Abstract
Coronavirus disease 2019 (COVID-19) has caused significant morbidity and mortality and new cases are on the rise globally, yet malaria-endemic areas report statistically significant lower incidences. We identified potential shared targets for an immune response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by immune determinants' shared identities with P. falciparum using the Immune Epitope Database and Analysis Resource Immune 9.0 browser tool. Probable cross-reactivity is suggested through HLA-A∗02:01 and subsequent CD8+ T-cell activation. The apparent immunodominant epitope conservation between SARS-CoV-2 (N and open reading frame (ORF) 1ab) and P. falciparum thrombospondin-related anonymous protein (TRAP) may underlie the low COVID-19 incidence in the malaria-endemic zone by providing immunity against virus infection to those previously infected with Plasmodium. Additionally, we hypothesize that the shared epitopes which lie within antigens that aid in the establishment of the P. falciparum erythrocyte invasion may be an alternative route for SARS-CoV-2 via the erythrocyte CD147 receptor, although this remains to be proven.
Keywords: CD147 receptor, epitope, homology, Plasmodium falciparum, SARS-CoV-2
Highlights
-
•
Malaria endemic zones are characterized by apparent low incidences of coronavirus disease 2019 (COVID-19).
-
•
On the basis of experimental and predicted in silico study, possible shared immunodominant epitopes between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and Plasmodium falciparum antigens were found.
-
•
Probable cross-reactivity is hypothesized through HLA-A∗02:01 and subsequent CD8+ T-cell activation, which may eventually lead to protection against SARS-CoV-2 in malaria-endemic zones.
-
•
The immunodominant epitope conservation between some of the SARS-CoV-2 antigens and P. falciparum thrombospondin-related anonymous protein may indicate a novel route for the virus.
Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, formerly HCoV-19), has led to significant morbidity and mortality in addition to adversely affecting healthcare systems and the global economy [1,2]. This acute respiratory illness ranges from a self-limiting acute upper respiratory tract infection to severe pneumonia, multiorgan failure and death [3]. There is currently no approved treatment or vaccine.
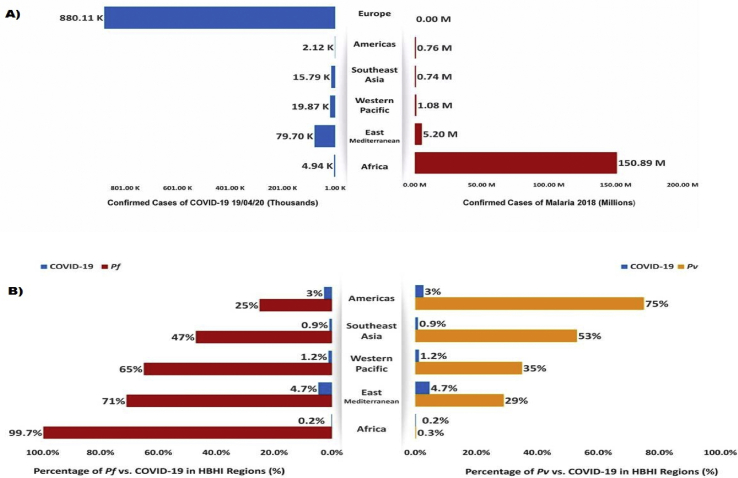
Most of the confirmed cases are confined to subtropical and temperate zones; countries in the equatorial and tropical zones seem to have the lowest incidences of COVID-19 (Supplementary Fig. S1). Interestingly, those countries have a high burden–high incidence (HBHI) of malaria infection [4]. According to World Health Organization (WHO) reports, the African region is characterized by a high prevalence of malaria (∼150.9 million in 2018), with dominant infection caused by Plasmodium falciparum (99% compared to ∼0.7% of Plasmodium vivax in 2018), and have the lowest number of cases of confirmed COVID-19 (∼187625 as of 17 June 2020) compared to other regions [[4], [5], [6]].
Malaria infection and its severity have shown an association with the ABO blood group system. Individuals with the O blood group are less susceptible to the infection compared to individuals with the A blood group [7]. The mechanism for this preferential protection is attributed to the rosette phenomenon, which is initially induced by the parasite proteins thrombospondin-related adhesive protein (MTRAP) and RH5. MTRAP and RH5 interact with the CD147 receptor on the erythrocyte surface, which results in making infected red blood cells (RBCs) stickier and therefore facilitate their binding to healthy erythrocytes, forming RBC clusters termed rosettes [7,8]. These rosettes assist parasites to escape recognition by phagocytosis. Clump formation is weakly seen in the case of individuals with O blood group.
The relationship between the ABO blood groups and susceptibility to COVID-19 infection has already been documented and is shown to follow a pattern similar to that of malaria [9]. Blood group A is associated with a high risk for acquiring the disease, whereas O blood group individuals have the lowest risk [10]. Several recent reports have highlighted the notably lower incidence of COVID-19 in malaria-endemic belts [11,12]. Though no mechanistic laboratory-based clarifications of this lower number of COVID-19 cases in the African malaria zone have been explained, several hypotheses have been put forward to answer the open query. The following factors have been widely suggested to be relevant: (a) warmer climate [13], (b) limited amount of international air traffic to and from Africa compared to the other continents, (c) public socioeconomic conditions, (d) early lockdown measures, (e) demographic factors, (f) protection provided by repeated use of antimalarial drugs and finally (g) factors related to host susceptibility, which might include immunologic and genetic factors [[14], [15], [16], [17]].
The genome of coronaviruses encodes 16 nonstructural proteins and four major structural proteins. A number of them are being targeted therapeutically and are being considered for candidates for vaccine development for COVID-19 [18,19]. In this study, we computationally and statistically analysed cases of COVID-19 in malaria-endemic regions and investigated possible shared immunogenic regions between dominant proteins of P. falciparum and SARS-CoV-2.
Methods
Statistical analysis of malaria-endemic regions and COVID-19 outbreak
We collected and analysed data from WHO reports and countries' status profiles for malaria HBHI in 2018 and the daily reports covering the COVID-19 outbreak [5,20]. Two-tailed tests of logistic regression coupled with chi-square, odds ratio and confidence interval analyses were performed by GraphPad Prism 5.0 software (GraphPad Software, La Jolla, CA, USA). Correlation confidence was used, and one-way ANOVA was performed with Bonferroni multiple comparison tests.
Defining immunodominant epitopes from SARS-CoV-2
SARS-CoV-2 B- and T-cell major histocompatibility complex (MHC)-restricted immunodominant regions and their corresponding epitopes, which are potential targets for immune responses, were identified by Grifoni et al. [21]. These potential epitopes were determined on the basis of sequence-shared identities with the closely related SARS-CoV and by parallel bioinformatic prediction approaches. Epitope sequences from SARS-CoV-2 were then retrieved and used to identify common immunodominant epitopes for P. falciparum.
Immunodominant regions of P. falciparum and homology with SARS-CoV-2
The Immune Epitope Database and Analysis Resource (IEDB, https://www.iedb.org) is a comprehensive repository of epitope data reported from the scientific literature. It includes antibody and T-cell epitopes for infectious disease, allergy, autoimmunity and transplantation. IEDB was used to search the immunome of Plasmodium falciparum (ID 5833) to identify the most immunogenic proteins (i.e. 23 proteins were determined) and retrieve their immunodominant epitopes. These proteins are apical membrane proteins 1 (AMP-1), circumsporozoite protein (CSP), merozoite surface protein 1 (MSP-1), thrombospondin-related anonymous protein (TRAP), liver stage antigen 3 (LSA3), circumsporozoite-related antigen (EXP1), sexual stage–specific protein 16 (Pfs16), sporozoite threonine and asparagine-rich protein (STARP), uncharacterized protein (UniProtQ8I5P1), DNAJ protein, putative (UniProt: Q8I0U6), serine-repeat antigen protein, RING finger protein (PFF0165c), Plasmodium exported protein (PHISTc), erythrocyte binding antigen 175, reticulocyte-binding protein homolog 5 (RPH5), rhoptry-associated protein 1 (RAP1), merozoite surface protein 3 (MSP-3), heat-shock 70 kDa protein, S antigen protein, early transcribed membrane protein 13 (ETRAMP13), Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP-1), erythrocyte binding antigen 140 (EBL-140) and erythrocyte binding antigen-181 (EBL-181).
These proteins were mapped back to the P. falciparum representative reference sequence using the IEDB's Immune 9.0 browser tool to display graphs and a data table that depict and list all the epitopes by their position and response frequency (RF, or how frequently a residue was found in a positive epitope) [22,23]. All B- and T-cell epitopes for the 23 proteins were selected on the basis of their sequence RF ˃ 0.0. For extra refinement of the results, we went further to identify T-cell epitopes with recognition restricted by human leukocyte antigen (HLA) MHC (class I (HLA-A, -B) and class II (HLA-DR, -DP, -DQ)); only positive assays were selected. Next, we aligned the P. falciparum B-cell, T-cell and T-cell MHC restriction epitopes to the SARS-CoV-2 immunodominant epitopes in order to calculate the percentage identity between each of the P. falciparum–dominant epitopes and SARS-CoV-2. Four or five amino acid shared residues were considered significant [24].
Prediction of T-cell MHC restricted epitopes
To identify and predict potential T-cell MHC restricted epitopes by alternative methods for selected P. falciparum antigenic targets (AMP-1, MSP-1, CSP, TRAP, SSP-2) and SARS-CoV-2 open reading frame (ORF) proteins (ORF3a, ORF1ab, ORF7a, ORF8, ORF10), we used the IEDB MHCI and MHCII online prediction tools (respectively http://tools.iedb.org/mhcI and http://tools.iedb.org/mhcII). These tools use different methods to determine the ability of the submitted sequence to bind to specific MHCI and –II molecules. The artificial neural network method was used to calculate the half-maximal inhibitory concentration (IC50) values of the peptide binding. For both frequent and nonfrequent alleles, the peptide length was set to 9 amino acids before the prediction. For MHCI epitopes, the alleles with affinity binding at IC50 of ≤500 nM were considered [25], while for MHCII, all epitopes that bind to many alleles with ≤1000 nM at IC50 were selected for further homology analysis [26].
Results
COVID-19 and malaria-endemic regions
We obtained results of malaria (year 2018) versus COVID-19 (up through 12 April 2020) cases worldwide in different regions including Africa (respectively 150887242 vs. 4943), Eastern Mediterranean (5202933 vs. 79695), West Pacific (1080872 vs. 19868), South-East Asia (742114 vs. 15735), the Americas (764980 vs. 46417) and Europe (0 vs. 880106) (Fig. 1). To examine the relative risk of exposure to COVID-19 and malaria coinfection, HBHI regions of malaria were assigned as cases and compared to Europe (which was considered the control), with zero cases of local malaria infection in 2018. The odds ratio indicated that all regions of malaria-endemic areas had a statistically significant lower relative risk of COVID-19 infection (p < 0.0001). Our results also demonstrated a significant reverse correlation between P. falciparum and COVID-19 death rates (r2 = −0.218, p < 0.001). Bonferroni multiple comparison tests were used to compare the mean of death rates and showed a significant variation between the death rate caused by P. falciparum and P. vivax against COVID-19 (p < 0.005) (Table 1, Table 2).
Fig. 1.
Coronavirus disease 2019 (COVID-19) vs. malaria. (A) Comparison between COVID-19 and malaria case numbers within different high burden–high incidence (HBHI) malaria regions. (B) Comparison between Plasmodium falciparum (Pf), P. vivax (Pv) and COVID-19 case percentages within HBHI malaria regions.
Table 1.
Logistic regression analysis associated with reduction factor of COVID-19 outbreak
| WHO region | No. of malaria cases in 2018 | No. of COVID-19 casesa | OR | 95% CI | p |
|---|---|---|---|---|---|
| African | 150887242 | 4943 | 179.1 | 174.1–184.1 | <0.0001 |
| Eastern Mediterranean | 5202933 | 79695 | 12.04 | 11.96–12.12 | <0.0001 |
| Western Pacific | 1080872 | 19868 | 45.30 | 44.68–45.93 | <0.0001 |
| South-East Asia | 742114 | 15735 | 56.93 | 56.06–57.82 | <0.0001 |
| Americas | 764980 | 46417 | 19.18 | 19.01–19.34 | <0.0001 |
| Europe | 0 | 880106 | — | — |
CI, confidence interval; COVID-19, coronavirus disease 2019; OR, odds ratio; WHO, World Health Organization.
As of 12 April 2020.
Table 2.
Average risk factor analysis by one-way ANOVA and Bonferroni multiple comparison test
| Death rate | Means (all regions) |
95% CI | Mean difference | r2 | t | p | |
|---|---|---|---|---|---|---|---|
| Malaria cases in 2018 | COVID-19 casesa | ||||||
| COVID-19 vs. malaria | 30370 | 65890 | −117300 to 188300 | 35520 | −0.21 | 0.7331 | 0.6780224 |
| COVID-19 vs. Pf | 30370 | 25860000 | −693000 to 17650000 | −25830000 | −0.218 | 1.946 | 0.6781664 |
| COVID-19 vs. Pv | 30370 | 590500 | −440300 to 42910000 | −560100 | −0.75413 | 0.04221 | 0.0832416 |
CI, confidence interval; COVID-19, coronavirus disease 2019; Pf, Plasmodium falciparum; Pv, Plasmodium vivax.
As of 12 April 2020.
Plasmodium falciparum immunodominant regions
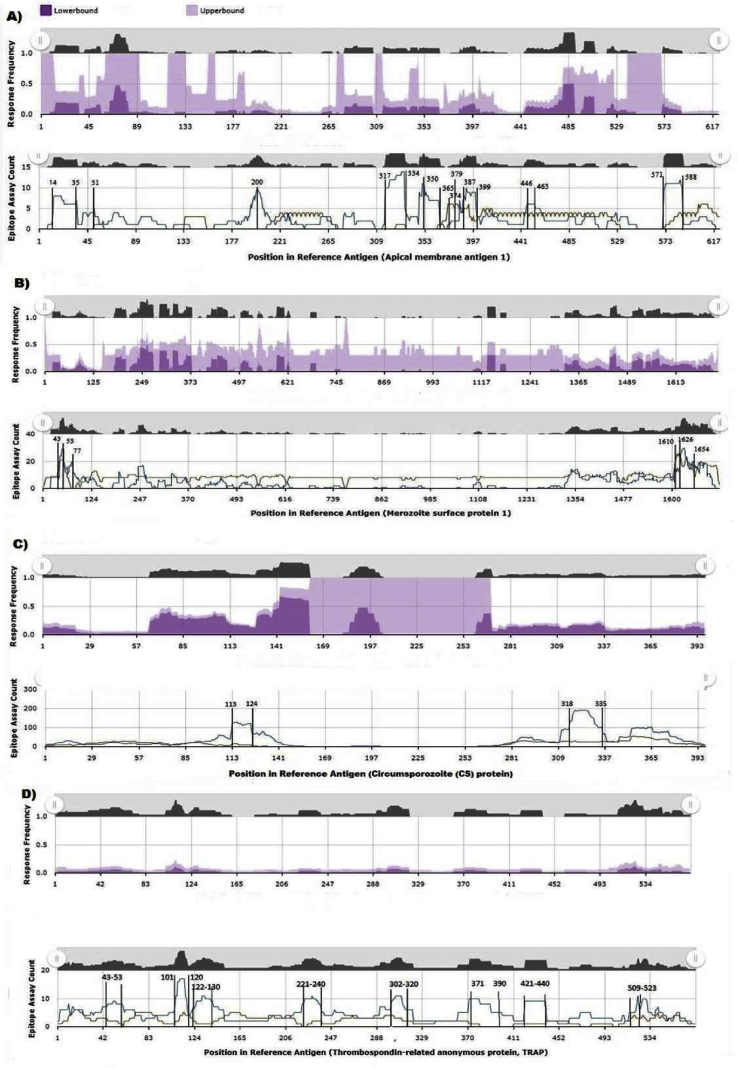
We screened the P. falciparum immunome to identify immunodominant regions. Twenty-three proteins were selected as promising immunogenic proteins. A total of 763 B-cell–immunodominant epitopes and 1084 T-cell–immunodominant epitopes were identified with RF ˃ 0.0. Many of these epitopes clustered in four proteins (AMP-1, MSP-1, CSP, TRAP) with a total of 190 and 918 B- and T-cell epitopes respectively. The analysis showed that AMP-1 has ten immune-conserved regions (residues 14–35, 51, 200, 317–334, 350–365, 374, 397, 387–399, 446–463, 571–588). MSP-1 has six regions located at both the N and C terminals of the protein; CSP has only two regions with potential interest. Residues 113–318, 335; and 43–53, 101, 120, 122–130, 221–240, 302–320, 371, 390, 421–440, 509–523 were identified for TRAP (Fig. 2). For SARS-CoV-2, ten immunodominant regions which were identified in reference to SARS-CoV by Grifoni et al. [21] via experimental data and confirmed with prediction approaches were used to define conserved shared regions with Plasmodium's tested immunogenic proteins.
Fig. 2.
T-cell–immunodominant regions based on Plasmodium falciparum–targeted proteins. (A) Apical membrane protein 1 (AMP-1). (B) Merozoite surface protein 1 (MSP-1). (C) Circumsporozoite protein (CSP). (D) Thrombospondin-related anonymous protein (TRAP). Specific epitope mapping response frequency (RF) score for each amino acid position was calculated and plotted over severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) consensus sequences. Cross-immune reactivity between P. falciparum and SARS-CoV-2 was identified.
Homology of Plasmodium B- and T-cell–immunodominant epitopes with SARS-CoV-2
All tested B-cell epitopes shared no significant homology with SARS-CoV-2. As such, no antibodies to Plasmodium could be proposed as eliciting an immune response against infection with SARS-CoV-2 through cross-reactivity. On the other hand, ≥40% of shared identities were noted between SARS-CoV-2 N protein (215–227 aa) and Plasmodium TRAP epitopes located at (509–523 aa) and also between ORF1ab (3661–3669 aa) and TRAP (101–130 aa). Although the phylogenetic distance between the two organisms would be expected, four or five shared amino acids in a single immunodominant epitope would be considered significant (Table 3).
Table 3.
Experimental T-cell–immunodominant epitopes from Plasmodium falciparum sharing homology with SARS-CoV-2 T-cell epitopes
| P. falciparum |
SARS-CoV-2 |
|||||||
|---|---|---|---|---|---|---|---|---|
| ID | Sequence | Mapped start–end | Epitope name | RF | Sequence | Protein name | Mapped start–end | Identity (%) |
| 55033 | RNNENRSYNRKHNNTPKHPE | 471–490 | TRAP | 0.03 | ALNTPKDHI | N | 138–146 | 44.4 |
| 31137 | KHNNTPKHPEREEHEKPDNN | 481–500 | 0.02 | 44.4 | ||||
| 34480 | KYKIAGGIAGGLALL | 509–520 | TRAP | 0.1 | GDAALALLLL | N | 215–224 | 40 |
| 20213 | GIAGGLALL | 515–523 | 0.37 | 44.4 | ||||
| 34480 | KYKIAGGIAGGLALL | 509–520 | TRAP | 0.1 | LALLLLDRL | N | 219–227 | 44.4 |
| 20213 | GIAGGLALL | 515–523 | 0.37 | 44.4 | ||||
| 28326 | IRLHSDASKNKEKALIIIKS | 101–120 | TRAP | 0.05 | SMWALIISV | ORF1ab | 3661–3669 | 44.4 |
| 7640 | DASKNKEKALIIIKS | 106–120 | 1 | 44.4 | ||||
| 32526 | KNKEKALII | 109–117 | 0.37 | 44.4 | ||||
| 30453 | KEKALIIIKSLLSTNLPYGK | 111–130 | 0.02 | 44.4 | ||||
| 549167 | KEKALIIIRSLLSTNLPYGR | 111–130 | 1 | 44.4 | ||||
ORF, open reading frame; RF, response frequency; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TRAP, thrombospondin-related anonymous protein.
Experimental and predicted T-cell MHC restriction homology with SARS-CoV-2
T-cell epitopes with recognition restricted by HLAs were also analysed. Typically, polymorphisms associated with MHC molecules result in the recognition of different epitopes. P. falciparum whole immunome was screened for T-cell MHC-immunodominant epitopes. The pool of MHCI- and MHCII-positive assays contained about 1610 immunodominant epitopes, most of them recognized in humans (n = 1575) and the rest reported in mice (n = 35) and rhesus macaques (n = 11). The same T-cell epitopes reported above have MHC restriction and equal identities with the same shared SARS-CoV-2 N and ORF1ab quarto-immunodominant epitopes. Interestingly, the N protein epitope located at 219–227 aa is extremely conserved in SARS-CoV (100% identity, RF = 0.29) and recognized by HLA-A∗02:01. This sequence is also partially shared by TRAP (504–513 aa) (44.4% identity, RF = 0.37), which is recognized by the same HLA-A∗02:01. Similarly, the ORF1ab epitope (3661–366aa) (89% shared SARS-CoV identity, RF = 0.42, recognized by HLA-A∗02:01 identified in HLA-transgenic mice) is 44.4%, homologous to TRAP precursor (114–122 aa, RF = 1) and also recognized by the same HLA molecule. Experimentally, HLA-A∗02:01 was assayed using interferon gamma enzyme-linked immunospot assay and reported to be restricted to CD8+ T-lymphocyte response to malaria in endemic areas (Table 4).
Table 4.
Experimental T-cell MHC restriction immunodominant epitopes from Plasmodium falciparum that share homology with SARS-CoV-2 T-cell MHC epitopes
| P. falciparum |
SARS-CoV-2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | Sequence | Mapped start–end | Epitope antigen name | Epitope parent name | MHC allele | Sequence | Protein name | Mapped start–end | Identity |
| 34480 | KYKIAGGIAGGLALL | 509–523 | Sporozoite surface protein 2 | TRAP | HLA-DRB1∗01:01, HLA-DRB1∗04:01, HLA-DRB1∗07:01, HLA-DRB1∗08:02, HLA-DRB1∗09:01, HLA-DRB1∗11:01, HLA-DRB1∗12:01, HLA-DRB1∗13:02, HLA-DRB5∗01:01 | GDAALALLLL | N | 215–224 | 40 |
| 34480 | KYKIAGGIAGGLALL | 509–523 | Sporozoite surface protein 2 | TRAP | HLA-DRB1∗01:01, HLA-DRB1∗04:01, HLA-DRB1∗07:01, HLA-DRB1∗08:02, HLA-DRB1∗09:01, HLA-DRB1∗11:01, HLA-DRB1∗12:01, HLA-DRB1∗13:02, HLA-DRB5∗01:01 | LALLLLDRL | N | 219–227 | 44.4 |
| 20762 | GLALLACAGL | 504–513 | TRAP precursor | TRAP | HLA-A∗02:01 | GDAALALLLL | N | 215–224 | 40 |
| 20762 | GLALLACAGL | 504–513 | TRAP precursor | TRAP | HLA-A∗02:01 | LALLLLDRL | N | 219–227 | 44.4 |
| 35388 | LEDIINLSKKKKKSINDTSF | 2557–2576 | Putative erythrocyte binding protein EBL-1 | EBL-140 | HLA-DRB1∗11:01 | WLMWLIINL | ORF1ab | 2292–2300 | 44.4 |
| 32526 | KNKEKALII | 109–117 | Sporozoite surface protein 2 | TRAP | HLA-B8 | SMWALIISV | ORF1ab | 3661–3669 | 44.4 |
| 2632 | ALIIIRSLL | 114–122 | TRAP precursor | TRAP | HLA-A∗02:01 | ||||
HLA, human leukocyte antigen; MHC, major histocompatibility complex; ORF, open reading frame; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TRAP, thrombospondin-related anonymous protein.
We then analysed the epitopes generated from the prediction approach, which resulted in the same sequences shared between TRAP epitopes and SARS-CoV-2 N and ORF1ab proteins with other MHCI and MHCII restrictions. Additionally, there was a novel immunodominant epitope identified in sporozoite surface protein 2 (SSP-2) located at 80–90 which shared a pentapeptide (55.6%) with SARS-CoV-2 S protein (1192–1200 aa) (100% identity with SARS-CoV, RF = 0.29). Both sequences also bind the HLA-A∗02:01 molecule and can be another potential candidate for the cellular immune response. Upon testing the sequence homology between SARS-CoV-2 predicted T-cell MHC epitopes for the accessory ORF proteins against the experimentally identified T-cell MHC epitopes from the P. falciparum immunome, we found that ORF3a epitopes (IVDEPEEHV, 236–244 aa; ALLAVFQSA, 51–59) share 44.4% homology with TRAP epitopes (DLDEPEQFRL, 543–552 aa; GLALLACAGL, 504–513 aa) respectively. These epitopes are again restricted by the HLA-A∗02:01 molecule (Table 5).
Table 5.
Experimental T-MHC restriction epitopes from Plasmodium falciparum that share homology with predicted SARS-CoV-2 ORF T-cell MHC epitopes
| P. falciparum |
SARS-CoV-2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | Sequence | Mapped start–end | Epitope antigen name | Epitope parent name | MHC allele | Sequence | Protein name | Mapped start–end | Identity (%) |
| 9041 | DLDEPEQFRL | 543–552 | TRAP precursor | TRAP | HLA-A∗02:01 | IVDEPEEHV | ORF3a | 236–244 | 44.4 |
| 20762 | GLALLACAGL | 504–513 | TRAP precursor | TRAP | HLA-A∗02:01 | ALLAVFQSA | ORF3a | 51–59 | 44.4 |
| 68914 | VICSFLVFL | 9–17 | Hypothetical protein PFL0800c | Uncharacterized protein | HLA-A∗02:03, HLA-A∗02:06 | LVFLGIITTV | ORF8 | 4–13 | 44.4 |
| 68915 | VICSFLVFLV | 9–18 | HLA-A∗02:03, HLA-A∗02:06 | 40 | |||||
| 16939 | FLVFLVFSNV | 13–22 | HLA-A∗02:06 | 40 | |||||
| 16939 | FLVFLVFSNV | 13–22 | Hypothetical protein PFL0800c | Uncharacterized protein | HLA-A∗02:01, HLA-A∗02:03 | FLVFLGIITT | ORF8 | 3–12 | 50 |
| 68914 | VICSFLVFL | 9–17 | HLA-A∗02:01, HLA-A∗02:03 | 55.6 | |||||
| 68915 | VICSFLVFLV | 9–18 | HLA-A∗02:01, HLA-A∗02:03 | 50 | |||||
| 16939 | FLVFLVFSNV | 13–22 | Hypothetical protein PFL0800c | Uncharacterized protein | HLA-A∗02:06 | LVFLGIITT | ORF8 | 4–12 | 44.4 |
| 68914 | VICSFLVFL | 9–17 | HLA-A∗02:06 | ||||||
| 68915 | VICSFLVFLV | 9–18 | HLA-A∗02:06 | ||||||
HLA, human leukocyte antigen; MHC, major histocompatibility complex; ORF, open reading frame; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TRAP, thrombospondin-related anonymous protein.
Discussion
The outbreak of COVID-19 caused by SARS-CoV-2 has caused significant devastation on multiple fronts, with 8061550 total confirmed cases and 440290 deaths globally as of 17 June 2020 [27]. This pandemic has adversely affected social practices, healthcare systems and economies. Despite the incremental increase in incidence in various parts of the world, Africa has remained the continent with the lowest number of confirmed cases and deaths. The poor socioeconomic status of Africa makes it one of the most vulnerable regions in the world as a result of malnutrition, endemic tropical infections, fragile healthcare systems, poverty and social practices that encourage close gatherings [4]. However, the consensus is that the actual situation of the pandemic in Africa remains unclear. According to the Director of the Africa Centres for Disease Control and Prevention (Africa CDC, UN), only 1.3 million COVID-19 tests were conducted by mid-May across the continent (one test for every 1000 individuals), compared to 3.6 million tests performed in Italy by the 27 May 2020 [28]. The social distancing applied by many governments in Africa to mitigate the pandemic is now creating extra financial hardship and food insecurity among African women and may not be effective in the future [29]. Despite the higher relative youth percentage in the African population (43% under 15 years old) compared to the European population (14% under 15 years old), the majority of the cases now are reported among individuals younger than 65 [30]. Nevertheless, regardless of the abovementioned challenges, as of 17 June 2020, it is apparent that African countries located within the HBHI of malaria (42 countries) reported fewer numbers of confirmed cases (n = 111852) and mortality (0.95%) due to SARS-CoV-2 compared to the 12 African countries outside the malaria zone (n = 149089 and 1.72% respectively). These findings are supported by our statistical analysis, which indicated a significantly lower risk of COVID-19 in malaria-endemic areas (p < 0.0001) with emphasis on P. falciparum–endemic areas (p < 0.001) and a reverse correlation between P. falciparum and COVID-19 death rates (r2 = −0.218, p < 0.001).
In malaria-endemic areas, the net immune response of one disease can be influenced by the predominant pathogen. The immune response to SARS-CoV-2 infection in malaria-endemic areas could be affected by prior exposure to the Plasmodium parasite [31]. However, considering the immune response through cross-reactivity could also be of value. The concept of ‘original antigenic sin’ is well known and had been used to explain many immunologic phenomena. Essentially, an adaptive immune response against one antigen can be used to combat another exposure by an unrelated antigen [32], with the second antigen relying on the memory established by the first antigen to initiate response [33,34]. In T-cell cross-reactivity, closely related sequences recognition is common and typically occurs between genetically related organisms. This is well documented for the same epitope isolated from different hepatitis C virus strains. Each epitope had a minor variation constricted to a single amino acid change. In this case, cross-reactivity occurs, but with substantial differences in the priming and magnitude of response [35]. Nonetheless, cross-reactivity can also be seen among random peptides or peptides with no shared sequences, or between those bearing relativity low homology [36,37]. The mechanism of recognition between T cell Receptor (TCR) and the epitope–MHC complex, where binding is not governed by the chemical principles of the epitope sequence, is well documented [38]. Searching the T-cell–immunodominant epitopes as well as T-cell MHC-restricted epitopes for shared sequences between P. falciparum and SARS-CoV-2 revealed several conserved tetrapeptides and pentapeptides between the two organisms. Because both organisms are evolutionarily distant, the four or five amino acids shared were considered to be of significance. These similarities were observed among immunodominant sequences N protein from SARS-CoV-2/SARS-CoV and TRAP from P. falciparum; and S protein–SARS-CoV-2/SARS-CoV and the predicted epitope in SSP-2 from P. falciparum. Separately, both epitopes can stimulate CD8+ T-lymphocyte response through HLA-A∗02:01 recognition.
In this argument, we have carefully applied the doctrine of original antigenic sin to explain possible cross-reactivity between SARS-CoV-2 and P. falciparum. TRAP is a type 1 membrane protein essential for sporozoite motility and cellular invasion. Twenty-one percent of MHCI-restricted CD8+ T-cell epitopes are found in this protein, and 28.6% to 100% of subjects from malaria-endemic areas responded to 504-GLALLACAGL-513–immunodominant epitope with restriction to HLA-A∗02:01 [39]. Four amino acid determinants found in this epitope were shared by SARS-CoV-2 nucleocapsid protein 219-LALLLLDRL-227, the recognition of which is also restricted by HLA-A∗02:01 [21]. Our assumption here is that the memory of the cellular adaptive immunity mounted against the abovementioned TRAP-immunodominant epitope could recognize the 219-LALLLLDRL-227–HLA-A∗02:01 complexes originating from SARS-CoV-2 infection in malaria-endemic regions and trigger an immune response. Of course, such an assumption needs further empirical testing to prove its validity and ascertain the strength of the primed response.
Although we could not find any shared immunodominant epitopes between P. falciparum and SARS-CoV-2 B-cell epitopes, many recent letters to the editors have suggested that antibodies against glycol immunodeterminants on P. falciparum infection could recognize SARS-CoV-2 envelope glycoproteins and induce activation of the complement system and proinflammatory cytokines [40,41].
Death due to malaria is attributed in part to Plasmodium invasion of erythrocytes, a process mediated by RH5-CD147 interactions. Recent findings have indicated infection of the host cell by SARS-CoV-2 through spike protein–CD147 interaction [42]. Though the CD147 receptor is also expressed on the erythrocyte surface, these authors focused only on the lung's receptor. Ulrich and Pillat [43] proposed the CD147 receptor as a target for COVID-19 treatment. In a recent study, homology modelling and molecular docking revealed that SARS-CoV-2 could attack haemoglobin and inhibit haeme metabolism. The authors also showed that the viral nonstructural proteins ORF3a and ORF1ab are assisted with the viral haemoglobin attack [44]. Of COVID-19 patients admitted to the intensive care unit with acute pneumonic complications, ∼49% presented with coagulopathy and thrombotic events [45]. High levels of d-dimer, or fibrinolysis-degraded fragments of fibrin, are often associated with COVID-19 infection and are used to detect in-hospital mortality [46,47], a situation that is also common with P. falciparum and P. vivax malaria [48]. Although not yet conclusively demonstrated, it is alleged that mature and immature erythrocytes could be implicated in SARS-CoV-2 entry into the body [49]. Both cells have CD147 and sialic acid receptors but lack angiotensin-converting enzyme 2 (ACE2) receptors. Unlike nucleated erythrocytes, mature RBCs lack the required machinery to support virus replication and provoke an immune response through MHC molecules. Therefore, erythrocyte-mediated infections are often fatal, as they go undiscovered by the immune clearance system. Nevertheless, viruses attacking erythrocytes would attract circulating antibodies, leading to antibody clumping. This would initiate a cascade of inflammatory responses that eventually leads to blood clotting. Given that P. falciparum antigens (TRAP and SSP-2) share immunodominant epitopes with SARS-CoV-2 antigens N, S, ORF1ab and ORF3a (Table 3, Table 4, Table 5), in addition to the involvement of the CD147 receptor in the invasion process of the virus to the erythrocyte, a receptor that is commonly used by P. falciparum in the blood stage, and the subsequent finding that the nanolipid Metadichol could Moderately inhibit both SARS-CoV-2 by blocking the ACE2 receptor and the malaria parasite [50], could indicate a plausible SARS-CoV-2 infection route related to the blood system.
Conclusion
The shared immunodominant epitopes with cross-immunogenic reactivity between SARS-CoV-2 antigens S, N, ORF1ab and ORF3a to that of the P. falciparum antigens TRAP and SSP-2 which are reported in this investigation could suggest an answer for the ambiguous reason why the lowest number of COVID-19 infections and mortality rates exist in malaria-endemic regions compared to the rest of the world. These results support other recently published data that relate to erythrocyte CD147 receptor and surrogate entry for the virus. This, in addition to the several shared epitopes between SARS-CoV-2 with those antigens from P. falciparum which are related to the later RBC invasion, are collectively suggested as a probable alternative route for SARS-CoV-2 via RBCs, although this remains to be practically demonstrated and warrants future investigations to confirm their validity.
Acknowledgements
The authors thank T. Zangeneh, D. Mahadevan and A. Kraft for their reviews. Dr. T. Zangeneh, Department of Medicine, Banner University Medical Center, AZ, USA, Dr. D. Mahadevan, University of Texas Health, San Antonio, TX, USA, Dr. A. Kraft, Emeritus University of Arizona Cancer Center, AZ, USA. We would like to thank the Federation of Arab Scientific Research Councils (FASRC), for funding the publication fee of this article.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nmni.2020.100817.
Conflicts of interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fauci A.S., Lane H.C., Redfield R.R. Covid-19—navigating the uncharted. N Engl J Med. 2020;382:1268–1269. doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization (WHO) 4 December 2019. World malaria report 2019.https://www.who.int/publications/i/item/9789241565721 Available at: [Google Scholar]
- 5.World Health Organization (WHO) Malaria country profile 2018. https://www.who.int/malaria/publications/world-malaria-report-2018/en/ Available at:
- 6.World Health Organization (WHO) Coronavirus disease (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 Available at:
- 7.Cserti C.M., Dzik W.H. The ABO blood group system and Plasmodium falciparum malaria. Blood. 2007;110:2250–2258. doi: 10.1182/blood-2007-03-077602. [DOI] [PubMed] [Google Scholar]
- 8.Rowe J.A., Handel I.G., Thera M.A., Deans A.M., Lyke K.E., Koné A. Blood group O protects against severe Plasmodium falciparum malaria through the mechanism of reduced rosetting. Proc Natl Acad Sci U S A. 2007;104:17471–17476. doi: 10.1073/pnas.0705390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taha A., Osman M., Abdolelkarim E., Holi M., Elbasheir M., Abuzeid N. Individuals with ‘A’ Rh-positive but not Rh-negative blood group are more vulnerable to SARS-CoV-2 infection: demographics and trend study on COVID-19 cases in Sudan. New Microbes New Infect. 2020 doi: 10.1016/j.nmni.2020.100763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J., Yang Y., Huang H., Li D., Gu D., Lu X. 27 March 2020. Relationship between the ABO blood group and the COVID-19 susceptibility. medRxiv. [DOI] [Google Scholar]
- 11.Sargin G., Yavaşoğlu S., Yavasoglu I. Is coronavirus disease 2019 (COVID-19) seen less in countries more exposed to malaria? Med Hypotheses. 2020;140:109756. doi: 10.1016/j.mehy.2020.109756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed A.E. Incidence of coronavirus disease (COVID-19) and countries affected by malarial infections. Travel Med Infect Dis. 2020 doi: 10.1016/j.tmaid.2020.101693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Alvarez M., Jarde A., Usuf E., Brotherton H., Bittaye M., Samateh A.L. COVID-19 pandemic in west Africa. Lancet Glob Health. 2020;8:e631–e632. doi: 10.1016/S2214-109X(20)30123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mbow M., Lell B., Jochems S.P., Cisse B., Mboup S., Dewals B.G. COVID-19 in Africa: dampening the storm? Science. 2020;369:624–626. doi: 10.1126/science.abd3902. [DOI] [PubMed] [Google Scholar]
- 15.Njenga M.K., Dawa J., Nanyingi M., Gachohi J., Ngere I., Letko M. Why is there low morbidity and mortality of COVID-19 in Africa? Am J Trop Med Hyg. 2020;103:564–569. doi: 10.4269/ajtmh.20-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubota Y., Shiono T., Kusumoto B., Fujinuma J. Multiple drivers of the COVID-19 spread: the roles of climate, international mobility, and region-specific conditions. PLoS One. 2020;15 doi: 10.1371/journal.pone.0239385. e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lalaoui R., Bakour S., Raoult D., Verger P., Sokhna C., Devaux C. What could explain the late emergence of COVID-19 in Africa? New Microbe. New Infect. 2020;38:100760. doi: 10.1016/j.nmni.2020.100760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct Target Ther. 2020;5:237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization (WHO) April 1 2020. Coronavirus disease (COVID-19) situation report 72.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Available at: [Google Scholar]
- 21.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vita R., Overton J.A., Greenbaum J.A., Ponomarenko J., Clark J.D., Cantrell J.R. The immune epitope Database (IEDB) 3.0. Nucleic Acid Res. 2015;43:D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R. The immune epitope Database (IEDB): 2018 update. Nucleic Acid Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanduc D., Shoenfeld Y. Inter-pathogen peptide sharing and original antigenic sin: solving a paradox. Open Immunol J. 2018;8:16–27. [Google Scholar]
- 25.Kim Y., Ponomarenko J., Zhu Z., Tamang D., Wang P., Greenbaum J. Immune epitope Database analysis Resource. Nucleic Acid Res. 2012;40:W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q., Wang P., Kim Y., Haste-Andersen P., Beaver J., Bourne P.E. Immune epitope Database analysis Resource (IEDB-AR) Nucleic Acid Res. 2008;36:W513–W518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization (WHO) June 17 2020. Coronavirus disease (COVID-19) situation report 149.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Available at: [Google Scholar]
- 28.Soy A. BBC News; 27 May 2020. Lack of COVID-19 testing undermines Africa’s ‘sucess’.https://www.bbc.com/news/world-africa-52801190 Available at: [Google Scholar]
- 29.Wenham C., Smith J., Morgan R. COVID-19: the gendered impacts of the outbreak. Lancet. 2020;395:846–848. doi: 10.1016/S0140-6736(20)30526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCaffrey D. Euronews; 12 May 2020. Analysis: Africa’s unexpected COVID-19 figures.https://www.euronews.com/2020/05/12/analysis-africa-s-unexpected-covid-19-figures Available at: [Google Scholar]
- 31.Clark I.A., Alleva L.M., Budd A.C., Cowden W.B. Understanding the role of inflammatory cytokines in malaria and related diseases. Travel Med Infect Dis. 2008;6:67–81. doi: 10.1016/j.tmaid.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Muraille E. The unspecific side of acquired immunity against infectious disease: causes and consequences. Front Microbiol. 2015;6:1525. doi: 10.3389/fmicb.2015.01525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank S. Princeton University Press; Princeton: 2002. Immunology and evolution of infectious disease. [PubMed] [Google Scholar]
- 34.Vatti A., Monsalve D.M., Pacheco Y., Chang C., Anaya J.M., Gershwin M.E. Original antigenic sin: a comprehensive review. J Autoimmun. 2017;83:12–21. doi: 10.1016/j.jaut.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Ziegler S., Skibbe K., Walker A., Ke X., Heinemann F.M., Heinold A. Impact of sequence variation in a dominant HLA-A∗02-restricted epitope in hepatitis C virus on priming and cross-reactivity of CD8+ T cells. J Virol. 2014;88:11080–11090. doi: 10.1128/JVI.01590-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evavold B.D., Sloan-Lancaster J., Wilson K.J., Rothbard J.B., Allen P.M. Specific T cell recognition of minimally homologous peptides: evidence for multiple endogenous ligands. Immunity. 1995;2:655–666. doi: 10.1016/1074-7613(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 37.Ishizuka J., Grebe K., Shenderov E., Peters B., Chen Q., Peng Y. Quantitating T cell cross-reactivity for unrelated peptide antigens. J Immunol. 2009;183:4337–4345. doi: 10.4049/jimmunol.0901607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin Y., Mariuzza R.A. The multiple mechanisms of T cell receptor cross-reactivity. Immunity. 2009;31:849–851. doi: 10.1016/j.immuni.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Heide J., Vaughan K.C., Sette A., Jacobs T., Schulze Zur Wiesch J. Comprehensive review of human Plasmodium falciparum–specific CD8+ T cell epitopes. Front Immunol. 2019;10:397. doi: 10.3389/fimmu.2019.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parodi A., Cozzani E. Coronavirus disease 2019 (COVID 19) and malaria: have anti glycoprotein antibodies a role? Med Hypotheses. 2020;143:110036. doi: 10.1016/j.mehy.2020.110036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panda A.K., Tripathy R., Das B.K. Plasmodium falciparum infection may protect a population from severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K., Chen W., Zhou Y.S., Lian J.Q., Zhang Z., Du P. bioRxiv; 14 March 2020. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. [DOI] [Google Scholar]
- 43.Ulrich H., Pillat M.M. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev Rep. 2020;16:434–440. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W., Li H. ChemRxiv; 13 July 2020. COVID-19: attacks the 1-beta chain of hemoglobin and captures the porphyrin to inhibit human heme metabolism.https://chemrxiv.org/articles/COVID-19_Disease_ORF8_and_Surface_Glycoprotein_Inhibit_Heme_Metabolism_by_Binding_to_Porphyrin/11938173 Available at: [Google Scholar]
- 45.Wise J. COVID-19 and thrombosis: what do we know about the risks and treatment? BMJ. 2020;369:m2058. doi: 10.1136/bmj.m2058. [DOI] [PubMed] [Google Scholar]
- 46.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L., Yan X., Fan Q., Liu H., Liu X., Liu Z. d-Dimer levels on admission to predict in-hospital mortality in patients with COVID-19. J Thromb Haemost. 2020;18:1324–1329. doi: 10.1111/jth.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dasgupta A., Rai S., Das Gupta A. Persistently elevated laboratory markers of thrombosis and fibrinolysis after clinical recovery in malaria points to residual and smouldering cellular damage. Indian J Hematol Blood Transfus. 2012;28:29–36. doi: 10.1007/s12288-011-0106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cavezzi A., Troiani E., Corrao S. COVID-19: hemoglobin, iron, and hypoxia beyond inflammation. A narrative review. Clin Pract. 2020;10:1271. doi: 10.4081/cp.2020.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raghavan P. Social science research network (SSRN) 7 July 2020. Metadichol®, a novel nano lipid formulation that inhibits SARS-CoV-2 and a multitude of pathological viruses in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.