Abstract
The disease called severe acute respiratory syndrome (SARS) is a lifestyle intimidating viral contamination affected by a positive, single stranded novel RNA virus (COVID-2019) from the enveloped coronaviruse family. The COVID-2019 virus has affected many people, scattering promptly, and researchers are attempting to find out medicines for its effectual cure in all over the globe. Chloroquine (ChQ) and its derivatives, an older drug used for the cure of malaria, is exposed to encompass a perceptible feasibility and commendable well-being in opposition to SARS CoV-2 associated pneumonia clinical trials conducted in China. Later on, a few investigations have been directed to find and present SARS CoV-2 antiviral medications. The aim of this present work deals with the potential binding interactions of some imidazolium salts with Nsp9 (Nonstructural protein 9) RNA binding protein of SARS CoV-2.
Keywords: Computational chemistry, Organic chemistry, Systems biology, Bioinformatics, Biotechnology, Biochemistry, Imidazole, Imidazolium salt, Benzoate, Protein-ligand interaction, SARS COV-2, Docking
Computational chemistry; Organic chemistry; Systems biology; Bioinformatics; Biotechnology; Biochemistry; Imidazole; Imidazolium salt; Benzoate; Protein-ligand interaction; SARS COV-2; DOCKING.
1. Introduction
A novel COVID-19 outbreak [1, 2], developed during the late December 2019, has influenced numerous individuals in Wuhan city, China [2], that has later on swayed and spread rapidly to around 200 nations around the world. Associated with fever, cough, and respiratory complications, the illness causes more than 25% mortality global wide. So far, there is no remedy for this dangerous viral sickness except some supportive healing processes. However, the key viral proteinase have recently been observed as an appropriate target for drug devise against SARS contamination due to its vital role in polyproteins processing indispensable for coronavirus imitation.
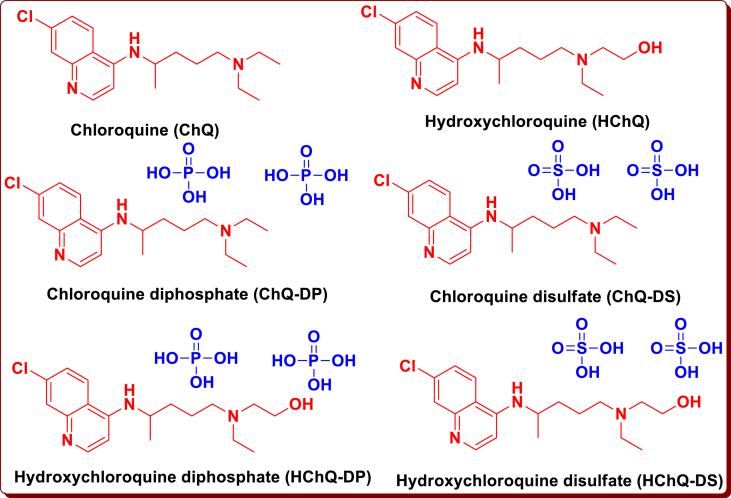
As a rule, COVID-19 is a penetrating developed malady, yet, it may equally be destructive, with a 2% case victim pace. Severe illness beginning may bring about mortality because of monstrous alveolar harm and forceful respiratory failure [2, 3]. As of March 26, 2020, the infection has caused around 4,62,684 cases affirmed and more than 20,834 deaths [4]. The infection has caused large numbers of cases to around 30,90,445 cases confirmed and more than 2,17, 769 deaths, around 59,34,936 cases confirmed and more than 3,67,166 deaths, and 1,01,85,374 cases confirmed and more than 5,03,862 deaths at the end of April, May and June 2020 respectively [5]. At that time (July 14, 2020), now the confirmed cases and mortality have been reached to 1,65,58,289 and 6,56,093 respectively [6]. However, no pathology has been accounted for because of barely accessible itemization or biopsy [2, 3]. On 17th February, 2020, the State Council of China released a newsflash preparation showing that chloroquine phosphate, an ancient medication employed for curing malaria, had revealed and checked sufficiency and praiseworthy wellbeing in curing the novel COVID-19 related pneumonia in multicenter medical preliminaries directed at China [7]. At the time in vitro investigations, chloroquine (ChQ) was discovered to block COVID-19 disease at a little-micromolar fixation, with a half-maximal in effect concentration (EC50) of 1.13 μM and a half-cytotoxic concentration (CC50) more noteworthy than 100 μM [8]. Chloroquine (ChQ) or Hydroxychloroquine (HChQ) is exploited to treat deadly diseases like malaria, rheumatoid arthritis and lupus erythematosus [9, 10]. Results also indicates that it contains a potentially wide range antiviral property by rising endosomal pH needed for infection/cell combination, just as meddling with the glycosylation of cell receptors of SARS-CoV [11, 12]. The anti-viral and anti-inflammatory potency of ChQ symbolize for its latent competence in treating COVID-19 patients. Now, Quinoline moiety containing drugs is used to hamper COVID-19 contagion (Figure 1) [13,14]. The discovery of new drug to inhibit and treat COVID-19 becomes absolutely essential and now, all scientists and researchers are in search of designing new potent drugs with the help of molecular docking tools and other pathways [15, 16, 17, 18, 19, 20, 21, 22, 23].
Figure 1.
Schematic representation of Quinoline moiety containing drugs is used to block COVID-19 infection.
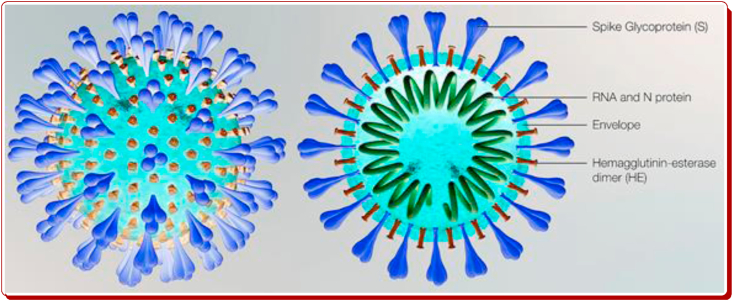
Coronaviruses (CoVs) are noticeably comprising a solitary-stranded positive-sense RNA genome compressed inside a membrane cover. The viral film is speckled with glycoprotein spikes that provide coronaviruses, their crown-like expression (Figure 2). Although coronaviruses infect each human beings and animals, sure forms of animals which include bats that have the biggest form of coronaviruses seem, by all accounts, to be invulnerable to coronavirus-actuated illness. Researchers have identified four types of coronaviruses namely alpha- (α-CoV), beta- (β-CoV), gamma- (γ-CoV), and delta-coronaviruses (δ-CoV). The betacoronavirus elegance consists of SARS-CoV, middle east respiratory syndrome coronavirus (MERS-CoV), mouse hepatitis coronavirus (MH-CoV) and bovine coronavirus (B-CoV) and the COVID-19 causative agent SARS-CoV-2. Just like SARS-CoV, MERS-CoV and SARS-CoV-2 are studded the lower respiratory system to motive viral pneumonia, yet it might furthermore influence the gastrointestinal system, coronary heart, liver, kidney and focal sensory system prompting various organ failures. Contemporary facts suggest that SARS-CoV-2 is far more spreadable/transmittable in contrast to SARS-CoV [24, 25, 26].
Figure 2.
Schematic representation of novel crown-like SARS-CoV-2.
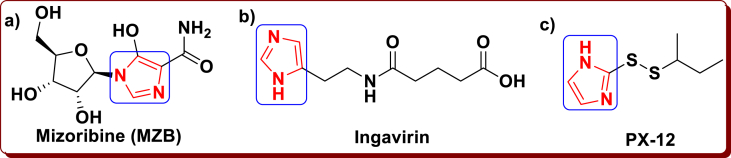
The immune-suppressant drug Mizoribine (MZB) is an imidazole nucleoside (Figure 3a), which inhibits SARS-CoV [26, 27] and it is isolated from the fungus Penicillium brefeldianum [28]. The imidazole moiety containing Russian antiviral medication is Ingavirin® (Figure 3b), which is used for the remedy and anticipation of the grave breathing viral contagions by methods for flu and non-flu with an utterly extraordinary mechanism of performance. This medication is given an early notoriety of contagions and the development of the anti-viral ceremonial of cells that forestalls the infection from replicating and dispersal in the animal corpse. It is groundbreaking against A and B type viruses, parainfluenza viruses, adenoviruses, breathe syncytial viruses, enteroviruses, metapneumoviruses, coronaviruses, which incorporates Coxsackie virus and rhinovirus [29, 30, 31]. A small molecule 1-methylpropyl 2-imidazolyl disulfide (PX-12, Figure 3c) is an inhibitor of stimulates apoptosis, down-regulates HIF-1α and vascular endothelial growth factor (VEGF), Trx-1 (Thioredoxin-1) and represses tumor growth in animal simulations. The PX-12, which is a preclinical or clinical-drug candidate and it is able to bind through the covalent to C-145 of the channel dyad and completely modify in SARS-CoV-2 Mpro [25, 32]. The several imidazole derivatives and imidazolium-based ionic salts were examined to identify their interaction and inhibition activity against coronavirus as antiviral agents [33, 34].
Figure 3.
Schematic representation of imidazole moiety containing drugs a) Mizoribine (MZB), b) Ingavirin, c) PX-12.
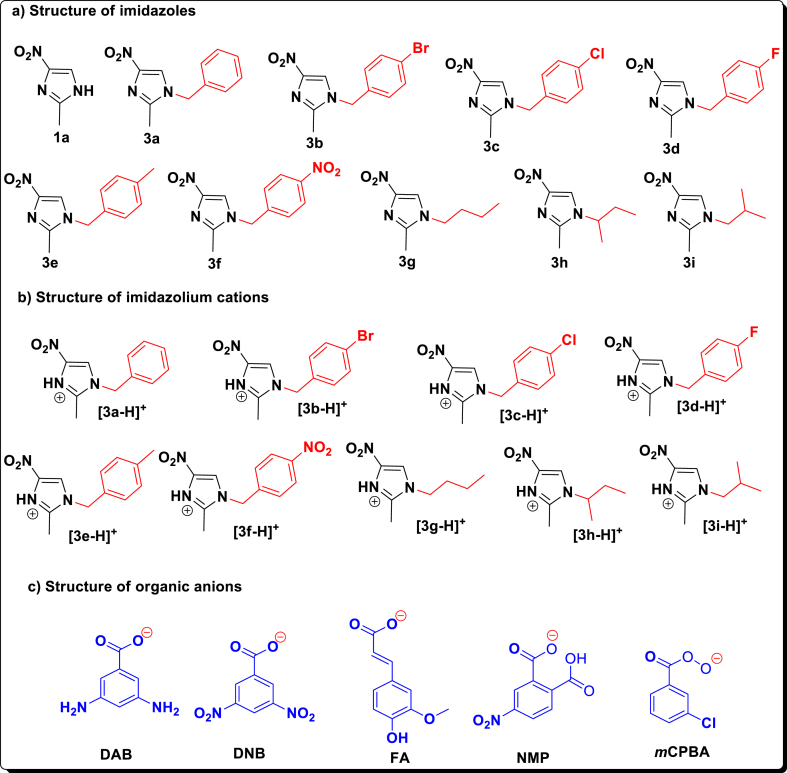
In our previous work, we reported the synthesis of N1-(4-substitutedbenzyl)-2-methyl-4-nitro-1H-imidazoles and N1-butyl-2-methyl-4-nitro-1H-imidazoles (Figure 4) [35] and thirty six Brønsted acidic based imidazolium salts incorporated with aromatic organic anions (Figure 4) [36, 37]. In continuation of our previous work, we would like to report, in the present article, a discovery of small molecules as new potential drugs for the inhibition of SARS CoV-2 contamination. In this line, we extensively carried out the molecular docking studies of N1-(4-substitutedbenzyl)/butyl-2-methyl-4-nitro-1H-imidazoles and their protic 4-nitroimidazolium salts with aromatic organic anions for the better understanding of the drug-receptor interaction with 6W4B. We strongly believe and hope that our results will accelerate the discernment of the binding affinities of imidazolium salts with SARS CoV-2 (Nsp9 RNA binding protein; PDB code: 6W4B) [38] as inhibitors and improve clinical strategies against the deadly carnivorous disease.
Figure 4.
The structure of a) imidazoles (3a-i) [35,37], b) imidazolium cations and c) organic anions [36, 37].
2. Methodology
2.1. Materials and methods
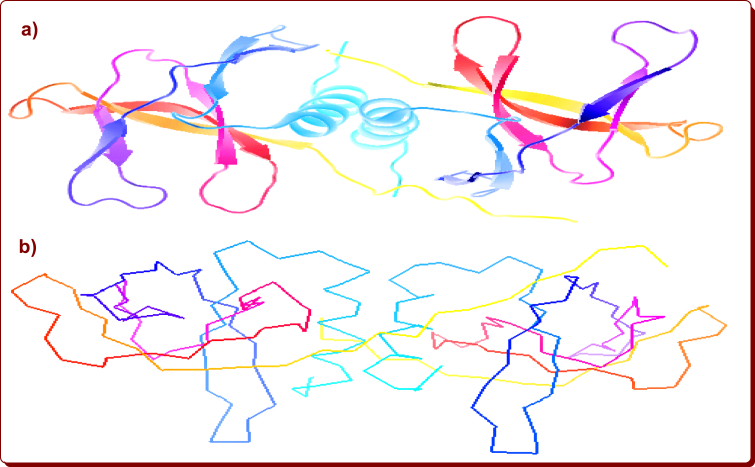
N1-(4-substituted benzyl)-2-methyl-4-nitro-1H-imidazoles, N1-butyl-2-methyl-4-nitro-1H-imidazoles and their protic 4-nitroimidazolium salts with aromatic organic anions were produced in our laboratory (Figure 4) [35, 36, 37]. Molecular docking simulations were performed using the 1-Click docking program to ascertain the binding nature and associative interactions with the Nsp9 RNA binding protein of SARS CoV-2, which was released from the Protein Data Bank (PDB code: 6W4B, Figure 5) [38]. After docking, the docking poses and binding modes were visualized by using Protein-Ligand Interaction Profile (PLIP) [39] and the results are summarized in Tables 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10.
Figure 5.
Schematic representation of structure of Nsp9 RNA binding protein of SARS CoV-2 a) cartoon and b) stick line.
Table 1.
Hydrophobic interactions of imidazoles (1a, 3a-i).
| Imidazole | Binding affinity (kcal/mol) | Index | Residue | AA | Distance (Å) | Ligand atom | Protein atom |
|---|---|---|---|---|---|---|---|
| 1a | -2.5 | - | - | - | - | - | - |
| 3a | -4.1 | 1 | 41A | PHE | 3.68 | 1697 | 247 |
| 3b | -3.7 | 1 | 36A | THR | 3.77 | 1694 | 218 |
| 2 | 41A | PHE | 3.78 | 1686 | 250 | ||
| 3c | -3.8 | 1 | 36A | THR | 3.57 | 1699 | 218 |
| 2 | 41A | PHE | 3.72 | 1695 | 252 | ||
| 3 | 41A | PHE | 3.54 | 1697 | 250 | ||
| 3d | -3.7 | 1 | 36A | THR | 3.66 | 1699 | 218 |
| 2 | 41A | PHE | 3.74 | 1695 | 252 | ||
| 3 | 41A | PHE | 3.60 | 1697 | 250 | ||
| 3e | -4.2 | 1 | 36A | THR | 3.61 | 1694 | 218 |
| 2 | 41A | PHE | 3.63 | 1697 | 252 | ||
| 3 | 41A | PHE | 3.61 | 1695 | 250 | ||
| 3f | -3.9 | 1 | 8A | VAL | 3.61 | 1701 | 20 |
| 2 | 36A | THR | 3.84 | 1700 | 218 | ||
| 3g | -3.0 | 1 | 36A | THR | 3.72 | 1695 | 218 |
| 2 | 41A | PHE | 3.60 | 1693 | 252 | ||
| 3h | -2.8 | 1 | 34A | ASN | 3.82 | 1695 | 203 |
| 2 | 41A | PHE | 3.69 | 1695 | 250 | ||
| 3i | -3.0 | 1 | 41A | PHE | 3.72 | 1695 | 248 |
Table 2.
Hydrophobic interactions of the Brønsted acidic based imidazolium cations.
| Imidazolium cation | Binding affinity (kcal/mol) | Index | Residue | AA | Distance (Å) | Ligand atom | Protein atom |
|---|---|---|---|---|---|---|---|
| [3a-H]+ | -3.8 | 1 | 36A | THR | 3.61 | 1695 | 218 |
| 2 | 41A | PHE | 3.71 | 1698 | 252 | ||
| 3 | 41A | PHE | 3.64 | 1695 | 250 | ||
| [3b-H]+ | -3.9 | 1 | 36A | THR | 3.62 | 1695 | 218 |
| 2 | 41A | PHE | 3.71 | 1698 | 252 | ||
| 3 | 41A | PHE | 3.59 | 1696 | 250 | ||
| [3c-H]+ | -3.7 | 1 | 36A | THR | 3.71 | 1695 | 218 |
| 2 | 41A | PHE | 3.78 | 1686 | 250 | ||
| [3d-H]+ | -3.9 | 1 | 8A | VAL | 3.74 | 1694 | 20 |
| 2 | 36A | THR | 3.92 | 1700 | 218 | ||
| [3e-H]+ | -4.0 | 1 | 36A | THR | 3.47 | 1695 | 218 |
| 2 | 41A | PHE | 3.71 | 1698 | 252 | ||
| 3 | 41A | PHE | 3.61 | 1696 | 250 | ||
| [3f-H]+ | -3.8 | 1 | 36A | THR | 3.64 | 1695 | 218 |
| 2 | 41A | PHE | 3.70 | 1698 | 252 | ||
| 3 | 41A | PHE | 3.58 | 1696 | 250 | ||
| [3g-H]+ | -3.0 | 1 | 34A | ASN | 3.91 | 1694 | 203 |
| 2 | 36A | THR | 3.73 | 1695 | 218 | ||
| 3 | 41A | PHE | 3.66 | 1695 | 250 | ||
| [3h-H]+ | -3.0 | 1 | 75B | ARG | 3.98 | 1686 | 1386 |
| [3i-H]+ | -3.3 | 1 | 7A | PRO | 3.70 | 1694 | 12 |
| 2 | 77B | VAL | 3.54 | 1686 | 1408 | ||
| 3 | 114B | GLN | 3.77 | 1695 | 1685 |
Table 3.
Hydrophobic interactions of organic anions.
| Organic anion | Binding affinity (kcal/mol) | Index | Residue | AA | Distance (Å) | Ligand atom | Protein atom |
|---|---|---|---|---|---|---|---|
| DAB | -3.1 | 1 | 8A | VAL | 3.67 | 1691 | 20 |
| 2 | 36A | THR | 3.66 | 1689 | 218 | ||
| DNB | -3.2 | 1 | 36A | THR | 3.70 | 1687 | 218 |
| 2 | 41A | PHE | 3.59 | 1689 | 252 | ||
| 3 | 41A | PHE | 3.76 | 1687 | 250 | ||
| FA | -3.5 | 1 | 7A | PRO | 3.67 | 1689 | 12 |
| 2 | 77B | VAL | 3.61 | 1692 | 1410 | ||
| 3 | 77B | VAL | 3.68 | 1689 | 1408 | ||
| NMP | -3.2 | 1 | 7A | PRO | 3.58 | 1691 | 12 |
| 2 | 77B | VAL | 3.64 | 1690 | 1408 | ||
| mCPBA | -3.3 | 1 | 36A | THR | 3.72 | 1692 | 218 |
| 2 | 41A | PHE | 3.69 | 1692 | 247 |
Table 4.
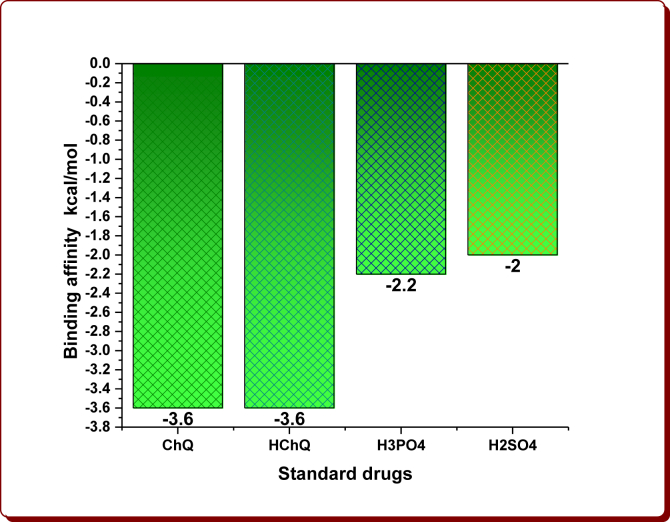
Hydrophobic interactions of ChQ, HChQ, H3PO4 and H2SO4.
| Standard drug | Binding affinity (kcal/mol) | Index | Residue | AA | Distance (Å) | Ligand atom | Protein atom |
|---|---|---|---|---|---|---|---|
| ChQ | -3.6 | 1 | 41A | PHE | 3.71 | 1704 | 250 |
| 2 | 41A | PHE | 3.63 | 1703 | 252 | ||
| 3 | 41A | PHE | 3.63 | 1707 | 248 | ||
| HChQ | -3.6 | 1 | 36A | THR | 3.58 | 1710 | 218 |
| 2 | 41A | PHE | 3.36 | 1705 | 250 | ||
| H3PO4 | -2.2 | - | - | - | - | - | - |
| H2SO4 | -2.0 | - | - | - | - | - | - |
Table 5.
Hydrogen bond interactions of imidazoles (1a, 3a-i).
| Imidazole | Index | Residue | AA | Distance (Å) |
Donor Angle | Donor atom | Acceptor atom | |
|---|---|---|---|---|---|---|---|---|
| H-A | D-A | |||||||
| 1a | 1 | 34A | ASN | 3.06 | 3.44 | 104.91 | 206 [Nam] | 1695 [O2] |
| 2 | 35A | THR | 2.49 | 2.94 | 106.66 | 1688 [Npl] | 210 [O2] | |
| 3a | - | - | - | - | - | - | - | - |
| 3b | 1 | 96A | ASN | 2.92 | 3.40 | 111.16 | 688 [Nam] | 1702 [O2] |
| 2 | 99A | ASN | 2.83 | 3.56 | 131.70 | 712 [Nam] | 1702 [O2] | |
| 3c | 1 | 6A | SER | 3.05 | 3.89 | 145.79 | 6 [O3] | 1688 [N2] |
| 2 | 37A | LYS | 3.01 | 3.89 | 148.87 | 219 [Nam] | 1702 [O2] | |
| 3d | 1 | 6A | SER | 3.16 | 4.02 | 148.65 | 6 [O3] | 1688 [N2] |
| 2 | 37A | LYS | 2.89 | 3.78 | 150.07 | 219 [Nam] | 1701 [O-] | |
| 3e | 1 | 6A | SER | 2.99 | 3.79 | 140.68 | 6 [O3] | 1688 [N2] |
| 2 | 37A | LYS | 3.20 | 4.05 | 144.83 | 219 [Nam] | 1701 [O-] | |
| 3f | 1 | 34A | ASN | 3.45 | 3.93 | 111.97 | 206 [Nam] | 1704 [O2] |
| 2 | 37A | LYS | 3.02 | 3.42 | 105.61 | 219 [Nam] | 1695 [O2] | |
| 3g | 1 | 6A | SER | 2.99 | 3.80 | 142.35 | 6 [O3] | 1688 [N2] |
| 2 | 37A | LYS | 3.22 | 4.07 | 145.80 | 219 [Nam] | 1698 [O2] | |
| 3h | 1 | 6A | SER | 3.08 | 3.87 | 139.59 | 6 [O3] | 1688 [N2] |
| 3i | - | - | - | - | - | - | - | - |
Table 6.
Hydrogen bond interactions of imidazolium cations.
| Imidazolium cation | Index | Residue | AA | Distance (Å) |
Donor Angle | Donor atom | Acceptor atom | |
|---|---|---|---|---|---|---|---|---|
| H-A | D-A | |||||||
| [3a-H]+ | 1 | 6A | SER | 3.10 | 3.89 | 139.60 | 6 [O3] | 1688 [N2] |
| 2 | 6A | SER | 3.35 | 3.89 | 115.13 | 1688 [N2] | 6 [O3] | |
| [3b-H]+ | 1 | 6A | SER | 3.04 | 3.84 | 140.17 | 6 [O3] | 1688 [N2] |
| 2 | 6A | SER | 3.32 | 3.84 | 113.27 | 1688 [N2] | 6 [O3] | |
| 3 | 37A | LYS | 3.17 | 4.02 | 145.11 | 219 [Nam] | 1702 [O-] | |
| [3c-H]+ | - | - | - | - | - | - | - | - |
| [3d-H]+ | 1 | 7A | PRO | 2.00 | 2.80 | 133.87 | 1688 [N2] | 10 [O2] |
| [3e-H]+ | 1 | 6A | SER | 3.17 | 4.00 | 144.69 | 6 [O3] | 1688 [N2] |
| 2 | 6A | SER | 3.57 | 4.00 | 107.49 | 1688 [N2] | 6 [O3] | |
| 3 | 37A | LYS | 3.10 | 3.96 | 147.19 | 219 [Nam] | 1703 [O2] | |
| [3f-H]+ | 1 | 6A | SER | 3.04 | 3.84 | 139.86 | 6 [O3] | 1688 [N2] |
| 2 | 6A | SER | 3.31 | 3.84 | 113.83 | 1688 [N2] | 6 [O3] | |
| 3 | 37A | LYS | 3.23 | 4.09 | 146.04 | 219 [Nam] | 1705 [O2] | |
| [3g-H]+ | 1 | 6A | SER | 3.06 | 3.89 | 144.21 | 6 [O3] | 1688 [N2] |
| 2 | 6A | SER | 3.47 | 3.89 | 106.63 | 1688 [N2] | 6 [O3] | |
| [3h-H]+ | 1 | 75B | ARG | 2.80 | 3.52 | 130.66 | 1392 [Ng+] | 1688 [N2] |
| 2 | 75B | ARG | 2.05 | 3.01 | 157.03 | 1688 [N2] | 1389 [Ng+] | |
| [3i-H]+ | 1 | 77B | VAL | 2.66 | 3.28 | 121.36 | 1404 [Nam] | 1688 [N2] |
Table 7.
Hydrogen bond interactions of organic anions.
| Organic anion | Index | Residue | AA | Distance (Å) |
Donor Angle | Donor atom | Acceptor atom | |
|---|---|---|---|---|---|---|---|---|
| H-A | D-A | |||||||
| DAB | 1 | 6A | SER | 2.32 | 3.04 | 129.74 | 6 [O3] | 1692 [Npl] |
| 2 | 6A | SER | 2.28 | 3.04 | 130.54 | 1692 [Npl] | 6 [O3] | |
| 3 | 34A | ASN | 3.21 | 3.78 | 118.19 | 206 [Nam] | 1697 [O.co2] | |
| 4 | 35A | THR | 2.45 | 3.01 | 114.88 | 1698 [Npl] | 210 [O2] | |
| DNB | 1 | 96A | ASN | 3.23 | 4.08 | 145.59 | 688 [Nam] | 1700 [O2] |
| 2 | 99A | ASN | 2.59 | 3.56 | 167.95 | 712 [Nam] | 1700 [O2] | |
| FA | - | - | - | - | - | - | - | - |
| NMP | - | - | - | - | - | - | - | - |
| mCPBA | 1 | 34A | ASN | 3.61 | 3.93 | 101.76 | 206 [Nam] | 1696 [O-] |
Table 8.
Hydrogen bond interactions of ChQ, HChQ, H3PO4 and H2SO4.
| Standard drug | Index | Residue | AA | Distance (Å) |
Donor Angle | Donor atom | Acceptor atom | |
|---|---|---|---|---|---|---|---|---|
| H-A | D-A | |||||||
| ChQ | 1 | 6A | SER | 3.04 | 3.78 | 134.72 | 6 [O3] | 1693 [N3] |
| HChQ | 1 | 34A | ASN | 3.64 | 4.03 | 106.32 | 206 [Nam] | 1703 [N2] |
| 2 | 35A | THR | 2.48 | 3.02 | 114.53 | 212 [O3] | 1698 [O3] | |
| H3PO4 | 1 | 34A | ASN | 3.31 | 3.66 | 103.60 | 1688 [O3] | 205 [O2] |
| 2 | 35A | THR | 2.15 | 2.90 | 132.34 | 212 [O3] | 1692 [O3] | |
| 3 | 35A | THR | 2.26 | 2.99 | 131.83 | 1692 [O3] | 210 [O2] | |
| H2SO4 | 1 | 35A | THR | 2.10 | 2.88 | 136.45 | 212 [O3] | 1689 [O3] |
| 2 | 35A | THR | 2.41 | 2.89 | 109.89 | 1689 [O3] | 210 [O2] | |
Table 9.
π-Stacking interaction of 3a, [3c-H]+ and mCPBA.
| Imidazole/Imidazolium cation/organic anion | Index | Residue | AA | Distance (Å) | Angle | Offset | Type | Ligand atom(s) |
|---|---|---|---|---|---|---|---|---|
| 3a | 1 | 41A | PHE | 3.78 | 0.49 | 1.13 | P | 1693–1698 |
| [3c-H]+ | 1 | 41A | PHE | 4.15 | 12.71 | 1.59 | P | 1694-1698 & 1700 |
| mCPBA | 1 | 41A | PHE | 3.77 | 4.57 | 0.97 | P | 1686- 1688, 1690- 1692 |
Table 10.
Salt bridge interaction of Ferulate anion.
| Organic anion | Index | Residue | AA | Distance (Å) | Ligand group | Ligand atom(s) |
|---|---|---|---|---|---|---|
| FA | 1 | 75B | ARG | 4.17 | Carboxylate | 1695 & 1696 |
2.2. Technical elements in bio-informatics studies
2.2.1. Receptors
In bio-informatics studies, receptors are chemical assemblies, created from protein, that get and transduce signs that might be composed into biological frameworks [40], which stick to ligands and induce responses inside the invulnerable gadget, such as cytokine receptors, development factor receptor and Fc receptor. Receptors are generally discovered in different insusceptible cells like B cells, T cells, NK cells, Monocytes and stem cells. A molecule that links to a receptor is known as a ligand, and can be a peptide (brief-protein) or every other small molecule together with a neurotransmitter, hormone, pharmaceutical-drug, toxin, or parts of the outside of a virus or microbe. While a ligand sticks to its matching receptor, it turns on or restrains the receptor's associated-biochemical track. Receptors can result in mobile increase, division and dying; influences membrane channels or alter cell binding. Receptors offer a vital function in sign transduction, immunotherapy and immune retorts.
2.2.2. Ligands
In pharmacology, a ligand is a small molecule which frames a macro-compound with a biomolecule to serve a biological motive. In ligand-protein linking, the ligand is typically a molecule which creates an indication via linking to a site of a target protein. This communication usually brings about a difference in conformation of the aimed protein [41, 42].
2.2.3. Docking
Within the area of molecular modeling, docking is a technique which forecasts the favored direction of one molecule to a second while certain to each different to shape a solid macro-compound [43]. Information about the desired direction in flip can be utilized to envisage the power of association or binding ability among organic compounds the usage of, as an instance, scoring functions.
2.2.4. Binding mode
The direction of the ligands is comparative with the receptors in addition to the conformation of the receptor and ligand while inevitable to each different.
2.2.5. Scoring
In computational science and molecular modeling, scoring capacities (arithmetic capacities) used to roughly foresee the linking ability between two molecules after they had been simulated. In general, one of the molecules is a diminutive organic compound, for example, a medication, and the second is the medication's biological objective, for example, a protein receptor [44]. Scoring capacities have additionally been created to anticipate the quality of inter-molecular communications between the two proteins [45] or among protein and DNA [46].
2.2.6. Protein-ligand interactions
Protein–ligand binding is crucial for all approaches occurring in residing organisms. Ligand-intervened sign diffusion through molecular balancing is crucial to all existence tactics; those chemical communications incorporates biological appreciation at molecular stage. The progression of the protein capabilities depends on the improvement of precise websites which might be intended to connect with ligands. Ligand fastening ability is essential to the law of life tasks of any creature. Protein-Ligand binding takes place via molecular mechanics which includes conformational adjustments among low and high attraction states. Ligand sticking communication alters the protein kingdom and its feature. The protein-ligand composite is a reversible non-covalent interplay between two biological (macro-)molecules. In non-covalent binding, there is no distribution of electrons like in covalent interactions or bonds. Non-covalent interaction relies upon Vander Waals forces, hydrogen bonds, hydrophobic forces, electrostatic interactions, π-π interactions in which no electrons are distributed among the two or more noteworthy included molecules [47].
2.2.7. Binding affinity
Binding affinity or bonding energy is delivered when a medication molecule is associated with a target, prompting a bringing down of the general vitality of the complex. The delivery of binding energy likewise makes up for any transformation of the ligand from its energy minimum to its bound conformation with the protein [48].
3. Results and interpretation
3.1. Protein-ligand binding communications
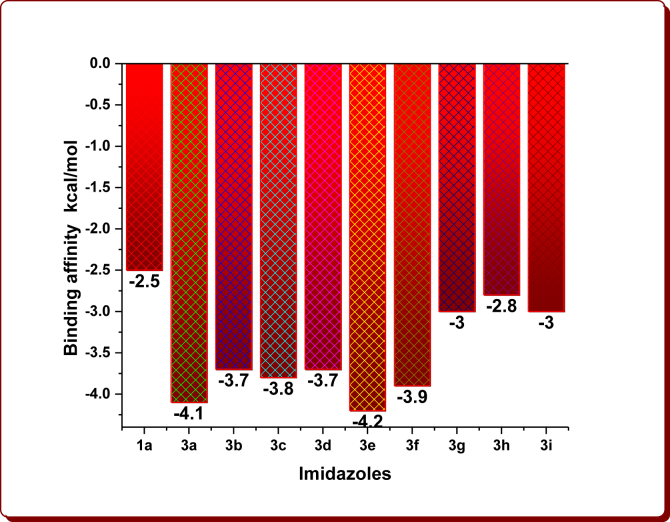
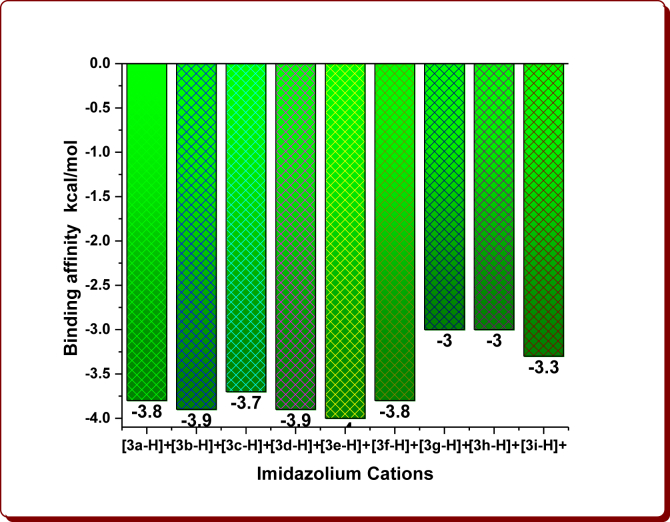
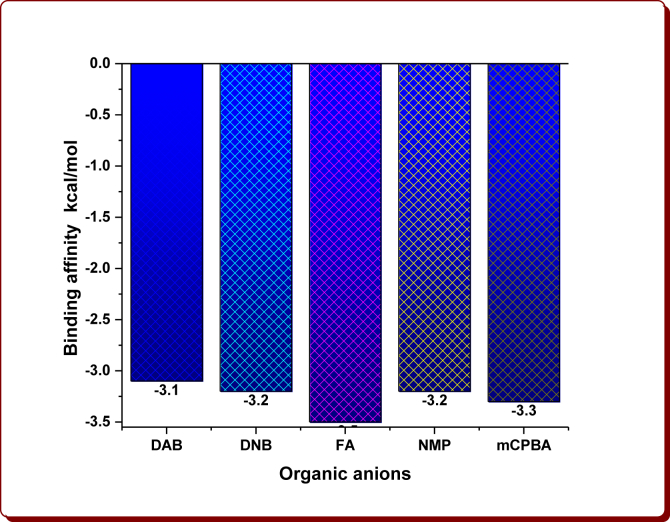
Molecular docking studies were simulated to examine the linking site and binding energies of N1-(4-substitutedbenzyl)/butyl-2-methyl-4-nitro-1H-imidazoles (3a-i), nine Brønsted acidic based imidazolium cations and aromatic organic anions (DAB: 3,5-Diaminobenzoate; DNB: 3,5-Dinitrobenzoate; FA: Ferulate; NMP: 5-Nitromonophthalate; mCPBA: m-Chloroperoxybenzoate) with 6W4B. The binding affinity values are given in Tables 1, 2, 3, and 4 and are displayed in Figures 6, 7, 8, and 9. The optimum binding interactions and binding sites are displayed in Figures 11, 12, 13, 14, 15, and 16 and are summarized in Tables 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10.
Figure 6.
The binding affinity of imidazoles.
Figure 7.
The binding affinity of imidazolium cations.
Figure 8.
The binding affinity of organic anions.
Figure 9.
The binding affinity of standard drugs.
Figure 11.
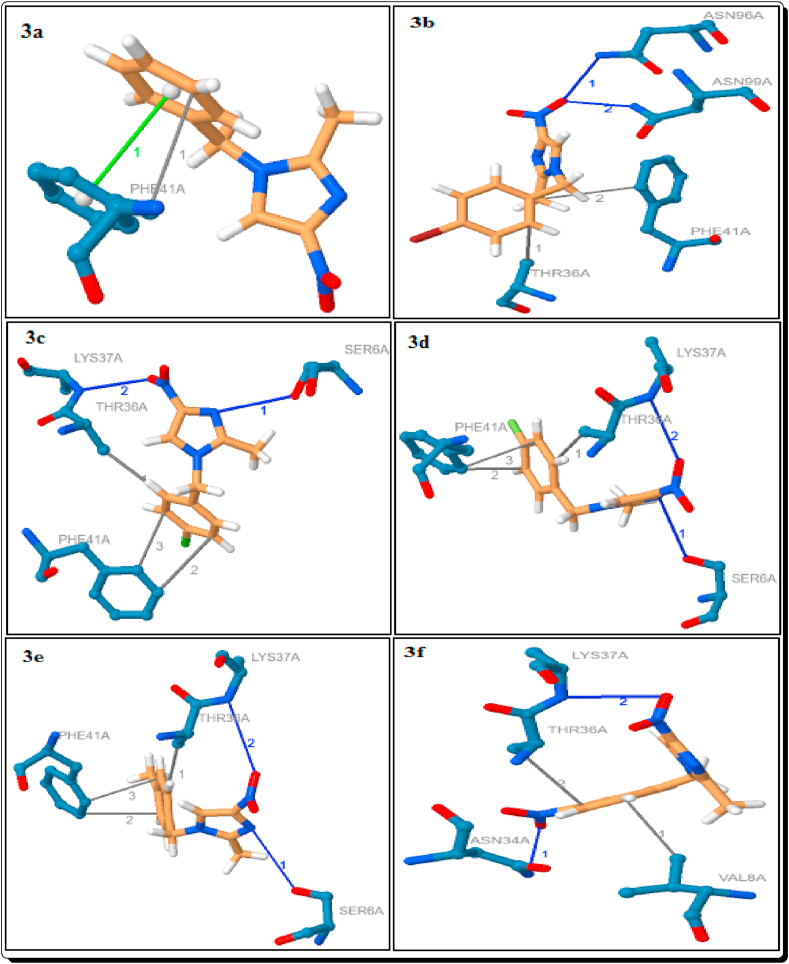
Docking poses and binding modes of 3a-f.
Figure 12.
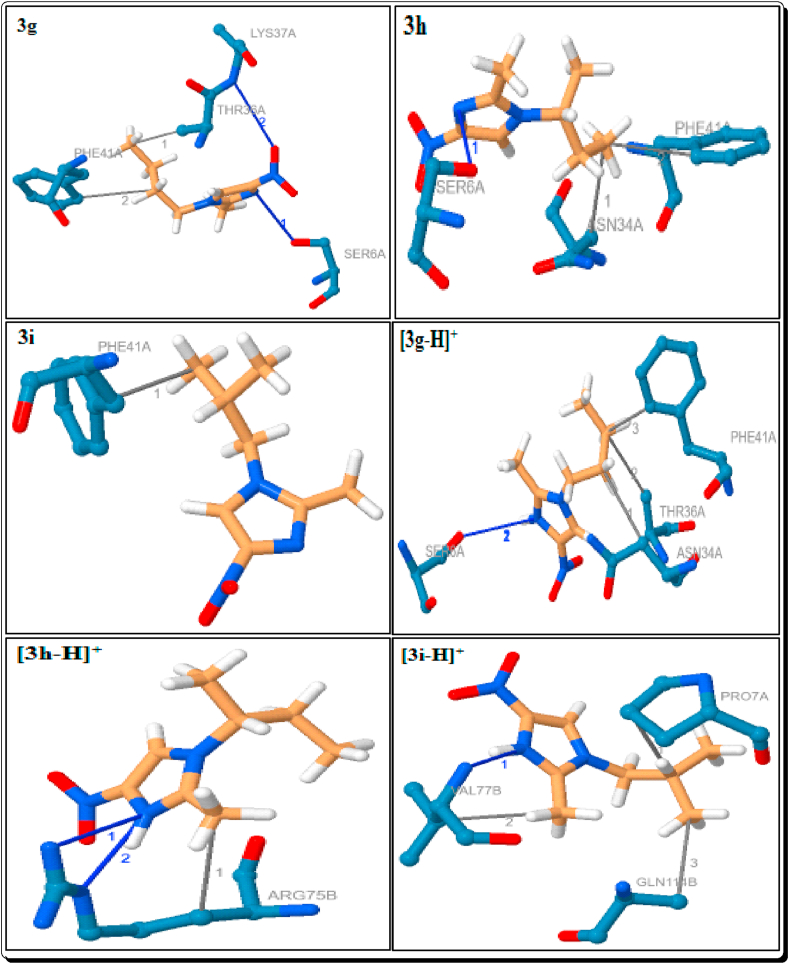
Docking poses and binding modes of 3g-i and their imidazolium cations.
Figure 13.
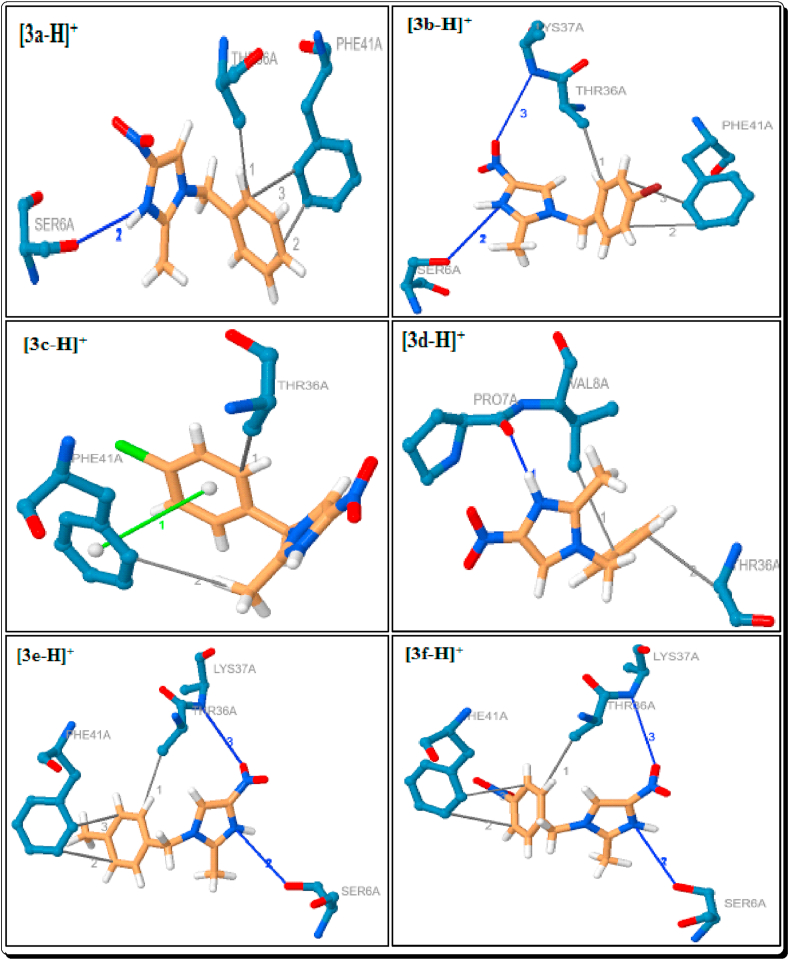
Docking poses and binding modes of [3a-H]+-[3f-H]+.
Figure 14.
Docking poses and binding modes of organic anions.
Figure 15.
Docking poses and binding modes of ChQ and HChQ
Figure 16.
Docking poses and binding modes of H2SO4 and H3PO4.
3.1.1. Binding affinity
The docking results clearly demonstrate that fact that the docking energies for the starting compound 1a has -2.5 kcal/mol, the 1-substituted imidazoles 3a-i have in the range from -2.8 to -4.2 kcal/mol and their imidazolium cations in the range from -3.0 to -4.0 kcal/mol (Tables 1 and 2, Figures 6 and 7). Among them, the compound 3e (-4.2 kcal/mol) and its imidazolium cation (-4.0 kcal/mol) have been shown the highest binding affinity, which is owing to the existence of the 4-Methylbenzyl moiety in the parent imidazolyl moiety. After the protonation, the imidazolium cations 3h-H+ and 3i-H+ alone showed remarkable binding affinities while others exhibited slightly less affinities (Figure 7). The cation 3h-H+ remained unchanged. The binding affinity of organic anions was found to be -3.1 kcal/mol for DAB, -3.2 kcal/mol for DNB, -3.5 kcal/mol for FA, -3.2 kcal/mol for NMP and -3.3 kcal/mol for mCPBA (Table 3, Figure 8). The binding affinities of ChQ, HChQ, H3PO4 and H2SO4 were found to be -3.6, -3.6, -2.2 and -2.0 kcal/mol respectively (Table 4, Figure 9).
3.1.2. Hydrophobic interactions
Among them docked compounds and standard drugs, the parent imidazole 1a, H3PO4 and H2SO4 only, have not shown the hydrophobic interactions (Tables 1 and 4, Figures 10 and 16). Except 3f, all other eight imidazoles have been showed 1–2 hydrophobic interactions with the residue PHE-41A which are due to the distance in the range from 3.54 Å to 3.78 Å. The imidazoles 3b-e and 3g showed a hydrophobic interaction with THR-36A (3.57–3.77 Å). The imidazole 3f has the two interactions with the residue VAL-8A and THR-36A with the distance of 3.61 and 3.84 Å respectively (Figure 11). The imidazole 3h has a hydrophobic interaction with ASN-34A (3.82 Å) (Table 1, Figure 12). The [3a-H]+, [3b-H]+, [3e-H]+ and [3f-H]+ have been shown the two hydrophobic interactions, and [3c-H]+ & [3g-H]+ showed a hydrophobic interaction with PHE-41A. The imidazolium cations [3a-H]+-[3g-H]+ have been shown a hydrophobic interaction with THR-36A. The cation [3d-H]+, [3g-H]+ and [3h-H]+ have been shown a hydrophobic interaction with the residue of VAL-8A (3.74 Å), ASN-34A (3.91 Å) and ARG-75B (3.98 Å) respectively. The cation [3i-H]+ showed the three hydrophobic interactions with the residue of PRO-7A (3.70 Å), VAL-77B (3.54 Å) and GLN-114B (3.77 Å) (Table 2, Figures 12 and 13).
Figure 10.
Docking pose and binding modes of 1a and representative modes of interactions.
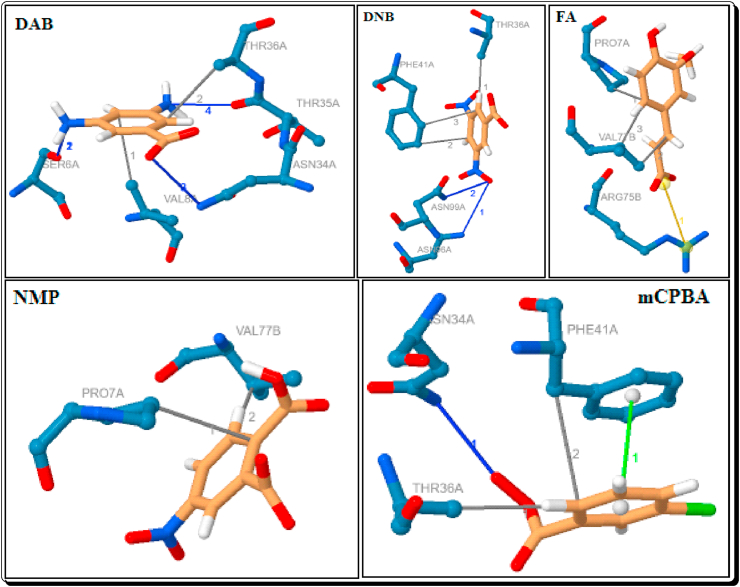
Among them docked organic anions, DAB, DNB and mCPBA have been shown a hydrophobic interaction with the residue of THR-36A (3.66–3.72 Å). The anion DNB and FA have been shown the two interactions with PHE-41A (3.59 & 3.76 Å) and VAL-77B (3.61 & 3.68 Å) respectively. The anion FA (3.67 Å) and NMP (3.58 Å) showed a hydrophobic interaction with the residue of PRO-7A. The anion DAB, NMP and mCPBA have been shown a hydrophobic interaction with the residue of VAL-8A (3.67 Å), VAL-77B (3.64 Å) and PHE-41A (3.69 Å) respectively (Table 3, Figure 14).
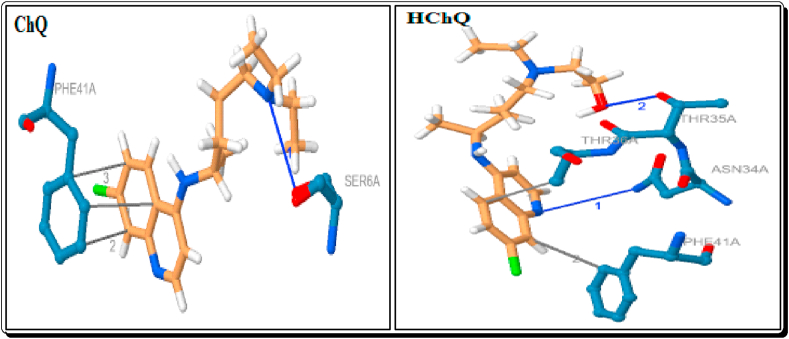
The ChQ has three hydrophobic interactions with the residue of PHE-41A (3.63–3.71 Å). The HChQ showed a hydrophobic interactions with THR-36A (3.58 Å) and PHE-41A (3.36 Å) (Figure 15). Compared to the docked compounds, six imidazoles (3b-g), seven imidazolium cations ([3a-H]+-[3g-H]+) and three organic anions (DAB, DNB and mCPBA) have been showing a strong hydrophobic interactions with THR-36A. The eight imidazoles (except 3f), six imidazolium cations (except [3d-H]+, [3h-H]+ and [3i-H]+), three organic anions DAB, NMP and mCPBA showed a strong hydrophobic interactions with PHE-41A (Table 4, Figure 14).
3.1.3. Hydrogen bond interactions
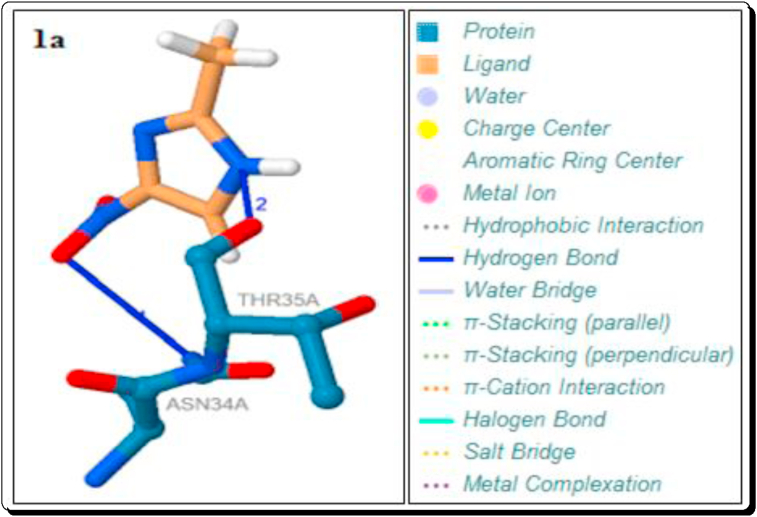
The hydrogen bond (H-bond) interactions, residues, H-A and D-A distance, donor angle, donor and acceptor atoms are summarized in Tables 5, 6, 7, and 8. Among them imidazoles, except 3a and 3i, all imidazoles are showing the strong 1–2 hydrogen bond interactions with the protein 6W4B. The parent imidazole 1a showed two strong hydrogen bond interactions with the residue of ASN-34A (3.06Å) and THR-35A (2.49Å). The imidazoles 3c-e, g and h showed a strong hydrogen bond interaction with the residue SER-6A with the distance in the range of 2.99–3.08Å (Table 5, Figures 11 and 12). The imidazoles 3c-g showed a strong hydrogen bond interaction with the residue LYS-47A (H-A distance in the range 2.89–3.22Å) and 3f has a hydrogen bond interaction with the residue ASN-34A (H-A distance in the range 3.45Å). The imidazole 3b has shown two strong hydrogen bond interactions with the residues ASN-96A (H-A distance is 2.92Å) and ASN-99A (H-A distance is 2.83Å) (Table 5, Figure 11).
Except [3c-H]+, all the Brønsted acidic based imidazolium cations have been shown a strong hydrogen bond interaction with the protein (Table 6, Figure 13). The cations [3a-H]+, [3b-H]+, [3e-H]+, [3f-H]+ and [3g-H]+ showed two hydrogen bond interactions with the residue SER-6A, which are due to the H-A distance in the range of 3.04–3.57Å. The cations [3b-H]+, [3e-H]+ and [3f-H]+ are showing a well-built hydrogen bond communication with the residue LYS-37A and the H-A distance 3.17, 3.10 and 3.23Å respectively. The cations [3h-H]+ has showed two hydrogen bond interactions with the residue ARG-75B and [3i-H]+ showed a hydrogen bond interaction with VAL-75B. Among them, the cation [3d-H]+ has a strong hydrogen bond interaction with the residue PRO-7A which is due to very short bond distance of 2.00Å and [3i-H]+ also showed strongest hydrogen bond interaction (with the residue VAL-75B) with the distance of 2.05Å (Table 6, Figures 12 and 13).
Among the five organic anions, anions FA and NMP have not shown any H-bond connections with the target protein (Table 7, Figure 14). The anion DAB has showed four H-bond interactions with the target protein, the two interactions with the residue SER-6A (one H-A distance is 2.32 Å and another one is 2.28 Å), one interaction with the residue ASN-34A (3.21 Å) and one interaction with THR-35A (2.45 Å). The anion DNB showed two interactions with ASN-96A (3.23 Å) and ASN-99A (2.59 Å). The anion mCPBA has an interaction with ASN-34A (3.61 Å). Among them anions, the anion DAB showed strong interactions with the protein (Table 7, Figure 14).
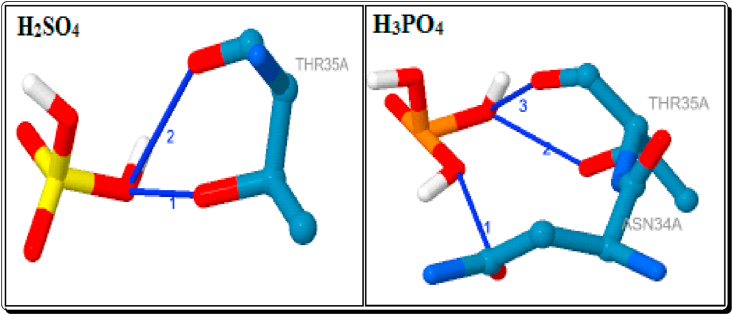
The currently used standard drugs for the COVID-2019 treatment such as Chloroquine (ChQ) and Hydroxy Chloroquine (HChQ) sulphate and phosphate derivatives also docked. The ChQ showed a hydrogen bond interaction with the residue SER-6A and its bond distance is 3.04 Å (Table 8, Figure 15). This type of the strong and short hydrogen bond interaction is also showing the five imidazoles (3c-e, g & h, 2.99–3.08Å), the five Brønsted acidic based imidazolium cations ([3a-H]+, [3b-H]+, [3e-H]+, [3f-H]+ & [3g-H]+) and the organic anion DAB (2.32 & 2.28 Å). A hydrogen bond interaction with ASN-34A showed by the standard drug HChQ (3.64 Å) and H3PO4 (3.31 Å). In our docked compounds, the imidazoles 1a (3.06Å) and 3f (3.45Å), the organic anions DAB (3.21 Å) and mCPBA (3.61 Å) also showed better binding the similar type of interaction with protein (Table 7, Figures 10, 11, and 15). The H3PO4 (2.15 & 2.26 Å) and H2SO4 (2.10 & 2.41 Å) are shown the two strong interactions and HChQ (2.48 Å) showed a strong interactions with the residue THR-35A (Table 8, Figures 15 and 16). Among them docked compounds, the imidazole 1a (2.49Å) and the anion DAB (2.45 Å) showed the similar type of hydrogen bond interaction (Tables 5 and 7, Figure 14).
3.1.4. π-Stacking interactions of 3a, [3c-H]+ and mCPBA
Among them simulated imidazoles, the imidazole 3a only showed the π-stacking interaction with the residue PHE-41A of the protein. The distance between ligand and protein is 3.78Å with the angle of 0.49 (Table 9, Figure 11). In the imiazolium cations and organic anions, the cation [3c-H]+ and the anion mCPBA have been shown the π-stacking interactions with the residue PHE-41A of the protein, which is due to the distance of 4.15 and 3.77 Å respectively (Table 9, Figures 13 and 14).
3.1.5. Salt bridge interactions of FA
Among them Imidazoles, Protic Imidazolium cations and Organic anions, the Ferulate anion (FA) only shown a salt bridge interaction with the residue ARG-75B of 6W4B, which is due to the carboxylate group with the distance of 4.17Å (Table 10, Figure 14).
4. Conclusion
In summary, the entire globe is screaming about the deadly novel Corona Virus and the ill effects that create in human beings. Presently, more than physical illness, this disease has developed a kind of fear among the general public all over the globe. Notwithstanding, no affirmed sedate at present exists to treat the malady. Currently accessible medications for COVID-19 treatment basically following up on the main protease (Mpro). In our present research, molecular docking experiments were applied to examine the effect of inhibitors on coronavirus proteinase. The major aim of this investigation was to look at the efficacy of a few restorative imidazoles, the Brønsted acidic based imidazolium cations and five organic anions that might be utilized to repress the COVID-19 disease pathway bio-informatically. Among them docked compounds, six imidazoles (3b-g), seven imidazolium cations ([3a-H]+-[3g-H]+) and three organic anions (DAB, DNB and mCPBA) have been showing a strong hydrophobic interactions with THR-36A. The eight imidazoles (except 3f), six imidazolium cations (except [3d-H]+, [3h-H]+, [3i-H]+), three organic anions DAB, NMP and mCPBA showed a strong hydrophobic interactions with PHE-41A. The strong and short hydrogen bond interactions are also showing the five imidazoles (3c-e, g & h, 2.99–3.08Å), the five Brønsted acidic based imidazolium cations ([3a-H]+, [3b-H]+, [3e-H]+, [3f-H]+ & [3g-H]+) and the organic anions DAB & mCPBA with the residues (ASN-34A & THR-35A) of COVID-2019 protein and their activity and efficiency are in accordance with the standard drugs (ChQ & HChQ derivatives). Yet, additional investigations are essential to explore in the future, the prospective usages and clinical trial of the compounds to restrain viral infections and will be accounted for in due course.
Declarations
Author contribution statement
D. Satheesh: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
A. Rajendran, K. Chithra: Analyzed and interpreted the data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Dr. AR, Mr. DS and Ms. KC, thank the HOD of Chemistry, the Principal and the management of Sir Theagaraya College, Chennai-21 and Loganatha Narayanaswamy Government College (Autonomous), Ponneri for their constant support and encouragement.
Contributor Information
Dhurairaj Satheesh, Email: satheeshvdm@gmail.com.
Annamalai Rajendran, Email: annamalai_rajendran2000@yahoo.com.
References
- 1.Wu F., Zhao S., Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan J.F., Yuan S., Kok K.H. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Notification of 2019-nCoV Infection. World Health Organization (WHO); 2019. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200326-sitrep-66-covid-19.pdf?sfvrsn=81b94e61_2 [Google Scholar]
- 5..; (a) Notification of 2019-nCoV Infection. World Health Organization (WHO); 2019. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200430-sitrep-101-covid-19.pdf?sfvrsn=2ba4e093_2 [Google Scholar]; (b) Notification of 2019-nCoV Infection. World Health Organization (WHO); 2019. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200531-covid-19-sitrep-132.pdf?sfvrsn=d9c2eaef_2 [Google Scholar]; (c) Notification of 2019-nCoV Infection. World Health Organization (WHO); 2019. https://www.who.int/docs/default-source/coronaviruse/20200630-covid-19-sitrep-162.pdf?sfvrsn=e00a5466_2 [Google Scholar]
- 6.Notification of 2019-nCoV Infection. World Health Organization (WHO); 2019. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200729-covid-19-sitrep-191.pdf?sfvrsn=2c327e9e_2 [Google Scholar]
- 7.Audio transcripts of the news briefing held by the State Council of China on February 17, 2020. The National health Commission of the People's Republic of China. http://www.nhc.gov.cn/xcs/yqfkdt/202002/f12a62d10c2a48c6895cedf2faea6e1f.shtml In Chinese.
- 8.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper Ross G., Magwere Tapiwanashe. Chloroquine has not disappeared. Afr. Health Sci. 2007;7:185–186. doi: 10.5555/afhs.2007.7.3.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox R.I. Mechanism of action of Hydroxychloroquine as an antirheumatic drug. Semin. Arthritis Rheum. 1993;23:82–91. doi: 10.1016/s0049-0172(10)80012-5. [DOI] [PubMed] [Google Scholar]
- 11.Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases? Lancet Infect. Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan Y., Zou Z., Sun Y., Li X., Xu K.F. Y.Wei, N. Jin, C. Jiang, Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 2013;23:300–302. doi: 10.1038/cr.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keyaerts E., Vijgen L., Maes P., Neyts J., Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;55:105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y.W., Yiu C.P.B., Wong K.Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000 Research. 2020;9:129. doi: 10.12688/f1000research.22457.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.John S.E.S., Tomar S., Stauffer S.R., Mesecar A.D. Targeting zoonotic viruses: structure-based inhibition of the 3C-like protease from bat coronavirus HKU4-The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS) Bioorg. Med. Chem. 2015;23:6036–6048. doi: 10.1016/j.bmc.2015.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Zhang J., Wang N., Li H., Shi Y., Guo G., Zou Q. Therapeutic drugs targeting 2019-nCoV main protease by high-throughput screening. BioRxiv. 2020 [Google Scholar]
- 21.Nosrati M., Behbahani M. Molecular docking study of HIV-1 protease with triterpenoides compounds from plants and Mushroom. J. Arak Uni. Med. Sci. 2015;18:67–79. [Google Scholar]
- 22.Yu R., Chen L., Lan R., Shen R., Li P. Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int. J. Antimicrob. Agents. 2020:106012. doi: 10.1016/j.ijantimicag.2020.106012. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Z., Peng C., Shi Y., Zhu Z., Mu K., Wang X., Zhu W. Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. BioRxiv. 2020 [Google Scholar]
- 24.Li Fang, Receptor Recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89(4):1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 26.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses — drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abuo-Rahma Gamal El-Din A., Mohamed Mamdouh F.A., Ibrahim Tarek S., Shoman Mai E., Samir Ebtihal, Abd El-Baky Rehab M. Potential repurposed SARS-CoV-2 (COVID-19) infection drugs. RSC Adv. 2020;10:26895–26916. doi: 10.1039/d0ra05821a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa H. Mizoribine and mycophenolate mofetil. Curr. Med. Chem. 1999;6(7):575–597. [PubMed] [Google Scholar]
- 29.Kolobukhina L.V., Merkulova L.N., Shchelkanov M., Burtseva E.I., Isaeva E.I., Malyshev N.A. Efficacy of ingavirin in adults with influenza. Ter. Arkh. 2009;81(3):51–54. [PubMed] [Google Scholar]
- 30.Loginova S., Borisevich S.V., Maksimov V.A., Bondarev V.P., Nebol’sin V.E. Therapeutic efficacy of Ingavirin, a new domestic formulation against influenza A virus (H3N2) Antib. Khimioterap. 2008;53(7–8):27–30. [PubMed] [Google Scholar]
- 31.Ingavirin New - well forgotten old? Evaluation of real effectiveness. https://ostit.ru/en/coronary-artery-disease/ingavirin-novoe-horosho-zabytoe-staroe-ocenka-realnoi-effektivnosti/ Available at:
- 32.Pawełczyk A., Zaprutko L. Anti-COVID drugs: repurposing existing drugs or search for new complex entities, strategies and perspectives. Future Med. Chem. 2020 doi: 10.4155/fmc-2020-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauro F.V., Francisco D.C., Marcela R.N., Maria L.R., Maria Virginia M.A., Alejandara G.E., Regina C.C. Design and synthesis of some imidazole derivatives: theoretical evaluation of interaction with a coronavirus (HCoV-NL63) Biointerf. Res. Appl. Chem. 2020;10(4):5869–5874. [Google Scholar]
- 34.Fister S., Mester P., Sommer J., Witte A.K., Kalb R., Wagner M., Rossmanith P. Virucidal influence of ionic liquids on phages P100 and MS2. Front. Microbiol. 2017;8:1608. doi: 10.3389/fmicb.2017.01608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satheesh D., Rajendran A., Saravanan R., Kannan S., Chithra K. An efficient room temperature synthesis of N1-(4-substitutedbenzyl)-2-methyl-4-nitro-1H-imidazoles and N1-butyl-2-methyl-4-nitro-1H-imidazoles. Iranian J. Org. Chem. 2018;10:2325–2331. [Google Scholar]
- 36.Satheesh D., Rajendran A., Chithra K., Saravanan R. Synthesis of some new protic N1-Benzyl/Butyl-2-methyl-4-nitro-1H-imidazol-3-ium salts with 3,5-Diaminobenzoate, 3,5-Dinitrobenzoate, (E)-3-(4-Hydroxy-3-methoxyphenyl) acrylate and 2-Carboxy-5-nitrobenzoate as organic anions. Res. Chem. 2020;2:100033. [Google Scholar]
- 37.Satheesh D., Rajendran A., Chithra K., Saravanan R. Synthesis and antimicrobial evaluation of N1 -benzyl/butyl-2-methyl-4-nitro-3-imidazolium 3 ´-chloroperoxy benzoates. Chem. Data Collect. 2020;28:100406. [Google Scholar]
- 38.Tan K., Kim Y., Jedrzejczak R., Maltseva N., Endres M., Michalska K., Joachimiak A. The crytal structure of Nsp 9 RNA binding protein of SARS CoV-2. RCSB Protein Data Bank. 2020 [Google Scholar]
- 39.Salentin Sebastian, Schreiber Sven, Joachim Haupt V., Adasme Melissa F., Schroeder Michael. PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015;43:W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall J.E. Elsevier Saunders; Philadelphia, PA: 2016. Guyton and Hall Textbook of Medical Physiology. pp. 930–937. [Google Scholar]
- 41.Teif V.B. Ligand-induced DNA condensation: choosing the model. Biophys. J. 2005;89(4):2574–2587. doi: 10.1529/biophysj.105.063909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teif V.B., Rippe K. Statistical-mechanical lattice models for protein-DNA binding in chromatin. J. Phys. Condens. Matter. 2010;22(41):414105. doi: 10.1088/0953-8984/22/41/414105. [DOI] [PubMed] [Google Scholar]
- 43.Lengauer T., Rarey M. Computational methods for biomolecular docking, current opinion in structural Biology. 1996;6(3):402–406. doi: 10.1016/s0959-440x(96)80061-3. [DOI] [PubMed] [Google Scholar]
- 44.Jain A.N. Scoring functions for protein-ligand docking, current protein & peptide science. 2006;7(5):407–420. doi: 10.2174/138920306778559395. [DOI] [PubMed] [Google Scholar]
- 45.Lensink M.F., Méndez R., Wodak S.J. Docking and scoring protein complexes: CAPRI 3rd Edition. Proteins. 2007;69(4):704–718. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 46.Robertson T.A., Varani G. An all-atom, distance-dependent scoring function for the prediction of protein-DNA interactions from structure. Proteins. 2007;66(2):359–374. doi: 10.1002/prot.21162. [DOI] [PubMed] [Google Scholar]
- 47.Bongrand P. Ligand-receptor interactions. Rep. Prog. Phys. 1999;62(6):921–968. [Google Scholar]
- 48.Zhou H.X., Rivas G., Minton A.P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.