Abstract
Background
Salivary tests for the new coronavirus (SARS-CoV-2) diagnosis have been suggested as alternative methods for the nasopharyngeal and oropharyngeal tests.
Method
Two reviewers independently performed a search in the following electronic databases: PubMed, Medline, Cochrane Library, Web of Science, Embase and Scopus to identify cross-sectional and cohort studies that used saliva samples for SARS-CoV-2 detection. The search strategy was: (“saliva”) and (“SARS-CoV-2” or “coronavirus” or “COVID-1”).
Results
A total of 363 studies were identified and 39 were selected for review. Salivary samples for SARS-CoV-2 detection was as consistent and sensitive as the nasopharyngeal swabs in most studies, having been effective in detecting asymptomatic infections previously tested negative in nasopharyngeal samples. Viral nucleic acids found in saliva obtained from the duct of the salivary gland may indicate infection in that gland. Live viruses could be detected in saliva by viral culture.
Conclusions
Salivary samples show great potential in SARS-CoV-2 detection and may be recommended as a simple and non-invasive alternative.
Keywords: Saliva, Coronavirus infections, Review
Highlights
-
•
It is a literature review reporting the saliva as tool for SARS-CoV-2 detection.
-
•
Viral nucleic acids was found in saliva obtained from duct of salivary gland.
-
•
Saliva samples are as consistent as nasopharyngeal samples in SARS-CoV-2 detection.
-
•
Saliva sample is low cost and non-invasive, being a viable alternative.
1. Introduction
In December 2019, an infection outbreak of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) emerged in Wuhan, Hubei province, China, and rapidly spread around the world [1], having been declared a pandemic by the WHO on March 11, 2020. More than 37.8 million cases were reported by October 14, 2020 around the world, resulting in 1,081,868 deaths [2]. The new 2019 coronavirus (2019-nCoV) is easily transmitted between humans through aerosol generation from infected people coughing, speaking or sneezing in close contact with others, and has an incubation period that ranges from 1 to 14 days [3,4].
The genetic sequencing done to 2019-nCoV, on January 7, 2020, allowed for fast tool-development for diagnostic tests through RT-PCR (reverse transcription polymerase chain reaction) [1]. Besides preventing transmission, its early and rapid detection is essential in controlling the virus spread [3,5]. Nasopharyngeal swabs (NPS) are widely used and recommended as a standard sample for the respiratory virus diagnosis, including SARS-CoV-2. However, this approach requires close contact with health professionals, increasing the cross-infection risk and may cause discomfort, coughing and even bleeding in patients, not being so desirable for serial viral load monitoring [6,7].
Salivary use for viral infection diagnosis has produced interest in recent years, mainly because it is a non-invasive technique, easy to collect and has a low cost [8]. Due to the absence of a standard protocol, saliva collection can be obtained from: a) stimulated or unstimulated saliva t or through oral swabs. Several viral infections can be detectable in saliva, as Epstein Barr virus, HIV, Hepatitis C virus, Rabies virus, Human papillomavirus, Herpes simplex virus and Norovirus [9]. In addition, saliva has also been reported as a positive detection means for coronavirus nucleic acid associated with severe acute respiratory syndrome [10] and, more recently, SARS-CoV-2 [3].
The advantages of using saliva samples for SARS-CoV-2 diagnosis, such as self-collection and collection outside hospitals, are that multiple samples can be easily obtained and there is a reduced need for health care professional handling during the sample collection, reduced nosocomial transmission risk, reduced test waiting time, and reduced PPE, transport and storage costs [7]. Another benefit for this non-invasive and economical collection method is a better perspective as community monitoring, both for asymptomatic infections and to guide end of quarantine [5,11]. Therefore, this work aims to conduct an integrative literature review reporting saliva use as a sample for the SARS-CoV-2 diagnosis.
2. Materials and methods
2.1. Research question
Can saliva be used as a diagnostic sample for SARS-CoV-2?
2.2. Search strategy
It is a current literature integrative review on saliva's use as a diagnostic tool for the new coronavirus discovered in 2019. Searches were carried out in the main national and international databases (PubMed, Medline, Cochrane Library, Web of Science, Embase and Scopus), in addition to a manual search. The search strategy used in all databases mentioned included the keywords and the terms MeSH (Medical Subject Headings): (“saliva”) and (“SARS-CoV-2” or “coronavirus” or “COVID-19″). The analyzed articles were published in English, Spanish or Portuguese.
For this integrative review, cross-sectional and cohort studies that corresponded the inclusion criteria dated from January to October 2020 were selected. Inclusion was based on two blind reviewers reading the manuscripts and confirming the eligibility criteria. The inclusion criteria referred to articles in English, Spanish or Portuguese who utilized saliva tests for detection of SARS-CoV-2. The exclusion criteria referred to articles that did not consider saliva as a possible diagnostic strategy for 2019-nCoV, besides literature reviews, letters to the editor, protocols and case reports.
Assessments were carried out by two independent reviewers in order to verify if the selected articles met the inclusion criteria. In case of disagreements, a third reviewer was consulted regarding the decision to include or not the article. Initially, studies were identified by reading title and articles abstracts. After that, the references selected were analyzed, manual searches for new studies were performed. Nineteen cross-sectional and cohort studies were considered in this research. After reading the selected articles, relevant information was collected and typed into a database according to the following criteria: authors, study design, study population, sample size, age range, test, saliva type, saliva collection method, respiratory or another sample and results (Table A.1, Table A.2, Table A.3).
Table A.1.
Studies that directly compared saliva use in SARS-CoV-2 detection with respiratory samples and that demonstrated good saliva results. Natal/RN, 2020.
| Authors | Study design | Study population | Sample size [Gender] | Age range [Mean] (Years) | Test | Saliva type | Saliva collection method | Respiratory sample | Results |
|---|---|---|---|---|---|---|---|---|---|
| Wyllie et al., 20206 | Cohort study | Patients who tested positive for SARS-CoV-2. Asymptomatic health professionals. |
44 [23 M, 21 F] 98 [16 M, 82 F] |
23-92 [61] 22-67 [36] |
RT-PCR | Saliva | Self-collection by spit. | OPS or NPS | 37 samples (84%) of saliva tested positive for SARS-CoV-2. SARS-CoV-2 detection in the saliva from two health professionals who tested negative by nasopharyngeal swab. |
| To et al., 20207 | Cohort study | Patients hospitalized with COVID-19 confirmed in the laboratory. | 12 [7 M, 5 F] | 37-75[62,5] | RT-PCR and viral culture | POS | Saliva with cough. | NPS | 2019-nCoV was detected in the initial saliva samples from 11 patients (91.7%). Live viruses were found in the saliva from 3 patients. |
| Azzi et al., 202012 | Cohort study | Patients with severe or very severe COVID-19. | 25 [17 M, 8 F] | 39-85 [61,5] | RT-qPCR | Saliva | Drooling technique. | NPS | All 25 initial samples showed positive results for the SARS-CoV-2 presence. |
| To et al., 202013 | Cohort study | Patients hospitalized with COVID-19 confirmed in the laboratory. | 23 [13 M, 10 F] | 37-75 [62] | RT-qPCR and EIA | POS | Saliva with cough. | NPS | 20 cases (87%) in which 2019 RNA-nCoV was detectable in saliva. |
| Iwasaki et al., 202014 | Cohort study | Patients suspicious of COVID-19 and patients with the diagnosis of COVID-19. | 76 [-] | 30-97∗[69] | RT-qPCR | Saliva | Self-collection by spit. | NPS | SARS-CoV-2 was detected in 8 (among 10 patients) with COVID-19 in nasopharyngeal and saliva samples. The overall agreement rate for virus detection was 97.4%. |
| Yokota et al., 202015 | Cohort study | Asymptomatic persons who have close contact with clinically confirmed COVID-19 patients. Asymptomatic travellers arriving at Tokyo and Kansai international airports |
161 [44 M, 26 F, 91U] 1763 [927 M, 832 F, 4U] |
29.8–66.4 [44.9] 22.6–47.4 [33.5] |
qRT-PCR and RT-LAMP | Saliva | Self-collection. | NPS | The nasopharynx and saliva samples obtained high sensitivity, with 86% and 92%, respectively, and specificity (greater than 99%) to the nucleic acid amplification test. |
| Wyllie et al., 202016 | Cohort study | COVID-19 inpatients at Yale-New Haven Hospital. | 70 [41 M, 29 F] | 13-91[61,4] | RT-PCR | Saliva | Self-collection by spit. | NPS | From 1 to 5 days after diagnosis, saliva samples (81%) were positive, while nasopharyngeal smear samples (71%) were positive, suggesting that both samples have at least similar sensitivity in the initial detection of SARS-CoV-2. |
| Han et al., 202017 | Cohort study | Mildly symptomatic and asymptomatic children with coronavirus disease. | 12 [5 M, 7 F] | 27 (days)-16 [5,6] | – | – | – | NPS | Saliva was collected from 11 children, with 8 tested positive for SARS-CoV-2. Positivity in saliva samples decreased from 80% at week 1–33% at week 2 and 11% at week 3. |
| Mao et al., 202018 | Cohort study | Patients with asymptomatic disease Patients with mild disease Patients with moderate disease |
6 [4 M, 2 F] 6 [6 M, 0 F] 22 [10 M, 12 F] |
28–48 [37] 21-57 [7,38] 21-64 [4,44] |
RT-qPCR | – | – | OPS and sputum | Saliva sample sensitivity, efficiency and specificity were only 74.10%, 83.90% and 94.40%, respectively, whereas for saliva-expectoration they were 93.40%, 94.00% and 95.20%, respectively, being a more effective diagnostic method. |
| Kojima et al., 202019 | Cross-sectional study | Symptomatic individuals not hospitalized recently tested for SARS-CoV-2 infection. | 45 [-] | 31-52 [42] | RT-qPCR | Saliva | Self-collected oral swab with and without doctor's supervision. | NPS and nasal swab | Oral swab samples self-collected and supervised by the physician detected 26 (90%) of the 29 infected individuals. Non-monitored self-collected oral fluid swab samples detected 19 (66%). |
| Pasomsub et al., 202020 | Cross-sectional study | Patients who experience fever or acute respiratory symptoms, along with a 14-day travel history from a COVID-19 endemic area or contact with an individual who has been confirmed or COVID-19 suspected. | 200 [69 M, 131 F] | 28-48 [36] | RT-PCR | Saliva | Saliva without cough. | NPS and throat swab | The COVID-19 prevalence diagnosed by saliva RT-PCR was 9.0%, showing high sensitivity and performance 84.2% and 98.9%, respectively. |
| Chen et al., 202021 | Cross-sectional study | Positive patients for COVID-19. | 58 [28 M, 30 F] | 31-52 [38] | RT-PCR POCT | POS | Saliva with cough. | NPS | Some patients (84.5%) had a positive result in both the nasopharyngeal swab and saliva, 10.3% had a positive result only in the nasopharyngeal swab and 5.2% had a positive result only in saliva. |
| McCormick-Baw et al., 202022 | Cross-sectional study | Patients with suspected COVID-19 and hospitalized patients positive for COVID-19 without the need for mechanical ventilation. | 156 [90 M, 66 F] | [8,47] | POCT | Saliva | Saliva without cough. | NPS | 49 positive tests by nasopharyngeal swab (47 also had positive saliva samples). A single sample demonstrated detectable SARS CoV-2 nucleic acid levels in saliva, but the nasopharyngeal swab was negative. |
| Leung et al., 202023 | Cross-sectional study | Patients admitted to the Prince of Wales Hospital in Hong Kong. | 62 [26 M, 36 F] | 19-85 [42] | RT-PCR | POS | Saliva with cough. | NPS | 95 sample pairs, 75 were positive for both nasopharyngeal and saliva samples; 13 positive saliva samples had corresponding negative nasopharyngeal samples and 7 positive nasopharyngeal samples had negative saliva samples. |
| Cheuk et al., 202024 | Cross-sectional study | Tested Patients for COVID-19. | 95 [38 M, 57 F] | 4-92 [39] | RT-PCR | Saliva | Self-collection by spit. | NPS | Saliva and nasopharyngeal samples positivity was 61.6% and 53.3%, respectively. Among the 6 discordant results, 4 presented positive saliva samples and negative nasopharyngeal samples. |
| Güçlü et al., 202025 | Cross-sectional study | Hospitalized patients with and without laboratory-confirmed Covid-19, with a finding consistent with COVID-19 in the Lung Computed Tomography (CT), and patients with complaints compatible with COVID-19 but normal CT. | 64 [37 M, 27 F] | -[51,04] | RT-PCR | Saliva | Self-collection. | OP/NP swab | Among 64 patients, 23 (35.9%) obtained positive saliva and OP-NP swab samples, 4 (6.25%) had only positive saliva sample and 4 (6.25%) only the OP/N swab. NP was positive. In general, saliva sensitivity and specificity was 85.19% and 89.19%, respectively. |
| Vaz et al., 202026 | Cross-sectional study | Health professionals with signs/symptoms suggestive of COVID-19, and infirmary patients with confirmed infection. | 155 [46 M, 109 F] | 33-48,5 [40] | RT-PCR | Saliva | Saliva without cough. | NPS or OPS | RT-PCR sensitivity and specificity for saliva were 94.4% and 97.62%, respectively. In addition, there was a high general agreement (96.1%) comparing the salivary sample with the gold standard test. |
| Rao et al., 202027 | Cross-sectional study | Asymptomatic adult male participants in a COVID-19 quarantine center. | 160 [160 M] | 18-36 [27] | RT-PCR | POS | Saliva with cough. | NPS | A higher detection rate for SARS-CoV-2 was found in saliva compared to NPS, being 93.1% and 52.5%, respectively. E and RdRp genes Ct values from the 73 concordant samples were significantly lower in saliva than in NPS (p < 0.05). |
| Byrne et al., 202028 | Cross-sectional study | Patients with COVID-19 symptoms. | 110 [49 M, 61 F] | – | RT-qPCR | Saliva | Self-collection. | Nasal and throat swab | Among 110 paired samples, 12 saliva samples tested positive for SARS-CoV-2, while 14 nasal and throat swab samples tested positive. The general viral loads were similar among all positive samples, ranging from 36 to 3.3 × 10 6 copies/mL. |
| Hanson et al., 202029 | Cross-sectional study | Adult patients with symptoms suggestive of COVID-19. | 354 [195 M, 173 F] | 18-75 [35] | RT-PCR | Saliva | Self-collection by spit. | NPS and anterior nasal swab | NPS and saliva samples had the highest positivity rates (22.5% and 22.9%) compared to the ANS (19.7%). The average Ct values for positive samples only for NPS was 27.0, and 28.2 for positive samples only for saliva. |
| Aita et al., 202030 | Cross-sectional study | Inpatients with COVID-19. | 49 [33 M, 16 F] | 28-86 [64] (M) 25-94 [60] (F) |
rRT-PCR | Saliva | Swab with absorbed saliva. | NPS | Among 43 patients with NPS and saliva samples paired, 7 cases tested positive in both samples and 35 tested negative for both NPS and saliva. One patient tested positive for saliva (Ct = 26), but not for NPS. |
| Uwamino et al., 202031 | Cross-sectional study | Hospitalized patients with COVID-19. Symptomatic university staff. |
32 [-] 115 [-] |
– | RT-PCR | Saliva | Saliva without cough. | NPS | From 196 samples collected, 32 tested positive for SARS-CoV-2 by both NPS and saliva, 15 by NPS but negative by saliva, and 11 samples that tested positive for saliva had NPS negative. The results obtained in the first 10 days of symptom onset were 96.4%. It was possible to detect viable viruses in two saliva samples. |
| Migueres et al., 202032 | Cross-sectional study | Hospitalized and ambulatory patients. | 123 [49 M, 74 F] | - [43] | RT-PCR | Saliva | Self-collection by spit. | NPS | Thirty-four patients tested positive in both samples, three only for saliva and 7 only for NPS. Saliva samples sensitivity was high for asymptomatic and symptomatic patients tested early, with 88.2% and 94.7% respectively, and lowest (50%) for symptomatic patients tested late after symptoms onset. |
| Senok et al., 202033 | Cross-sectional study | Adult patients undergoing COVID-19 testing. | 401 [329 M, 72 F] | [5,35] | RT-PCR | Saliva | Saliva without cough. | NPS | Saliva sensitivity and specificity were 73.1% and 97.6%, respectively. The general SARS-CoV-2 detection prevalence by NPS was 6.5%, while, by saliva, it was 7%. |
| Altawalah et al., 202034 | Cross-sectional study | COVID-19 suspected patients. | 891 [-] | – | RT-PCR | POS | Saliva with cough. | NPS | The general agreement between the NPS and saliva samples was 91.25%. Saliva sensitivity and diagnostic specificity were 83.43% and 96.71%, respectively, and the detection rate was 83.43%. The median Ct values did not differ significantly between NPS and saliva. |
POS: Posterior Oropharyngeal Saliva; NPS: Nasopharyngeal swab; OPS: Oropharyngeal swab; RT-PCR: Reverse Transcription Polymerase Chain Reaction; rRT-PCR: Real-time Reverse Transcription Polymerase Chain Reaction; RT-qPCR: Quantitative Reverse Transcription Polymerase Chain Reaction; EIA: Enzyme Immunoassays; RT-LAMP: Reverse-Transcription Loop Mediated Isothermal Amplification; U: unknown; POCT: Point-of-care testing.
Age group (10 patients with COVID-19).
Table A.2.
Studies that directly compared saliva use in SARS-CoV-2 detection with respiratory samples and that demonstrated inaccurate saliva results. Natal/RN, 2020.
| Authors | Study design | Study population | Sample size [Gender] | Age range [Mean] (Years) | Test | Saliva type | Saliva collection method | Respiratory sample | Results |
|---|---|---|---|---|---|---|---|---|---|
| Jamal et al., 202035 | Cohort study | Positive patients for COVID-19 with nasopharyngeal, midturbinate or nasal swab. | 91 [52 M, 39 F] | 23-106 [66] | RT-PCR | Saliva | Self-collection by spit. | NPS | 72 patients had at least one positive specimen (nasopharyngeal swab or saliva). 61% of these 72 patients, both were positive, 28% only the nasopharyngeal swab was positive, and in 11% only saliva was positive. Nasopharyngeal swabs were 17% more sensitive than saliva overall. |
| Fang et al., 202036 | Cohort study | COVID-19 patients admitted to Central Hospital of Xiangtan. | 32 [16 M, 16 F] | 34-54 [41] | RT-PCR | – | – | Nasal swab. | Nasal swab samples showed 100.0% positivity, while the positive rate for saliva was 78.1%. The viral shedding time of SARS-CoV-2 of nasal swab was significantly longer than that of blood and saliva. |
| Kim et al., 202037 | Cohort study | Patients with SARS-CoV-2 infection. | 15 [5 M, 10 F] | 17-91 [59] | rRT-PCR | Saliva | Self-collection by spit. | NPS/OPS and sputum. | General rRT-PCR sensitivity for saliva compared to the sensitivity for naso/oropharyngeal samples was lower, being 64% and 77% respectively. |
| Chong et al., 202038 | Cohort study | COVID-19-infected children. | 18 [10 M, 8 F] | 1,8–11,1 [6,6] | rRT-PCR | Saliva | Self-collection by spit or through syringe. | NPS | The Ct values had statistically significant differences between saliva and NPS samples (1–3, 4–7 and 8–10 days after onset symptoms), and did not differ significantly in the period of 11–15 days. In five children, saliva samples tested negative on day 1–3 and became positive on day 4–7. |
| Williams et al., 202039 | Cross-sectional study | Outpatients who come to a COVID-19 screening clinic. | 622 [-] | – | RT-PCR | Saliva | Self-collection by spit. | NPS | 39 positive tests for SARS-CoV-2 by nasopharyngeal swab (33 tested positive for saliva). Nucleic acid was detected in the saliva from 1 out of 50 patients with negative test for nasopharyngeal swab. |
| Kam et al., 202040 | Cross-sectional study | Pediatric hospitalized patients confirmed for COVID-19. | 11 [-] | Symptomatic 2,1–12,5 [4,8] Asymptomatic 0,3–11,8 [3,8] |
RT-qPCR | Saliva | Oral swab. | NPS | SARS-CoV-2 was detected in at least 1 oral swab sample in 9 of the 11 children (81.8%). Two children with positive nasopharyngeal tests had negative results in the saliva samples in two collection days. In general, oral samples produced lower viral loads and had low sensitivity (25–71.4%) compared to nasopharyngeal samples. |
| Skolimowska et al., 202041 | Cross-sectional study | Symptomatic healthcare workers and household contacts presenting to a COVID-19 outpatient clinic. | 132 [43 M, 89 F] | 30-51 [39] | RT-PCR | Saliva | Saliva without cough. | NPS/OPS | Among the paired samples, 18 NP/OP swab samples tested positive, with 15 tested positive also for saliva. Saliva obtained sensitivity and specificity of 83.3% and 99.1%, respectively. Saliva Ct values were significantly higher than for swabs. |
| Lai et al., 202042 | Cross-sectional study | Patients with SARS-CoV-2 infection confirmed. | 50 [23 M, 27 F] | 16-72 [-] | RT-PCR | Saliva | Clearing the throat gargling saliva. | NPS and throat swab and sputum. | Saliva samples obtained RT-PCR positivity lower rates (68.7%) and lower viral RNA concentrations (mean log copy/mL 3.54) compared to sputum (89.4%, 5.03) and swabs (80.4%, 4.63). |
| Landry et al., 202043 | Cross-sectional study | COVID-19 suspected symptomatic outpatients. | – | – | RT-PCR | Saliva | Saliva without cough. | NPS | Among the 35 positive samples, 33 were positive for NPS, while 30 were positive for saliva. The general sensitivity for saliva was 85.7% (95% CI 70.6%–93.7%). The median Ct value was significantly lower for NPS than for saliva (p = 0.0331). |
NPS: Nasopharyngeal swab; OPS: Oropharyngeal swab; RT-PCR: Reverse Transcription Polymerase Chain Reaction; rRT-PCR: Real-time Reverse Transcription Polymerase Chain Reaction; RT-qPCR: Quantitative Reverse Transcription Polymerase Chain Reaction.
Table A.3.
Studies that did not directly compare saliva use in SARS-CoV-2 detection with respiratory samples. Natal/RN, 2020.
| Authors | Study design | Study population | Sample size [Gender] | Age range [Mean] (Years) | Test | Saliva type | Saliva collection method | Another samples | Results |
|---|---|---|---|---|---|---|---|---|---|
| Zhang et al., 202044 | Cohort study | Patients at Wuhan Pulmonar Hospital. | 15 [-] | – | RT-qPCR | Saliva | Oral swab. | Anal swab and blood. | 8 patients (53.3%) were positive for 2019-nCoV in oral swab on day 0, while there were only 4 (25%) positive oral swab on day 5. |
| Hung et al., 202045 | Cohort study | Patients confirmed with SARS-CoV-2 infection. | 18 [8 M, 10 M] | 18-61 [53] | RT-PCR | POS | Saliva with cough. | – | Higher viral loads were found in saliva samples collected in the morning, showing statistically significant differences when compared to the samples collected at night. |
| Chen et al., 202046 | Cross-sectional study | Patients whose 2019-nCoV nucleic acid detection remained positive before or on the sample collection day. | 31 [15 M, 16 F] | 18-86 [60,6] | RT-PCR | Saliva | Swab at the salivary gland canal opening. | OPS | 13 positive cases for 2019-nCoV nucleic acid detection by oropharyngeal swab (4 cases with positive saliva detection and 3 in serious condition). |
| Nagura-Ikeda et al., 202047 | Cross-sectional study | Patients with laboratory-confirmed COVID-19. | 103 [66 M, 37 F] | 18-87 [46] | LDT RT-qPCR, cobas SARS-CoV-2 test, direct RT-qPCR, RT-LAMP, RAT. | Saliva | Self-collection. | – | The viral RNA detection was significantly higher in saliva samples collected from symptomatic patients within 9 days after the symptoms onset than in samples collected 10 days after symptoms onset or in the saliva from asymptomatic patients. |
| Randad et al., 202048 | Cross-sectional study | Patients with RT-PCR confirmed prior SARS-CoV-2 infection | 28∗ [−] | [−] | RT-PCR, IgG, IgA, IgM | Saliva | Brushing the gum line. | Serum sample. | 22 amostras combinadas obtiveram resultado positivo para detecção de SARS-CoV-2 pela saliva e pelo soro, e 6 resultados foram negativos para ambas amostras. A detecção salivar específica de IgG para SARS-CoV-2 obteve alta sensibilidade e especificidade. |
POS: Posterior Oropharyngeal Saliva; OPS: Oropharyngeal swab; RT-PCR: Reverse Transcription Polymerase Chain Reaction; RT-qPCR: Quantitative Reverse Transcription Polymerase Chain Reaction; LDT: Laboratory developed tests; RT-LAMP: Reverse-Transcription Loop Mediated Isothermal Amplification; RAT: Rapid Antigen Test.
Participants with matched saliva-serum samples.
3. Results
3.1. Study selection
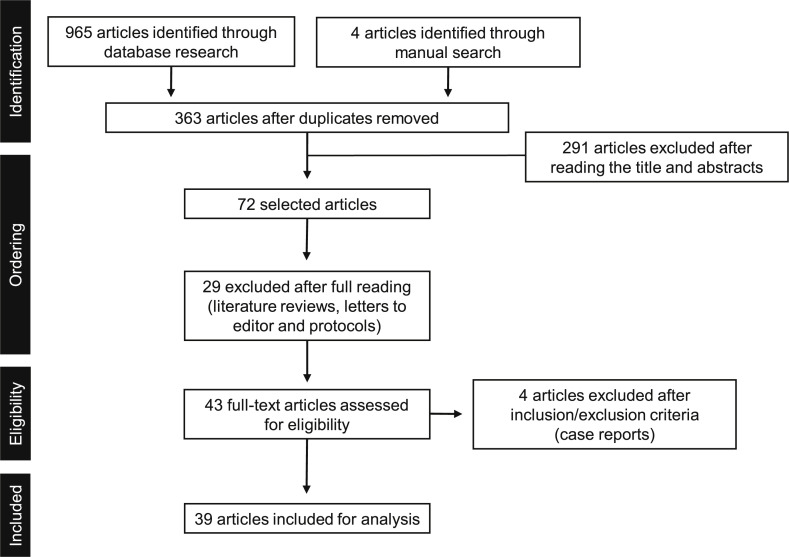
Nine hundred and sixty five studies published between January and October 2020 were identified after an initial electronic search in the main databases, in addition to four identified through manual search. Three hundred and sixty-three articles were evaluated after duplicates removed. After reading the titles and abstracts, 291 articles were excluded. Of the 72 articles selected for reading in full text, 29 were excluded (literature reviews, letters to the editor and protocols). Forty-three were fully evaluated, four being excluded when the inclusion/exclusion criteria were applied once they were case reports, totaling 39 manuscripts included in the analysis (Fig. A.1). No randomized controlled clinical trial was found. All manuscripts included were cross-sectional or cohort studies [6,7,[12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48]] (Table A.1, Table A.2, Table A.3).
Fig. A.1.
Article selection flowchart.
Nine hundred and sixty five studies identified through electronic search and 4 through manual search, totaling 363 articles after duplicates removed, published between January and October 2020 were evaluated. After reading the titles and abstracts, 291 articles were excluded. Of the 72 articles selected for reading in full text, 29 were excluded. Forty three were fully evaluated, four being excluded when the inclusion / exclusion criteria were applied, totaling thirty nine articles included for the current review .
In samples whose posterior oropharyngeal saliva is collected, other secretions are added besides the saliva that is secreted by larger or smaller salivary glands, which may come from the upper or lower respiratory tract and gingival fluid [7,55]. All studies that used this collection method demonstrated good performance in SARS-CoV-2 detection, including asymptomatic and pre-symptomatic infections [7,13,21,23,27,34].
3.2. Study characteristics
According to the methodology used and the results obtained, the studies were subdivided into three sessions: studies that directly compared saliva use in SARS-CoV-2 detection with respiratory samples and that demonstrated good saliva results; those who directly compared saliva use in SARS-CoV-2 detection with respiratory samples and who demonstrated inaccurate saliva results; and those who did not directly compare saliva use in SARS-CoV-2 detection with respiratory samples.
3.2.1. Studies that directly compared saliva use in SARS-CoV-2 detection with respiratory samples and that demonstrated good results from saliva
In total, 25 articles compared saliva use in relation to the other respiratory sample for the SARS-CoV-2 detection and obtained promising results regarding the salivary samples use, both for the initial diagnosis and for monitoring the course of the disease (Table A.1). The approaches reported in these studies for the saliva collection included: spit, cough, drooling technique, oral swab and swab with absorbed saliva.
Pasomsub et al., (2020) evaluated two hundred pairs of nasopharyngeal and oropharyngeal swab samples, and saliva samples from potentially infected patients, demonstrated that saliva tests had high sensitivity (84.2%) and specificity (98.9%), with good diagnostic performance when compared to standard nasopharyngeal and oropharyngeal swab tests.
High sensitivity and specificity were also found in a mass screening study, evaluating 1924 people possibly infected, both for saliva samples (86% and 99.96%) and for NPS (92% and 99.93%) [15]. In this study, three samples negative for saliva were positive for NPS, while six samples negative for NPS were positive for saliva. Observing the median cycle threshold (Ct) values, being 40 in the qRT-PCR test for NPS and between 33.7 and 37.2 per qRT-PCR for saliva, it was demonstrated that the viral load was equivalent between the samples.
Similarly, Wyllie et al., (2020) [6], Azzi et al., (2020), Kojima et al., (2020) and Pasomsub et al., (2020) were able to detect SARS-CoV-2 in saliva samples, while negative NPS were observed in 11% (2/18), 20% (6/29) 8% (2/25) and 21% (8/38) of infected patients, respectively, the last two after treatment days. In addition, viral RNA was found in the saliva from asymptomatic patients who presented negative NPS in three studies (2/98 [6]; 1/50 [39]; 1/106 [20]). Wyllie et al., (2020) [6] also demonstrated five nasopharyngeal sample cases (22.7%) in which there was a negative test followed by a positive result during the next collection, without any occurrences being seen for the saliva samples.
In contrast, To et al., (2020) [7] demonstrated that all patients whose nasopharyngeal samples showed negative results for 2019-nCoV, obtained negative results for saliva samples. Meanwhile, Vaz et al., (2020) found four participants with the virus detected in NPS or oropharyngeal swab (OPS) but not in the saliva. However, the difference in the SARS-CoV-2 detection rate found between saliva and swabs did not reach statistical significance.
It was detected a general decline in the saliva samples viral load after hospitalization in most patients [6,7,16]. A case with viral spillage in saliva was found by To et al., (2020) [7] even after 11 days on hospitalization. In another study by To et al., (2020) [13], 2019-nCoV RNA was detected for 20 days or more in recovered and symptom-free patients, and one patient tested positive for SARS-CoV-2 in saliva even after two days of negative results. In that study, the mean viral load of salivary or other respiratory samples was 5.2 log10 copies/mL.
When comparing the viral load between nasopharyngeal and salivary samples, Iwasaki et al., (2020) did not identify significant differences in viral loads between samples, with a mean of 5.4 ± 2.4 and 4.1 ± 1.4 log10 copies of the gene/ml in nasopharynx and saliva samples, respectively (p = 0.184), while Wyllie et al., (2020) [6], using a SARS-CoV-2 detection limit of 5610 virus copies/mL, found significantly higher SARS-CoV-2 titers in saliva than nasopharyngeal smears for the corresponding samples (p = 0.0001).
Kojima et al. (2020) evaluated the performance of the saliva swab samples self-collected by the individuals recruited, who had been tested positive for SARS-CoV-2 and compared to nasopharyngeal swabs, The four testing groups were as follows: a) with and b) without a doctor's supervision, c) self-collected nasal swab samples and d) nasopharyngeal swab collected by the doctor. For both saliva tests, all participants received the same instructions. The unsupervised group performed the collection only following these instructions, while the supervised group had instant feedback. The supervised saliva swab collection group was able to detect 90% of the infected individuals, being the highest rate when compared to the saliva swab group performed without supervision (66%), which was also the worst performance in the study sample. Meanwhile, posterior nasopharyngeal swab samples detected 79% of those infected.
3.2.2. Studies that directly compared saliva use in SARS-CoV-2 detection with respiratory samples and that demonstrated inaccurate saliva results
Nine studies demonstrated less saliva sensitivity as a diagnostic sample or that there was less time of viral shedding compared to respiratory samples when comparing the saliva use in relation to the other respiratory sample for the SARS-CoV-2 detection (Table A.2). The saliva collection approaches were by spitting, using a syringe, oral swab and clearing the throat and gargling the saliva itself.
Kam et al., (2020) and Chong et al., (2020) demonstrated low sensitivity in saliva samples use for SARS-CoV-2 detection in infected children, varying from 25 to 71.4% on different collection days in the first week after diagnosis [40] and reaching a sensitivity peak of 52.9% in the period of 4–7 days [38]. Kam et al., (2020) still found statistically significant differences (p < 0.001) between the median Ct values of the oral and nasopharyngeal samples of infected patients, with an average difference of 10.7 (range 6.1–16, 1).
Likewise, Williams et al., (2020) showed lower Ct values in nasopharyngeal samples than in saliva samples, suggesting lower viral loads in saliva. In addition, Jamal et al., (2020) observed greater sensitivity in nasopharyngeal swab samples compared to saliva samples in SARS-CoV-2 detection, especially in disease with advanced stages. Both samples showed greater sensitivity in the first week, with less difference in sensitivity between them.
Meanwhile, Kim et al., (2020) observed a greater difference in sensitivity between naso/oropharyngeal and saliva samples in the initial stage of symptoms (93% and 53%, respectively), and saliva sensitivity was especially lower in patients who had no sputum (55%). Despite this, the median Ct value of saliva did not differ significantly from the naso/oropharyngeal samples (p = 0.7531), being 32 (IQR 28–38) and 33 (27–35), respectively.
3.2.3. Studies that did not directly compare saliva use in SARS-CoV-2 detection with respiratory samples
A total of 5 non-comparative studies between saliva and respiratory samples were evaluated (Table A.3). These included saliva samples analyzed at different times of the day, in different tests, investigating the specific antibodies responses to salivary SARS-CoV-2, evaluating the saliva collected directly from the salivary duct and comparing it with samples and blood and anal swabs.
Evaluating saliva samples at different times during the day, Hung et al., (2020) suggested a trend for higher viral loads in the morning (median Ct value = 34,5) compared to the other four times: before lunch (38,2), before tea time (36,3), before dinner (41) and before bed (41), occurring in 8 out of 13 patients who had detectable viral loads [45].
When analyzing saliva samples in different tests, Nagura-Ikeda et al. (2020) showed sufficient sensitivity in clinical use to detect SARS-CoV-2 from tests such as LDT RT-qPCR, cobas SARS-CoV-2 high-throughput system and RT-LAMP. However, it was observed that rapid antigen tests using saliva are not recommended for the early disease diagnosis due to their low sensitivity [47].
However, when it comes to serological testing using saliva collected 10 days or more after the onset of symptoms, Randad et al. (2020) demonstrated that the anti-SARS-CoV-2 IgG assay detects SARS infection -CoV-2 with high sensitivity and specificity, reaching 100% and 99%, respectively, for the GenScript N antigen, and 89% and 100% for Mt. Sinai RBD [48].
2019-nCoV nucleic acid was detected in pure saliva [46]. The samples were collected from the salivary gland canal opening after cleaning the oral cavity, avoiding contamination by other respiratory tract secretions. Among the 13 patients positive for oropharyngeal swab, four tested positive also for saliva, being three critical cases with ventilatory support. In addition, the authors assessed the oral symptoms reported by patients with COVID-19, and reported that main symptoms were dry mouth (46.3%) and dysgeusia (47.2%).
Zhang et al., (2020) [44] collected blood samples, oral and anal swabs from patients at the Wuhan Lung Hospital who had nCoV 2019 positive oral swabs on admission. For the virus molecular detection, it was demonstrated that, of the positive tests in the initial phase of the disease, the majority were oral swabs (50%), while the minority were anal swabs (25%). In the later phase, this relationship was reversed, with more positive tests of anal swabs (37.5%) than of oral swabs (25%).
4. Discussion
Diagnostic techniques based on nasopharyngeal and oropharyngeal samples were recommended for the COVID-19 detection in outpatients by WHO on January 9, 2020 [49]. These tests for the virus detection have strong evidence, with studies indicating they are more sensitive than other respiratory samples. This is because it has been proven that there is active viral replication in the tissues of the upper respiratory tract, with higher viral loads being seen in the first week of symptoms in the pharynx samples [50]. However, its performance can cause pain and discomfort in patients, in addition to bleeding in patients with thrombocytopenia, making them uninteresting for serial monitoring about the viral load [7,51].
Saliva is a hypotonic liquid secreted by the parotid, submandibular, sublingual and minor salivary glands that are distributed throughout the oral cavity. These glands are very permeable and surrounded by blood capillaries, allowing the molecules and biomarkers exchange, which can be secreted together with saliva. Thus, these biomarkers in saliva have been analyzed and used to detect local and systemic diseases, such as caries, periodontitis, oral and lung cancer, diabetes, cardiovascular diseases and viral infections [11]. Oral fluid samples can indicate the virus infection presence by screening for viral nucleic acids, antigens and antibodies [9].
Thus, saliva samples have been suggested as tools for the respiratory viruses’ detection, such as influenza A virus, influenza B virus, parainfluenza virus and respiratory syncytial virus (RSV), in order to reduce costs and time associated with collections [52,53]. A cohort study demonstrated that the detection of respiratory viruses in salivary samples, by an automated multiplex Clinical Laboratory Improvement Amendments-waived point-of-care molecular assay, has high sensitivity and specificity [53].
In 2003, the coronavirus from severe acute respiratory syndrome (SARS-CoV) spread rapidly from China to more than 30 countries, being a highly contagious disease [10]. It is known that both SARS-CoV and 2019-nCoV, a coronavirus similar to SARS-CoV, can be transmitted efficiently between humans through the droplets generation when speaking, coughing or sneezing [11]. In addition, they interact with the angiotensin II-converting enzyme receptor (ACE2) in host cells, found expressed in the lungs, esophagus, ileum, colon, liver, bladder and in the salivary gland and tongue [3,54].
The results found by Chen et al., (2020) [46] may suggest that epithelial cells that line the minor salivary glands ducts, which express ACE2, were infected, generating infected saliva. In addition, viable viruses could be identified in saliva samples in two studies, using collecting saliva method with and without cough [7,31]. Thus, the oral cavity can be a host for 2019-nCoV.
Viral nucleic acid's presence after treatment days seen in some studies [7,13] may indicate low excretion levels in saliva, even after resolution symptoms. Thus, reliable tests are necessary, including different tests combinations, at the time of hospital discharge for patients admitted with SARS-CoV-2, avoiding the possibility that these recovered patients continue to transmit the virus through direct or indirect contact through the generation of droplets from infected saliva.
Although most studies in this review have shown good results utilizing saliva as a diagnostic sample, compared to the gold standard of nasopharyngeal samples, some limitations have been reported. Among them, the faster decrease in viral load than NPS in hospitalized patients [14,17], less sensitivity compared to nasopharyngeal samples during the one week collection period in children [38,40] and the saliva characteristics that could making sample processing difficult [43].
Given that saliva collection is a simpler technique, it is strongly suggested as an alternative screening procedure in places with limited resources. In addition, the ease in salivary sample collection for the SARS-CoV-2 diagnosis allows for self-sample collection by the patients themselves, not necessarily in a hospital environment or with the health professional presence. This system would make it possible, in addition to epidemiological control and self-surveillance, to reduce the exposure risk for health professionals and to reduce the need for personal protective equipment.
On May 7, 2020, the U.S. Food and Drug Administration (FDA) approved the first in home saliva collection test for the SARS-CoV-2 diagnosis [56]. Thus, saliva's self-collection may allow for more specific results due to the possibility of performing it when symptoms arise, especially in the first week, and in the morning, when the saliva has higher viral loads [6,19,45].
A study evaluated the general acceptability of North American adults in relation to the collecting process, packaging and sending self-collected saliva samples, oropharyngeal swab and dried blood card [57]. It was possible to observe high acceptability and confidence in relation to the self-collection of both saliva and other samples by patients, which reinforces the possibility of facilitating mass screening processes, reducing the costs associated with collections.
The possibility of utilizing this simple technique, with lower cost and more comfort in detecting 2019-nCoV in non-hospital environments would bring advantages to dental care, reducing the waiting period or even allowing immediate intervention based on positive results.
5. Conclusions
Saliva seems to be a promising resource in the SARS-CoV-2 detection, having demonstrated similar performance to nasopharyngeal swabs, plus advantages such as low cost, disease course monitoring, not being invasive and avoiding close contact with health professionals. In addition, salivary samples are a good alternative for epidemiological studies and asymptomatic or pre-symptomatic infections detection and can be useful in screening systems and dental care.
Conflicts of interest
A conflicting interest exists when professional judgement concerning a primary interest (such as patient's welfare or the validity of research) may be influenced by a secondary interest (such as financial gain or personal rivalry). It may arise for the authors when they have financial interest that may influence their interpretation of their results or those of others. Examples of potential conflicts of interest include employment, consultancies, stock ownership, honoraria, paid expert testimony, patent applications/registrations, and grants or other funding.
Funding source
All sources of funding should also be acknowledged and you should declare any involvement of study sponsors in the study design; collection, analysis and interpretation of data; the writing of the manuscript; the decision to submit the manuscript for publication. If the study sponsors had no such involvement, this should be stated. Please state any sources of funding for your research.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgments
The authors would like to thank the Universidade Federal do Rio Grande do Norte (UFRN, Brazil) for allowing this study and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazil) for supporting this manuscript.
References
- 1.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/s0140-6736-(20)-30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization WHO coronavirus disease (COVID-19) dashboard. https://covid19.who.int/
- 3.Xu R., Cui B., Duan X., Zhang P., Zhou X., Yuan Q. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int J Oral Sci. 2020;12:11. doi: 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma S, Kumar V, Chawla A, Logani A. Rapid detection of SARS‐CoV‐2 in saliva: can an endodontist take the lead in point‐of‐care COVID‐19 testing? Int Endod J. 10.1111/iej.13317. [DOI] [PMC free article] [PubMed]
- 5.Sullivan P.S., Sailey C., Guest J.L. Detection of SARS-CoV-2 RNA and antibodies in diverse samples: protocol to validate the sufficiency of provider-observed, home-collected blood, saliva, and oropharyngeal samples. JMIR Public Health and Surveillance. 2020 Apr-Jun;6(2) doi: 10.2196/19054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyllie A.L., Fournier J., Casanovas-Massana A. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. April 22, 2020. [cited 2020 July 15]. medRxiv [Preprint]. Avaliable from: [DOI]
- 7.To KKW, Tsang OTY, Yip CCY, et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clinical Infectious Diseases, ciaa149, 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed]
- 8.Corstjens P.A.M., Abrams W.R., Malamud D. Detecting viruses by using salivary diagnostics. J Am Dent Assoc. 2012 Oct;143(10 0):12S–18S. doi: 10.14219/jada.archive.2012.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corstjens P.A.M., Abrams W.R., Malamud D. Saliva and viral infections. Periodontol 2000. 2016 Feb;70(1):93–110. doi: 10.1111/prd.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W.K., Chen S.Y., Liu I.J. Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg Infect Dis. 2004 Jul;10(7):1213–1219. doi: 10.3201/eid1007.031113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santosh T.S., Parmar R., Anand H., Srikanth K., Saritha M. A review of salivary diagnostics and its potential implication in detection of covid-19. Cureus. 2020 Apr;12(4) doi: 10.7759/cureus.7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azzi L., Carcano G., Gianfagna F. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. July 2020;81(Issue 1):e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.To K.K.W., Tsang O.T.Y., Leung W.S. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(Issue 5):p565–574. doi: 10.1016/S1473-3099(20)30196-1. May 01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwasaki S., Fujisawa S., Nakakubo S. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect. 2020 Jun 4 doi: 10.1016/j.jinf.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokota I., Shane P.Y., Okada K. Mass screening of asymptomatic persons for SARS-CoV-2 using saliva. Clin Infect Dis. 2020 Sep 25:ciaa1388. doi: 10.1093/cid/ciaa1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyllie A.L., Fournier J., Casanovas-Massana A. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020 Aug 28 doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han M.S., Seong M.W., Kim N. Viral RNA load in mildly symptomatic and asymptomatic children with COVID-19, Seoul, South Korea. Emerg Infect Dis. 2020 Oct;26(10):2497–2499. doi: 10.3201/eid2610.202449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao M.H., Guo J.J., Qin L.Z., Han Z.X., Wang Y.J., Yangc D. Serial semiquantitative detection of SARS-CoV-2 in saliva samples. J Infect. 2020 Oct 6 doi: 10.1016/j.jinf.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojima N., Turner F., Slepnev V. Self-collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for covid-19 detection. April 15, 2020. [cited 2020 July 15]. medRxiv [Preprint]. Avaliable from: [DOI] [PMC free article] [PubMed]
- 20.Pasomsub E., Watcharananan S.P., Boonyawat K. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019 (COVID-19): a cross-sectional study. Clin Microbiol Infect. 2020 May 15 doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J.K.H., Yip C.C.Y., Poon R.W.S. Evaluating the use of posterior oropharyngeal saliva in a point-of-care assay for the detection of SARS-CoV-2. Emerg Microb Infect. 2020;9(2021) doi: 10.1080/22221751.2020.1775133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mccormick-Baw C, Morgan K, Gaffney D, et al. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using cepheid xpert xpress SARS-CoV-2. J Clin Microbiol. doi:10.1128/JCM.01109-20. [DOI] [PMC free article] [PubMed]
- 23.Leung E.C., Chow V.C., Lee M.K., Lai R.W. Deep throat saliva as an alternative diagnostic specimen type for the detection of SARS‐CoV‐2. J Med Virol. 2020 Jul 14 doi: 10.1002/jmv.26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheuk S, Wong Y, Tse H, et al. Posterior oropharyngeal saliva for the detection of SARS-CoV-2. Clinical Infectious Diseases, ciaa797, 10.1093/cid/ciaa797. [DOI] [PMC free article] [PubMed]
- 25.Güçlü E., Koroglu M., Yürümez Y. Comparison of saliva and oro-nasopharyngeal swab sample in the molecular diagnosis of COVID-19. Rev Assoc Med Bras. 2020;66(8) doi: 10.1590/1806-9282.66.8.1116. São Paulo Aug. 2020 Epub Sep. 11. [DOI] [PubMed] [Google Scholar]
- 26.Vaz S.N., Santana D.S., Netto E.M. Saliva is a reliable, non-invasive specimen for SARS-CoV-2 detection. Braz J Infect Dis. 2020 Aug 31 doi: 10.1016/j.bjid.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao M., Rashid F.A., Sabri F.S.A.H. Clinical infectious diseases. An Official Publication of the Infectious Diseases Society of America; 2020 August 6. Comparing nasopharyngeal swab and early morning saliva for the identification of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byrne R.L., Kay G.A., Kontogianni K. Saliva alternative to upper respiratory swabs for SARS-CoV-2 diagnosis. Emerg Infect Dis. 2020 Nov doi: 10.3201/eid2611.203283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanson K.E., Barker A.P., Hillyard D.R. Self-collected anterior nasal and saliva specimens versus healthcare worker-collected nasopharyngeal swabs for the molecular detection of SARS-CoV-2. J Clin Microbiol. 2020 Aug doi: 10.1128/JCM.01824-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aita A., Basso D., Cattelan A.M. SARS-CoV-2 identification and IgA antibodies in saliva: one sample two tests approach for diagnosis. Clin Chim Acta. 2020 Nov;510:717–722. doi: 10.1016/j.cca.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uwamino Y., Nagata M., Aoki W. Accuracy and stability of saliva as a sample for reverse transcription PCR detection of SARS-CoV-2. J Clin Pathol Month. 2020 doi: 10.1136/jclinpath-2020-206972. 0 No 0. [DOI] [PubMed] [Google Scholar]
- 32.Migueres M., Mengelle C., Dimeglio C. Saliva sampling for diagnosing SARS-CoV-2 infections in symptomatic patients and asymptomatic carriers. J Clin Virol. 2020 Sep;130:104580. doi: 10.1016/j.jcv.2020.104580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senok A., Alsuwaidi H., Atrah Y. Saliva as an alternative specimen for molecular COVID-19 testing in community settings and population-based screening. Infect Drug Resist. 1 October 2020;2020:3393–3399. doi: 10.2147/IDR.S275152. 13 Pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altawalah H., AlHuraish F., AlkandariWA Ezzikouri S. Saliva specimens for detection of severe acute respiratory syndrome coronavirus 2 in Kuwait: a cross-sectional study. J Clin Virol. Nov 2020;132 doi: 10.1016/j.jcv.2020.104652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jamal A.J., Mozafarihashjin M., Coomes E. Clinical Infectious Diseases; June: 2020. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang Z., Zhang Y., Hang C., Ai J., Li S., Zhang W. Comparisons of viral shedding time of SARS-CoV-2 of different samples in ICU and non-ICU patients. J Infect. 2020 Jul;81(1):147–178. doi: 10.1016/j.jinf.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S.E., Lee J.Y., Lee A. Viral load kinetics of SARS-CoV-2 infection in saliva in Korean patients: a prospective multi-center comparative study. J Kor Med Sci. 2020 Aug 10;35(31):e287. doi: 10.3346/jkms.2020.35.e287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chong C.Y., Kam K.Q., Li J. Saliva is not a useful diagnostic specimen in children with Coronavirus Disease 2019. Clin Infect Dis. 2020 Sep 14:ciaa1376. doi: 10.1093/cid/ciaa1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol. doi:10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed]
- 40.Kam K., Yung C.F., Maiwald M. Clinical utility of buccal swabs for SARS-CoV-2 detection in covid-19-infected children. Journal of the Pediatric Infectious Diseases Society. July 2020;9(Issue 3):370–372. doi: 10.1093/jpids/piaa068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skolimowska K., Rayment M., Jones R. Non-invasive saliva specimens for the diagnosis of COVID-19: caution in mild outpatient cohorts with low prevalence. Clin Microbiol Infect. 2020 Jul 18 doi: 10.1016/j.cmi.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai C.K.C., Chen Z., Lui G. Prospective study comparing deep-throat saliva with other respiratory tract specimens in the diagnosis of novel coronavirus disease (COVID-19) J Infect Dis. 2020 Aug 1:jiaa487. doi: 10.1093/infdis/jiaa487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landry M.L., Criscuolo J., Peaper D.R. Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J Clin Virol. 2020 Sep;130 doi: 10.1016/j.jcv.2020.104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W., Du R.H., Li B. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microb Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung D.L.L., Li X., Chiu K.H.Y. Early-morning vs spot posterior oropharyngeal saliva for diagnosis of SARS-CoV-2 infection: implication of timing of specimen collection for community-wide screening. Open Forum Infectious Diseases. 2020 Jun;7(6) doi: 10.1093/ofid/ofaa210. Published online 2020 Jun 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L., Zhao J., Peng J. Detection of 2019-nCoV in saliva and characterization of oral symptoms in COVID-19 patients. March 14, 2020. Available at SSRN: [DOI] [PMC free article] [PubMed]
- 47.Nagura-Ikeda M., Imai Z., Tabata S. Clinical evaluation of self-collected saliva by RT-qPCR, direct RT-qPCR, RT-LAMP, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol. 2020 Jul 7 doi: 10.1128/JCM.01438-20. JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Randad P.R., Pisanic N., Kruczynski K. Version 1. medRxiv. Preprint. 2020 May 26. COVID-19 serology at population scale: SARS-CoV-2-specific antibody responses in saliva. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.World Health Organization Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117
- 50.Wölfel R., Corman V.M., Guggemos W. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 51.Khurshid Z., Asiri F.Y.I., Wadaani H.A. Human saliva: non-invasive fluid for detecting novel coronavirus (2019-nCoV) Int J Environ Res Publ Health. 2020 Apr;17(7):2225. doi: 10.3390/ijerph17072225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson J., Lee B.E., Kothapalli S., Craig W.R., Fox J.D. Use of throat swab or saliva specimens for detection of respiratory viruses in children. Clin Infect Dis. 2008 Apr 1;46(7):e61–e64. doi: 10.1086/529386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.To K.K.W., Yip C.C.Y., Lai C.Y.W. Saliva as a diagnostic specimen for testing respiratory virus by a point-of-care molecular assay: a diagnostic validity study. Clin Microbiol Infect. 2019;25:372e378. doi: 10.1016/j.cmi.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 54.Liu L., Wei Q., Alvarez X. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J Virol. 2011 Apr;85(8):4025–4030. doi: 10.1128/JVI.02292-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabino-Silva R., Jardim A.C.G., Siqueira W.L. Coronavirus COVID-19 impacts to dentistry and potential salivary diagnosis. Clin Oral Invest. 2020;24:1619–1621. doi: 10.1007/s00784-020-03248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Live Science First at-home saliva test for COVID-19 earns FDA approval. https://www.livescience.com/at-home-saliva-test-for-covid19.html
- 57.Valentine-Graves M., Hall E., Guest J.L. At-home self-collection of saliva, oropharyngeal swabs and dried blood spots for SARS-CoV-2 diagnosis and serology: post-collection acceptability of specimen collection process and patient confidence in specimens. PLoS One. 2020;15(8) doi: 10.1371/journal.pone.0236775. Aug 5. [DOI] [PMC free article] [PubMed] [Google Scholar]