Abstract
BACKGROUND
Prediction of survival after the treatment of hepatocellular carcinoma (HCC) has been widely investigated, yet remains inadequate. The application of artificial intelligence (AI) is emerging as a valid adjunct to traditional statistics due to the ability to process vast amounts of data and find hidden interconnections between variables. AI and deep learning are increasingly employed in several topics of liver cancer research, including diagnosis, pathology, and prognosis.
AIM
To assess the role of AI in the prediction of survival following HCC treatment.
METHODS
A web-based literature search was performed according to the Preferred Reporting Items for Systemic Reviews and Meta-Analysis guidelines using the keywords “artificial intelligence”, “deep learning” and “hepatocellular carcinoma” (and synonyms). The specific research question was formulated following the patient (patients with HCC), intervention (evaluation of HCC treatment using AI), comparison (evaluation without using AI), and outcome (patient death and/or tumor recurrence) structure. English language articles were retrieved, screened, and reviewed by the authors. The quality of the papers was assessed using the Risk of Bias In Non-randomized Studies of Interventions tool. Data were extracted and collected in a database.
RESULTS
Among the 598 articles screened, nine papers met the inclusion criteria, six of which had low-risk rates of bias. Eight articles were published in the last decade; all came from eastern countries. Patient sample size was extremely heterogenous (n = 11-22926). AI methodologies employed included artificial neural networks (ANN) in six studies, as well as support vector machine, artificial plant optimization, and peritumoral radiomics in the remaining three studies. All the studies testing the role of ANN compared the performance of ANN with traditional statistics. Training cohorts were used to train the neural networks that were then applied to validation cohorts. In all cases, the AI models demonstrated superior predictive performance compared with traditional statistics with significantly improved areas under the curve.
CONCLUSION
AI applied to survival prediction after HCC treatment provided enhanced accuracy compared with conventional linear systems of analysis. Improved transferability and reproducibility will facilitate the widespread use of AI methodologies.
Keywords: Deep learning, Artificial neuronal network, Recurrence, Liver transplantation, Resection, Hepatocellular cancer
Core Tip: Prediction of survival after the treatment of hepatocellular carcinoma (HCC) has been widely investigated yet remains inadequate. The application of artificial intelligence (AI) is an emerging adjunct to traditional statistics due to its ability to process vast amounts of data and find hidden interconnections between variables. The current study aimed to assess the role of various methodologies of AI in the prediction of survival after treatment of HCC by performing a systematic review of the literature.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the most common primary liver malignancy and the third most common cause of cancer-related death worldwide. Surgery, in the form of liver transplantation and resection, is the mainstay of treatment as the only potentially curative treatment option. Ablation has emerged as an alternative treatment to resection for small tumors. In contrast, intra-arterial treatments and chemotherapy can offer disease control and be used as part of a multimodal therapeutic strategy[1].
Many factors affect survival following the treatment of HCC. Among them, we can consider background liver condition, radiologic and histologic characteristics of the tumor, biologic markers, and comorbidities.
Traditionally, conventional linear models, such as the survival analysis and the Cox proportional hazard models, have been used to evaluate the prognosis of HCC[2-4]. Nevertheless, linear systems can have considerable limitations and often fail to capture the complexity of the interactions among clinicopathological characteristics[5]. With the intent to overcome such constraints, artificial intelligence (AI) has been employed with growing interest in healthcare research during the last decade, in particular applying deep learning (DL) techniques in artificial neural networks (ANN)[6]. ANN is a mathematical model that resembles the structure and function of a biological neural system using computer technology. It consists of a highly interconnected set of units, beginning with an input layer (the data to be analyzed), one or more hidden layers that process the data, and an output layer that provides the outcomes. The peculiarity of ANN is that it can be trained by exposing the network to examples of input/output pairs, thus improving its reliability[7]. During DL, the model reassigns a different weight to the connections within each hidden layer. ANN can learn from errors by comparing any generated output with desired outputs. The error is backpropagated, and the existing weights between connections are modified accordingly. Once learning is complete, ANN can create connections and make predictions on datasets that have not been observed before.
AI has been used to build models to predict a variety of outcomes related to HCC, such as tumor diagnosis, pathology characteristics, response to treatment, and survival[7,8]. With the growing availability of big data from fields such as genomics, AI can unravel otherwise hidden connections between tumor elements because of the increasing computational power of modern technology[9].
The objective of the current study was to systematically review the application of AI and DL in the prediction of survival among patients who were treated for HCC, as well as compare the performance of AI methods relative to linear prediction models.
MATERIALS AND METHODS
Search sources and study design
A systematic review of the published literature focused on the prognostic impact of AI in the management of HCC was undertaken. The search strategy was performed following the Preferred Reporting Items for Systemic Reviews and Meta-Analysis (PRISMA) guidelines[10].
The specific research question formulated in the present study includes the following PICO components: (1) Patient: Patient with a confirmed HCC; (2) Intervention: Evaluation of HCC treatment using AI; (3) Comparison: Evaluation of HCC treatment without using AI; and (4) Outcome: Patient death and/or tumor recurrence. A search of the PubMed and Cochrane Central Register of Controlled Trials Databases was conducted using the following terms: (Artificial intelligence OR deep learning) AND (HCC OR hepatocellular carcinoma OR hepatocellular cancer). The search period was from "1985/01/01" to "2020/02/29".
The systematic qualitative review included only English studies that included human patients. Published reports were excluded based on several criteria: (1) Data on animal models; (2) Lacked enough clinical details; and (3) Had non-primary source data (e.g., review articles, non-clinical studies, letters to the editor, expert opinions, and conference summaries). In the case of studies originating from the same center, possible overlapping of clinical cases was examined, and the most informative study was considered eligible.
Data extraction and definitions
Following a full-text review of the eligible studies, two independent authors (Lai Q and Larghi Laureiro Z) performed the data extraction and crosschecked all outcomes. During the selection of articles and extraction of the data, potential discrepancies were resolved following a consensus with a third reviewer (Mennini G). Collected data included the first author of the publication, year of publication, country, number of reported cases, research question/purpose, the method used, and key findings.
Quality assessment
Selected studies were systematically reviewed with the intent to identify potential sources of bias. The quality of the papers was assessed using the Risk of Bias In Non-randomized Studies of Interventions tool[11].
RESULTS
Search results and study characteristics
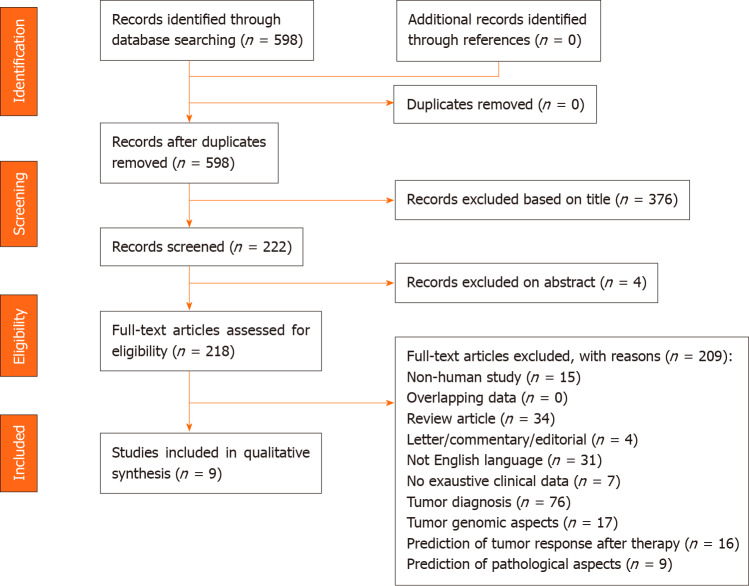
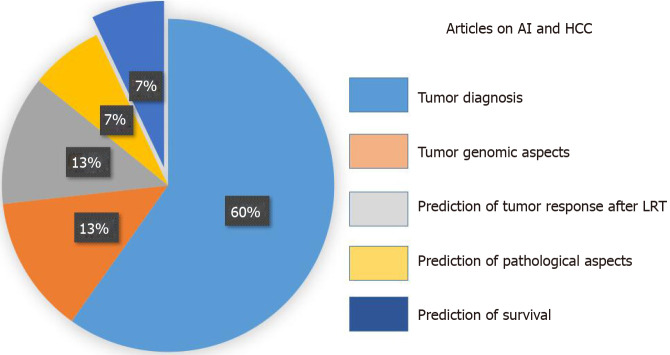
The PRISMA flow diagram schematically depicts the article selection process (Figure 1). Among the 598 articles screened, a total of 127 studies reported on the use of AI in HCC. Among these articles, only 9 (7.1%) studies referred to the use of AI in the prediction of survival among patients with HCC and were included in this review[12-20]. Other studies using AI in HCC were excluded; specifically, these studies reported on the use of AI for the diagnosis of the tumor (n = 76, 59.8%), identification of specific genes or pathways (n = 17, 13.4%), prediction of tumor response after therapy (n = 16, 12.6%), and the prediction of pathological aspects (n = 9, 7.1%) (Figure 2). All studies included in the analytic cohort were published in the last decade except for one that was published in 1995[12]. All articles were from Asia; five studies were based on a population from Taiwan[13-17], two from China[18,20], one from Japan[12], and one from India[19].
Figure 1.
Preferred Reporting Items for Systemic Reviews and Meta-Analysis flowchart of the literature search and study selection.
Figure 2.
Different articles exploring the impact of artificial intelligence as diagnostic or prognostic tool in the setting of hepatocellular carcinoma management. AI: Artificial intelligence; HCC: Hepatocellular carcinoma; LRT: Locoregional therapy.
Qualitative assessment of the included studies
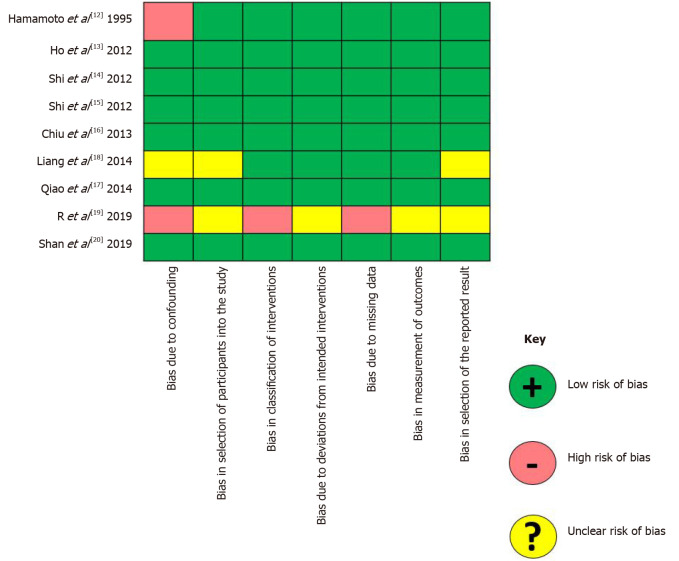
Results from the qualitative assessment of the included studies are depicted in Figure 3. Six studies had a low risk of bias, while two studies were at high risk for bias, mainly due to the presence of potential confounders. In one study, due to the absence of clear data explaining the characteristics of the comparison groups, the risk of bias was unclear.
Figure 3.
Results of the Risk of Bias In Non-randomized Studies of Interventions tool for the extracted articles.
Review of the eligible studies
Data extracted from the nine eligible articles are reported in detail in Table 1. The largest studies were based on the same population of patients coming from the Taiwan Bureau of National Health Insurance. All patients had a diagnosis of a malignant neoplasm of the liver and underwent a hepatectomy between 1998-2009 (n = 22926)[14,15]. In all other studies, the sample size was smaller than 1000 cases, and in two cases, the sample size was smaller than 100[12,17].
Table 1.
Articles focused on the role of artificial intelligence in the prediction of survival
| Ref. | Country/region | n | Research question/purpose | Method used | Key findings |
| Hamamoto et al[12], 1995 | Japan | 11 | ANN for the prediction of survival after HCC resection. | ANN was trained with the data of 54 resected patients and then prospectively used. | The outcomes in the prospective cohort were successfully predicted in all the cases (10 successful, 1 died). |
| Ho et al[13], 2012 | Taiwan | 482 | To validate the use of ANN model for predicting 1-, 3-, and 5-yr disease-free survival after hepatic resection, and to compare it with LR and decision tree model. | Training set: 80% of the cases; validation set: Remaining 20% of the cases. | The ANN model outperformed the other models in terms of prediction accuracy (AUC for 5-yr disease-free survival: 0.864 vs 0.627-0.736). |
| Shi et al[14], 2012 | Taiwan | 22926 | ANN model for predicting in-hospital mortality in HCC surgery patients and to compare it with LR models. | This study analyzed administrative claims data obtained from the Taiwan Bureau of National Health Insurance. | Compared to the LR models, the ANN models had a better accuracy rate in 97.28% of cases, and a better ROC curve in 84.67% of cases. |
| Shi et al[15], 2012 | Taiwan | 22926 | To validate the ANN models for predicting 5-yr mortality in HCC resected patients, and to compare them with LR models. | This study analyzed administrative claims data obtained from the Taiwan Bureau of National Health Insurance. | Compared to the LR models, the ANN models had a better accuracy rate in 96.57% of cases, and a better receiver operating characteristic curves in 88.51% of cases. |
| Chiu et al[16], 2013 | Taiwan | 434 | To compare significant predictors of mortality for HCC resected patients between ANN and LR models, and to evaluate the predictive accuracy of ANN and LR in different survival year estimation models. | Training set: 80% of the cases; validation set: Remaining 20% of the cases. | The results indicated that ANN had double to triple numbers of significant predictors at 1-, 3-, and 5-yr survival models as compared with LR models. Scores of accuracy, sensitivity, specificity, and AUC using ANN were superior to those of LR. |
| Qiao et al[17], 2014 | China | 543; 182; 104 | ANN for the prediction of survival in early HCC cases following partial hepatectomy. | Training set: 75% of the cases; internal validation set: Remaining 25% of the cases; external validation set. | In the training cohort, the AUC of the ANN was larger than that of the Cox model (0.855 vs 0.826, P = 0.0115). These findings were confirmed with the internal and external validation cohorts. |
| Liang et al[18], 2014 | Taiwan | 83 | Use of support vector machine for the development of recurrence predictive models for HCC patients receiving RFA treatment. | Five feature selection methods including genetic algorithm, simulated annealing algorithm, random forests and hybrid methods were utilized. | The developed support vector machine-based predictive models using hybrid methods had averages of the sensitivity, specificity, and AUC as 67%, 86%, and 0.69. |
| R et al[19], 2019 | India | 152 | To use artificial plant optimization algorithm to select optimal features and parameters of classifiers to improve the effectiveness and efficiency of prediction of HCC recurrence. | Different methods tested. | The sampling based multiple measurement artificial plant optimized random forest classifier with statistical measure showed the best results (balanced accuracy: 0.955). |
| Shan et al[20], 2019 | China | 156 | Peritumoral radiomics for the prediction of early recurrence after HCC curative resection or ablation. | Training cohort (n = 109) and validation cohort (n = 47). Using CT images, two regions of interest were delineated around the lesion for feature extraction o tumoral radiomics and peritumoral radiomics. | In the validation cohort, the ROC curves, calibration curves and decision curves indicated that the CT-based peritumoral radiomics model had better calibration efficiency and provided greater clinical benefits. |
ANN: Artificial neural network; HCC: Hepatocellular carcinoma; AUC: Area under the curve; LR: Logistic regression; RFA: Radiofrequency ablation; CT: Computed tomography; ROC: Receiver operating characteristic.
The use of ANN in populations of patients who underwent surgery was reported in six articles[12-16,18]. The outcomes investigated included in-hospital postoperative mortality[14], long-term overall survival[12,15,16,18], and disease-free survival after hepatic resection[13]. Several other studies used different AI systems rather than ANN. Specifically, a support vector machine was used for the development of predictive models relative to the recurrence of HCC following radiofrequency ablation[17]. Besides, an Artificial Plant Optimization algorithm was used to assess the effectiveness and efficiency to predict HCC recurrence[19]. Peritumoral radiomics was used to predict early recurrence after HCC curative-intent resection or ablation[20].
A cohort was used in the majority of studies to train the AI network[12-16,18,20]; in one study, a double five-fold cross-validation loop method was adopted[17]. In all studies, AI demonstrated superior predictive performance compared with other traditional models. In several studies, the ANN outperformed logistic regression or Cox regression models[13-16,18]. In all cases, the prediction accuracy of the AI models expressed as the areas under the curve was significantly improved compared with traditional statistical techniques[13-16,18].
DISCUSSION
The use of AI in healthcare began in the early 1970s and has gained increased acceptance over the last decades. In particular, the development of AI in medical research and its clinical applications have gained popularity, in part because of the widespread use of AI in almost all fields of human life[21]. The current literature search revealed that many AI studies focused on diagnosis, and the application of AI to distinguish the radiological features of HCC. The identification and diagnostic discrimination of benign vs malignant liver masses has been the objective of a previous systematic review that noted AI could differentiate liver cancer and, in particular, HCC from other lesions better compared with other methods such as Bayesian models and expert radiologists image inspection[8]. The present systematic review is important because it is the first to summarize the ability of AI systems to predict patient survival following treatment of HCC. Our results revealed that different types of AI methods have been employed in the existing studies with heterogeneous patient sample sizes. The majority of the included studies (n = 6/9) utilized ANN for the analysis of predictors of post-treatment survival, which is in line with the results of other systematic reviews on the prediction of outcomes[22,23]. Considering the need for more accurate prediction, investigators have compared AI techniques with traditional linear models to optimize treatment decision-making. Although several prediction models have utilized both pre- and postoperative variables, these models have not proved useful in clinical decision-making since they require information that can only be available after resection or other treatment. In contrast, models with only preoperative variables can help guide treatment strategies in the preoperative setting[24,25].
Importantly, our systematic review revealed that the prediction of survival using AI methodology was highly accurate and remained robust in studies with limited sample sizes, although current knowledge in prediction modeling using AI has noted that AI performs better when applied to larger sample sizes[26]. Although the reason for the consistent high predictive accuracy of AI models is multifactorial, the complexity of AI models (e.g., a higher number of events per variable) further reinforces the superiority of their performance, which might explain the outstanding results even when used in smaller size studies[27].
Reproducibility and applicability of AI models in clinical practice and across different centers might be questioned due to the difficulties in acquiring and utilizing a dedicated software to process the data. In addition, as ANN learns from examples, one may argue that ANN needs to be trained before it can be applied to varying datasets that are different from the one it was initially built on. Nevertheless, what emerged from this systematic review was that AI could be an outstanding adjunct to conventional linear systems of analysis to predict post-treatment survival. Cucchetti et al[7] made their ANN available online so that other centers can test and possibly enrich their model aiming to predict HCC tumor grade and micro-vascular invasion preoperatively. Besides, when applied to other aspects of HCC, AI is particularly useful for exploring interconnections of big data such as in genomics. ANN combined with genotyping for microsatellite mutations/deletions was able to predict HCC recurrence after liver transplantation with an 85% accuracy in the center where the model was developed, and with 89.5% accuracy when examined in data from another center[28]. AI applied to radiomics is increasingly investigated: Machine learning has been used to provide a quantitative interpretation of computed tomography scans to reclassify indeterminate nodules and potentially avoid biopsy and improve patients safety[29]. Similarly, neural network algorithms have been built with the intent to objectively and reproducibly provide liver imaging reporting and data system categories concordant with the expert radiologists classifcation[30].
One of the downsides associated with the application of ANN in clinical practice might be the disproportionate number of input factors per patient (too many, e.g., thousands of proteins for gene expression) relative to the number of patients (too little). The risk of overfitting the dataset can be mitigated by strictly filtering out potentially irrelevant variables[31]. In particular, selecting the variables to use as input factors in ANN using traditional statistics has been employed as a strategy to improve efficiency and reduce redundancy of the AI model, as confirmed by all of the studies using ANNs included in this systematic review. When analyzing cancer patient data (i.e., too many dimensions for a relatively small number of samples), combining DL with other techniques of machine learning have been used to identify prognostic gene signatures and differentiate between better and worse prognosis in patients with various types of tumors including HCC[32].
CONCLUSION
Artificial intelligence can provide an enhanced prediction of survival following treatment of HCC compared with conventional linear models. The use of AI can be particularly helpful to process large amounts of data, as well as help identify patterns and associations that are not evident with traditional techniques given the complexity of the biological systems. AI has a promising role in health-care research and its application to HCC. While an increasing amount of data becomes available per patient, it is important to identify to what extent AI can help guide clinical decision-making and optimize the prediction of long-term outcomes based on the unique characteristics of each patient.
ARTICLE HIGHLIGHTS
Research background
Prediction of survival after the treatment of hepatocellular carcinoma (HCC) has been widely investigated, yet remains inadequate. The application of artificial intelligence (AI) is emerging as a valid adjunct to traditional statistics due to the ability to process vast amounts of data and find hidden interconnections between variables. AI and deep learning are increasingly employed in several topics of liver cancer research, including diagnosis, pathology, and prognosis.
Research motivation
AI applied to survival prediction after HCC treatment should provide enhanced accuracy compared with conventional linear systems of analysis.
Research objectives
Improved transferability and reproducibility will facilitate the widespread use of AI methodologies.
Research methods
A web-based literature search was performed according to the Preferred Reporting Items for Systemic Reviews and Meta-Analysis guidelines using the keywords “artificial intelligence”, “deep learning” and “hepatocellular carcinoma” (and synonyms).
Research results
Among the 598 articles screened, nine papers met the inclusion criteria, six of which had low-risk rates of bias. Eight articles were published in the last decade; all came from eastern countries. Patient sample size was extremely heterogenous (n = 11-22926). AI methodologies employed included artificial neural networks (ANN) in six studies, as well as support vector machine, artificial plant optimization, and peritumoral radiomics in the remaining three studies. All the studies testing the role of ANN compared the performance of ANN with traditional statistics. Training cohorts were used to train the neural networks that were then applied to validation cohorts. In all cases, the AI models demonstrated superior predictive performance compared with traditional statistics with significantly improved areas under the curve.
Research conclusions
AI applied to survival prediction after HCC treatment provided enhanced accuracy compared with conventional linear systems of analysis.
Research perspectives
Improved transferability and reproducibility will facilitate the widespread use of AI methodologies.
Footnotes
Conflict-of-interest statement: The authors have no conflict of interest to declare.
PRISMA 2009 Checklist statement: The authors have read the PRISMA 2009 Checklist, and the manuscript was prepared and revised in accordance with this checklist.
Manuscript source: Invited manuscript
Peer-review started: August 29, 2020
First decision: September 12, 2020
Article in press: October 1, 2020
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lerut JP S-Editor: Chen XF L-Editor: A P-Editor: Liu JH
Contributor Information
Quirino Lai, Hepato-biliary and Organ Transplant Unit, Department of Surgery, Sapienza University of Rome, Rome 00161, Italy. quirino.lai@uniroma1.it.
Gabriele Spoletini, General Surgery and Liver Transplantation Unit, Fondazione Policlinico Universitario A Gemelli IRCCS, Rome 00100, Italy.
Gianluca Mennini, Hepato-biliary and Organ Transplant Unit, Department of Surgery, Sapienza University of Rome, Rome 00161, Italy.
Zoe Larghi Laureiro, Hepato-biliary and Organ Transplant Unit, Department of Surgery, Sapienza University of Rome, Rome 00161, Italy.
Diamantis I Tsilimigras, Wexner Medical Center, The Ohio State University, Columbus, OH 43210, United States.
Timothy Michael Pawlik, Wexner Medical Center, The Ohio State University, Columbus, OH 43210, United States.
Massimo Rossi, Hepato-biliary and Organ Transplant Unit, Department of Surgery, Sapienza University of Rome, Rome 00161, Italy.
References
- 1.Vitale A, Trevisani F, Farinati F, Cillo U. Treatment of hepatocellular carcinoma in the Precision Medicine era: from treatment stage migration to therapeutic hierarchy. Hepatology. 2020:Online ahead of print. doi: 10.1002/hep.31187. [DOI] [PubMed] [Google Scholar]
- 2.Vitale A, Lai Q, Farinati F, Bucci L, Giannini EG, Napoli L, Ciccarese F, Rapaccini GL, Di Marco M, Caturelli E, Zoli M, Borzio F, Sacco R, Cabibbo G, Virdone R, Marra F, Felder M, Morisco F, Benvegnù L, Gasbarrini A, Svegliati-Baroni G, Foschi FG, Missale G, Masotto A, Nardone G, Colecchia A, Bernardi M, Trevisani F, Pawlik TM Italian Liver Cancer (ITA. LI.CA) group. Utility of Tumor Burden Score to Stratify Prognosis of Patients with Hepatocellular Cancer: Results of 4759 Cases from ITA.LI.CA Study Group. J Gastrointest Surg. 2018;22:859–871. doi: 10.1007/s11605-018-3688-y. [DOI] [PubMed] [Google Scholar]
- 3.Lai Q, Vitale A, Halazun K, Iesari S, Viveiros A, Bhangui P, Mennini G, Wong T, Uemoto S, Lin CC, Mittler J, Ikegami T, Zhe Y, Zheng SS, Soejima Y, Hoppe-Lotichius M, Chen CL, Kaido T, Lo CM, Rossi M, Soin AS, Finkenstedt A, Emond JC, Cillo U, Lerut J. Identification of an Upper Limit of Tumor Burden for Downstaging in Candidates with Hepatocellular Cancer Waiting for Liver Transplantation: A West-East Collaborative Effort. Cancers (Basel) 2020;12:452. doi: 10.3390/cancers12020452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai Q, Nicolini D, Inostroza Nunez M, Iesari S, Goffette P, Agostini A, Giovagnoni A, Vivarelli M, Lerut J. A Novel Prognostic Index in Patients With Hepatocellular Cancer Waiting for Liver Transplantation: Time-Radiological-response-Alpha-fetoprotein-INflammation (TRAIN) Score. Ann Surg. 2016;264:787–796. doi: 10.1097/SLA.0000000000001881. [DOI] [PubMed] [Google Scholar]
- 5.Huang S, Yang J, Fong S, Zhao Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020;471:61–71. doi: 10.1016/j.canlet.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Cleophas TJ, Cleophas TF. Artificial intelligence for diagnostic purposes: principles, procedures and limitations. Clin Chem Lab Med. 2010;48:159–165. doi: 10.1515/CCLM.2010.045. [DOI] [PubMed] [Google Scholar]
- 7.Cucchetti A, Piscaglia F, Grigioni AD, Ravaioli M, Cescon M, Zanello M, Grazi GL, Golfieri R, Grigioni WF, Pinna AD. Preoperative prediction of hepatocellular carcinoma tumour grade and micro-vascular invasion by means of artificial neural network: a pilot study. J Hepatol. 2010;52:880–888. doi: 10.1016/j.jhep.2009.12.037. [DOI] [PubMed] [Google Scholar]
- 8.Azer SA. Deep learning with convolutional neural networks for identification of liver masses and hepatocellular carcinoma: A systematic review. World J Gastrointest Oncol. 2019;11:1218–1230. doi: 10.4251/wjgo.v11.i12.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen B, Garmire L, Calvisi DF, Chua MS, Kelley RK, Chen X. Harnessing big 'omics' data and AI for drug discovery in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17:238–251. doi: 10.1038/s41575-019-0240-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutton B, Salanti G, Caldwell DM, Chaimani A, Schmid CH, Cameron C, Ioannidis JP, Straus S, Thorlund K, Jansen JP, Mulrow C, Catalá-López F, Gøtzsche PC, Dickersin K, Boutron I, Altman DG, Moher D. The PRISMA extension statement for reporting of systematic reviews incorporating network meta-analyses of health care interventions: checklist and explanations. Ann Intern Med. 2015;162:777–784. doi: 10.7326/M14-2385. [DOI] [PubMed] [Google Scholar]
- 11.Sterne JA, Hernán MA, Reeves BC, Savović J, Berkman ND, Viswanathan M, Henry D, Altman DG, Ansari MT, Boutron I, Carpenter JR, Chan AW, Churchill R, Deeks JJ, Hróbjartsson A, Kirkham J, Jüni P, Loke YK, Pigott TD, Ramsay CR, Regidor D, Rothstein HR, Sandhu L, Santaguida PL, Schünemann HJ, Shea B, Shrier I, Tugwell P, Turner L, Valentine JC, Waddington H, Waters E, Wells GA, Whiting PF, Higgins JP. ROBINS-I: a tool for assessing risk of bias in non-randomised studies of interventions. BMJ. 2016;355:i4919. doi: 10.1136/bmj.i4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamamoto I, Okada S, Hashimoto T, Wakabayashi H, Maeba T, Maeta H. Prediction of the early prognosis of the hepatectomized patient with hepatocellular carcinoma with a neural network. Comput Biol Med. 1995;25:49–59. doi: 10.1016/0010-4825(95)98885-h. [DOI] [PubMed] [Google Scholar]
- 13.Ho WH, Lee KT, Chen HY, Ho TW, Chiu HC. Disease-free survival after hepatic resection in hepatocellular carcinoma patients: a prediction approach using artificial neural network. PLoS One. 2012;7:e29179. doi: 10.1371/journal.pone.0029179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi HY, Lee KT, Lee HH, Ho WH, Sun DP, Wang JJ, Chiu CC. Comparison of artificial neural network and logistic regression models for predicting in-hospital mortality after primary liver cancer surgery. PLoS One. 2012;7:e35781. doi: 10.1371/journal.pone.0035781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi HY, Lee KT, Wang JJ, Sun DP, Lee HH, Chiu CC. Artificial neural network model for predicting 5-year mortality after surgery for hepatocellular carcinoma: a nationwide study. J Gastrointest Surg. 2012;16:2126–2131. doi: 10.1007/s11605-012-1986-3. [DOI] [PubMed] [Google Scholar]
- 16.Chiu HC, Ho TW, Lee KT, Chen HY, Ho WH. Mortality predicted accuracy for hepatocellular carcinoma patients with hepatic resection using artificial neural network. ScientificWorldJournal. 2013;2013:201976. doi: 10.1155/2013/201976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiao G, Li J, Huang A, Yan Z, Lau WY, Shen F. Artificial neural networking model for the prediction of post-hepatectomy survival of patients with early hepatocellular carcinoma. J Gastroenterol Hepatol. 2014;29:2014–2020. doi: 10.1111/jgh.12672. [DOI] [PubMed] [Google Scholar]
- 18.Liang JD, Ping XO, Tseng YJ, Huang GT, Lai F, Yang PM. Recurrence predictive models for patients with hepatocellular carcinoma after radiofrequency ablation using support vector machines with feature selection methods. Comput Methods Programs Biomed. 2014;117:425–434. doi: 10.1016/j.cmpb.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 19.R D, P R. An Optimized HCC Recurrence Prediction Using APO Algorithm Multiple Time Series Clinical Liver Cancer Dataset. J Med Syst. 2019;43:193. doi: 10.1007/s10916-019-1265-x. [DOI] [PubMed] [Google Scholar]
- 20.Shan QY, Hu HT, Feng ST, Peng ZP, Chen SL, Zhou Q, Li X, Xie XY, Lu MD, Wang W, Kuang M. CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging. 2019;19:11. doi: 10.1186/s40644-019-0197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yasnitsky LN. Artificial Intelligence and Medicine: History, Current State and Forecasts for the Future. Curr Hypertens Rev. 2020 doi: 10.2174/1573402116666200714150953. [DOI] [PubMed] [Google Scholar]
- 22.Senanayake S, White N, Graves N, Healy H, Baboolal K, Kularatna S. Machine learning in predicting graft failure following kidney transplantation: A systematic review of published predictive models. Int J Med Inform. 2019;130:103957. doi: 10.1016/j.ijmedinf.2019.103957. [DOI] [PubMed] [Google Scholar]
- 23.Abbod MF, Catto JW, Linkens DA, Hamdy FC. Application of artificial intelligence to the management of urological cancer. J Urol. 2007;178:1150–1156. doi: 10.1016/j.juro.2007.05.122. [DOI] [PubMed] [Google Scholar]
- 24.Tsilimigras DI, Mehta R, Moris D, Sahara K, Bagante F, Paredes AZ, Farooq A, Ratti F, Marques HP, Silva S, Soubrane O, Lam V, Poultsides GA, Popescu I, Grigorie R, Alexandrescu S, Martel G, Workneh A, Guglielmi A, Hugh T, Aldrighetti L, Endo I, Pawlik TM. Utilizing Machine Learning for Pre- and Postoperative Assessment of Patients Undergoing Resection for BCLC-0, A and B Hepatocellular Carcinoma: Implications for Resection Beyond the BCLC Guidelines. Ann Surg Oncol. 2020;27:866–874. doi: 10.1245/s10434-019-08025-z. [DOI] [PubMed] [Google Scholar]
- 25.Tsilimigras DI, Mehta R, Moris D, Sahara K, Bagante F, Paredes AZ, Moro A, Guglielmi A, Aldrighetti L, Weiss M, Bauer TW, Alexandrescu S, Poultsides GA, Maithel SK, Marques HP, Martel G, Pulitano C, Shen F, Soubrane O, Koerkamp BG, Endo I, Pawlik TM. A Machine-Based Approach to Preoperatively Identify Patients with the Most and Least Benefit Associated with Resection for Intrahepatic Cholangiocarcinoma: An International Multi-institutional Analysis of 1146 Patients. Ann Surg Oncol. 2020;27:1110–1119. doi: 10.1245/s10434-019-08067-3. [DOI] [PubMed] [Google Scholar]
- 26.van der Ploeg T, Austin PC, Steyerberg EW. Modern modelling techniques are data hungry: a simulation study for predicting dichotomous endpoints. BMC Med Res Methodol. 2014;14:137. doi: 10.1186/1471-2288-14-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gauglitz G. Artificial vs. human intelligence in analytics : Do computers outperform analytical chemists? Anal Bioanal Chem. 2019;411:5631–5632. doi: 10.1007/s00216-019-01972-2. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Luna H, Vargas HE, Byrne T, Rakela J. Artificial neural network and tissue genotyping of hepatocellular carcinoma in liver-transplant recipients: prediction of recurrence. Transplantation. 2005;79:1737–1740. doi: 10.1097/01.tp.0000161794.32007.d1. [DOI] [PubMed] [Google Scholar]
- 29.Mokrane FZ, Lu L, Vavasseur A, Otal P, Peron JM, Luk L, Yang H, Ammari S, Saenger Y, Rousseau H, Zhao B, Schwartz LH, Dercle L. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol. 2020;30:558–570. doi: 10.1007/s00330-019-06347-w. [DOI] [PubMed] [Google Scholar]
- 30.Yamashita R, Mittendorf A, Zhu Z, Fowler KJ, Santillan CS, Sirlin CB, Bashir MR, Do RKG. Deep convolutional neural network applied to the liver imaging reporting and data system (LI-RADS) version 2014 category classification: a pilot study. Abdom Radiol . 45:24–35. doi: 10.1007/s00261-019-02306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartosch-Härlid A, Andersson B, Aho U, Nilsson J, Andersson R. Artificial neural networks in pancreatic disease. Br J Surg. 2008;95:817–826. doi: 10.1002/bjs.6239. [DOI] [PubMed] [Google Scholar]
- 32.Choi J, Oh I, Seo S, Ahn J. G2Vec: Distributed gene representations for identification of cancer prognostic genes. Sci Rep. 2018;8:13729. doi: 10.1038/s41598-018-32180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]