × 10−3 | 3 |
| GO:0019233 | Sensory perception of pain | 2.17 × 10−3 | 5 |
| GO:0030282 | Bone mineralization | 3.11 × 10−3 | 5 |
| GO:0050805 | Negative regulation of synaptic transmission | 4.09 × 10−3 | 4 |
| GO:0043409 | Negative regulation of MAPK cascade | 4.39 × 10−3 | 6 |
| GO:0019371 | Cyclooxygenase pathway | 6.38 × 10−3 | 2 |
| GO:0031392 | Regulation of prostaglandin biosynthetic process | 7.40 × 10−3 | 2 |
| GO:0006925 | Inflammatory cell apoptotic process | 2.02 × 10−2 | 2 |
3.4. KEGG Enrichment
We obtained 148 pathways in total, which belonged to several categories, including tumor, inflammation, infection, and other pathways. After analyzing the results of KEGG enrichment, we found that GTF mainly focused on the regulation of proinflammatory cytokines, damage associated molecular pattern (DAMP), neural sensitization, and pain-related protein kinase (Table 2).
Table 2.
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment result.
| ID | KEGG pathway | p value | Count |
|---|---|---|---|
| hsa04657 | IL-17 signaling pathway | 4.64 × 10−18 | 21 |
| hsa04668 | TNF signaling pathway | 9.66 × 10−18 | 22 |
| hsa04620 | Toll-like receptor signaling pathway | 4.63 × 10−12 | 16 |
| hsa04010 | MAPK signaling pathway | 3.76 × 10−11 | 24 |
| hsa04621 | NOD-like receptor signaling pathway | 2.40 × 10−7 | 15 |
| hsa04630 | JAK-STAT signaling pathway | 3.94 × 10−7 | 14 |
| hsa04726 | Serotonergic synapse | 2.15 × 10−5 | 10 |
| hsa04622 | RIG-I-like receptor signaling pathway | 2.35 × 10−5 | 8 |
| hsa04062 | Chemokine signaling pathway | 6.33 × 10−5 | 12 |
| hsa04728 | Dopaminergic synapse | 2.40 × 10−2 | 6 |
| hsa04750 | Inflammatory mediator regulation of TRP channels | 2.84 × 10−2 | 5 |
GTF can regulate the synthesis and secretion of inflammatory factors by regulating IL-17 (hsa04657), TNF (hsa04668), and chemokine signaling pathways (hsa04062). IL-17, secreted by CD4+ T cells, can induce epithelial cells and endothelial cells to synthesize IL-6, PG, and other cytokines, and combination with the TNF signaling pathway to promote inflammation [33]. Inhibition of IL-17 can effectively relieve pain and inhibit peripheral nerve sensitization [34]. C-X3-C motif chemokine ligand 1 (CX3CL1) is a kind of chemokine. It was found that CX3CL1 and its receptor CX3CR1 were very important in the development of neuropathic pain [35]. Soluble CX3CL1 bound to CX3CR1 on the surface of microglia, which lead to the increase of intracellular calcium concentration and the occurrence of neuropathic pain [36, 37]. In the study of network pharmacology of GTF, it was shown that the application of GTF can affect those pathways and inhibit the occurrence of pain. At the same time, we found that GTF can regulate the response of the body to DAMP by regulating Toll-like receptor (hsa04620), NOD-like receptor (hsa04621), and RIG-I-like receptor signaling pathways (hsa04622). It had been proven that Toll-like receptors were highly expressed in microglia of mice with neuropathy. When the Toll-like receptor signaling pathway was inhibited, it can effectively inhibit the occurrence of pain [38].
In the aspect of neurotransmitter transmission and central sensitization, GTF can regulate the release of neurotransmitters and maintain the stability of neuron membrane potential by influencing serotonergic (hsa04726) and dopaminergic synapse (hsa04728). Transient receptor potential (TRP) was widely distributed in nociceptive neurons and was related to the persistence of pain [39]. GTF regulated this pathway (hsa04750) to affect pain duration.
p38 MAPK was widely distributed in the spinal dorsal horn and can be activated by various external stimuli such as trauma stimulation and inflammatory factors [40] and participated in the occurrence of pain through phosphorylated voltage-gated sodium channels. The JAK-STAT pathway was activated by interleukin-6 (IL-6) involving in the occurrence of inflammation and pain [41]. GTF can regulate the release of PG and excitatory amino acids by regulating these two pathways, to reduce cancer pain.
3.5. Molecular Docking
POCASA v1.1 was used to predict the most likely docking pocket of protein, which was sorted according to the volume of the pocket (Figure S1, docking pocket from large to small: a–e). In addition, compared with the active sites reported in the PubMed database, the docking sites of protein targets were obtained. The detailed information for core protein is listed in Table S7.
Figure 4 shows the mapping of the strongest affinity of 10 key drug molecules and 10 core target proteins. We found that the binding energy between the molecule and the target protein was approximately between −3.59 and −9.43 kcal·mol−1. It can be seen from Figure 4 that AVPR2, CX3CL1, p38 MAPK, prostaglandin-endoperoxide synthase 2 (PTGS2), and TRPV1 have stronger docking energy. It means that the compounds in GTF bind well to the above target proteins.
Figure 4.
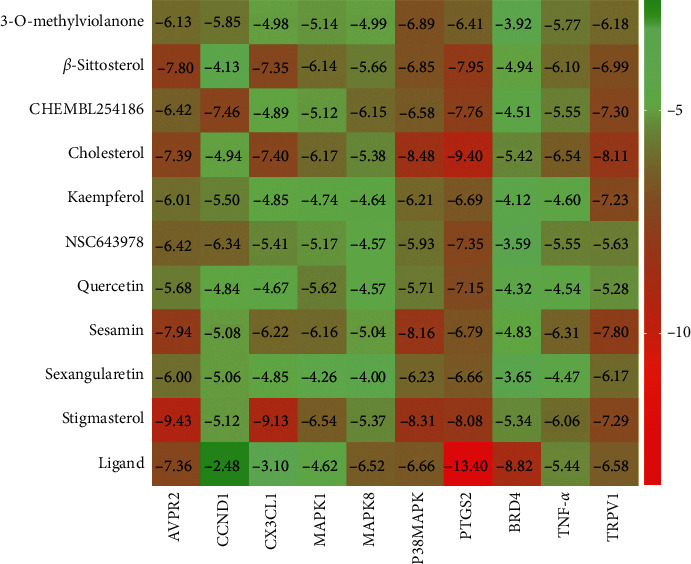
The binding energy of 10 key drug molecules and 10 target proteins.
Arginine vasopressin (AVP), an important analgesic substance, can be synthesized and secreted by the paraventricular nucleus of the hypothalamus [42]. After external stimulation, the expression of AVP increased and transported to the midbrain aqueduct gray matter, nucleus raphe magnus, caudate nucleus, and other related nuclei, resulting in the secretion of endogenous opioid peptide, 5-hydroxytryptamine and acetylcholine, and activating vasopressin receptors in central and peripheral tissues, which played a crucial role in nociception. Studies had shown that high dose of AVP can increase the action potential of C-type nociceptive fibers and produce analgesic effect [43]. The receptors of AVP, including V1a, V1b, and V2, belong to G protein-coupled receptor [44]. The compounds contained in GTF can stably bind to G protein-coupled receptor, producing analgesic effect.
CX3CL1 and its receptor CX3CR1 were both expressed in the nervous system [35] and played a role in promoting the occurrence of pain, which had been confirmed in many experiments. The researchers believed that the painful behavior caused by CX3CL1 was achieved by exciting CX3CR1 on microglia and activating the p38 MAPK signaling pathway [45]. Interestingly, our molecular docking results showed that GTF had better binding effect with CX3CL1 and p38 MAPK. In the pain model of bone tumor in rats made by injecting Walker 256 breast cancer cells into the tibial marrow cavity, intrathecal injection of CX3CR1 neutralizing antibody can reduce the development of pain and hyperalgesia, and blocking CX3CR1 can inhibit the activation of spinal microglia and the phosphorylation level of p38 MAPK [46]. It indicated the role of CX3CL1/CX3CR1/p38 MAPK pathway in the formation and development of cancer pain. This further showed that GTF can affect the occurrence and development of pain by regulating the above pathways.
PTGS2, also known as cyclooxygenase-2 (COX-2), was the second isozyme of cyclooxygenase. COX was an important rate-limiting enzyme for PG. COX-2 can be induced by a variety of inflammatory mediators and cytokines and participated in tissue inflammation and cell differentiation and proliferation. Upregulation of COX-2 was also related to antiapoptosis and tumor angiogenesis [47]. The results of molecular docking showed that the pharmacodynamic molecules selected in GTF had stronger binding energy to COX-2, which meant that the active ingredients in GTF can regulate the development of inflammatory pain and tumors through COX-2.
TRPV1 was a heat-activated cation channel protein, which was expressed on primary afferent neurons and upregulated after inflammation and nerve injury and was closely related to inflammation and acute and chronic pain [48]. Inhibition of TRPV1 activity was one of the pain treatment methods. In the preclinical study, it has the potential to be a receptor of nonopioid analgesics [49]. The results of molecular docking showed that the compounds contained in GTF can better combine with TRPV1 and played an analgesic role.
According to the docking energy results, we selected the 2 compounds with the strongest protein docking results from the 5 proteins, respectively. Stigmasterol had the strongest affinity with AVPR2, CX3CL1, and cholesterol with PTGS2, p38 MAPK, and TRPV1. The binding patterns of compounds and proteins were plotted, and then the interaction between compounds and binding sites and surrounding amino acid residues was observed (Figure 5). Cholesterol and stigmasterol both occupied the docking pocket of the protein and bound stably. The analysis of the docking effect of compounds and receptor proteins is shown in Table 3.
Figure 5.
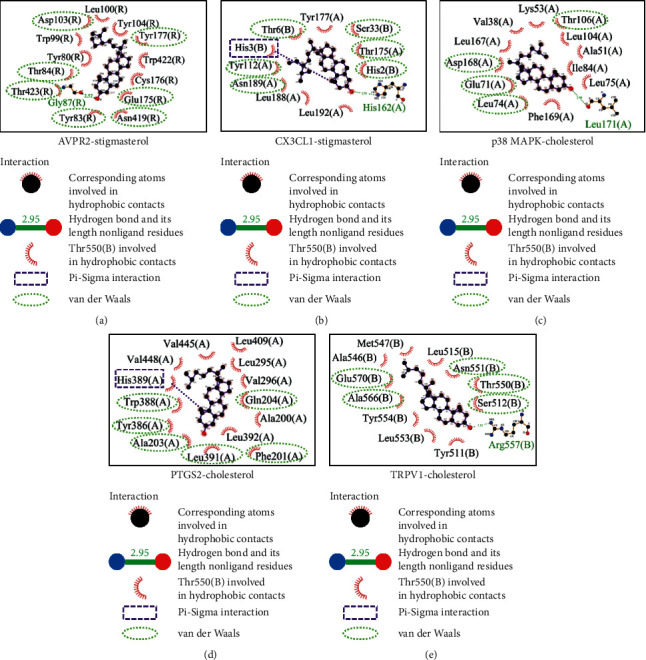
Binding mode of protein and compounds.
Table 3.
Docking effect analysis of compounds and targets.
| Compound | Target | Binding energy (kcal/mol) | Hydrogen bonding interaction | Hydrophobic interaction |
|---|---|---|---|---|
| Stigmasterol | AVPR2 | −9.43 | Cly87 | Tyr80, Tyr83, Tyr84, Trp99, Leu100, Asp103, Tyr104, Glu175, Cys176, Tyr177, Asn419, Trp422, Thr423 |
| CX3CL1 | −9.13 | His162 | Chain (A): Tyr112, Thr175, Tyr177, Leu188, Asn189, Leu192; China (B): His2, His3, Thr6, Ser33 |
|
| Cholesterol | p38 MAPK | −8.48 | Leu171 | Val38, Ala51, Lys53, Glu71, Leu74, Leu75, Ile84, Leu104, Thr106, Leu167, Asp168, Phe169 |
| PTGS2 | −9.4 | Not formed | Ala200, Phe201, Ala203, Gln204, Leu295, Val296, Tyr386, Tyr388, His389, Leu391, Leu392, Leu409, Val445, Val448 | |
| TRPV1 | −8.11 | Arg557 | Tyr511, Ser512, Leu515, Ala546, Met547, Thr550, Asn551, Leu553, Tyr554, Ala566, Glu570 |
All the five protein targets are able to form hydrophobic interaction and van der Waals force with compounds and stable binding. Stigmasterol (with AVPR2 and CX3CL1) and cholesterol (with p38 MAPK and TRPV1) can form hydrogen bonds on Gly87, His162, Leu171, and Arg557 residues, respectively, which made the binding more robust. CX3CL1 (with stigmasterol) and PTGS2 (with cholesterol) can form Pi-Sigma interaction on His3 and His389 residues, respectively, which increased the binding stability.
Stigmasterol is a common phytosterol. Modern pharmacological studies showed that it can effectively relieve acute and chronic pain and had neuroprotective effect on ischemic/reperfusion injury and good anti-inflammatory effect [50, 51]. In the process of molecular docking, the stigmasterol also had strong binding activity with AVPR2 and CX3CL1. It also suggested that stigmasterol in GTF was also an effective component in relieving cancer pain.
Cholesterol, widely consisted in vivo, a precursor of neurosteroid biosynthesis, is an endogenous produced molecule that inhibits TRPV1 activity and plays an important role in the regulation of nervous system injury and disorder [52]. Little is known about its role in neuropathic pain. Recent studies have shown that in inflammatory animal models, transcutaneous cholesterol delivery can alleviate allergic reactions and cholesterol homeostasis can help regulate inflammation and pain [53]. Increasing cholesterol content can inhibit the expression of PTGS2 and the secretion of PG [54], while decreasing cholesterol content can enhance p38 MAPK and inflammatory activity [55]. In molecular docking research, we found that cholesterol contained in GTF can stably target PTGS2, p38 MAPK, and TRPV1, which may play an analgesic role through MAPK and chemokine pathways.
4. Conclusion
To sum up, we revealed the potential pharmacological mechanism of GTF in the treatment of cancer pain from a systematic perspective, which may involve the secretion of inflammatory cytokines, membrane potential, bone protection, and other biological processes by regulating chemokine, MAPK, and TRP channels. The cholesterol and stigmasterol in GTF can be the key pharmacodynamic molecules for analgesia in molecular docking screening. This study provides clues for understanding the synergistic effect of GTF in relieving cancer pain. However, considering that this study is mainly based on data analysis and molecular docking, further experiments are necessary to verify the results.
Acknowledgments
This research was supported by the Talent Development Fund Award plan of Cancer Hospital of Chinese Academy of Medical Science (Grant no. RC2016007) and Research and Development of Ten Diseases and Ten Drugs—Preclinical Study of Yishen Gukang Formula in the Treatment of Cancer-Related Pain (Z171100001717017).
Abbreviations
- GTF:
Gu-tong formula
- JUN:
Jun proto-oncogene
- MAPK:
Mitogen-activated protein kinase
- RELA:
RELA proto-oncogene
- AVPR2:
Arginine vasopressin receptor 2
- CX3CL1:
C-X3-C motif chemokine ligand 1
- PTGS2:
Prostaglandin-endoperoxide synthase 2
- TRPV1:
Transient receptor potential vanilloid-1
- RALP:
Radix Aconiti Lateralis Preparata
- CC:
Cortex Cinnamomi
- RC:
Rhizoma Curculiginis
- HA:
Herba Asari
- RZ:
Rhizoma Zingiberis
- CCO:
Radix Clematidis
- PCSP:
Pseudobulbus Cremastrae seu Pleiones
- BMK:
Scorpio
- FC:
Flos Caryophylli
- TCMSP:
Traditional Chinese Medicine Systems Pharmacology
- OB:
Bioavailability
- DL:
Drug-likeness
- TCMID:
Traditional Chinese Medicine Information Database
- BATMAN-TCM:
A Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine
- OMIM:
Online Mendelian Inheritance in Man
- PPI:
Protein-protein interaction
- GO:
Gene Ontology
- KEGG:
Kyoto Encyclopedia of Genes and Genomes
- PI3K/AKT:
Phosphoinositide 3-kinase/protein kinase B
- PG:
Prostaglandin
- DSI:
Disease-specific index
- NPR1:
Natriuretic peptide receptor 1
- OPRM1:
Opioid receptor mu 1
- CXCL8:
Interleukin-8
- RB1:
Retinoblastoma-associated protein
- CDKN1A:
Cyclin-dependent kinase inhibitor 1
- FOS:
Proto-oncogene c-Fos
- TNF:
Tumor necrosis factor
- ESR1:
Estrogen receptor
- MYC:
Myc proto-oncogene protein
- DAMP:
Damage associated molecular pattern
- IL-6:
Interleukin-6
- JAK-STAT:
Janus kinase-signal transducer and activator of transcription
- COX-2:
Cyclooxygenase-2.
Data Availability
The data used to support the findings of this study are included within the article and the supplementary information files.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Li Feng conceived and designed the research. Jinyuan Chang collected the data and drafted the manuscript. Lixing Liu, Yaohan Wang, and Hao Li analyzed the data and provided valuable suggestions on the investigation. Yutong Sui critically reviewed the manuscript and assisted in the final write-up of the manuscript. All authors read and approved the final manuscript.
Supplementary Materials
Figure S1. The docking pocket of target proteins predicted by POCASA v1.1, and the order of pocket volume is from large to small: a–e. Table S1. The targets of cancer-related pain in DisGeNET. Table S2. The targets of cancer-related pain in GeneCards. Table S3. The targets of cancer-related pain in OMIM. Table S4. All the GTF compounds. Table S5. Potential targets of GTF compounds downloaded from TCMSP database. Table S6. Potential targets of BMK active compounds downloaded from PharmMapper database. Table S7. The PDB information for core protein.
References
- 1.Portenoy R. K. Treatment of cancer pain. The Lancet. 2011;377(9784):2236–2247. doi: 10.1016/s0140-6736(11)60236-5. [DOI] [PubMed] [Google Scholar]
- 2.Fallon M., Giusti R., Aielli F., et al. Management of cancer pain in adult patients: ESMO clinical practice guidelines. Annals of Oncology. 2018;29(4):iv166–iv191. doi: 10.1093/annonc/mdy152. [DOI] [PubMed] [Google Scholar]
- 3.Eisenberg E., Berkey C. S., Carr D. B., Mosteller F., Chalmers T. C. Efficacy and safety of nonsteroidal antiinflammatory drugs for cancer pain: a meta-analysis. Journal of Clinical Oncology. 1994;12(12):2756–2765. doi: 10.1200/jco.1994.12.12.2756. [DOI] [PubMed] [Google Scholar]
- 4.Gaertner J., Stamer U. M., Remi C., et al. Metamizole/dipyrone for the relief of cancer pain: a systematic review and evidence-based recommendations for clinical practice. Palliative Medicine. 2017;31(1):26–34. doi: 10.1177/0269216316655746. [DOI] [PubMed] [Google Scholar]
- 5.Tian J. Clinical Observation of Bone-Pain Plaster in the Treatment of Cancerous Somatic Pain of Yin Cold Stagnation Type Master. Beijing, China: Beijing University of Traditional Chinese Medicine; 2015. https://kns.cnki.net/kcms/detail/detail.aspx?FileName=1015386149.nh&DbName=CMFD2015. [Google Scholar]
- 6.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6(1):p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wishart D. S., Feunang Y. D., Marcu A., et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Research. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue R., Fang Z., Zhang M., Yi Z., Wen C., Shi T. TCMID: traditional Chinese medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Research. 2013;41:D1089–D1095. doi: 10.1093/nar/gks1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z., Guo F., Wang Y., et al. BATMAN-TCM: a bioinformatics analysis tool for molecular mechanism of traditional Chinese medicine. Scientific Reports. 2016;6(1):p. 21146. doi: 10.1038/srep21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., Shen Y., Wang S., et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Research. 2017;45(W1):W356–W360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piñero J., Ramírez-Anguita J. M., Saüch-Pitarch J., et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Research. 2020;48(D1):D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stelzer G., Rosen N., Plaschkes I., et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Current Protocols in Bioinformatics. 2016;54(1):p. 1. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 13.Hamosh A., Scott A. F., Amberger J., Valle D., McKusick V. A. Online mendelian inheritance in man (OMIM) Human Mutation. 2000;15(1):57–61. doi: 10.1002/(sici)1098-1004(200001)15:1<57::aid-humu12>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 14.The UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Research. 2019;47(D1):D506–D515. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otasek D., Morris J. H., Bouças J., Pico A. R., Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biology. 2019;20(1):p. 185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin C.-H., Chen S.-H., Wu H.-H., Ho C.-W., Ko M.-T., Lin C.-Y. CytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Systems Biology. 2014;8(4):p. S11. doi: 10.1186/1752-0509-8-s4-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szklarczyk D., Gable A. L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Y., Wang H., Pan Y., Liu T. Investigating the multi-target pharmacological mechanism of hedyotis diffusa willd acting on prostate cancer: a network pharmacology approach. Biomolecules. 2019;9(10):p. 591. doi: 10.3390/biom9100591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G., Wang L.-G., Han Y., He Q.-Y. Clusterprofiler: an R package for comparing biological themes among gene clusters. Omics: A Journal of Integrative Biology. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J., Zhou Y., Tanaka I., Yao M. Roll: a new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics. 2010;26(1):46–52. doi: 10.1093/bioinformatics/btp599. [DOI] [PubMed] [Google Scholar]
- 21.Morris G. M., Huey R., Lindstrom W., et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. Journal of Computational Chemistry. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauf A., Imran M., Khan I. A., et al. Anticancer potential of quercetin: a comprehensive review. Phytotherapy Research. 2018;32(11):2109–2130. doi: 10.1002/ptr.6155. [DOI] [PubMed] [Google Scholar]
- 23.Imran M., Salehi B., Sharifi-Rad J., et al. Kaempferol: a key emphasis to its anticancer potential. Molecules. 2019;24(12):p. 2277. doi: 10.3390/molecules24122277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bin Sayeed S. M., Ameen S. S. Beta-sitosterol: a promising but orphan nutraceutical to fight against cancer. Nutrition and Cancer. 2015;67(8):1214–1220. doi: 10.1080/01635581.2015.1087042. [DOI] [PubMed] [Google Scholar]
- 25.Li Q., Wei L., Lin S., Chen Y., Lin J., Peng J. Synergistic effect of kaempferol and 5fluorouracil on the growth of colorectal cancer cells by regulating the PI3K/Akt signaling pathway. Molecular Medicine Reports. 2019;20(1):728–734. doi: 10.3892/mmr.2019.10296. [DOI] [PubMed] [Google Scholar]
- 26.Leo M., Schulte M., Schmitt L. I., Schäfers M., Kleinschnitz C., Hagenacker T. Intrathecal resiniferatoxin modulates TRPV1 in DRG neurons and reduces TNF-induced pain-related behavior. Mediators of Inflammation. 2017;2017:8. doi: 10.1155/2017/2786427.2786427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li R., Zhao C., Yao M., Song Y., Wu Y., Wen A. Analgesic effect of coumarins from radix angelicae pubescentis is mediated by inflammatory factors and TRPV1 in a spared nerve injury model of neuropathic pain. Journal of Ethnopharmacology. 2017;195:81–88. doi: 10.1016/j.jep.2016.11.046. [DOI] [PubMed] [Google Scholar]
- 28.Zhao D., Han D. F., Wang S. S., Lv B., Wang X., Ma C. Roles of tumor necrosis factor-α and interleukin-6 in regulating bone cancer pain via TRPA1 signal pathway and beneficial effects of inhibition of neuro-inflammation and TRPA1. Molecular Pain. 2019;15:1–10. doi: 10.1177/1744806919857981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilron I., Baron R., Jensen T. Neuropathic pain: principles of diagnosis and treatment. Mayo Clinic Proceedings. 2015;90(4):532–545. doi: 10.1016/j.mayocp.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 30.Al-Olabi L., Polubothu S., Dowsett K., et al. Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. Journal of Clinical Investigation. 2018;128(11):p. 5185. doi: 10.1172/jci124649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Futakuchi M., Fukamachi K., Suzui M. Heterogeneity of tumor cells in the bone microenvironment: mechanisms and therapeutic targets for bone metastasis of prostate or breast cancer. Advanced Drug Delivery Reviews. 2016;99:206–211. doi: 10.1016/j.addr.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 32.Curtin P., Youm H., Salih E. Three-dimensional cancer-bone metastasis model using ex-vivo co-cultures of live calvarial bones and cancer cells. Biomaterials. 2012;33(4):1065–1078. doi: 10.1016/j.biomaterials.2011.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hot A., Lavocat F., Lenief V., Miossec P. Simvastatin inhibits the pro-inflammatory and pro-thrombotic effects of IL-17 and TNF-α on endothelial cells. Annals of the Rheumatic Diseases. 2013;72(5):754–760. doi: 10.1136/annrheumdis-2012-201887. [DOI] [PubMed] [Google Scholar]
- 34.Luo H., Liu H.-Z., Zhang W.-W., et al. Interleukin-17 regulates neuron-glial communications, synaptic transmission, and neuropathic pain after chemotherapy. Cell Reports. 2019;29(8):2384–2397. doi: 10.1016/j.celrep.2019.10.085. [DOI] [PubMed] [Google Scholar]
- 35.Old E. A., Nadkarni S., Grist J., et al. Monocytes expressing CX3CR1 orchestrate the development of vincristine-induced pain. Journal of Clinical Investigation. 2014;124(5):2023–2036. doi: 10.1172/jci71389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mecca C., Giambanco I., Donato R., Arcuri C. Microglia and aging: the role of the TREM2-DAP12 and CX3CL1-CX3CR1 axes. International Journal of Molecular Sciences. 2018;19(1):p. 318. doi: 10.3390/ijms19010318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindia J., McGowan E., Jochnowitz N., Abbadie C. Induction of CX3CL1 expression in astrocytes and CX3CR1 in microglia in the spinal cord of a rat model of neuropathic pain. The Journal of Pain. 2005;6(7):434–438. doi: 10.1016/j.jpain.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Peng Y., Zhang X., Zhang T., et al. Lovastatin inhibits Toll-like receptor 4 signaling in microglia by targeting its co-receptor myeloid differentiation protein 2 and attenuates neuropathic pain. Brain, Behavior, and Immunity. 2019;82:432–444. doi: 10.1016/j.bbi.2019.09.013. [DOI] [PubMed] [Google Scholar]
- 39.Basso L., Altier C. Transient receptor potential channels in neuropathic pain. Current Opinion in Pharmacology. 2017;32:9–15. doi: 10.1016/j.coph.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Tsuda M., Mizokoshi A., Shigemoto-Mogami Y., Koizumi S., Inoue K. Activation of p38 mitogen-activated protein kinase in spinal hyperactive microglia contributes to pain hypersensitivity following peripheral nerve injury. Glia. 2004;45(1):89–95. doi: 10.1002/glia.10308. [DOI] [PubMed] [Google Scholar]
- 41.Ding C.-P., Guo Y.-J., Li H.-N., Wang J.-Y., Zeng X.-Y. Red nucleus interleukin-6 participates in the maintenance of neuropathic pain through JAK/STAT3 and ERK signaling pathways. Experimental Neurology. 2018;300:212–221. doi: 10.1016/j.expneurol.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 42.Yang J., Li P., Zhang X.-Y., et al. Arginine vasopressin in hypothalamic paraventricular nucleus is transferred to the caudate nucleus to participate in pain modulation. Peptides. 2011;32(1):71–74. doi: 10.1016/j.peptides.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 43.Juif P.-E., Poisbeau P. Neurohormonal effects of oxytocin and vasopressin receptor agonists on spinal pain processing in male rats. Pain. 2013;154(8):1449–1456. doi: 10.1016/j.pain.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Decaux G., Soupart A., Vassart G. Non-peptide arginine-vasopressin antagonists: the vaptans. The Lancet. 2008;371(9624):1624–1632. doi: 10.1016/s0140-6736(08)60695-9. [DOI] [PubMed] [Google Scholar]
- 45.O’Sullivan S. A., Gasparini F., Mir A. K., Dev K. K. Fractalkine shedding is mediated by p38 and the ADAM10 protease under pro-inflammatory conditions in human astrocytes. Journal of Neuroinflammation. 2016;13(1):p. 189. doi: 10.1186/s12974-016-0659-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu J.-H., Yang J.-P., Liz L., et al. Involvement of CX3CR1 in bone cancer pain through the activation of microglia p38 MAPK pathway in the spinal cord. Brain Research. 2012;1465:1–9. doi: 10.1016/j.brainres.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 47.Desai S. J., Prickril B., Rasooly A. Mechanisms of phytonutrient modulation of cyclooxygenase-2 (COX-2) and inflammation related to cancer. Nutrition and Cancer. 2018;70(3):350–375. doi: 10.1080/01635581.2018.1446091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bujak J. K., Kosmala D., Szopa I. M., Majchrzak K., Bednarczyk P. Inflammation, cancer and immunity-implication of TRPV1 channel. Frontiers in Oncology. 2019;9:p. 1087. doi: 10.3389/fonc.2019.01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sapio M. R., Neubert J. K., LaPaglia D. M., et al. Pain control through selective chemo-axotomy of centrally projecting TRPV1+ sensory neurons. Journal of Clinical Investigation. 2018;128(4):1657–1670. doi: 10.1172/jci94331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morales-Lázaro S. L., Rosenbaum T. Cholesterol as a key molecule that regulates TRPV1 channel function. Advances in Experimental Medicine and Biology. 2019;1135:105–117. doi: 10.1007/978-3-030-14265-0_6. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y., Flores D., Carrisoza-Gaytán R., Rohatgi R. Cholesterol affects flow-stimulated cyclooxygenase-2 expression and prostanoid secretion in the cortical collecting duct. American Journal of Physiology-Renal Physiology. 2015;308(11):F1229–F1237. doi: 10.1152/ajprenal.00635.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin-Segura A., Casadome-Perales A., Fazzari P., et al. Aging increases hippocampal DUSP2 by a membrane cholesterol loss-mediated RTK/p38MAPK activation mechanism. Frontiers in Neurolofy. 2019;10:p. 675. doi: 10.3389/fneur.2019.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amsalem M., Poilbout C., Ferracci G., Delmas P., Padilla F. Membrane cholesterol depletion as a trigger of Nav1.9 channel-mediated inflammatory pain. The EMBO Journal. 2018;37(8) doi: 10.15252/embj.201797349.e97349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leitão M. M., Radai J. A. S., Ferrari I. C., et al. Effects of an ethanolic extract and fractions from Piper glabratum (Piperaceae) leaves on pain and inflammation. Regulatory Toxicology and Pharmacology. 2020;117 doi: 10.1016/j.yrtph.2020.104762.104762 [DOI] [PubMed] [Google Scholar]
- 55.Sun J., Li X., Liu J., Pan X., Zhao Q. Stigmasterol exerts neuro-protective effect against ischemic/reperfusion injury through reduction of oxidative stress and inactivation of autophagy. Neuropsychiatric Disease and Treatment. 2019;15:2991–3001. doi: 10.2147/ndt.s220224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The docking pocket of target proteins predicted by POCASA v1.1, and the order of pocket volume is from large to small: a–e. Table S1. The targets of cancer-related pain in DisGeNET. Table S2. The targets of cancer-related pain in GeneCards. Table S3. The targets of cancer-related pain in OMIM. Table S4. All the GTF compounds. Table S5. Potential targets of GTF compounds downloaded from TCMSP database. Table S6. Potential targets of BMK active compounds downloaded from PharmMapper database. Table S7. The PDB information for core protein.
Data Availability Statement
The data used to support the findings of this study are included within the article and the supplementary information files.


