To explore the mutational possibilities of insertions and deletions (indels) in the Klebsiella pneumoniae carbapenemase (KPC) beta-lactamase, we selected for ceftazidime-avibactam-resistant mutants. Of 96 screened mutants, we obtained 19 indels (2 to 15 amino acids), all located in the loops surrounding the active site. Three antibiotic susceptibility phenotypes emerged: an extended-spectrum-beta-lactamase-like phenotype, an activity restricted to ceftazidime, and a carbapenem-susceptible KPC-like phenotype.
KEYWORDS: KPC, carbapenemase, ceftazidime-avibactam, deletion, hydrolysis spectrum, insertion, mutation, mutational tolerance, omega loop
ABSTRACT
To explore the mutational possibilities of insertions and deletions (indels) in the Klebsiella pneumoniae carbapenemase (KPC) beta-lactamase, we selected for ceftazidime-avibactam-resistant mutants. Of 96 screened mutants, we obtained 19 indels (2 to 15 amino acids), all located in the loops surrounding the active site. Three antibiotic susceptibility phenotypes emerged: an extended-spectrum-beta-lactamase-like phenotype, an activity restricted to ceftazidime, and a carbapenem-susceptible KPC-like phenotype. Tolerance for indels reflects the evolvability of KPC beta-lactamase, which could challenge the therapeutic management of patients.
INTRODUCTION
Mutations are the substrate of natural selection and allow microorganisms to face environmental changes. Among mutations, insertions and deletions (indels) in the DNA are important sources of variation in nature.
The hydrolysis spectrum of antibiotics can be modified in cases of indels in antibiotic resistance genes, such as beta-lactamases genes (1–3). Recently, Klebsiella pneumoniae carbapenemase (KPC), a class A beta-lactamase, has emerged and is spreading worldwide, challenging the therapeutic management of infected patients. The core of the active site of KPC beta-lactamase contains a serine at position 70. This residue is surrounded by 4 loops (in Ambler numbering): one between the α3- and the α4-helices (Leu102 to Ser106), the omega-loop (Arg164 to Asp179), the loop between the β3- and β4-strands (Cys238 to Thr243; Loop238–243), and the loop between the β5-strand and the α11-helix (Ala267 to Ser275; Loop267–275) (4, 5). KPC enzymes confer resistance to penicillins, cephalosporins, carbapenems, and most beta-lactamase inhibitors. Avibactam, a non-beta-lactam beta-lactamase inhibitor, can inactivate KPC-type carbapenemases, offering new therapeutic alternatives (6). However, in vivo and in vitro resistance (7–12) to the ceftazidime-avibactam (CZA) combination has been reported worldwide and requires particular attention. The diversification of KPCs is very rapid, and at the time of writing, 54 clinical alleles (July 2020), including some selected during a ceftazidime-avibactam treatment, have been described (13). Although indels are rare in beta-lactamases, they are highly represented in KPC beta-lactamase clinical isolates (at least 10/54 isolates, 18.5%, including 8 insertions and 2 deletions) (13–16) and also have been described in vitro under ceftazidime-avibactam selective pressure (8, 9, 17).
Here, we further investigated the contribution and impact of indels in the evolution of KPC with ceftazidime-avibactam.
Anticancer chemotherapies have been shown to increase the global mutation rate of bacteria through the induction of the SOS system (18, 19). Therefore, we used 2 molecules of anticancer chemotherapies (dacarbazine, an alkylating drug, and the antimetabolite azacytidine) to increase the mutation rate and facilitate the selection of in vitro ceftazidime-avibactam-resistant mutants from clinical isolates of Enterobacteriaceae (an Enterobacter cloacae isolate harboring blaKPC-3 [RD26] and an Escherichia coli isolate harboring blaKPC-2 [RD29]). Mutants were selected by plating overnight cultures on Müeller-Hinton (MH) agar containing 4 times the MIC of CZA (4:1 ratio), and the frequency of emergence of antibiotic-resistant mutants was calculated by plating appropriate dilutions of overnight cultures on MH agar. Growing colonies with an indel in the coding region were screened by PCR amplification of the blaKPC gene and sequencing (18). KPC genes with indels were cloned in the pBR322 plasmid (Gibson assembly) and expressed in E. coli TOP10 (18). Antibiotic MICs were determined in duplicate according to the supplier's recommendations, either with the Etest method (for CZA and ertapenem) (bioMérieux, France) or by broth microdilution using the Sensititre ESB1F sensor plates for the other antibiotics (Thermo Scientific), and interpreted using EUCAST breakpoints (20). The reference strains E. coli ATCC 25922 and ATCC 35218 and K. pneumoniae ATCC 700603 were used as controls (21).
The frequency of the emergence of CZA-resistant mutants from E. cloacae RD26 was 10−6, 10−7, and 10−9 after incubation with azacitidine (0.5 mg/liter), dacarbazine (10 mg/liter), and no anticancer drug, respectively. That of E. coli RD29 was 10−8, 10−5, and 10−10 for azacitidine, dacarbazine, and no anticancer drug, respectively. Ninety-six CZA-resistant mutants, selected randomly from culture with or without anticancer drugs (16 colonies from each culture condition), were screened by PCR amplification and sequencing. We isolated 19 mutants with 17 unique indels, listed in Table 1, representing almost 20% (19/96) of CZA-resistant mutants selected in vitro. Eight were found in KPC-2 and 9 in KPC-3. Overall, eight mutants had insertions and nine had deletions. Five indels were also associated with nonsynonymous mutations. Indels were found under all culture conditions with no difference of frequencies depending on conditions.
TABLE 1.
Insertions and deletions and impact on MIC for various beta-lactams
| Isolate cloned in pBR322 and expressed in E. coli Top10 | Clinical allele | Deletion/insertion | Ancestral allele | Associated mutation | Site | Phenotypic profilea | MICb (mg/liter) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | TZP | CFZ | CEF | CRO | CPD | CAZ | CZA | CAZ-clavu | CTX | CTX-clavu | FEP | FOX | ETP | IPM | MEM | |||||||
| RD29 | KPC-2 | >16 | >64 | >16 | >16 | 128 | >32 | 16 | 0.5 | 16 | 16 | 2 | 8 | 16 | 1.5 | 2 | 4 | |||||
| RD26 | KPC-3 | >16 | >64 | >16 | >16 | 128 | >32 | 64 | 1 | 32 | 16 | 4 | 8 | 16 | 1 | 2 | 4 | |||||
| Insertions | ||||||||||||||||||||||
| D2 | ins165_EL | KPC-3 | Ω Loop | ESBL-like | >16 | <4 | >16 | >16 | 4 | 32 | >128 | 16 | 2 | 2 | <0.12 | 2 | 8 | 0.016 | <0.5 | <1 | ||
| E2 | KPC-25 | ins165_EL | KPC-2 | Ω Loop | ESBL-like | >16 | 8 | >16 | >16 | 4 | 16 | 128 | 4 | 1 | 2 | <0.12 | 2 | 8 | 0.023 | <0.5 | <1 | |
| D6 | ins168_LK | KPC-2 | Ω Loop | ESBL-like | >16 | <4 | >16 | >16 | 4 | 16 | >128 | 32 | 2 | 2 | <0.12 | 2 | 8 | 0.023 | <0.5 | <1 | ||
| D8 | ins168_LK | KPC-3 | A185T | Ω Loop | KPC-like | >16 | 64 | >16 | >16 | 8 | 32 | 32 | 2 | 4 | 4 | 0.5 | 2 | 8 | 0.25 | 1 | <1 | |
| mut-6 | ins175_SAIPG | KPC-2 | Ω Loop | KPC-like | >16 | >64 | >16 | >16 | 16 | >32 | >128 | 24 | 8 | 8 | 2 | 8 | 16 | 0.125 | <0.5 | <1 | ||
| 2 = C11 | KPC-44 | ins261_AVYTRAPNKDDKHSE | KPC-2 | Loop267–275 | KPC-like | >16 | 64 | >16 | >16 | 16 | >32 | 128 | 64 | 8 | 8 | 0.25 | 8 | 16 | 0.19 | 1 | <1 | |
| mut-1 = D1 | KPC-29 | ins269_KDD | KPC-3 | Loop267–275 | KPC-like | >16 | >64 | >16 | >16 | 32 | >32 | >128 | 32 | 32 | 16 | 1 | 16 | 16 | 0.094 | 2 | <1 | |
| C8 | ins274_SEAVI | KPC-3 | G147R | Loop267–275 | ESBL-like | >16 | 16 | >16 | >16 | 8 | 32 | 128 | 32 | 8 | 4 | 0.25 | 4 | 8 | 0.032 | <0.5 | <1 | |
| Deletions | ||||||||||||||||||||||
| C1 | del165-175_WELELNSAIPG | KPC-3 | Ω Loop | CAZase | <8 | <4 | <8 | <8 | 2 | 2 | 64 | 24 | 16 | 0.5 | <0.12 | <1 | 8 | 0.008 | <0.5 | <1 | ||
| C2 | del166-171_ELELNS | KPC-3 | A172D | Ω Loop | CAZase | <8 | <4 | <8 | <8 | 2 | 4 | 64 | 32 | 8 | 0.5 | <0.12 | <1 | 8 | 0.012 | <0.5 | <1 | |
| C6 | del168-171_ELNS | KPC-3 | Ω Loop | CAZase | <8 | 8 | <8 | 16 | 4 | 8 | >128 | >256 | 4 | 2 | <0.12 | <1 | 8 | 0.012 | <0.5 | <1 | ||
| C7 | del168-169_EL | KPC-2 | Ω Loop | CAZase | <8 | <4 | <8 | <8 | 2 | 8 | 64 | 1.5 | 0.5 | 1 | <0.12 | <1 | 16 | 0.012 | <0.5 | <1 | ||
| E1 | del167-170_LELN | KPC-2 | S171P | Ω Loop | CAZase | <8 | <4 | <8 | 16 | 8 | 16 | >128 | 24 | 2 | 4 | <0.12 | 2 | 16 | 0.023 | <0.5 | <1 | |
| C4 | del176-178_DA | KPC-3 | Ω Loop | ESBL-like | >16 | <4 | <8 | <8 | <1 | 4 | 32 | 8 | 2 | 0.5 | <0.12 | <1 | 8 | 0.006 | <0.5 | <1 | ||
| mut-7 | del_176-179_DAR | KPC-3 | Ω Loop | KPC-like | >16 | >64 | >16 | >16 | 16 | >32 | >128 | 32 | 8 | 8 | 0.25 | 8 | 16 | 0.094 | 1 | <1 | ||
| mut-11 | del239-240_GV | KPC-2 | Loop238–243 | ESBL-like | >16 | <4 | >16 | >16 | 16 | >32 | >128 | 32 | 4 | 8 | <0.12 | 8 | 8 | 0.012 | <0.5 | <1 | ||
| 5 | del240-241_VY | KPC-2 | G239D | Loop238–243 | ESBL-like | >16 | <4 | >16 | >16 | 8 | >32 | >128 | 48 | 4 | 8 | <0.12 | 8 | 8 | 0.023 | <0.5 | <1 | |
ESBL-like, extended-spectrum-beta-lactamase-like phenotype defined by AMP, MIC of >16 mg/liter, TZP, MIC of ≤16 mg/liter, and a > 4-fold decrease in MIC between CTX and CTX-clavulanate. KPC-like, KPC-like phenotype defined by a polyvalent resistance and TZP MIC of ≥64 mg/liter. CAZase, highly specialized activity on ceftazidime defined by ampicillin MIC of <8 mg/liter and resistance to CAZ and CZA.
AMP, ampicillin; TZP, piperacillin-tazobactam (1:4); CFZ, cefazolin; CEF, cephalothin; CRO, ceftriaxone; CPD, cefpodoxime; CAZ, ceftazidime; CZA, ceftazidime-avibactam; CAZ-clavu, ceftazidime-clavulanate (1:4); CTX, cefotaxime; CTX-clavu, cefotaxime-clavulanate (1:4); FEP, cefepime; FOX, cefoxitin; ETP, ertapenem; IPM, imipenem; MEM, meropenem.
Insertions.
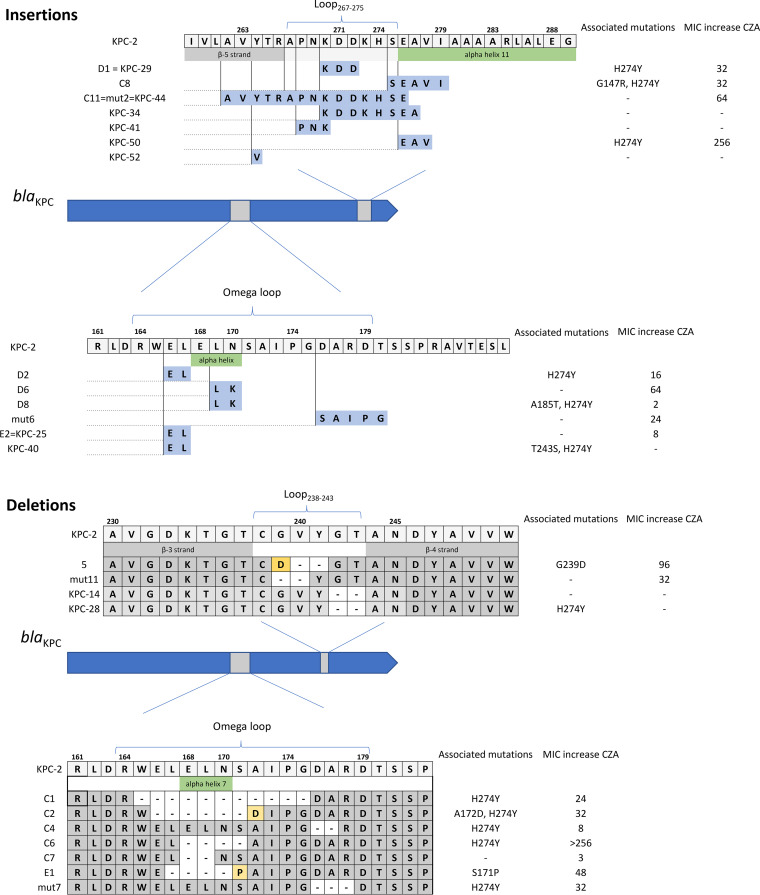
The eight insertions were tandem repeat (TR) mutations. The size of the repeat units of the TR mutations ranged from 6 bp to 45 bp (Table 1). The longest insertion occurred in Loop267–275. Some mutants were previously described in clinical isolates (KPC-25, KPC-29, and KPC-44 [13]). Of note, 2 mutants were found twice in our experiments, and these are the same mutants already described in clinical alleles (KPC-29 and KPC-44). This suggests that these mutants are more likely to emerge in vivo. Figure 1 represents alleles found in our study and those identified in clinical isolates.
FIG 1.
Representation of the protein with the insertion (A) and deletion (B) mutations and the fold increase in MIC to ceftazidime-avibactam (CZA) compared to that of KPC-2.
The resistance profile of the insertion mutants was highly heterogeneous for beta-lactams. In the omega-loop, we observed a decrease in MIC between cefotaxime and cefotaxime associated with clavulanate, suggesting an extended-spectrum beta-lactamase (ESBL)-like phenotypic profile. This phenotype was also evaluated by carrying out susceptibility tests by the disk diffusion method, with evidence of synergy between clavulanic acid and cefotaxime, cefepime, or aztreonam (22).
Insertions at positions 261 and 269 (previously described in the clinical alleles KPC-29 and KPC-44, respectively) showed high CZA MICs and tazobactam MICs of ≥64 mg/liter. Although their resistance to ertapenem and meropenem decreased, they retained a detectable activity on imipenem, revealing a polyvalent resistance of these mutants toward beta-lactams.
Deletions.
The nine deletions ranged from 6 bp to 33 bp. The longest deletion occurred in the omega-loop. No mutant observed in our study had yet been described in clinical isolates (10). Compared to the parent strains, all of the mutants with deletion mutations were more susceptible to carbapenems. We observed a consistent phenotype for mutants with deletions between positions 165 and 175, with a significant decrease in their resistance to all beta-lactams, including penicillins, except ceftazidime and CZA (Table 1). Thus, the mutated enzyme became highly specialized on ceftazidime. Mutants with deletions in Loop238–243 showed resistance to most penicillins and cephalosporins. They were strongly inhibited by clavulanic acid and tazobactam, showing an ESBL-like phenotype, with a decrease in MIC when clavulanate was combined with cefotaxime, compared to inhibition by cefotaxime alone. A phenotypic synergy between clavulanic acid and cefotaxime, cefepime, or aztreonam was also observed on antibiotic susceptibility testing by the disk diffusion method (22). Their carbapenemase activity was no longer detectable.
The tolerance to indel mutations depends on the region in which they occur in the protein. The 3 main loops surrounding the active site are affected either by insertions or deletions, reinforcing the idea that loops are more tolerant to indels, especially the omega-loop. Mutations in the omega-loop can modify salt bridges (specifically between Arg164 and Asp179) and, consequently, enhance the flexibility of the loop, widening the substrate spectrum (23, 24).
Altogether, the mutants with indels showed decreased susceptibility to ceftazidime-avibactam and a higher susceptibility toward some natural substrates of KPC, such as penicillins, cephalosporins (cefotaxime and cefepime), aztreonam, or carbapenems. The trade-off observed here in the KPC mutants is reported in other class A beta-lactamases (25). Within the class A beta-lactamases, a trade-off is described as an increased activity against cephalosporins (such as ceftazidime), generally resulting from both a loss of thermodynamic stability and a kinetic activity against their ancestral targets (25).
The variants with indels exhibited various resistance profiles (Table 1), with 3 different phenotypes being highlighted.
ESBL-like phenotype.
The ESBL-like phenotype is encountered with some insertions in the omega-loop and deletions in the Loop238–243 (Table 1). This resistance phenotype was defined by a resistance to ampicillin with a MIC of >16 mg/liter, a piperacillin-tazobactam MIC of ≤16 mg/liter, and a >4-fold decrease in MIC between cefotaxime and cefotaxime combined with clavulanate. Evidence of a synergy between clavulanic acid and cefotaxime, cefepime, or aztreonam was also observed on antibiotic susceptibility testing made by disk diffusion method (22).
Highly specialized activity on ceftazidime.
Deletions in the omega-loop section containing the alpha helix 7 show a consensus in the hydrolytic modifications of the protein. The mutants reported here are hypersensitive to most beta-lactams and, notably, ampicillin (MIC of <8 mg/liter) but not ceftazidime and ceftazidime-avibactam (Table 1). This suggests that for these variants, by the narrowing of the omega-loop, the deletion of the residues impacts not only the conformation of the substrate binding sites but also the catalytic regions of KPC beta-lactamase (26). The first clinical mutant with a deletion in the omega loop was recently described (11).
KPC-like phenotype with polyvalent activity on beta-lactams.
Even if phenotypical resistance profiles are modified (mainly on carbapenems), these mutants conserved polyvalent resistance on the same beta-lactams as their ancestral KPC-2 and KPC-3. In particular, piperacillin-tazobactam MICs were ≥64 mg/liter.
In our study, we explored and gave a more comprehensive view of the KPC beta-lactamase’s tolerance to indels that allows the protein to rapidly adapt in response to antibiotic treatments, such as ceftazidime-avibactam. The ability of the enzyme to modify its structure and tolerate indels while maintaining its activity reflects its plasticity. The thermodynamic stability of the KPC enzyme compared to those of other class A beta-lactamases (27) could explain their fascinating adaptability to the environment. If we add in their high tolerance for mutations, their evolutionary potential remains unpredictable and should be further evaluated. The clinical consequence of this evolutive capacity could challenge the therapeutic management of patients in the future.
Data availability.
blaKPC genes were submitted to GenBank under accession numbers MW077223 (KPC-C1), MW077224 (KPC-C2), MW077225 (KPC-C6), MW077226 (KPC-E1), MW077227 (KPC-C4), MW077228 (KPC-C7), MW077229 (KPC-D2), MW077230 (KPC-D6), MW077231 (KPC-D8), MW077232 (KPC-E2), MW077233 (KPC-D1), MW077234 (KPC-C8), MW077235 (KPC-C11), MW077236 (KPC-5-F), MW077237 (KPC-mut-6), MW077238 (KPC-mut-11), and MW077239 (KPC-mut-7).
ACKNOWLEDGMENTS
Claire Amaris Hobson is currently supported by funds from the Fondation ARC pour la Recherche sur le Cancer (grant no. DOC20190509066) for a Ph.D.
We have no conflicts to declare.
REFERENCES
- 1.Nijhuis RHT, Oueslati S, Zhou K, Bosboom RW, Rossen JWA, Naas T. 2015. OXY-2–15, a novel variant showing increased ceftazidime hydrolytic activity. J Antimicrob Chemother 70:1429–1433. doi: 10.1093/jac/dkv002. [DOI] [PubMed] [Google Scholar]
- 2.Arpin C, Labia R, Andre C, Frigo C, El Harrif Z, Quentin C. 2001. SHV-16, a β-lactamase with a pentapeptide duplication in the omega loop. Antimicrob Agents Chemother 45:2480–2485. doi: 10.1128/aac.45.9.2480-2485.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nukaga M, Haruta S, Tanimoto K, Kogure K, Taniguchi K, Tamaki M, Sawai T. 1995. Molecular evolution of a class C beta-lactamase extending its substrate specificity. J Biol Chem 270:5729–5735. doi: 10.1074/jbc.270.11.5729. [DOI] [PubMed] [Google Scholar]
- 4.Levitt PS, Papp-Wallace KM, Taracila MA, Hujer AM, Winkler ML, Smith KM, Xu Y, Harris ME, Bonomo RA. 2012. Exploring the role of a conserved class A residue in the Ω-Loop of KPC-2 β-lactamase. A mechanism for ceftazidime hydrolysis. J Biol Chem 287:31783–31793. doi: 10.1074/jbc.M112.348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galdadas I, Lovera S, Pérez-Hernández G, Barnes MD, Healy J, Afsharikho H, Woodford N, Bonomo RA, Gervasio FL, Haider S. 2018. Defining the architecture of KPC-2 Carbapenemase: identifying allosteric networks to fight antibiotics resistance. Sci Rep 8:12916. doi: 10.1038/s41598-018-31176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tooke CL, Hinchliffe P, Bragginton EC, Colenso CK, Hirvonen VHA, Takebayashi Y, Spencer J. 2019. β-Lactamases and β-lactamase inhibitors in the 21st century. J Mol Biol 431:3472–3500. doi: 10.1016/j.jmb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnes MD, Winkler ML, Taracila MA, Page MG, Desarbre E, Kreiswirth BN, Shields RK, Nguyen M-H, Clancy C, Spellberg B, Papp-Wallace KM, Bonomo RA. 2017. Klebsiella pneumoniae carbapenemase-2 (KPC-2), substitutions at ambler position Asp179, and resistance to ceftazidime-avibactam: unique antibiotic-resistant phenotypes emerge from β-lactamase protein engineering. mBio 8:e00528-17. doi: 10.1128/mBio.00528-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen MH, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. doi: 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livermore DM, Warner M, Jamrozy D, Mushtaq S, Nichols WW, Mustafa N, Woodford N. 2015. In vitro selection of ceftazidime-avibactam resistance in Enterobacteriaceae with KPC-3 carbapenemase. Antimicrob Agents Chemother 59:5324–5330. doi: 10.1128/AAC.00678-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oueslati S, Iorga BI, Tlili L, Exilie C, Zavala A, Dortet L, Jousset AB, Bernabeu S, Bonnin RA, Naas T. 2019. Unravelling ceftazidime/avibactam resistance of KPC-28, a KPC-2 variant lacking carbapenemase activity. J Antimicrob Chemother 74:2239–2246. doi: 10.1093/jac/dkz209. [DOI] [PubMed] [Google Scholar]
- 11.Antinori E, Unali I, Bertoncelli A, Mazzariol A. 2020. Klebsiella pneumoniae KPC producer resistant to ceftazidime-avibactam due to a deletion in the blaKPC3 gene. Clin Microbiol Infect 26:946.e1–946.e3. doi: 10.1016/j.cmi.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Humphries RM, Hemarajata P. 2017. Resistance to ceftazidime-avibactam in Klebsiella pneumoniae due to porin mutations and the increased expression of KPC-3. Antimicrob Agents Chemother 61:e00537-17. doi: 10.1128/AAC.00537-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, Retailleau P, Iorga BI. 2017. Beta-lactamase database (BLDB)–structure and function. J Enzyme Inhib Med Chem 32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Räisänen K, Koivula I, Ilmavirta H, Puranen S, Kallonen T, Lyytikäinen O, Jalava J. 2019. Emergence of ceftazidime-avibactam-resistant Klebsiella pneumoniae during treatment, Finland, December 2018. Euro Surveill 24:1900256. doi: 10.2807/1560-7917.ES.2019.24.19.1900256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naas T, Dortet L, Iorga BI. 2016. Structural and functional aspects of class A carbapenemases. Curr Drug Targets 17:1006–1028. doi: 10.2174/1389450117666160310144501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel L, Vuillemin X, Juhas M, Masseron A, Bechtel-Grosch U, Tiziani S, Mancini S, Nordmann P. 2020. KPC-50 confers resistance to ceftazidime-avibactam associated with reduced carbapenemase activity. Antimicrob Agents Chemother 64:e00321-20. doi: 10.1128/AAC.00321-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Göttig S, Frank D, Mungo E, Nolte A, Hogardt M, Besier S, Wichelhaus TA. 2019. Emergence of ceftazidime/avibactam resistance in KPC-3-producing Klebsiella pneumoniae in vivo. J Antimicrob Chemother 74:3211–3216. doi: 10.1093/jac/dkz330. [DOI] [PubMed] [Google Scholar]
- 18.Hobson CA, Bonacorsi S, Hocquet D, Baruchel A, Fahd M, Storme T, Tang R, Doit C, Tenaillon O, Birgy A. 2020. Impact of anticancer chemotherapy on the extension of beta-lactamase spectrum: an example with KPC-type carbapenemase activity towards ceftazidime-avibactam. Sci Rep 10:1–8. doi: 10.1038/s41598-020-57505-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meunier A, Nerich V, Fagnoni-Legat C, Richard M, Mazel D, Adotevi O, Bertrand X, Hocquet D. 2019. Enhanced emergence of antibiotic-resistant pathogenic bacteria after in vitro induction with cancer chemotherapy drugs. J Antimicrob Chemother 74:1572–1577. doi: 10.1093/jac/dkz070. [DOI] [PubMed] [Google Scholar]
- 20.European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2020. Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0. https://www.eucast.org/clinical_breakpoints/.
- 21.European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2020. Quality control. Version 10.0. https://www.eucast.org/ast_of_bacteria/quality_control/.
- 22.Drieux L, Brossier F, Sougakoff W, Jarlier V. 2008. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect 14(Suppl 1):90–103. doi: 10.1111/j.1469-0691.2007.01846.x. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez CE, Roberts P, Ostermeier M. 2019. Fitness effects of single amino acid insertions and deletions in TEM-1 β-lactamase. J Mol Biol 431:2320–2330. doi: 10.1016/j.jmb.2019.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathonet P, Deherve J, Soumillion P, Fastrez J. 2006. Active TEM-1 beta-lactamase mutants with random peptides inserted in three contiguous surface loops. Protein Sci 15:2323–2334. doi: 10.1110/ps.062303606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Minasov G, Shoichet BK. 2002. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol 320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 26.Yi H, Choi JM, Hwang J, Prati F, Cao T-P, Lee SH, Kim HS. 2016. High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A β-lactamase, PenL. Sci Rep 6:36527. doi: 10.1038/srep36527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehta SC, Rice K, Palzkill T. 2015. Natural variants of the KPC-2 carbapenemase have evolved increased catalytic efficiency for ceftazidime hydrolysis at the cost of enzyme stability. PLoS Pathog 11:e1004949. doi: 10.1371/journal.ppat.1004949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
blaKPC genes were submitted to GenBank under accession numbers MW077223 (KPC-C1), MW077224 (KPC-C2), MW077225 (KPC-C6), MW077226 (KPC-E1), MW077227 (KPC-C4), MW077228 (KPC-C7), MW077229 (KPC-D2), MW077230 (KPC-D6), MW077231 (KPC-D8), MW077232 (KPC-E2), MW077233 (KPC-D1), MW077234 (KPC-C8), MW077235 (KPC-C11), MW077236 (KPC-5-F), MW077237 (KPC-mut-6), MW077238 (KPC-mut-11), and MW077239 (KPC-mut-7).